Abstract
Neutralization-resistant simian-human immunodeficiency virus AD8 (SHIVAD8) variants that emerged in an infected macaque elite neutralizer targeting the human immunodeficiency virus type 1 (HIV-1) gp120 N332 glycan acquired substitutions of critical amino acids in the V3 region rather than losing the N332 glycosylation site. One of these resistant variants, carrying the full complement of gp120 V3 changes, was also resistant to the potent anti-HIV-1 monoclonal neutralizing antibodies PGT121 and 10-1074, both of which are also dependent on the presence of the gp120 N332 glycan.
TEXT
A major challenge for developing an effective human immunodeficiency virus type 1 (HIV-1) vaccine has been the identification of immunogens capable of eliciting broadly reacting neutralizing antibodies (NAbs). Although most HIV-1-infected individuals produce NAbs within several months or a year of exposure, this neutralizing activity is directed primarily against autologous virus (1–3). Levels of anti-HIV-1 antibodies able to neutralize viral strains from other infected individuals are generally much lower and delayed in their appearance (4–6). More-recent studies, employing pseudovirions in conjunction with high-throughput assays, have shown that approximately 20% of HIV-1-infected persons generate NAb responses against genetically diverse virus strains 2 to 4 years following HIV acquisition (7–10). Furthermore, so-called elite neutralizers, comprising approximately 1% of seropositive individuals, develop extremely potent cross-clade antiviral NAbs (11, 12). Analyses of HIV-1 envelope epitopes targeted by elite neutralizers have shown that a limited number of specificities are responsible for the broad and potent activities observed. These targets include the gp120 CD4 binding site, a gp120 V1/V2 glycan-dependent site (N160), the N332 glycan located in the C3 region of gp120, and the membrane-proximal external region (MPER) in gp41 (13–18).
We previously reported that one rhesus monkey (CE8J), inoculated with an uncloned preparation of the R5 simian-human immunodeficiency virus AD8-LN (SHIVAD8-LN) (19), developed potent cross-clade anti-HIV-1 NAbs similar to those observed for HIV-1-infected elite neutralizers (20). In that study and in the present one, we used pseudotyped (PS) viruses carrying the SHIVAD8 envelope protein (designated CK15), derived from the recently described pathogenic SHIVAD8 molecular clone SHIVAD8-EO (21), to monitor anti-SHIVAD8 neutralizing activity in the TZM-bl cell neutralization assay. CK15 PS virus preparations have been used to detect and quantitate anti-virus NAbs in plasma samples collected from macaques infected with uncloned or cloned SHIVAD8 inocula (20–22).
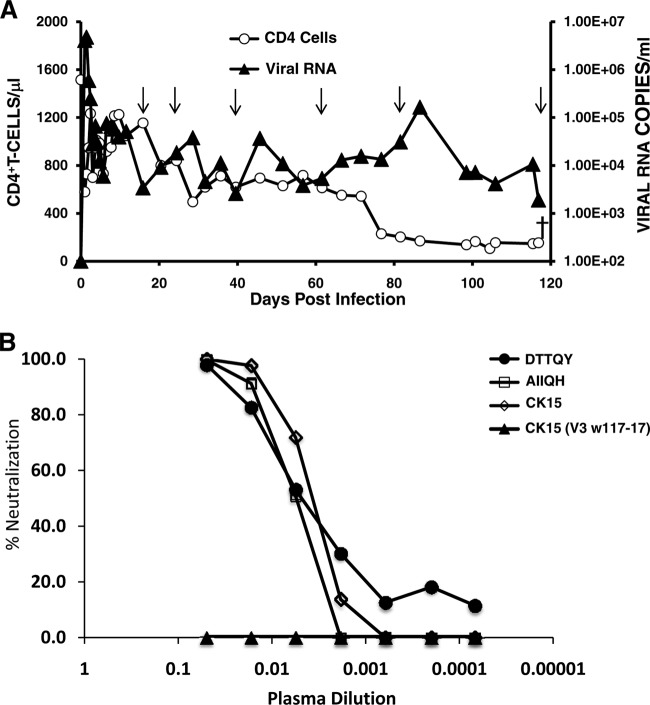
Plasma mapping studies revealed that NAbs in the elite neutralizer CE8J macaque exclusively targeted the HIV-1 gp120 N332 glycan, located immediately downstream of the V3 loop; removal of this glycosylation site from the Env of several HIV-1 isolates eliminated cross-reactive neutralization sensitivity (20). We also reported that this broadly reacting neutralizing activity persisted throughout the 2-year infection of monkey CE8J. Consistent with several studies reporting a positive correlation between the presence of anti-HIV-1 cross-reacting NAbs and plasma virus load (9, 10, 23), macaque CE8J, like HIV-1 elite neutralizers, who rarely derive any clinical benefit from the potent cross-reacting NAbs they generate (24, 25), ultimately succumbed to immunodeficiency and had to be euthanized at week 117 postinfection (p.i.) (Fig. 1) because of chronic Campylobacter coli enteritis. Notably, the virus recovered from this animal at the time of death was resistant to neutralization when tested with plasma specimens collected at weeks 50 and 87 p.i. (20).
Fig 1.
Neutralization variants emerge in elite neutralizer macaque CE8J inoculated with SHIVAD8. (A) Macaque CE8J was inoculated intravenously with 3.2 × 105 50% tissue culture infective doses (TCID50) of SHIVAD8#2-LN (19), and levels of plasma viral RNA and CD4+ T lymphocytes were determined. The arrows indicate plasma collection time points for determinations of neutralization sensitivities of circulating virus populations. (B) Sensitivities of the reference PS SHIVAD8 (CK15) and derivatives carrying gp120 V3 regions, amplified at various times during the infection of macaque CE8J, were determined using limiting dilutions of week 117 plasma in the TZM-bl cell assay. The V3 signature motifs shown are described in Fig. 3.
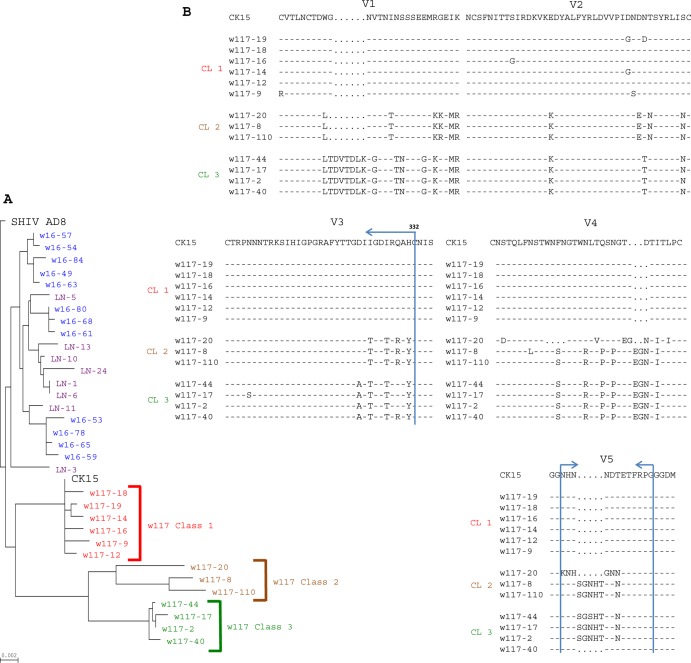
We have examined the env genes present in virus circulating in macaque CE8J at early and late times after its SHIVAD8 infection. As can be seen from the phylogenetic analysis shown in Fig. 2A, the virus population present at week 16 p.i. was closely related to the original uncloned SHIVAD8-LN inoculum (the LN series in the phylogenetic tree) used to infect monkey CE8J. At the time of euthanasia (week 117 p.i.), this analysis revealed that three classes of virus were present in the plasma. Each class had retained the N332 glycan but carried distinctive gp120 variable regions (Fig. 2B). It also should be noted that the CK15 env gene segment, present in the PS SHIVAD8 preparation used in neutralization assays, tracks with week 117 class 1 viruses in the phylogenetic tree (Fig. 2A). The alignments of the week 117 viruses revealed that the gp120 variable regions in the class 1 viruses were very similar to those present in the CK15 Env component of the PS SHIVAD8. In contrast, the week 117 class 3 viruses carried defining mutations and/or insertions located in all of the gp120 variable regions and were clearly different from the CK15 reference sequence (Fig. 2B). Class 2 viruses possessed an intermediate env gene sequence signature, containing a V1 region partially related to that present in class 1 viral RNAs and V2 to V5 segments with features found in the class 3 SHIV RNAs. No consistent amino acid changes were identified in gp41 coding regions of the week 117 SHIVs.
Fig 2.
Genetic analyses of SHIVAD8 env genes in macaque CE8J at weeks 16 and 117 postinfection. Viral RNA was amplified by RT-PCR from plasma collected at weeks 16 (blue) and 117 (red, class 1; brown, class 2; green, class 3) postinfection or from the SHIVAD8#2-LN inoculum (purple) (LN) and sequenced. Phylogenetic trees were generated by neighbor joining (A), and env gene sequences were aligned using ClustalW (B). The blue arrows and vertical lines demarcate the terminus of a gp120 variable region. The CK15 env gene is a component of both the recently described pathogenic SHIVAD8 molecular clone (21) and the reference SHIVAD8 PS virus used in TZM-bl neutralization assays.
We initially determined the neutralization sensitivity of the week 117 SHIVAD8 classes by preparing PS viruses using cytomegalovirus (CMV) plasmids expressing representative reverse transcription-PCR (RT-PCR)-amplified env genes. PS viruses were successfully obtained from 293T cells transfected with two class 1 (w117-16 and w117-19) and two class 3 (w117-2 and w117-17) amplicons (Fig. 2B). Neutralization assays utilized TZM-bl cells in combination with week 117 plasma diluted 1:20 as previously described (26). A >95% reduction of the input PS virus-induced luciferase activity was scored as neutralization sensitive, and a <30% reduction was rated as neutralization resistant. These assays revealed that the week 117 class 1 PS viruses and the CK15 PS virus control were sensitive to neutralization by week 117 plasma, whereas the two class 3 PS SHIVs were resistant (Table 1). PS viruses containing class 2 amplicons (w117-8 and w117-20) of sufficient titer to be evaluated for the neutralization phenotype were not able to be generated from transfected 293T cells. Further analyses revealed that similar amounts of envelope glycoproteins were (i) synthesized intracellularly and (ii) released as particle-associated proteins from the transfected cells by all three classes of week 117 PS viruses (see Fig. S1 in the supplemental material). However, the class 2 SHIVs were functionally defective in generating luciferase signals following infections of TZM-bl cells, and their neutralization sensitivities were not able to be determined.
Table 1.
Neutralization phenotypes for different SHIVAD8 PS virusesa
| PS virus group and strain | Neutralization phenotype |
|---|---|
| CK15 | Sensitive |
| Class 1 viruses | |
| w117-16 | Sensitive |
| w117-19 | Sensitive |
| Class 3 viruses | |
| w117-2 | Resistant |
| w117-17 | Resistant |
| Variable-region substitutions | |
| w117-17 (V1/V2 CK15) | Resistant |
| w117-17 (V1/V2+V4 CK15) | Resistant |
| w117-17 (V5 CK15) | Resistant |
| w117-17 (V3 CK15) | Sensitive |
| CK15 (V3 w117-17) | Resistant |
Pseudovirions carrying the entire gp120 coding region from week 117 class 1 or class 3 viruses were assayed for neutralization sensitivity using a 1:20 dilution of plasma collected from macaque CE8J at week 117 p.i. The neutralization phenotype of week 117 PS viruses with the indicated CK15 variable-region substitutions or the CK15 env gene with the class 3 w117-17 V3 variable-region insert was also determined with the same plasma sample.
Because the only consistent differences between the class 1 and class 3 SHIVAD8 derivatives mapped to the variable regions of gp120, our initial strategy was to identify variable-region determinants responsible for the neutralization resistance of the class 3 SHIVs. Individual or combinations of gp120 variable regions, derived from the neutralization-sensitive molecularly cloned CK15 env gene associated with the PS SHIVAD8, were inserted into the genetic background of the neutralization-resistant class 3 w117-17 env gene, and the persistence of neutralization resistance was evaluated. As shown in Table 1, the transfer of the V1/V2 segments, the V1/V2 plus V4 segments, or the V5 segment from the neutralization-sensitive CK15 env gene into the w117-17 env gene background, generating w117-17 (V1/V2 CK15), w117-17 (V1/V2+V4 CK15), and w117-17 (V5 CK15) pseudotyped viruses, respectively, had no effect on neutralization resistance. In contrast, the insertion of CK15 V3 sequences alone into the w117-17 env gene background, thereby generating w117-17 (V3 CK15), abrogated neutralization resistance. More importantly, as shown in Fig. 1B and indicated in Table 1, the reciprocal transfer of the w117-17 V3 region into the CK15 env gene, generating CK15 (V3 w117-17), conferred full neutralization resistance to the previously sensitive PS SHIVAD8, which was neutralized with a 50% effective concentration (EC50) titer of 1:316 using the week 117 plasma. Taken together, these results indicated that the gp120 V3 loop of the class 3 viruses was the principal determinant of resistance.
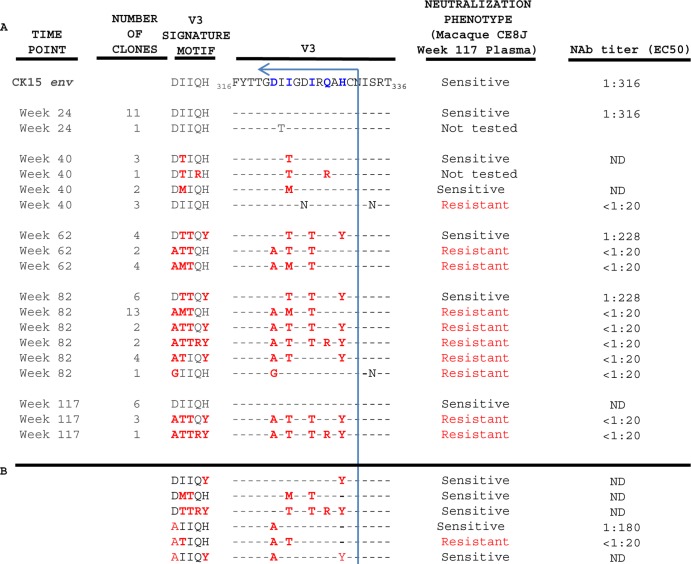
Because it seemed likely that the “resistant” V3 amino acid signature motif 321A-T–T-R/Q-Y330, present in the week 117 class 3 SHIVs (Fig. 2), may have emerged in a stepwise fashion, virus populations, circulating at various times during the two-plus years of infection of macaque CE8J, were evaluated by RT-PCR amplification of plasma viral RNA. The neutralization sensitivity and, in several instances, the EC50 neutralization titers of individual SHIVs from each time point were determined with CK15 PS viruses carrying specific gp120 V3 substitutions. As shown in Fig. 3A, the gp120 V3 region in circulating SHIVs at week 24 p.i. was essentially unchanged from that present in the neutralization-sensitive PS SHIVAD8 bearing theCK15 env gene, indicating that neutralization resistance had not arisen during this period.
Fig 3.
V3 neutralization-resistant SHIVAD8 variants emerge in a stepwise fashion beginning at week 40 p.i. (A) RT-PCR clones were prepared from plasma samples collected from macaque CE8J at various times postinfection, and the indicated gp120 V3 region substitutions were introduced into the CK15 env gene and expressed by a CMV vector. The resultant PS viruses were tested for neutralization sensitivity using week 117 plasma in a TZM-bl cell-based assay. Signature V3 amino acids present in the reference PS SHIVAD8 are indicated in violet, and those present in emerging variants are indicated in red. The latter changes have been incorporated into the V3 motifs listed. The arrow and blue vertical line demarcate the C terminus of the gp120 V3 region. ND, not done. (B) PS viruses carrying gp120 V3 substitutions not observed in vivo were also constructed, and their neutralization phenotypes were determined.
At week 40, the initial signs of V3 sequence evolution became evident (Fig. 3A). Six of nine env gene segments amplified at this time point contained V3 sequences with an I-to-T/M substitution at class 3 signature V3 residue 323. Nonetheless, when tested for neutralization sensitivity in the TZM-bl cell assay, SHIVAD8 pseudovirions carrying these changes remained neutralization sensitive. Three of the week 40 amplicons carried the S334N substitution, a change that eliminates the potential N-linked glycosylation site at N332 of gp120. Our previous study demonstrated that HIV-1 isolates lacking this glycan were not neutralized with plasma collected at multiple time points from macaque CE8J (20). When tested, a PS SHIVAD8 bearing the S334N change was, in fact, neutralization resistant.
At week 62 p.i., all 10 V3 gp120 segments amplified from circulating SHIVs carried amino acid substitutions associated with neutralization resistance to week 117 plasma, yet every segment had retained the N332 glycan required for neutralization sensitivity. The acquisition of the D321A change, plus at least two other signature V3 substitutions in six of these week 62 amplicons, conferred neutralization resistance (Fig. 3A). In contrast, pseudovirions bearing V3 regions with the V3 DTTQY signature motif remained neutralization sensitive, exhibiting an EC50 neutralization titer (1:228) slightly lower than that observed (1:316) for PS virions bearing the CK15 env gene (Fig. 1B). Thus, at week 62 p.i., 6 of 10 amplified env gene segments carried critical changes in gp120 V3 regions and exhibited a neutralization-resistant phenotype to week 117 plasma.
By week 82 p.i., 21 of the 28 plasma-derived env gene amplicons carried gp120 V3 regions with the D321A change in combination with at least two other signature V3 substitutions. All were neutralization resistant. A single amplicon carried the S334N substitution, eliminating the N332 glycosylation site. Interestingly, between weeks 60 and 86, the levels of circulating CD4+ T lymphocytes declined from 614 to 169 cells/μl and continued to fall between weeks 86 and 100 p.i. (Fig. 1). This decrease was associated with a striking decline in the titers of neutralizing antibody directed against the CK15 PS virus (from 1:5,193 [week 77] to 1:397 [week 101]) (20), with both parameters ostensibly reflecting a deteriorating and increasingly dysfunctional immune system. Consistent with these findings, several of the env gene amplicons recovered at week 117 carried gp120 V3 regions lacking signature neutralization-resistant motifs and resembled the neutralization-sensitive class 1 viruses (compare Fig. 3A with Fig. 2B). The latter finding may be indicative of the reemergence of a more fit archival virus in an immunocompromised animal.
To further investigate critical amino acid substitutions conferring resistance against the polyclonal NAbs present in macaque CE8J at week 117, env genes with V3 changes not observed in vivo were constructed, introduced into SHIVAD8 pseudovirions, and assayed for neutralization sensitivity. As shown in Fig. 3B, V3 regions with multiple signature amino acid substitutions, but not the D321A change, were all neutralization sensitive. A PS virus with a V3 region containing only the D321A change (AIIQH) was also neutralization sensitive (Fig. 1B), exhibiting an EC50 neutralization titer of 1:180 with week 117 plasma. The minimal gp120 V3 changes conferring resistance to week 117 plasma possessed the ATIQH signature motif (Fig. 3B).
A group of potent, cross-reactive monoclonal NAbs targeting gp120 carbohydrate epitopes has recently been described (16, 27, 28). The sensitivity of several of these MAbs is dependent on the presence of the gp120 N332 glycan, and some have also been reported to interact with residues mapping to the V3 loop. Because the broad polyclonal NAbs generated in macaque CE8J are also dependent on the presence of the N332 glycan, we wondered whether the resistant SHIV variants emerging in this animal had lost their sensitivity to these potent neutralizing MAbs. As shown in Table 2, a SHIVAD8 bearing a V3 region with the ATTQY motif was resistant to the week 117 CE8J plasma, PGT 121, and 10-1074 but not to the PGT126, PGT128, and PGT130 neutralizing MAbs. In contrast, the closely related SHIVAD8 variant, which carried a V3 region with the ATTQH motif, was resistant to the week 117 monkey plasma but not to any of the N332 glycan-dependent neutralizing MAbs. One might conclude from these results that a V3 region with only the H330Y substitution (DIIQY motif) might confer neutralization resistance to PGT 121 and 10-1074. However, when such a DIIQY-pseudotyped SHIVAD8 was tested, it was found to be sensitive to both PGT121 and 10-1074. The latter result is consistent with a previous report showing that an alanine substitution at residue 330 has no effect on PGT 121 sensitivity (16).
Table 2.
Neutralization sensitivity of SHIVAD8 V3 variants to potent neutralizing monoclonal antibodies
| V3 signature motif | V3 sequence | Neutralization phenotype for wk 117 CE8J plasma (1:20) | MAb concn (μg/ml)a |
||||
|---|---|---|---|---|---|---|---|
| PGT121 | 10-1074 | PGT126 | PGT128 | PGT130 | |||
| DIIQH (CK15) | 315FYTTGDIIGDIRQAHCNIS334 | Sensitive | 0.081 | 0.132 | 0.106 | 0.037 | 0.206 |
| ATTQH | 315FYTTGAITGDTRQAHCNIS334 | Resistant | 0.185 | 0.435 | 0.057 | 0.042 | 0.174 |
| ATTQY | 315FYTTGAITGDTRQAYCNIS334 | Resistant | >1,000 | >1,000 | 0.020 | 0.019 | 0.066 |
Values are the monoclonal antibody concentrations in μg/ml at which relative luminescence units were reduced 50% compared to virus control wells (no antibody).
Taken together, our findings show that stepwise substitutions of critical amino acids in the C-terminal one-third of the SHIVAD8 gp120 V3 region are sufficient to confer resistance to polyclonal plasma NAbs present in an infected macaque elite neutralizer as well as against two potent monoclonal NAbs, PGT121 and 10-1074, both of which target the N332 glycan. The resistance to these different antiviral neutralizing activities, measured in the context of a gp120 retaining the N332 glycan, was dependent on the location and number of gp120 V3 amino acid substitutions: resistance to the PGT121 and 10-1074 MAbs required 4 amino acid changes, whereas 2 critical amino acid substitutions were sufficient for resistance to week 117 plasma. It is worth noting that the V3 neutralization-resistant SHIVAD8 variants that arose in macaque CE8J were clinically significant. Unlike the resistant viruses emerging in an HIV-1-infected elite neutralizer, which lost replication fitness (29), they possessed robust in vivo replication properties and induced CD4+ T cell depletion and death from immunodeficiency (Fig. 1).
It was somewhat unexpected that the vast majority of neutralization escape variants that emerged in macaque CE8J had retained the N332 glycan and evolved by inducing amino acid substitutions in the C-terminal portion of the V3 loop. Although 3 of 9 amplicons recovered at week 40 p.i. had lost the N332 glycan and were found to be neutralization resistant, this SHIVAD8 variant remained a minor component of circulating virus and was not detected again until week 82 p.i., and then as only 1 of 28 amplicons recovered at that time point.
Crystal structures of PGT121 and PGT128 antigen binding domains interacting with HIV-1 JR-FL and HIV-1 YU-2 gp120 molecules, respectively, have revealed that both neutralizing MAbs bind to epitopes associated with the N332 glycan and amino acids located at the base of the V3 loop (27, 28). The resistance observed to PGT121 and 10-1074 but not to PGT126, PGT128, and PGT130 very likely reflects the derivation of the PGT121/10-1074 MAbs from the same HIV-1-infected African donor and the isolation of PGT126, PGT128, and PGT130 from other infected individuals (27). Differences in the orientations of respective heavy chain complementarity-determining regions of these N332-dependent neutralizing MAbs may affect their interactions with their gp120 targets and explain the variable neutralization sensitivities observed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ronald Plishka for performing nucleotide sequence analyses and are indebted to Dennis Burton, The Scripps Institute, and Michel Nussenzweig, Rockefeller University, for providing aniti-HIV-1 neutralizing monoclonal antibodies.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print 29 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00878-13.
REFERENCES
- 1. Albert J, Abrahamsson B, Nagy K, Aurelius E, Gaines H, Nystrom G, Fenyo EM. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4:107–112 [DOI] [PubMed] [Google Scholar]
- 2. Bosch KA, Rainwater S, Jaoko W, Overbaugh J. 2010. Temporal analysis of HIV envelope sequence evolution and antibody escape in a subtype A-infected individual with a broad neutralizing antibody response. Virology 398:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aasa-Chapman MM, Hayman A, Newton P, Cornforth D, Williams I, Borrow P, Balfe P, McKnight A. 2004. Development of the antibody response in acute HIV-1 infection. AIDS 18:371–381 [DOI] [PubMed] [Google Scholar]
- 5. Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 7. Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L, CAPRISA002 Study Team 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7:e1001251. 10.1371/journal.ppat.1001251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Euler Z, van den Kerkhof TL, van Gils MJ, Burger JA, Edo-Matas D, Phung P, Wrin T, Schuitemaker H. 2012. Longitudinal analysis of early HIV-1-specific neutralizing activity in an elite neutralizer and in five patients who developed cross-reactive neutralizing activity. J. Virol. 86:2045–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6:e1001028. 10.1371/journal.ppat.1001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishimura Y, Shingai M, Willey R, Sadjadpour R, Lee WR, Brown CR, Brenchley JM, Buckler-White A, Petros R, Eckhaus M, Hoffman V, Igarashi T, Martin MA. 2010. Generation of the pathogenic R5-tropic simian/human immunodeficiency virus SHIVAD8 by serial passaging in rhesus macaques. J. Virol. 84:4769–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walker LM, Sok D, Nishimura Y, Donau O, Sadjadpour R, Gautam R, Shingai M, Pejchal R, Ramos A, Simek MD, Geng Y, Wilson IA, Poignard P, Martin MA, Burton DR. 2011. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc. Natl. Acad. Sci. U. S. A. 108:20125–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shingai M, Donau OK, Schmidt SD, Gautam R, Plishka RJ, Buckler-White A, Sadjadpour R, Lee WR, LaBranche CC, Montefiori DC, Mascola JR, Nishimura Y, Martin MA. 2012. Most rhesus macaques infected with the CCR5-tropic SHIV(AD8) generate cross-reactive antibodies that neutralize multiple HIV-1 strains. Proc. Natl. Acad. Sci. U. S. A. 109:19769–19774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gautam R, Nishimura Y, Lee WR, Donau O, Buckler-White A, Shingai M, Sadjadpour R, Schmidt SD, LaBranche CC, Keele BF, Montefiori D, Mascola JR, Martin MA. 2012. Pathogenicity and mucosal transmissibility of the R5-tropic simian/human immunodeficiency virus SHIVAD8 in rhesus macaques: implications for use in vaccine studies. J. Virol. 86:8516–8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Euler Z, van Gils MJ, Bunnik EM, Phung P, Schweighardt B, Wrin T, Schuitemaker H. 2010. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J. Infect. Dis. 201:1045–1053 [DOI] [PubMed] [Google Scholar]
- 25. Mascola JR, Montefiori DC. 2010. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28:413–444 [DOI] [PubMed] [Google Scholar]
- 26. Willey R, Nason MC, Nishimura Y, Follmann DA, Martin MA. 2010. Neutralizing antibody titers conferring protection to macaques from a simian/human immunodeficiency virus challenge using the TZM-bl assay. AIDS Res. Hum. Retroviruses 26:89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. U. S. A. 109:E3268–E3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sather DN, Carbonetti S, Kehayia J, Kraft Z, Mikell I, Scheid JF, Klein F, Stamatatos L. 2012. Broadly neutralizing antibodies developed by an HIV-positive elite neutralizer exact a replication fitness cost on the contemporaneous virus. J. Virol. 86:12676–12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.