Introduction
During its lifetime, an RNA molecule is escorted by a cohort of RNA-binding protein (RBP) partners in ever-changing ribonucleoprotein (RNP) complexes. RBPs critically regulate the structure, localization, and function of both coding and non-coding RNAs [reviewed in (Glisovic et al., 2008)]. Since RNPs therefore play fundamental roles in normal and diseased cells, it is crucial to catalogue and functionally dissect their composition correctly (Khalil and Rinn, 2011). With the advent of new mass spectrometry and high-throughput sequencing methods, genome-wide data have enabled previously unprecedented views of the RNP world.
While these powerful technologies have yielded novel insights into RNP biology and transcript regulation, all experimental designs bear caveats. The “Observer Effect” is a term frequently applied in physics to describe the perturbations made by the act of observation on the phenomenon being investigated (Buks et al., 1998). Seemingly small perturbations affecting the cellular environment or buried within a purification scheme, as necessitated by an experimental protocol, can have global consequences. These concerns are relevant to the interpretation of recent large-scale screens and some specific issues have been systematically tested in independent experiments. Caution is therefore warranted in genome-wide studies of protein-RNA interactions.
Here we briefly review approaches currently used to obtain genome-wide profiles of RNA-protein interactions in living cells. We highlight recent studies of the mRNA-bound proteome and address pitfalls inherent in such investigations.
RIP-Chip
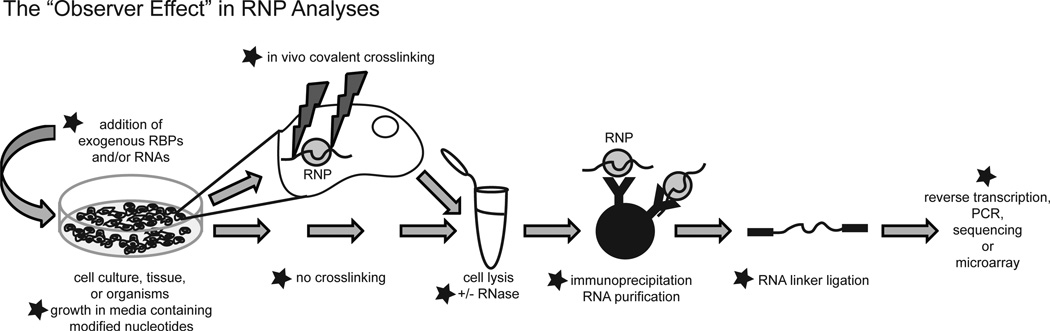
To define the in vivo composition of RNPs, many global studies of RBPs have employed RNA immunoprecipitation coupled with microarray analyses (RIP-Chip). In general, such protocols begin with creation of a lysate of cells or tissue that is then subjected to immunoprecipitation with an antibody directed against an RBP of interest. Formaldehyde or UV crosslinking may or may not be used to link protein-RNA complexes covalently before lysis. RNAs that coimmunoprecipitate with the protein are then subjected to microarray analyses for identification [Fig. 1; protocol for method: (Keene et al., 2006)]. RIP-Chip analyses have demonstrated the ubiquity of protein-RNA interactions and have laid the foundation for many structural and functional studies (Khalil and Rinn, 2011).
Figure 1.
The Observer Effect in RNP analysis. The diagram shows a generalized approach to the isolation and analysis of RNA bound to RBPs. In some experiments, exogenous RBPs and/or RNAs are expressed or transfected into cells. Cultured cells, tissues, or whole organisms are either subjected to in vivo covalent crosslinking (UV or formaldehyde) or lysed directly without crosslinking; in some cases, cells are grown in modified media to enhance crosslinking. The cell lysate is often treated with RNase to digest RNAs into workable fragments before the RBPs are immunoprecipitated. After RNA is purified from the immunoprecipitate, RNA linkers are ligated to both ends to facilitate reverse transcription, PCR, and sequencing. The stars indicate steps subject to documented occurrences of the Observer Effect, which are described in the text.
However, RIP-Chip has limitations. RIP-Chip without crosslinking has been used to select stable RNPs, often including noncoding RNAs, which survive the conditions of the immunoprecipitation protocol. Yet, transient interactions are not readily captured by this method. In analyses designed to characterize less stable RNPs, particularly those involving mRNAs, non-crosslinked RNAs and proteins reassociate upon cell lysis, yielding false-positive results that do not reflect in vivo interactions (Mili and Steitz, 2004; Riley et al., 2012). Predicting whether remodeling of an RNP will occur after cell lysis is not as simple as comparing protein-RNA binding constants, because the concentrations of both the RNA targets and competing RBPs contribute to the outcome. The demonstrated reproducibility of RIP-Chip experiments is ~60–75% (Khalil et al., 2009), complicating analyses and inarguably requiring many replicates, which are not always undertaken. Finally, data from RIP-Chip without crosslinking represent the sum of direct and indirect interactions of a protein with RNA (Keene et al., 2006), and binding sites cannot be mapped to nucleotide resolution.
CLIP
To address many of the shortcomings of RIP-Chip, a crosslinking and immunoprecipitation (CLIP) protocol was developed by the Darnell lab [Fig. 1; method first described: (Ule et al., 2003); applications of CLIP reviewed: (Darnell, 2010)] and its utility demonstrated in a pioneering study of the brain-specific splicing factor, Nova. In CLIP, UV light (254 nm) covalently couples specific amino acids in bound RBPs to photo-reactive nucleotide bases in RNAs in unperturbed live cells or tissue. Lysates are subjected to immunoprecipitation and stringent purification steps are used to isolate RNAs crosslinked to the protein of interest. RNA sequencing then identifies RNA regions directly bound to the RBP, background is very low, and a defined consensus sequence for binding can be derived [for a review and technical comparison of CLIP approaches, see (Konig et al., 2012; Milek et al., 2012)].
CLIP has been widely applied to many RBPs and adapted in several ways (Darnell, 2010; Konig et al., 2012). The addition of high-throughput sequencing of crosslinked RNA fragments (HITS-CLIP) permits genome-scale identification of direct RNA targets, largely overcomes the issue of UV crosslinking inefficiency (Licatalosi et al., 2008), and exhibits good reproducibility between biological replicates [for example, R2>0.8 for replicates of Argonaute-mRNA HITS-CLIP comparing results from individual mouse brains (Chi et al., 2009)]. However, multiple biological and technical replicates are still required to draw reliable global conclusions. While the advent of high-throughput sequencing has improved the depth of the CLIP approach significantly, inherent problems remain in generating accurate sequencing reads due to limitations in the biochemistry of high-throughput sequencing and in the mapping of sequencing reads to a reference genome (Li et al., 2011; Pickrell et al., 2012).
Photoactivatable-ribonucleotide-enhanced crosslinking (PAR-CLIP) is a related method developed to facilitate mapping of crosslinks at single-nucleotide resolution (Hafner et al., 2010). In PAR-CLIP, a modified nucleotide such as 4-thiouridine is added to cell media and incorporated into newly synthesized RNAs; UV light (365 nm) forges crosslinks between modified residues and protein or RNA molecules lying in close proximity. Moreover, the incorporation of photoactivatable ribonucleotides in the PAR-CLIP approach affords an internal control for crosslinking (Ascano et al., 2012). Direct comparisons of HITS-CLIP and PAR-CLIP data have demonstrated that the two methods yield similarly resolved genomic landscapes and specific binding sites for RBPs (Kishore et al., 2011). The reliability of global conclusions from CLIP surveys, and the identification of stable, functionally relevant RNA-protein interactions, also benefits from extensive experimental replication (Jungkamp et al., 2011).
All CLIP procedures are elaborate, multi-step procedures that require extensive optimization and proper controls. Bias can arise from several sources. The nucleotide composition of the RNA linkers that are ligated to the precipitated RNAs or RNA fragments to prepare them for reverse transcription, PCR, and sequencing has been documented to affect ligation efficiency in the creation of small RNA libraries (Hafner et al., 2011). The aforementioned 254 nm and 365 nm UV crosslinking chemistries exhibit differential sequence preferences (Castello et al., 2012). Sequence-specific RNase overdigestion can bias CLIP results as well (Kishore et al., 2011). Since any procedure involving immunoprecipitation is subject to background signal, reproducing CLIP experiments (>4 biological replicates) is necessary to reduce background significantly (Chi et al., 2009).
Perturbations to in vivo stoichiometry
To facilitate application of RIP-Chip or CLIP, many genome-wide studies rely upon the addition or expression of exogenous, sometimes tagged, RBPs or RNAs. When the levels of these exogenous proteins or RNAs are assessed (a crucial control)—typically by non-quantitative Western blot analyses, Northern blots, or qPCR—the entire cell population is examined. However, it should be appreciated that the levels of such artificially expressed molecules probably vary significantly from one cell to the next, creating a notable Observer Effect in at least some cells. Basic principles of chemical stoichiometry apply to RBPs and their targets inside cells; hence, the in vivo ratio of RNA to protein is often tightly controlled to encourage correct interactions and prevent non-specific binding events [see (Wright et al., 2011) for one such detailed analysis]. By probing the system with transfected RNA or proteins, cellular stoichiometry is perturbed. Both false-positive and false-negative results can be generated (Khan et al., 2009; Mili and Steitz, 2004; Riley et al., 2012).
The consequences of microRNA (miRNA) or small interfering RNA (siRNA) transfection in genome-wide target identification have been explored in detail. Comparison of >150 published genome-wide studies of transcript responses to mi/siRNA transfection showed that endogenous miRNA function is significantly impaired, as endogenous miRNA targets are de-repressed in transfected cells; the data also revealed time- and concentration-dependent alterations in the experimental output (Khan et al., 2009). These findings are consistent with the competing endogenous RNA hypothesis, which posits that the relative ratios of cellular RNAs are key to their functioning within large-scale regulatory networks (Salmena et al., 2011).
Subcellular localization
Another important consideration for RNA biology is the subcellular location of RNP complexes. In addition to respecting barriers imposed by membrane boundaries within the cell, RNPs often localize by assorting into functionally distinct subcompartments in a temporally appropriate manner. These aggregates or “RNA granules” include Cajal bodies and nucleoli within the nucleus, and neuronal granules, stress granules and processing (P−) bodies within the cytoplasm. RNA granules assemble from soluble components into dynamic RNP aggregates with hydrogel-like characteristics [reviewed in (Weber and Brangwynne, 2012)]. The identities of the protein and RNA components of RNA granules are of great interest, but have been technically difficult to define. In a pair of recent publications, the McKnight group identified protein and RNA components of RNA granules that were isolated by precipitation with a small molecule (Han et al., 2012; Kato et al., 2012). Mass spectrometry revealed that an overwhelming majority of the precipitated RBPs bear repetitive motifs of low-complexity sequences (LCS), which are intrinsically disordered. Further, certain LCS proteins were capable of forming hydrogel aggregates in vitro, similar to RNA granules, independent of the precipitant (Kato et al., 2012).
These studies represent a significant advance in our understanding of the complexity and subcellular organization of RNPs. The existence of RBP aggregates could explain the detection of indirectly associated mRNAs in immunoprecipitates where the analysis does not include generation of protein-RNA covalent bonds. Perhaps, some of the experimental variability in RIP-Chip data derives from association of secondary RBPs with direct RNA-binders.
From global to specific
A global profile of the mRNA-bound proteome of HeLa cells, which was obtained by variants of both HITS-CLIP (conventional crosslinking or “cCL”) and PAR-CLIP (“PAR-CL”) selection of polyadenylated RNAs coupled with quantitative mass spectrometry of RNA-bound RBPs, also revealed an overrepresentation of LCS in RBPs (Castello et al., 2012). Remarkably, >300 novel RBPs were discovered using this well-controlled, highly replicated approach. A similar study identified ~245 novel RBPs in human embryonic kidney cells using PAR-CLIP/mass spectrometry (Baltz et al., 2012). In both studies, a notable fraction of RBPs did not exhibit identifiable RNA binding motifs, emphasizing the importance of a purely biochemical approach. In this new catalogue of RBPs, Castello and colleagues detected a structural theme: that intrinsic disorder often correlates with the inclusion of short, repetitive amino acid motifs (Castello et al., 2012). Together these complementary studies reaffirm that there is still much to learn about the molecular basis of protein-RNA interactions.
Going forward, a methodology is needed to catalogue the complete complement of RBPs that associate with a particular individual RNA as it proceeds through the stages of its existence—from transcription to decay. Perhaps some variant of the CHART (capture hybridization analysis of RNA targets) procedure (Simon et al., 2011), devised to detect DNA sequences and proteins that are formaldehyde crosslinked to a selected RNA, will fill this gap.
Conclusions
Genome-wide studies of RNP complexes have recently led to momentous advances in our understanding of RBPs, including the characterization of novel binding sites for splicing factors, the definition of subcellular structures through which RNAs traffic, and miRNA target identification. They have also uncovered hundreds of previously uncharacterized RBPs. However, studies of individual RBPs will continue to play defining roles in our quest for mechanistic insights into RNA biology. As we move toward broader and higher resolution studies, it is important to bear in mind that the Observer Effect can influence the outcome of any study. It remains essential to use complementary methods of validation in the study of RBPs.
References
- Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Buks E, Schuster R, Heiblum M, Mahalu D, Umansky V. Dephasing in electron interference by a 'which-path' detector. Nature. 1998;391:871–874. [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. 2010 doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Jungkamp AC, Stoeckius M, Mecenas D, Grun D, Mastrobuoni G, Kempa S, Rajewsky N. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell. 2011;44:828–840. doi: 10.1016/j.molcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Rinn JL. RNA-protein interactions in human health and disease. Semin Cell Dev Biol. 2011;22:359–365. doi: 10.1016/j.semcdb.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2012;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, Cheung VG. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milek M, Wyler E, Landthaler M. Transcriptome-wide analysis of protein-RNA interactions using high-throughput sequencing. Semin Cell Dev Biol. 2012;23:206–212. doi: 10.1016/j.semcdb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Gilad Y, Pritchard JK. Comment on "Widespread RNA and DNA sequence differences in the human transcriptome". Science. 2012;335:1302. doi: 10.1126/science.1210484. author reply 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley KJ, Yario TA, Steitz JA. Association of Argonaute proteins and microRNAs can occur after cell lysis. RNA. 2012;18:1581–1585. doi: 10.1261/rna.034934.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J. 2011;30:533–545. doi: 10.1038/emboj.2010.334. [DOI] [PMC free article] [PubMed] [Google Scholar]