Abstract
Neuroectodermal signalling centres induce and pattern many novel vertebrate brain structures but are absent, or divergent, in invertebrate chordates. This has led to the hypothesis that signalling centre genetic programs were first assembled in stem vertebrates, which potentially drove morphological innovations. However, this scenario presumes that extant cephalochordates accurately represent ancestral chordate characters, which has not been tested using close chordate outgroups. Here, we report that genetic programs homologous to three vertebrate signalling centres; the anterior neural ridge, zona limitans intrathalamica, and isthmic organizer are present in the hemichordate Saccoglossus kowalevskii. Fgf8/17/18, sfrp1/5, hh, and wnt1 are expressed in vertebrate-like arrangements in hemichordate ectoderm, and homologous genetic mechanisms regulate ectodermal patterning in both animals. We propose these genetic programs were components of an unexpectedly complex, ancient genetic regulatory scaffold for deuterostome body patterning that degenerated in amphioxus and ascidians, but was retained to pattern divergent structures in hemichordates and vertebrates.
During vertebrate development, the brain arises from coarsely patterned planar neuroectoderm through successive refinement of regional identities, resulting after morphogenesis and growth, in a complex structure composed of highly specialized areas1-3. In contrast, invertebrate chordates have relatively simple nervous systems that lack unambiguous homologs of many vertebrate brain regions 4-6. These clear disparities in nervous system complexity indicate that key innovations in patterning mechanisms have attended the evolution of the vertebrate brain from a simpler CNS. However, identifying the presumed genetic regulatory novelties has been elusive, and comparative studies have largely revealed similarities, rather than differences, in the early transcriptional architectures of diverse bilaterian nervous systems7-12. Notable exceptions are CNS signalling centres, which have emerged as strong candidates for vertebrate genetic regulatory novelties involved in early vertebrate brain evolution8-10. These centres act as secondary organizers that mediate regional patterning in the CNS and are often necessary and sufficient for the establishment of vertebrate-specific brain structures1-3,13-18.
The anterior neural ridge (ANR), zona limitans intrathalamica (ZLI) and isthmic organizer (IsO) are the three primary signalling centres that direct anteroposterior (AP) patterning in the vertebrate anterior neural plate and then brain1-3. Homologous signalling centres and their molecular signatures are absent, or divergent, in amphioxus and ascidians8-10,19-25, consistent with the hypothesis that they are vertebrate novelties whose origins could have driven CNS innovations in stem vertebrates8-10. However, this hypothesis depends on amphioxus adequately representing ancestral states for chordate developmental genetic characters, and does not account for the possibility of secondary losses in cephalochordates. Here, we present developmental data from the hemichordate Saccoglossus kowalevskii to test an alternative scenario for signalling centre origins, namely, that ANR, ZLI, and IsO genetic programs predate chordate origins and were secondarily simplified or lost along the lineages leading to the invertebrate chordates.
Hemichordates are a deuterostome phylum closely related to chordates26 and are a promising outgroup for investigating chordate evolution. Previous studies established that despite significant body plan divergence between the two groups, hemichordates and vertebrates share a broadly conserved transcriptional regulatory architecture during early body patterning7,27-29, revealed by their similar maps of expression domains for genes involved in early AP patterning. This combination of morphological divergence and developmental genetic similarity makes hemichordates an informative outgroup for testing the proposed coupling of vertebrate morphological and developmental genetic innovations. Here, we present descriptive and functional evidence that genetic programs homologous to the ANR, ZLI, and IsO are present in S. kowalevskii, suggesting that they were elements of an ancient genetic and developmental toolkit for deuterostome body patterning that were subsequently modified and elaborated to regulate vertebrate brain development.
An ANR-like regulatory program in hemichordates
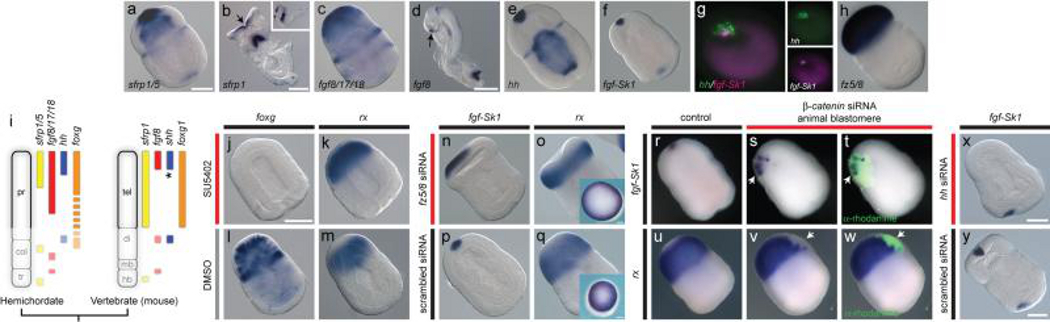
The ANR is located in the anterior neural plate of vertebrates and is a source of fibroblast growth factors (FGFs) and secreted frizzled-related proteins (SFRPs), which establish and pattern the telencephalon 1,2,15,16,30,31. Topologically consistent with vertebrate CNS expression domains, sfrp1/5 and fgf8/17/18 are expressed in S. kowalevskii anterior proboscis ectoderm (Fig. 1a-d). In vertebrates, FGFs and Wnt antagonists mediate ANR function and telencephalon patterning15,16,30-32. In mice, conditional fgfr1,2,3 knockout abolishes the telencephalon16, while fgf8 mutants have a posteriorized neocortex33. Similarly, treating S. kowalevskii embryos with the FGF receptor inhibitor SU540234 from late gastrula through double-groove stages (28-36 hrs) eliminated the morphological boundary between the proboscis and collar and resulted in altered gene expression domains indicating loss and/or posteriorization of anterior proboscis (Fig. 1j-m). Expression of foxg, which is an FGF target in vertebrates and marks S. kowalevskii proboscis ectoderm, was completely eliminated (Fig. 1j, l), while rx, which is normally excluded from the anterior proboscis, was expressed up to the anterior limit of the embryo (Fig. 1k, m).
Figure 1. An ANR-like signalling centre in S. kowalevskii.
a-h, S. kowalevskii and mouse in situ hybridizations for markers of ANR and telencephalon. S. kowalevskii embryos are at double groove stage (36 hrs), and are shown in dorsal view with anterior to top left, except where noted. Embryos are optically cleared except in o, q insets and r-w. Mouse embryos are at stage ~E8.5. a, S. kowalevskii sfrp1/5 expression. b, frontal view of mouse sfrp1 expression, arrow denotes ANR. Inset shows lateral view. c, S. kowalevskii fgf8/17/18 expression. d, mouse fgf8 expression, arrow denotes ANR. e, S. kowalevskii hh expression. f, S. kowalevskii fgf-Sk1 expression. g, frontal view of double fluorescence in situ hybridization for hh and fgf-Sk1. h, fz5/8 expression. i, AP expression topologies in S. kowalevskii and mouse embryos. Anterior to top. j-k, foxg (j) and rx (k) expression in embryos treated with SU5402. l, m, foxg (l) and rx (m) expression in embryos treated with DMSO. n, expanded apical fgf-Sk1 expression in an embryo injected with fz5/8 siRNA. o, retracted rx expression in an embryo injected with fz5/8 siRNA. p, fgf-Sk1 expression in a siRNA control embryo. q, rx expression in a siRNA control embryo. Insets in o, q show frontal views of uncleared embryos. r, wild type fgf-Sk1 expression. s, fgf-Sk1 expression in descendants of a blastomere injected with β -catenin siRNA. t, merged darkfield/fluorescence images showing clonal descendants of injected cell (green). u, wild-type rx expression. v, rx is not expressed in descendants of a blastomere injected with β -catenin siRNA. w, merged darkfield/fluorescence images showing location of the β-catenin deficient clone (green). x, fgf-Sk1 expression in an embryo injected with hh siRNA. y, fgf-Sk1 expression in a siRNA control embryo. Scale bars = 100 μm in S. kowalevskii, and for mice, 200 μm in (b) and 500 μm in (d). Abbreviations: col, collar; di, diencephalon; hb, hindbrain; mb, midbrain; pr, proboscis; tel, telencephalon; tr, trunk. *, note shh is expressed in the medial ganglionic eminence, near to the ANR.
Wnt antagonists expressed in the ANR and/or telencephalon are also critical for forebrain development in vertebrates2,15,35. To explore potential similarities in Wnt functions between the telencephalon and proboscis, we first suppressed Wnt activity in the developing proboscis by utilizing RNAi to knock down the Wnt receptor fz5/8, which is expressed with a sharp boundary at the proboscis base (Fig. 1h). Fz5/8 siRNA expanded apical identity at the expense of the posterior proboscis as demonstrated by expansion of the apical marker fgf-Sk1 (Fig. 1n, p) and contraction of rx expression to a more posterior domain (Fig. 1o, q). Embryos injected with a scrambled control siRNA were indistinguishable from wild type (Fig. 1p, q), and the effects of fz5/8 knockdown were limited to its endogenous expression domain (Supplementary Fig. 1). Second, we suppressed Wnt signalling in small patches of proboscis ectoderm by injecting β-catenin siRNA into single animal blastomeres at early cleavage stages. β-catenin-deficient clones in the posterior proboscis ectoderm expressed the apical marker fgf-Sk1 (Fig. 1r-t), but not the more posterior proboscis marker rx (Fig. 1u-w), which indicates a transformation of posterior to apical fate in the absence of local Wnt signalling.
Although not a genetic component of the ANR itself, shh is expressed in the nearby medial ganglionic eminence1,36 (see Fig. 2c, d) and interacts with FGFs to regulate telencephalon patterning37. The apical S. kowalevskii proboscis ectoderm expresses hh in a domain partially overlapping fgf-Sk1, with hh more broadly expressed dorsally and laterally (Fig. 1g). Embryos injected with hh siRNA lacked apical fgf-Sk1 expression (Fig. 1x, y) indicating that hh at least partially regulates anterior FGF signalling and apical/ventral patterning. This finding raises the hypothesis that an anterior signalling domain including FGFs, Wnt antagonists, and hh was present in stem deuterostomes and was spatially partitioned during vertebrate evolution.
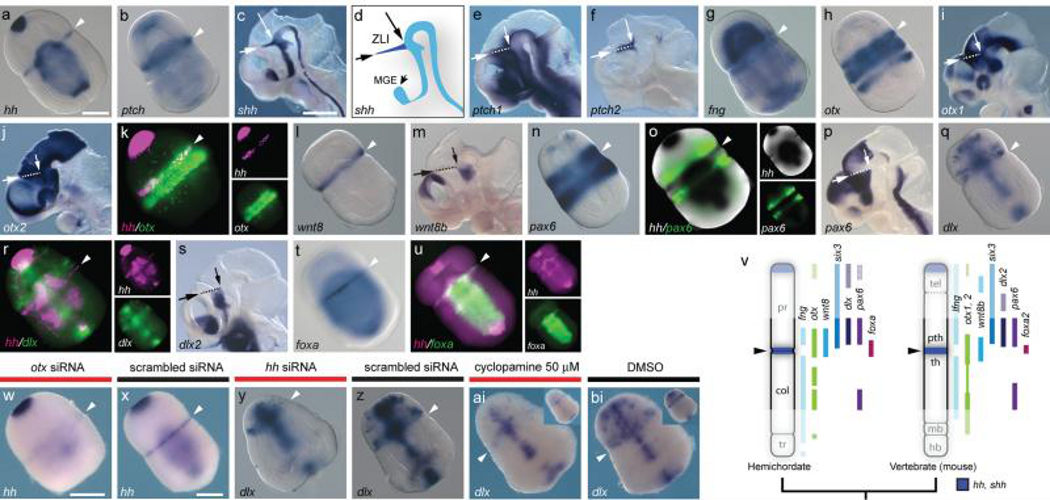
Figure 2. A ZLI-like signalling centre in S. kowalevskii.
a-u, in situ hybridizations for S. kowalevskii and mouse homologs of ZLI and diencephalon markers. Arrowheads mark the proboscis/collar boundary in S. kowalevskii. Mouse images show hemisected heads at E10.5, and dashed lines indicate the ZLI with arrows denoting its extent. a, S. kowalevskii hh expression. b, S. kowalevskii ptch expression (also see Supplementary fig. 2). c, mouse shh expression. d, diagram of shh expression showing ZLI in dark blue. e, mouse ptch1 expression. f, mouse ptch2 expression. g, S. kowalevskii fng expression. h, S. kowalevskii otx expression. i, mouse otx1 expression. j, mouse otx2 expression. k, hh (magenta) and otx (green) are co-expressed at the presumptive proboscis/collar boundary. l, S. kowalevskii wnt8 expression. m, mouse wnt8b expression. n, S. kowalevskii pax6 expression. o, double in situ hybridization showing pax6 expression (fluorescence, green) anterior to hh (colorometric, black). p, mouse pax6 expression. q, S. kowalevskii dlx expression. r, double FISH showing dlx expression (green) in the proboscis base anterior to hh (magenta). s, mouse dlx2 expression. t, S. kowalevskii foxa expression. u, double FISH showing foxa (green) and hh (magenta) expression. v, diagram of AP expression topologies of ZLI and forebrain marker homologs in S. kowalevskii and mice. Six3 expression based on previous data7. Anterior to top. w, x, otx siRNA downregulates hh expression at the proboscis/collar boundary (w) relative to a control siRNA (x). y, z, hh siRNA reduces dlx expression in the proboscis base (y) relative to a scrambled siRNA (z). ai, bi, cyclopamine treatment reduces dlx expression in the proboscis base (ai) relative to a control embryo treated with DMSO (bi). Insets show ventral views. Scale bars = 100 μm in S. kowalevskii embryos and 1 mm in mice. Abbreviations: mge, medial ganglionic eminence; pth, prethalamus; th, thalamus; others as described previously.
A ZLI-like regulatory program in hemichordates
The ZLI is located in the vertebrate diencephalon and specifies the flanking prethalamus and thalamus1,17,18,23. The molecular signature of the ZLI is a narrow, transverse domain of shh expression (Fig. 2C, D) that patterns the mid-diencephalon along its AP axis17,18,23. A homologous hh expression domain is absent in invertebrate chordates, supporting the hypothesis that the ZLI is a vertebrate genetic innovation, possibly associated with forebrain origins10,21-23. Surprisingly, we found that S. kowalevskii hh is expressed in a circumferential ectodermal band at the proboscis/collar boundary (Fig. 2a) in a transcriptional context of expression domains similar to the posterior vertebrate forebrain7. Broader ptch expression indicates that hh signals to adjacent ectoderm (Fig. 2b) similar to the ZLI (Fig. 2e, f). The proboscis/collar boundary region also expresses fng, otx, wnt8 (Fig. 2g-m, v) and six37 in relative spatial domains characteristic of the mid-diencephalon and ZLI. In vertebrates, lfng is expressed broadly in the diencephalon except for the ZLI38, while wnt8b and otx genes mark the ZLI itself in a combinatorial pattern that is unique to this region18 (Fig. 2i, j, m). S. kowalevskii hh is also expressed in a fng-negative, otx and wnt8-positive territory posterior to six37 (fig. 2 g, h, k, l, v). In vertebrates, otx genes are expressed in restricted domains in the mid-diencephalon at the stage when the ZLI forms (Fig. 2i, j), and otx1l and otx2 knockdown eliminates shh expression at the zebrafish ZLI39. Similarly, in S. kowalevskii, hh and otx are co-expressed in the presumptive proboscis/collar boundary ectoderm shortly after gastrulation (Fig. 2k), and otx siRNA knockdown reduced hh expression at the proboscis/collar boundary (Fig. 2w, x).
Homologs of diencephalic markers regulated by shh at the vertebrate ZLI are also expressed in similar topological arrangements at the S. kowalevskii proboscis/collar boundary. In vertebrates, pax6 and dlx2 are expressed anterior to the ZLI in the prethalamus (Fig. 2p, s), where their expression requires shh17,18. Similarly, S. kowalevskii pax6 and dlx are expressed anterior to hh at the proboscis base (Fig. 2n, o, q, r, v). In vertebrates, foxa2 is expressed in the ZLI itself17, and in S. kowalevskii, foxa is expressed at the proboscis/collar boundary (Fig. 2t, u). Targeting hh function in S. kowalevskii by injecting hh siRNA downregulated dlx at the proboscis/collar boundary (Fig. 2y, z). However, these embryos had other strong defects in AP and DV patterning, likely due to additional roles of hh, complicating the assessment of hh function at the proboscis/collar boundary (Supplementary Fig. 2). To reduce early pleiotropic effects, we treated embryos with the Hh signalling inhibitor cyclopamine40 from the end of gastrulation through double-groove stage. 50 μM cyclopamine downregulated dlx at the proboscis/collar boundary with limited effects on midline expression and general morphology (Fig. 2ai, bi), suggesting that Hh signalling from the proboscis/collar boundary regulates dlx. The prominent similarities in expression of ZLI marker homologs in S. kowalevskii and vertebrates and the conserved functions for otx and hh suggest that an ancestral signalling centre homologous to the ZLI was present in early deuterostomes and was independently lost in amphioxus and ascidians.
An IsO-like regulatory program in hemichordates
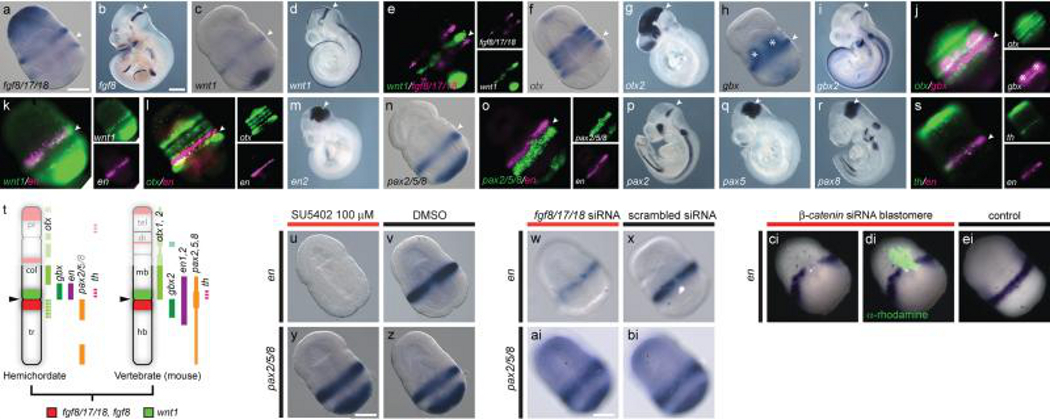
The IsO is located at the midbrain/hindbrain boundary (MHB) and is defined molecularly by abutting domains of FGF8 and WNT11,3, which induce and pattern adjacent neural structures1,3,13,14. The search for a pre-vertebrate IsO has focused on expression patterns of these ligands along with the transcription factors otx, gbx, en, and pax2/5/8, whose combinatorial expression patterns in vertebrates define a molecular territory unique to the MHB (Fig. 3b, d, g, i, m, p-r). In amphioxus, CNS expression of fgf8/17/18 is restricted to the anterior cerebral vesicle19,24, and wnt1 is not expressed in the CNS25. In C. intestinalis, fgf8/17/18 is expressed in larval visceral ganglion where it regulates en and pax2/5/8 to delineate the sensory vesicle and neck regions20 suggesting that at least a partial IsO-like signalling centre predates vertebrates. However, wnt1 is absent in the C. intestinalis genome, making it difficult to infer the full extent of this ancestral centre. In Drosophila melanogaster, otx and gbx orthologs are expressed in patterns similar to the MHB, but the absence of compelling similarities in the expression of fgf8/17/18-related genes and repeated expression of wnt1 and en at parasegmental boundaries12 weakens hypotheses of homology with the vertebrate IsO. To assess the presence of an IsO-like region in hemichordates, we investigated expression patterns for S. kowalevskii homologs of vertebrate MHB markers (Fig. 3a-t). We found that at double-groove stage (36 hrs), wnt1 and fgf8/17/18 are expressed in adjacent ectodermal bands in the anterior trunk with wnt1 expressed anterior to fgf8/17/18 (Fig. 3a, c, e); a topology similar to the vertebrate IsO (Fig. 3b, d). In vertebrates, abutting domains of otx and gbx genes position the IsO3, and opposing otx and gbx domains are also found in protostomes12 and amphioxus8,9 suggesting that this pattern is ancestral to bilaterians. A reassessment of otx and gbx expression patterns in S. kowalevskii7 revealed that they are also expressed in adjacent domains at the collar/trunk coelom boundary with gbx expressed in the ectoderm between the most posterior otx domains (Fig. 3f, h, j). However, the spatial arrangements of otx/wnt1 and gbx/fgf8/17/18 are reversed in S. kowalevskii and vertebrates (Fig. 3t) implying that divergent mechanisms position homologous IsO-like regions in these groups. In vertebrates, pax2,5,8 and en1,2 are co-expressed at the MHB (Fig. 3m, p-r). However, pax2/5/8 and en do not overlap in S. kowalevskii (Fig. 3n, o) or invertebrate chordates9,20, which suggests that regulation of en genes by pax2/5/8 genes is a novel feature of vertebrate development. Beyond similarities in expression of signalling molecules and transcription factors, we found that the catecholaminergic neuron marker th (tyrosine hydroxylase) is co-expressed with en in the S. kowalevskii posterior collar (Fig. 3s), raising the possibility that this neuronal population is homologous to vertebrate midbrain dopaminergic neurons.
Figure 3. An IsO-like signalling centre in S. kowalevskii.
a-s, in situ hybridizations for S. kowalevskii and mouse homologs of MHB markers. Arrowheads mark the S. kowalevskii collar/trunk coelom boundary the mouse IsO. a, S. kowalevskii fgf8/17/18 expression. b, mouse fgf8 expression. c, S. kowalevskii wnt1 expression. d, mouse wnt1 expression. e, double FISH showing S. kowalevskii wnt1 (green) expressed directly anterior to fgf8/17/18 (magenta). f, S. kowalevskii otx expression. g, mouse otx2 expression. h, S. kowalevskii gbx expression. Asterisks denote endodermal domains. i, mouse gbx2 expression. j, double FISH for S. kowalevskii otx (green) and gbx (magenta). k, double FISH for S. kowalevskii wnt1 (green) and en (magenta). l, double FISH for S. kowalevskii en and otx. m, mouse en2 expression. n, S. kowalevskii pax2/5/8 expression. o, double FISH showing S. kowalevskii pax2/5/8 and en expression. p-r, expression of mouse pax2 (p), pax5 (q), and pax8 (r). s, double FISH for S. kowalevskii th (green) and en (magenta). t, summary of AP expression topologies in hemichordates and mice. Anterior to top. u, en expression is reduced in an embryo treated with SU5402. v, en expression in a DMSO-treated control embryo. w, en expression in an embryo injected with fgf8/17/18 siRNA. x, en expression in an embryo injected with a control siRNA. y, pax2/5/8 expression in an embryo treated with SU5402. z, pax2/5/8 expression in a DMSO-treated control embryo. ai, pax2/5/8 expression in an embryo injected with fgf8/17/18 siRNA. bi, pax2/5/8 expression in an embryo injected with a control siRNA. ci, en is not expressed in descendants of a blastomere injected with β -catenin siRNA. di, merged darkfield/fluorescence images showing location of the β-catenin deficient clone (green). ei, wild-type en expression. Scale bars = 100 μm in S. kowalevskii embryos and 1 mm in mice. Abbreviations as described previously.
Functional assays further support the deep deuterostome ancestry of an MHB-like genetic module. In vertebrates, fgf8 mediates organizing abilities of the IsO and maintains expression of other MHB markers3,13,14. To assess similar requirements for FGFs in hemichordates, we first treated embryos with 100 μM SU5402 at the end of gastrulation to suppress FGF signalling without perturbing earlier patterning events. En expression was strongly reduced in SU5402 treated embryos (Fig. 3u, v), while pax2/5/8 was unaffected (Fig. y, z) compared to DMSO treated embryos. To specifically test for a role of fgf8/17/18, we injected fertilized oocytes with fgf8/17/18 siRNA. Fgf8/17/18 knockdown reduced en expression (Fig. 3w, x), but had no effect on pax2/5/8 (Fig. 3ai, bi) relative to embryos injected with a scrambled control siRNA. The absence of any effect on pax2/5/8 expression in these experiments highlights differences in gene regulation downstream of fgf8/17/18 between hemichordates and chordates3,20.
In vertebrates, wnt1 is required to maintain en expression at the MHB3,41. To assess local Wnt functions at the S. kowalevskii collar/trunk boundary, we injected single blastomeres with siRNA at early cleavage stages to suppress Wnt signalling in small patches of collar/trunk ectoderm. Clonal descendants of injected blastomeres failed to express en (Fig. 3ci-ei), demonstrating a similar role for Wnts in regulating en genes at the MHB and collar/trunk boundary.
Discussion
Extensive similarities in expression patterns of signalling centre markers and conserved functions for FGF, Wnt, and Hh signals in S. kowalevskii and vertebrates provide compelling evidence that signalling centres homologous to the ANR, ZLI, and IsO were parts of an ancient genetic regulatory scaffold that predate the morphological innovations of vertebrates (Fig. 4). Therefore, assembly of these genetic networks did not trigger morphological novelties of the brain. Rather, early vertebrate brain evolution involved modifying and elaborating existing signalling centres to pattern novel structures within a highly conserved gene regulatory framework for AP ectodermal patterning7.
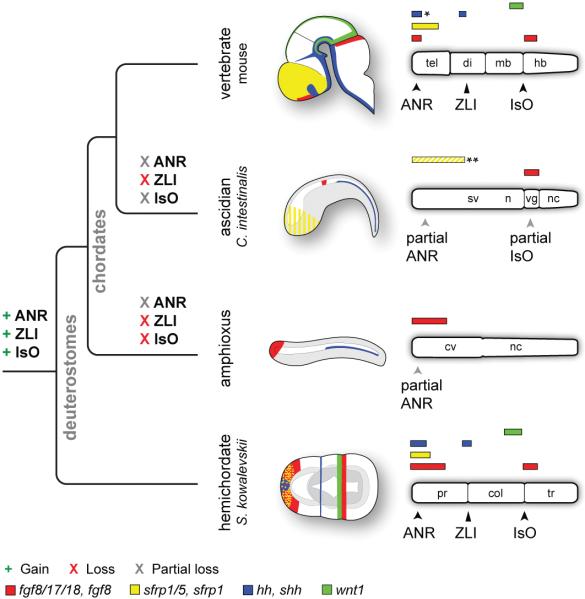
Figure 4. Evolutionary gain and loss of ANR, ZLI, and IsO-like genetic programs.
Schematic diagrams depicting expression of fgf8, sfrp1, shh, and wnt1 homologs in the mouse brain and ectoderm of C. intestinalis, amphioxus, and S. kowalevskii. Embryos are oriented with anterior to left and dorsal to top. Bar diagrams are oriented with anterior to the left. Diagrams depict only expression domains related to signalling components of vertebrate CNS signalling centres. Abbreviations: cv, cerebral vesicle; n, neck; nc, nerve cord; sv, sensory vesicle; vg, visceral ganglion; others as described previously. Diagrams not to scale. *Note that shh is expressed in the medial ganglionic eminence, adjacent to the ANR.**Sfrp1/5 is expressed in the C. intestinalis anterior ectoderm from 64-cells up to neurulation, but is then downregulated in the anterior ectoderm and CNS (depicted as yellow stripes).
Widespread losses of signalling centre components in invertebrate chordates and the unresolved nature of the ancestral deuterostome nervous system present a challenge for inferring when the ANR, ZLI, and IsO genetic circuits were first deployed to regulate CNS patterning specifically, and to what extent these integrations could have been associated with origins of vertebrate novelties. In S. kowalevskii, signalling centre programs are deployed at stages when the nervous system is still circumferentially organized7 and in body regions that have been described as containing components of the adult peripheral, rather than central, nervous system in the hemichordate Ptychodera flava42. Notably, similar to their roles in the vertebrate brain, the ZLI and IsO-like signalling centres are located at morphological boundaries in S. kowalevskii, suggesting ancient roles in demarcating ectodermal divisions. We propose that rather than having evolved for CNS patterning, the most ancient role for signalling centres was in general body plan regionalization.
This work also highlights that basal chordates have not retained all ancestral chordate characters and have undergone substantial independent evolutionary changes. This is generally accepted for urochordates, but based on available data, cephalochordates are often considered to be the most informative extant group for reconstructing ancestral chordate characters8,9,24. Our data provide clear evidence that secondary losses of complex developmental mechanisms have occurred in cephalochordates, and that overlooking this possibility can inflate the numbers of putative vertebrate novelties. Although difficult to test, divergence of ANR, ZLI, and IsO-like genetic programs in extant invertebrate chordate lineages may have been associated with the loss or modification of anterior ectodermal structures of the CNS and head. Based on these new observations, reanalyses of early chordate fossils accommodating scenarios of greater anterior complexity in stem chordates may be informative. However, in the absence of additional fossil data, predictions of the morphological consequences of developmental genetic losses based on molecular data alone are problematic.
This study provides a compelling example of the challenges associated with identifying key developmental genetic innovations responsible for morphological innovations at macroevolutionary scales. The unexpected presence of ANR, ZLI, and IsO-like programs in S. kowalevskii highlights the point that basing outgroup choice primarily on morphological criteria can lead to erroneous conclusions about links between morphological and developmental genetic characters: although by almost all morphological criteria amphioxus shares more similarities with vertebrates than do hemichordates, our data support the hypothesis that in certain cases hemichordates will be a more informative group than basal chordates for reconstructing stem chordate characters and understanding the origins of vertebrate developmental genetic processes. Additional data from protostomes, especially lophotrochozoans, will be required to assess whether the ANR, ZLI, and IsO genetic programs are unique deuterostome features or have even deeper bilaterian origins.
Our findings stress the importance of broad phylogenetic sampling, including morphologically divergent outgroups, to identify gene regulatory innovations responsible for evolutionary changes in body plans. With the expanding use of novel model organisms, it appears that secondary losses of complex developmental regulatory characters may occur commonly43-45 with as yet poorly understood consequences for morphological evolution. Conversely, the presence of complex developmental modules that regulate morphologically disparate structures in distantly related lineages suggests a loose coupling between morphological and gene regulatory evolution over macroevolutionary timescales, and highlights the difficulties of reconstructing ancestral morphological characters from molecular genetic data.
Methods
Embryo procurement
Gravid S. kowalevskii adults were collected from the intertidal near Woods Hole, MA and maintained in flow-through seawater tables at the Marine Biological Laboratory. Spawning and embryo rearing used established techniques7. Mouse embryos were obtained using standard protocols approved by the University of Chicago Institutional Animal Care and Use Committee.
Experimental manipulations
SU5402 (Calbiochem) and cyclopamine (Calbiochem) were resuspended in minimal volumes of DMSO and diluted in 0.2 μM filtered seawater for treatments. Embryos were raised at room temperature (RT). Control embryos were obtained from the same batches and raised in an equivalent concentration of DMSO. siRNA microinjections were performed as described previously27,29. siRNAs were resuspended at 100 mM and injected at 1/10 concentration in a suspension buffer27 including either calcein or lysinated 10,000 MW tetramethylrhodamine dextran (Molecular Probes). Unsuccessfully injected embryos, or embryos with abnormal cleavage patterns, were discarded. siRNAs were ordered from Ambion or Qiagen, and sequences are provided in Supplementary table 1.
In situ hybridizations
Colorimetric in situ hybridizations were performed as described previously7,31. Clonal descendants of injected blastomeres were identified after in situ hybridization using α-tetramethylrhodamine (Molecular Probes) immunofluorescence. For fluorescence in situ hybridization (FISH), pre-hybridization steps were performed using our standard protocol. For double FISH, embryos were simultaneously hybridized with antisense RNA probes for both genes labeled separately with digoxigenin-11-UTP (Roche) or fluorescein-12-UTP (Roche). Due to reduced sensitivity of FISH, we typically utilized two to three times more probe compared to colorimetric methods. Post-hybridization washes and blocking steps utilized our standard protocol. Embryos were then incubated in 1:200 α-digoxigenin-POD or α-fluorescein-POD antibodies (Roche) in blocking solution (2% Roche blocking reagent, 1x MAB) for four hours at RT on a rotary shaker. Embryos were washed 4 times in 1x MAB for 30 minutes and once for 1 hour at RT, or overnight at 4°C. Embryos were then washed in 0.1 M imidazole (Sigma) in 1x PBS for 10 minutes at RT and probes detected using a Tyramide Signal Amplification (TSA) Plus kit (Perkin-Elmer). Embryos were rinsed in 1x amplification diluent and incubated in cyanine-3 or cyanine-5 tyramide diluted 1:50 in 1x amplification diluent for 45 minutes on a rotary shaker at RT. Embryos were washed three times in detergent solution (1% triton x-100, 1% SDS, 0.5% sodium deoxycholate, 50 mM Tris-HCL pH 8.0, 1 mM EDTA pH 8.0, 150 mM sodium chloride) for 20 minutes at 60°C. If detecting a second gene, embryos were washed once for 20 minutes in solution X (50% formamide, 2x SSC, 1% SDS) at 60°C. Embryos were then washed three times in 1x MAB for five minutes and returned to the blocking step.
For genes with low signal using standard FISH methods, we performed an additional DNP amplification step. After primary antibody incubation and 1x MAB washes, embryos were incubated in DNP amplification reagent diluted 1:50 in 1x amplification diluent (Perkin-Elmer) for 5-10 minutes at RT on a rotary shaker. Embryos were washed 4 times in 1x MAB for 30 minutes and once for 1 hour at RT, re-blocked for one hour, and incubated in α-DNP-HRP antibody (Perkin-Elmer) diluted 1:200 in blocking solution for four hours at RT on a rotary shaker. Embryos were washed 4 times in 1x MAB for 30 minutes and once for 1 hour at RT. Expression was then detected using a TSA Plus kit (Perkin-Elmer) as described above.
Photography
Embryos were photographed using Zeiss Axiocam MRc5 or MRm cameras on Zeiss AxioImager.Z1 or Discovery.V12 microscopes. For optical clearing, S. kowalevskii embryos were dehydrated into methanol, cleared using Murray's clearing reagent (1:2 benzyl alcohol to benzyl benzoate), and mounted in permount (Fisher Scientific). Cleared embryos were imaged under DIC optics, and uncleared embryos were photographed in 1x PBS or 1x MAB on agarose-coated dishes. Images were acquired using Zeiss Axiovision 4.8 software and adjusted for color balance and/or levels or gamma using Zeiss Axiovision 4.8 or Adobe Photoshop CS3-5 software. Images of experimental and control embryos were processed utilizing the same parameters.
Gene identification
S. kowalevskii homologs of vertebrate genes were identified in an EST library screen46, Amino acid sequences were aligned using ClustalW47, and putative homologies were confirmed by constructing gene trees using MrBayes 3.1.248,49 with parameters optimized for each gene (Supplementary Figs. 3, 4). See Supplementary Table 2 for accession numbers.
Supplementary Material
Acknowledgements
The authors would like to acknowledge John Gerhart and Marc Kirschner for assistance and support, Robert Freeman for bioinformatics assistance, Ellyn Farrelly, Mark Terasaki, and Sebastien Darras for technical guidance, the members of the Lowe lab for stimulating discussions, and Gregory Wray and John Gerhart for comments on drafts of the manuscript. We also thank the staff of the Marine Biological Laboratory, the Waquoit Bay National Estuarine Research Reserve, Carl Zeiss, and Nikon, USA for assistance during our field season. This work was funded by the Searle Kinship Foundation (C.J.L), Brain Research Foundation (C.J.L.), NSF grant 1049106 (C.J.L), National Institutes of Health grant R01 HD42330 (E.A.G.), and The University of Chicago Hinds Fund (A.M.P). A.M.P. was supported by a Marine Biological Laboratory Frank R. Lillie Fellowship, National Institute of Child Health and Development institutional training grant 1T32HD055164-01A1, and National Institute of Neurological Disorders and Stroke predoctoral fellowship 1F31NS074738-01A1.
Footnotes
Author contributions
A.M.P., C.J.L., and J.A. conceived the project. A.M.P. and C.J.L. performed the hemichordate experiments and wrote the paper. E.E.M. and S.A. performed mouse experiments, and E.A.G. edited the paper. All authors discussed and commented on the data.
Author information
S. kowalevskii gene sequences have been deposited in Genbank, and accession numbers are provided in Supplementary Table 2. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Echevarria D, Vieira C, Gimeno L, Martinez S. Neuroepithelial secondary organizers and cell fate specification in the developing brain. Brain Res Brain Res Rev. 2003;43:179–191. doi: 10.1016/j.brainresrev.2003.08.002. doi:S0165017303002078 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. doi:S1534580704000279 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. doi:10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- 4.Wicht H, Lacalli TC. The nervous system of amphioxus: structure, development, and evolutionary significance. Canadian Journal of Zoology. 2005;83:122–150. doi:10.1139/z04-163. [Google Scholar]
- 5.Lacalli TC. Prospective protochordate homologs of vertebrate midbrain and MHB, with some thoughts on MHB origins. International journal of biological sciences. 2006;2:104–109. doi: 10.7150/ijbs.2.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meinertzhagen IA, Lemaire P, Okamura Y. The neurobiology of the ascidian tadpole larva: recent developments in an ancient chordate. Annu Rev Neurosci. 2004;27:453–485. doi: 10.1146/annurev.neuro.27.070203.144255. doi:10.1146/annurev.neuro.27.070203.144255. [DOI] [PubMed] [Google Scholar]
- 7.Lowe CJ, et al. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. doi:S0092867403004690 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Holland LZ, Short S. Gene duplication, co-option and recruitment during the origin of the vertebrate brain from the invertebrate chordate brain. Brain Behav Evol. 2008;72:91–105. doi: 10.1159/000151470. doi:000151470 [pii] 10.1159/000151470. [DOI] [PubMed] [Google Scholar]
- 9.Holland LZ. Chordate roots of the vertebrate nervous system: expanding the molecular toolkit. Nat Rev Neurosci. 2009;10:736–746. doi: 10.1038/nrn2703. doi:nrn2703 [pii] 10.1038/nrn2703. [DOI] [PubMed] [Google Scholar]
- 10.Irimia M, et al. Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the Zona Limitans Intrathalamica (ZLI) brain organizer. Evodevo. 2010;1:7. doi: 10.1186/2041-9139-1-7. doi:2041-9139-1-7 [pii] 10.1186/2041-9139-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomer R, Denes AS, Tessmar-Raible K, Arendt D. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell. 2010;142:800–809. doi: 10.1016/j.cell.2010.07.043. doi:S0092-8674(10)00891-3 [pii] 10.1016/j.cell.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 12.Urbach R. A procephalic territory in Drosophila exhibiting similarities and dissimilarities compared to the vertebrate midbrain/hindbrain boundary region. Neural Dev. 2007;2:23. doi: 10.1186/1749-8104-2-23. doi:1749-8104-2-23 [pii] 10.1186/1749-8104-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crossley PH, Martinez S, Martin GR. Midbrain development induced by FGF8 in the chick embryo. Nature. 1996;380:66–68. doi: 10.1038/380066a0. doi:10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- 14.Reifers F, et al. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- 15.Houart C, et al. Establishment of the telencephalon during gastrulation by local antagonism of Wnt signaling. Neuron. 2002;35:255–265. doi: 10.1016/s0896-6273(02)00751-1. doi:S0896627302007511 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Paek H, Gutin G, Hebert JM. FGF signaling is strictly required to maintain early telencephalic precursor cell survival. Development. 2009;136:2457–2465. doi: 10.1242/dev.032656. doi:136/14/2457 [pii] 10.1242/dev.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiecker C, Lumsden A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat Neurosci. 2004;7:1242–1249. doi: 10.1038/nn1338. doi:nn1338 [pii] 10.1038/nn1338. [DOI] [PubMed] [Google Scholar]
- 18.Scholpp S, Wolf O, Brand M, Lumsden A. Hedgehog signalling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development. 2006;133:855–864. doi: 10.1242/dev.02248. doi:dev.02248 [pii] 10.1242/dev.02248. [DOI] [PubMed] [Google Scholar]
- 19.Meulemans D, Bronner-Fraser M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS One. 2007;2:e787. doi: 10.1371/journal.pone.0000787. doi:10.1371/journal.pone.0000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai KS, Stolfi A, Levine M, Satou Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development. 2009;136:285–293. doi: 10.1242/dev.026419. doi:dev.026419 [pii] 10.1242/dev.026419. [DOI] [PubMed] [Google Scholar]
- 21.Shimeld SM. The evolution of the hedgehog gene family in chordates: insights from amphioxus hedgehog. Dev Genes Evol. 1999;209:40–47. doi: 10.1007/s004270050225. [DOI] [PubMed] [Google Scholar]
- 22.Takatori N, Satou Y, Satoh N. Expression of hedgehog genes in Ciona intestinalis embryos. Mech Dev. 2002;116:235–238. doi: 10.1016/s0925-4773(02)00150-8. doi:S0925477302001508 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Scholpp S, Lumsden A. Building a bridal chamber: development of the thalamus. Trends Neurosci. 2010;33:373–380. doi: 10.1016/j.tins.2010.05.003. doi:S0166-2236(10)00077-9 [pii] 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertrand S, et al. Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1014235108. doi:10.1073/pnas.1014235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holland LZ, Holland NN, Schubert M. Developmental expression of AmphiWnt1, an amphioxus gene in the Wnt1/wingless subfamily. Dev Genes Evol. 2000;210:522–524. doi: 10.1007/s004270000089. doi:10.1007/s004270050342. [DOI] [PubMed] [Google Scholar]
- 26.Bourlat SJ, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. doi:nature05241 [pii] 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- 27.Darras S, Gerhart J, Terasaki M, Kirschner M, Lowe CJ. {beta}-Catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii. Development. 2011;138:959–970. doi: 10.1242/dev.059493. doi:138/5/959 [pii] 10.1242/dev.059493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillis JA, Fritzenwanker JH, Lowe CJ. A stem-deuterostome origin of the vertebrate pharyngeal transcriptional network. Proceedings. Biological sciences / The Royal Society. 2011 doi: 10.1098/rspb.2011.0599. doi:10.1098/rspb.2011.0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe CJ, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. doi:06-PLBI-RA-0385R2 [pii] 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimamura K, Rubenstein JL. Inductive interactions direct early regionalization of the mouse forebrain. Development. 1997;124:2709–2718. doi: 10.1242/dev.124.14.2709. [DOI] [PubMed] [Google Scholar]
- 31.Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. doi:10.1126/science.10642521064252 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Walshe J, Mason I. Unique and combinatorial functions of Fgf3 and Fgf8 during zebrafish forebrain development. Development. 2003;130:4337–4349. doi: 10.1242/dev.00660. [DOI] [PubMed] [Google Scholar]
- 33.Garel S, Huffman KJ, Rubenstein JL. Molecular regionalization of the neocortex is disrupted in Fgf8 hypomorphic mutants. Development. 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- 34.Mohammadi M, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 35.Lagutin OV, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. doi:10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crossley PH, Martinez S, Ohkubo Y, Rubenstein JL. Coordinate expression of Fgf8, Otx2, Bmp4, and Shh in the rostral prosencephalon during development of the telencephalic and optic vesicles. Neuroscience. 2001;108:183–206. doi: 10.1016/s0306-4522(01)00411-0. doi:S0306-4522(01)00411-0 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Hebert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. doi:10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeltser LM, Larsen CW, Lumsden A. A new developmental compartment in the forebrain regulated by Lunatic fringe. Nat Neurosci. 2001;4:683–684. doi: 10.1038/89455. doi:10.1038/89455 [pii] [DOI] [PubMed] [Google Scholar]
- 39.Scholpp S, et al. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development. 2007;134:3167–3176. doi: 10.1242/dev.001461. doi:dev.001461 [pii] 10.1242/dev.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. doi:10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMahon AP, Joyner AL, Bradley A, McMahon JA. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. doi:0092-8674(92)90222-X [pii] [DOI] [PubMed] [Google Scholar]
- 42.Nomaksteinsky M, et al. Centralization of the deuterostome nervous system predates chordates. Curr Biol. 2009;19:1264–1269. doi: 10.1016/j.cub.2009.05.063. doi:S0960-9822(09)01234-2 [pii] 10.1016/j.cub.2009.05.063. [DOI] [PubMed] [Google Scholar]
- 43.Gavino MA, Reddien PW. A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Current biology : CB. 2011;21:294–299. doi: 10.1016/j.cub.2011.01.017. doi:10.1016/j.cub.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grande C, Patel NH. Nodal signalling is involved in left-right asymmetry in snails. Nature. 2009;457:1007–1011. doi: 10.1038/nature07603. doi:10.1038/nature07603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campo-Paysaa F, Marletaz F, Laudet V, Schubert M. Retinoic acid signaling in development: tissue-specific functions and evolutionary origins. Genesis. 2008;46:640–656. doi: 10.1002/dvg.20444. doi:10.1002/dvg.20444. [DOI] [PubMed] [Google Scholar]
- 46.Freeman RM, Jr., et al. cDNA sequences for transcription factors and signaling proteins of the hemichordate Saccoglossus kowalevskii: efficacy of the expressed sequence tag (EST) approach for evolutionary and developmental studies of a new organism. Biol Bull. 2008;214:284–302. doi: 10.2307/25470670. doi:214/3/284 [pii] [DOI] [PubMed] [Google Scholar]
- 47.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. doi:btm404 [pii] 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 48.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 49.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.