Abstract
A mouse- and human-brain-abundant, nuclear factor (NF)-κB-regulated, micro RNA-146a (miRNA-146a) is an important modulator of the innate immune response and inflammatory signaling in specific immunological and brain cell types. Levels of miRNA-146a are induced in human brain cells challenged with at least five different species of single- or double-stranded DNA or RNA neurotrophic viruses, suggesting a broad role for miRNA-146a in the brain’s innate immune response and antiviral immunity. Upregulated miRNA-146a is also observed in pro-inflammatory cytokine-, Aβ42 peptide- and neurotoxic metal-induced, oxidatively stressed human neuronal-glial primary cell cocultures, in murine scrapie and in Alzheimer’s disease (AD) brain. In AD, miRNA-146a levels are found to progressively increase with disease severity and co-localize to brain regions enriched in inflammatory neuropathology. This study provides evidence of upregulation of miRNA-146a in extremely rare (incidence 1–10 per 100 million) human prion-based neurodegenerative disorders, including sporadic Creutzfeldt–Jakob disease (sCJD) and Gerstmann–Straussler-Scheinker syndrome (GSS). The findings suggest that an upregulated miRNA-146a may be integral to innate immune or inflammatory brain cell responses in prion-mediated infections and to progressive and irreversible neurodegeneration of both the murine and human brain.
Micro RNA (miRNA) are small, noncoding regulatory molecules that are important epigenetic, posttranscriptional regulators of messenger RNA (mRNA) complexity. Their major mode of action is to interact, via base-pair complementarity, with the 3′ untranslated region (3′UTR) of their target messenger RNA (mRNA), and in doing so decrease the capability of that specific mRNA to be expressed (Barbato et al. 2009; De Smaele et al. 2010; Li et al. 2010a; Provost 2010; Roshan et al. 2009; Saudstad 2010). Of the approximately 1250 known miRNA in human tissues (miR-Base version 16; Cambridge, UK), the central nervous system (CNS) utilizes only a discrete subset of these, numbering probably less than 100 different major miRNA species (Burmistrova et al. 2007; Lukiw 2007; Sethi and Lukiw 2010). A 22-nucleotide mouse-and human-brain abundant miRNA-146a that are identical in nucleotide sequence, originally described as a mediator of inflammatory signaling in human monocytes (Taganov et al., 2006) has been specifically associated with (a) upregulation of inflammatory neurodegeneration in cortical and hippocampal regions in AD and in transgenic murine models of AD (Lukiw et al., 2008; Li et al., 2010); (b) prion-induced neuro-degeneration (Saba et al., 2008); (c) the down-regulation of interleukin-1 receptor associated kinase-1 (IRAK-1) in endotoxin-and cytokine-challenged human monocytes and astroglial cells (Taganov et al., 2006; Cui et al., 2010); and (d) the down-regulation of IRAK-1 and complement factor H (CFH), an important repressor of complement signaling in AD and in AD brain cell models (Lukiw et al., 2008) (Figure1). This inducible miRNA-146a was found to be further upregulated in pro-inflammatory cytokine-, amyloid beta 42 amino acid (Aβ42) peptide-, oxidation-, or neurotoxic-metal-stressed primary human neuronal–glial coculture cell models of neurodegenerative disease (Alexandrov et al. 2005; Cui et al. 2010; Lukiw et al. 2008; Pogue et al. 2009). This study expanded and advanced our investigations into miRNA-146a–mediated signaling in murine and human prion disease to further understand the involvement of this brain-enriched, stress-induced miRNA-146a in the molecular-genetic mechanism that drives the process of prion-mediated inflammatory neurodegeneration.
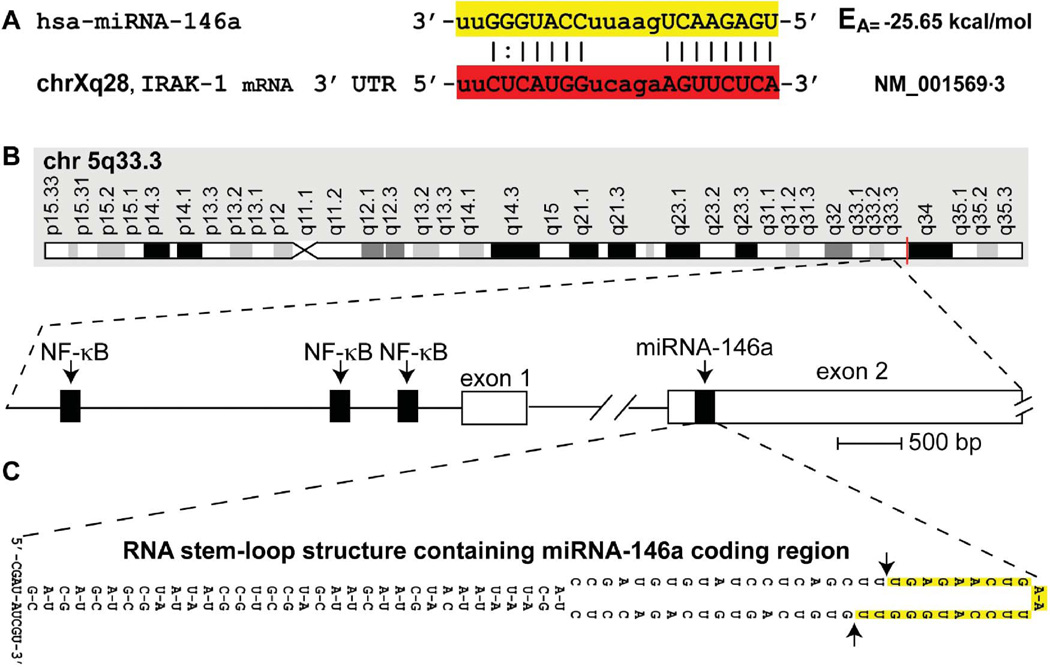
FIGURE 1.
(A) Homo sapiens micro RNA-146a (hsa-miRNA-146a) is a 22-nucleotide small RNA (highlighted in yellow) abundant in mouse and human immune cells and the limbic system, with known mRNA 3′ UTR targets that include the interleukin-1 receptor associated kinase 1 (IRAK-1; highlighted in red); uppercase ribonucleotides involved in fully (|) or partially (:) complementary hydrogen bonding (Cui et al. 2010; Taganov et al. 2006); chromosome location of the human IRAK-1 gene (chrXq28) and Genbank accession (NM_01569.3) indicated; EA = predicted energy of association for miRNA-mRNA interaction (Sethi and Lukiw 2009; miRBASE, Cambridge, UK); the mouse and human miRNA-146a are identical in sequence (Li et al. 2010a; 2010b; Sethi and Lukiw 2009). (B) Structural features of the miRNA-146a encoding DNA locus at chromosome 5q33.3 and details of the NF-κB-sensitive miRNA146a gene showing three upstream (5′) regulatory NF-КB binding sites; miRNA-146a transcription is highly NF-КB sensitive (Cui et al. 2010; Lukiw et al. 2008; Taganov et al. 2006). (C) The pre-miRNA-146a transcribed from the miRNA146a locus (chr 5q33.3) has strong potential to form a highly stable 35-base-pair stem, 60-nucleotide loop RNA structure (stem-loop EA= −49.5 kcal/mol); other predicted secondary structures of alternate stem-loop configurations are possible and the 5′ and 3′ ends of pre-miRNA-146a may be significantly extended. In several preferential EA models, such as the one depicted in (C), the stem-loop structure containing the miRNA-146a sequence is consistently located in the very distal end of the predicted loop (highlighted in yellow; delineated by arrows), after which Dicer (RNase III) processing of this pre-miRNA-146a (Yokota 2009) yields the mature miRNA-146a highlighted in yellow in (A) (color figure available online).
MATERIALS AND METHODS
Mouse and Human Brain Total RNA
Mouse and human brain samples used in these studies were selected from archived tissue or total RNA extract sources at the Louisiana State University (LSU) Neuroscience Center, New Orleans, the University of California at Irvine, and the Oregon Health Sciences Center, Portland, OR. Human brain RNA tissues extracts were used in accordance with the institutional review board and biosafety guidelines at the LSU Health Sciences Center and donor institutions (Cui et al. 2008; Lukiw 2007). Archived total RNA isolated from the short incubation model of murine scrapie (strain 139A in Compton White [CW] mice) used here have been previously analyzed for chromatin structural aberrations (McLachlan et al. 1986). Due to their extreme rarity and limited availability, only sCJD and GSS small RNA, and no brain tissues protein extracts, were available for the current investigation. The current study focused on human brain 5S RNA, miRNA-125b, and miRNA-146a abundance; miRNA-125b and miRNA-146a are brain-enriched miRNA known to be associated with the regulation of gliosis, glial cell proliferation, and the innate immune and inflammatory response (Cui et al. 2010; Hill et al. 2009; Lukiw et al. 2008; Pogue et al. 2009 2010). To assess relative miRNA-125b and miRNA-146a abundance levels, all brain samples were from the murine brain cortex or human temporal lobe neocortex, and were age-matched to neurologically normal murine cortex or human neocortex controls. The average ages of sCJD (n = 3) and the controls (n = 3) were 65 ± 9 yr and the mean age of the GSS (n = 2) and controls (n = 6) was 61 yr. There were no significant differences in total RNA yield or purity between any murine or human control, or prion-affected brain samples (Sethi and Lukiw 2009).
Small RNA Isolation, Northern Dot Blot Analysis, DNA and miRNA Arrays
A guanidine isothiocyanate- and silica gel-based membrane total RNA purification system and miRNA isolation kit (PureLink, Invitrogen, Carlsbad, CA) were used to isolate total small RNA (5S RNA, tRNA and miRNA), and the concentration and quality of these total small RNA were determined using RNA 6000 Nano LabChips and a 2100 Bioanalyzer (Caliper Technologies, Mountainview, CA; Agilent Technologies, Palo Alto, CA). Mammalian brain tissues typically yield about 1 µg of total RNA per mg wet weight of tissue (Cui et al. 2010; Lukiw et al. 1992; Pogue and Lukiw 2004; Sethi and Lukiw 2009). Total small RNA was either (a) analyzed using Cy3/Cy5 fluorescent labeling and human miRNA array panels (containing 856 individual human miRNAs and 56 controls) as previously described (LC Sciences, Houston, TX; Pogue et al. 2010), or (b) small RNA samples (25 µg) were run out on 15% Tris–borate–EDTA (ethylenediamine tetraactic acid)–urea polyacrylamide denaturing gels (TBE-urea; Invitrogen, Carlsbad, CA), and after ethidium bromide staining, total miRNA species (<25 nucleotides) were excised and end-labeled using [γ-32P]-δATP (6000 Ci/mmol; Amersham-GE Healthcare) according to the manufacturer’s protocols (Invitrogen; Cui et al. 2008; Li et al. 2010b; Lukiw 2007; Pogue et al. 2010; Riazanskaia et al. 2002) (Figure 2). Total radiolabeled small RNA from control or prion-associated brain tissues were then probed against miRNA dot-blot panels containing fixed amounts of synthetic human 5SRNA and miRNA-146a targets (Cui et al. 2005; Hill et al. 2009) (Figure 2).
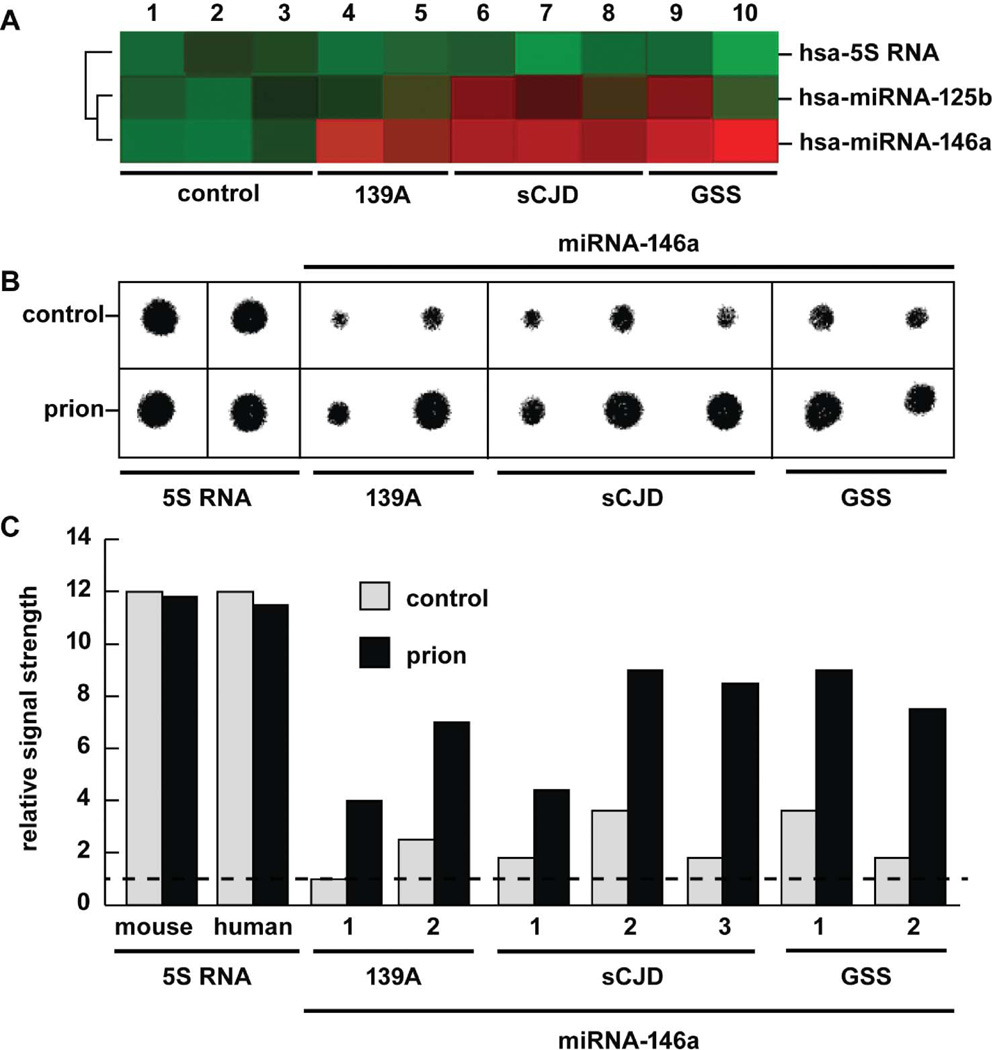
FIGURE 2.
Abundance of hsa-miRNA-125b and/or hsa-miRNA-146a in comparison to an internal control hsa-5S RNA marker in murine scrapie 139a, sporadic Creutzfeldt-Jakob disease (sCJD), and Gerstmann-Straussler-Scheinker syndrome (GSS) compared to age-matched controls (control) in the same brain region as analyzed by (A) fluorescent miRNA array cluster analysis (LC Sciences, Houston TX) and (B) concentrated Northern dot-blot analysis of 5S RNA and miRNA-146a using high-specific-activity radiolabeled probes (Cui et al. 2005; Sethi and Lukiw 2009); (C) quantified results from Northern dot-blot analysis; dashed horizontal line indicates control 139A levels relative to 5S RNA for ease of comparison. In (A) panel 1 is the age-matched control for murine 139a (n= 2; lanes 4 and 5); lanes 2 and 3 are the mean age-matched control for the individual cases for sCJD (n = 3; lanes 6–8) and GSS (n= 2, lanes 9 and 10), respectively; by convention, green colors indicate no change and red colors indicate upregulation (LC Sciences; Lukiw et al. 2008). Squared black lines to the left of panel (A) indicate that a mean signal intensity comparison between hsa-5S–RNAand hsa-miRNA-125b yielded a significance of p > .07 (ANOVA), and between 5S–RNA and hsa-miRNA-146a yielded a p< .01 (ANOVA). miRNA-125b was found to be significantly upregulated in one of two GSS cases and two of three CJD cases; miRNA-146a was found to be consistently elevated in all sCJD or GSS cases using miRNA array analysis; and this was further confirmed using quantitative Northern dot blot analysis (B and C). The 22 nucleotide hsa-5S–RNA probe was derived from the 5′ end of the 107 nucleotide human 5S ribosomal RNA (5S RNA) and in panel (B) was loaded at 1/10 the concentration of miRNA-146a (Sethi and Lukiw 2009) (color figure available online).
Using a vacuum dot blot apparatus human brain miRNA of interest including miRNA-125b, miRNA-146a, human 5S ribosomal RNA (5S RNA), or other miRNA controls were spotted onto GeneScreen Plus nylon membranes using a Biomek 2000 lab automation workstation (Beckman, Fullerton, CA). Blots were cross-linked, baked, hybridized, and probed using the radiolabeled probes described earlier according to the manufacturer’s protocol (NEN Research Products, Boston; Hill et al. 2001; Lukiw et al. 1998; 2008). As a preliminary screen and for normalization purposes, Northern dot blot arrays were probed with total labeled miRNA isolated from control human brain tissues (Cui et al. 2010). Specific miRNA showing the strongest hybridization signals were studied further. This [γ-32P]-δATP radiolabel-based miRNA analytical assay was found to be at least as sensitive as fluorometrically based mRNA and miRNA detection (Alexandrov et al. 2005; Cui et al. 2005; Sethi and Lukiw 2009).
Data Analysis and Interpretation
In Northern dot blots 5S RNA, an abundant 107-ribonucleotide marker, was used as an internal small RNA abundance control and relative miRNA signal strengths were quantified against 5S RNA in each sample using data-acquisition software provided with a GS250 molecular imager (Bio-Rad, Hercules, CA). MiRNA ribonucleotide sequence analysis was performed using visual inspection, miR-BASE (http://microrna.sanger.ac.uk/sequences), the DBD transcription factor database search (Medical Research Council, Cambridge, UK), and/or GeneSpring algorithms (GeneSpring Redwood City, CA). Graphic presentations were performed using Excel algorithms (Microsoft, Seattle, WA) and Adobe Photoshop 6.0 (Adobe Systems, San Jose CA). Statistical significance was analyzed using a two-way factorial analysis of variance (p, ANOVA; SAS Institute, Cary, NC); a p < .05 was deemed as being statistically significant.
RESULTS
Figure 1 shows the sequence of the 22 nucleotide mature miRNA-146a and a preferred miRNA-mRNA highly stable interaction involving miRNA-146a and the 3′ untranslated region (3′ UTR) of IRAK-1 (Lukiw et al. 2008; Taganov et al. 2006), the chromosome locus for human miRNA-146a located at chr 5q33, and a highly probable RNA pre-miRNA-146a stem-loop structure containing the mature miRNA-146a coding region in the loop domain (Krol and Krzyzosiak 2006; Lukiw 2007; Roshan et al. 2009). As depicted in Figure 2, the abundance of miRNA-146a was analyzed using two different methods; miRNA fluorescent array analysis of 1213 human miRNA targets (Sethi and Lukiw 2008; LC Sciences, Houston, TX) and Northern dot blot analysis using high-specific-activity radiolabeled small RNA probes onto highly concentrated immobilized small RNA targets (Lukiw et al. 2008) (Figure 2). A high vacuum filtration method was found to concentrate small RNA onto dot blots approximately 2 mm in diameter using a Biomek 2000 laboratory automation workstation yields highly quantitative small RNA determinations (Cui et al. 2010; Lukiw 2007). While no significant changes were observed in the abundance of 5S RNA in any mouse or human total RNA examined, miRNA-146a showed an increase in scrapie 139A of 2.5- to 3.5-fold over age-matched controls, an rise of miRNA-146a in sCJD of 2.4- to 2.7-fold over controls, and an elevation of miRNA-146a of 2.3- to 4-fold in GSS. Interestingly, a brain-abundant miRNA-125b known to be associated with gliosis and glial cell proliferation (Pogue et al. 2010) was found to be significantly increased in three of the five human prion cases examined; both CJD and GSS neuropathology are associated with varying degrees of reactive astrogliosis (Prusiner 1994; Reimer et al. 2009).
DISCUSSION
The involvement of small RNA signaling in both common and rare human neurological disorders associated with astrogliosis, memory loss, and progressive dementia, including AD and prion disease, has been known for some time (Lukiw et al. 1992). Chromatin structural and transcriptional changes and the upregulated abundance of small RNA that include miRNA-146a are observed in human brain cells in response to pathophysiological and oxidative stress (Cui et al. 2004; Lukiw et al. 2005; Barbato et al. 2009; Lukiw 2007; Pogue et al. 2009), as a response to viral and prion infection (Hill et al. 2010; McLachlan et al. 1986; Saba et al. 2008), in AD (Cui et al. 2010; Lukiw 2007; Lukiw et al. 2008), and in several other progressive neurological disorders including schizophrenia (Burmistrova et al. 2007) and temporal-lobe epilepsy wherein neuroinflammatory signaling is activated (Aronica et al. 2010). More specifically, host miRNA-146a levels are progressively upregulated in human brain cells challenged with herpes simplex-1 (HSV-1; Higaki et al. 2002; Hill et al. 2001; 2009; Lukiw et al. 2010), human immunodeficiency virus-1 (HIV-1; Rom et al. 2010), Kaposi’s sarcoma-associated herpes virus (KSHV; Punj et al. 2010), Epstein–Barr virus (EBV; Motsch et al. 2007), and vesicular stomatitis virus (VSV; Lian et al. 2010), indicating that a host-generated miRNA-146a plays important immune response roles in viral pathogenesis, or viral evasion from host immune signaling, or both (Hill et al. 2009). In a recent large study of AD brains, miRNA-146a was found to be increased to an average of 2.6-fold over age-matched controls in the temporal lobe neocortex (n = 66; Cui et al. 2010). Interestingly, the inducible expression of miRNA-146a is significantly upregulated in primary cocultures of human astroglial cells stressed using the pro-inflammatory cytokine interleukin-1beta (IL-1β) and amyloid beta 42 (Aβ42) peptide, and this inducible upregulation is significantly quenched using specific nuclear factor (NF)-κB inhibitors including curcumin, pyrrolidine dithiocarbamate (PDTC), and the resveratrol analog CAY10512 (Cui et al. 2010). The relatively rare transmissible spongiform encephalopathies including sCJD and GSS, occurring at an incidence of 1–10 per 100 million population, are characterized by reactive gliosis, altered immune responses, and subsequent inflammatory degeneration of neuronal tissue (Collins et al. 2001; Prusiner 1994). Interference with inflammatory signaling may represent a novel therapeutic approach to slow down the course of prion disease development (Reimer et al. 2009). In vitro, NF-κB inhibition and anti-miRNA-146a strategies demonstrated potential pharmacological value in treating pathological conditions wherein miRNA-146a is overexpressed (Cui et al. 2010; Lukiw et al. 2008). Of further interest is that expression of miRNA-146a correlates with amyloid plaque density, synaptic pathology, and cognition in aging Tg2576 and in 5xFAD amyloid overexpressing transgenic mouse models used in the study of Aβ42 peptide-mediated aspects of AD-type neurodegeneration (Li et al. 2010b).
These findings have several interesting implications: (a) an NF-κB-driven, miRNA-146a-mediated inflammatory neurodegenerative response appears to share an underlying commonality in viral infection, AD, prion disease and temporal-lobe epilepsy (Aronica et al. 2010; Hill et al. 2010; Li et al 2010a; Lukiw 2007; Saba et al. 2008); (b) known miRNA-146a mRNA targets, namely, components of the innate immune response and pro-inflammatory signaling factors such as complement factor H (CFH) and the Tolllike receptor/interleukin-1 receptor (TLR/IL1-R) interleukin-1 receptor associated kinase-1 (IRAK-1; Cui et al. 2010; Taganov et al. 2006), may also be targeted in prion disease; and (c) highly specific miRNA-146a-regulated epigenetic immune and inflammatory neurodegenerative processes are conserved over evolution between the brains of mouse and humans that through comparative genomics show a species divergence of at least 75 million years (Zhu et al. 2007). These indicate an underlying common and highly conserved inflammatory response to neurological insults induced by viral infections, AD, and prion disease when compared to healthy, aging murine or human controls that exhibit no overtly abnormal immunopathology.
CONCLUSION
In summary, these data further support the observation than an inducible, NF-κB-regulated miRNA-146a is upregulated in a wide range of murine and human viral and prion infections and in neurological disorders associated with inflammatory neurodegeneration including AD. It will be of interest to see (a) whether miRNA-146a is similarly overexpressed in other relatively common animal prion diseases such as bovine spongiform encephaolopathy (BSE) and in chronic wasting disease (CWD) of mule deer and elk, or in other rare human transmissible spongiform encephalopathies such as fatal familial insomnia (FFI), and (b) whether in BSE, CWD, or FFI specific miRNA-146a targets such as CFH and IRAK-1 mRNA and innate immune or inflammatory signaling are similarly affected. Data indicate that significant upregulation of miRNA-146a coupled to downregulation of pathogenic target mRNAs may contribute to an altered innate immune or inflammatory response to both viral- and prion-mediated infection that also accompanies progressive, irreversible inflammatory neurodegeneration in a surprisingly wide range of murine and human brain diseases. Anti-miRNA-146a strategies may have novel therapeutic benefit in the treatment of these insidious, progressive, and lethal neurological disorders.
Acknowledgments
Thanks are extended to the families, physicians, and researchers who contributed to the murine and human brain bank tissue and total RNA resources used in these studies, and especially to Drs. P. Alexandrov, R. I. Carp, L. Carver, C. Chen, S. Gettner, E. Head, H. LeBlanc, W. Poon, T. Saing, and Jian Zhang at donor institutions. Some of the control and pathological brain tissues used in these studies were provided by the Memory Impairments and Neurological Disorders (MIND) Institute at the University of California, Irvine, Alzheimer’s Disease Research Center (UCI-ADRC); funding for the UCI-ADRC was provided by NIH/NIA grant P50 AG16573. Thanks are also extended to Drs. Hilary Thompson, Yuan Yuan Li, and Darlene Guillot for expert statistical analysis and technical assistance. These studies were supported in part by a Translational Research Initiative grant from Louisiana State University (WJL), by an Alzheimer Association Investigator-Initiated Research Grant IIRG-09-131729 (WJL), NIH NIA AG18031 (WJL), and by NIH NIA AG038834 (WJL).
Footnotes
Publisher's Disclaimer: Full terms and conditions of use: http://www.tandfonline.com/page/terms-and-conditions
This article may be used for research, teaching, and private study purposes. Any substantial or systematic reproduction, redistribution, reselling, loan, sub-licensing, systematic supply, or distribution in any form to anyone is expressly forbidden.
The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae, and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand, or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
REFERENCES
- Alexandrov PN, Zhao Y, Pogue AI, Tarr MA, Kruck TP, Percy ME, Cui JG, Lukiw WJ. Synergistic effects of iron and aluminum on stress-related gene expression in primary human neural cells. J. Alzheimers Dis. 2005;8:117–127. doi: 10.3233/jad-2005-8204. [DOI] [PubMed] [Google Scholar]
- Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- Barbato C, Ruberti F, Cogoni C. Searching for MIND: microRNAs in neurodegenerative diseases. J. Biomed Biotechnol. 2009;2009:871313. doi: 10.1155/2009/871313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmistrova OA, Goltsov AY, Abramova LI, Kaleda VG, Orlova VA, Rogaev EI. MicroRNA in schizophrenia: Genetic and expression analysis of miR-130b (22q11) Biochemistry (Moscow) 2007;72:578–582. doi: 10.1134/s0006297907050161. [DOI] [PubMed] [Google Scholar]
- Collins S, McLean CA, Masters CL. Gerstmann–Sträussler–Scheinker syndrome, fatal familial insomnia, and kuru: A review of these less common human transmissible spongiform encephalopathies. J. Clin. Neurosci. 2001;8:387–397. doi: 10.1054/jocn.2001.0919. [DOI] [PubMed] [Google Scholar]
- Cui JG, Kuroda H, Chandrasekharan NV, Pelaez RP, Simmons DL, Bazan NG, Lukiw WJ. Cyclooxygenase-3 gene expression in Alzheimer hippocampus and in stressed human neural cells. Neurochem Res. 2004;29:1731–1737. doi: 10.1023/b:nere.0000035809.70905.8a. [DOI] [PubMed] [Google Scholar]
- Cui JG, Zhao Y, Lukiw WJ. Isolation of high spectral quality RNA using run-on gene transcription; Application to gene expression profiling of human brain. Cell. Mol. Neurobiol. 2005;25:789–794. doi: 10.1007/s10571-005-4035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ. Differential regulation of interleukin-1-receptor-associated kinase-1 (IRAK-1) and IRAK-2 by micro RNA-146a and NF-κB in stressed human astroglial cells and in Alzheimer’s disease. J. Biol. Chem. 2010;285:38951–38960. doi: 10.1074/jbc.M110.178848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smaele E, Ferretti E, Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010;1338:100–111. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- Higaki S, Gebhardt BM, Lukiw WJ, Thompson HW, Hill JM. Effect of immunosuppression on gene expression in the HSV-1 latently infected mouse trigeminal ganglion. Invest. Ophthalmol. Vis. Sci. 2002;43:1862–1869. [PubMed] [Google Scholar]
- Hill JM, Lukiw WJ, Gebhardt BM, Higaki S, Loutsch JM, Myles ME, Thompson HW, Kwon BS, Bazan NG, Kaufman HE. Gene expression analyzed by microarrays in HSV-1 latent mouse trigeminal ganglion following heat stress. Virus Genes. 2001;23:273–280. doi: 10.1023/a:1012517221937. [DOI] [PubMed] [Google Scholar]
- Hill JM, Zhao Y, Clement C, Neumann DM, Lukiw WJ. HSV-1 infection of human brain cells induces miRNA-146a and Alzheimer-type inflammatory signaling. Neuroreport. 2009;20:1500–1505. doi: 10.1097/WNR.0b013e3283329c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Krzyzosiak WJ. Structure analysis of microRNA precursors. Methods Mol. Biol. 2006;342:19–32. doi: 10.1385/1-59745-123-1:19. [DOI] [PubMed] [Google Scholar]
- Li L, Chen XP, Li YJ. MicroRNA-146a and human disease. Scand. J. Immunol. 2010a;71:227–231. doi: 10.1111/j.1365-3083.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. Increased expression of miRNA-146a in Alzheimer’s disease transgenic mouse models. Neurosci. Lett. 2010b Oct 8; doi: 10.1016/j.neulet.2010.09.079. [Epub ahead of print] PubMed PMID: 20934487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H, Liu W, Liu Q, Jin H, Sun Y, Li J, Xia Z, Gao H. A laboratory-attenuated vesicular stomatitis virus induces apoptosis and alters the cellular miRNA expression profile in BHK cells. Arch. Virol. 2010;155:1643–1653. doi: 10.1007/s00705-010-0749-2. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Bazan NG. Cyclooxygenase 2 RNA message abundance, stability, and hypervariability in sporadic Alzheimer neocortex. J. Neurosci Res. 1997;50:937–945. doi: 10.1002/(SICI)1097-4547(19971215)50:6<937::AID-JNR4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Handley P, Wong L, McLachlan DRC. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD) Neurochem. Res. 1992;17:591–597. doi: 10.1007/BF00968788. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, LeBlanc HJ, Carver LA, McLachlan DR, Bazan NG. Run-on gene transcription in human neo-cortical nuclei. Inhibition by nanomolar aluminum and implications for neurodegenerative disease. J. Mol. Neurosci. 1998;11:67–78. doi: 10.1385/JMN:11:1:67. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Percy ME, Kruck TP. Nanomolar aluminum induces pro-inflammatory and pro-apoptotic gene expression in human brain cells in primary culture. J Inorg Biochem. 2005;99:1895–1898. doi: 10.1016/j.jinorgbio.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a–mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J. Biol. Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Cui JG, Yuan LY, Bhattacharjee PS, Corkern M, Clement C, Kammerman EM, Ball MJ, Zhao Y, Sullivan PM, Hill JM. Acyclovir or Abeta42 peptides attenuate HSV-1-induced miRNA-146a levels in human primary brain cells. Neuroreport. 2010;21:922–927. doi: 10.1097/WNR.0b013e32833da51a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan DRC, Lukiw WJ, Cho HJ, Carp RI, Wisniewski H. Chromatin structure in scrapie and Alzheimer’s disease. Can. J. Neurol. Sci. 1986;13:427–431. doi: 10.1017/s0317167100037057. [DOI] [PubMed] [Google Scholar]
- Motsch N, Pfuhl T, Mrazek J, Barth S, Grässer FA. Epstein-Barr virus-encoded latent membrane protein 1 induces the expression of the cellular miRNA miR-146a. RNA Biol. 2007;4:131–137. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- Pogue AI, Lukiw WJ. Angiogenic signaling in Alzheimer’s disease. Neuroreport. 2004;15:1507–1510. doi: 10.1097/01.wnr.0000130539.39937.1d. [DOI] [PubMed] [Google Scholar]
- Pogue AI, Li YY, Cui JG, Zhao Y, Kruck TP, Percy ME, Tarr MA, Lukiw WJ. Characterization of an NF-κB-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J. Inorg. Biochem. 2009;103:1591–1595. doi: 10.1016/j.jinorgbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, Lukiw WJ. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci. Lett. 2010;476:18–22. doi: 10.1016/j.neulet.2010.03.054. [DOI] [PubMed] [Google Scholar]
- Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer’s and prion diseases. Brain Res. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Molecular biology and genetics of prion diseases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994;343:447–463. doi: 10.1098/rstb.1994.0043. [DOI] [PubMed] [Google Scholar]
- Punj V, Matta H, Schamus S, Tamewitz A, Anyang B, Chaudhary PM. Kaposi’s sarcoma-associated herpesvirus-encoded viral FLICE inhibitory protein (vFLIP) K13 suppresses CXCR4 expression by up-regulating miRNA-146a. Oncogene. 2010;29:1835–1844. doi: 10.1038/onc.2009.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazanskaia N, Lukiw WJ, Grigorenko A, Korovaitseva G, Dvoryanchikov G, Moliaka Y, Nicolaou M, Farrer L, Rogaev E. Regulatory region variability in the human presenilin-2 (PSEN2) gene: Potential contribution to the gene activity and risk for AD. Mol. Psychiatry. 2002;7:891–898. doi: 10.1038/sj.mp.4001101. [DOI] [PubMed] [Google Scholar]
- Riemer C, Gültner S, Heise I, Holtkamp N, Baier M. Neuroinflammation in prion diseases: Concepts and targets for therapeutic intervention. CNS Neurol. Disorders Drug Targets. 2009;8:329–341. doi: 10.2174/187152709789542014. [DOI] [PubMed] [Google Scholar]
- Rom S, Rom I, Passiatore G, Pacifici M, Radhakrishnan S, Del Valle L, Piña-Oviedo S, Khalili K, Eletto D, Peruzzi F. CCL8/MCP-2 is a target for mir-146a in HIV-1-infected human microglial cells. FASEB J. 2010;24:2292–2300. doi: 10.1096/fj.09-143503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshan R, Ghosh T, Scaria V, Pillai B. MicroRNAs: novel therapeutic targets in neurodegenerative diseases. Drug Discov. Today. 2009;14:1123–1129. doi: 10.1016/j.drudis.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh JI. MicroRNAs and their therapeutic potential for human diseases: Aberrant microRNA expression in Alzheimer’s disease brains. J. Pharmacol. Sci. 2010 Oct 9; doi: 10.1254/jphs.10r11fm. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Saugstad JA. MicroRNAs as effectors of brain function with roles in ischemia and injury, neuroprotection, and neurodegeneration. J. Cereb. Blood Flow Metab. 2010;30:1564–1576. doi: 10.1038/jcbfm.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: Specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem. Soc. Trans. 2008;36:1211–1215. doi: 10.1042/BST0361211. [DOI] [PubMed] [Google Scholar]
- Yokota T. MicroRNA and the central nervous system. Brain Nerve. 2009;61:167–176. [PubMed] [Google Scholar]
- Zhu J, Sanborn JZ, Diekhans M, Lowe CB, Pringle TH, Haussler D. Comparative genomics search for losses of long-established genes on the human lineage. PLoS Comput Biol. 2007;3:e247. doi: 10.1371/journal.pcbi.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]