Abstract
The substantial morbidity and mortality associated with invasive fungal infections constitute undisputed tokens of their severity. The continued expansion of susceptible population groups (such as immunocompromised individuals, patients undergoing extensive surgery, and those hospitalized with serious underlying diseases especially in the intensive care unit) and the limitations of current antifungal agents due to toxicity issues or to the development of resistance, mandate the development of novel antifungal drugs. Currently, drug discovery is transitioning from the traditional in vitro large-scale screens of chemical libraries to more complex bioassays, including in vivo studies on whole animals; invertebrates, such as Caenorhabditis elegans, are thus gaining momentum as screening tools. Key pathogenesis features of fungal infections, including filament formation, are expressed in certain invertebrate and mammalian hosts; among the various potential hosts, C. elegans provides an attractive platform both for the study of host-pathogen interactions and the identification of new antifungal agents. Advantages of compound screening in this facile, relatively inexpensive and not as ethically challenged whole-animal context, include the simultaneous assessment of antifungal efficacy and toxicity that could result in the identification of compounds with distinct mechanisms of action, for example by promoting host immune responses or by impeding fungal virulence factors. With the recent advent of using predictive models to screen for compounds with improved chances of bioavailability in the nematode a priori, high-throughput screening of chemical libraries using the C. elegans-c. albicans antifungal discovery assay holds even greater promise for the identification of novel antifungal agents in the near future.
Keywords: Invertebrate model hosts, non-mammalian animal models, systemic mycoses, Candida albicans, high-throughput in vivo screening, drug bioaccumulation model, Caenorhabditis elegans
INTRODUCTION
The incidence of invasive fungal infections that are associated with significant morbidity and mortality has increased in recent years (reviewed in [1, 2]). The mortality rate of invasive candidiasis, which is currently the fourth most common nosocomial blood-stream infection, can reach 40% [3-5]. Factors postulated to have contributed to the rise of Candida infections include the increase in invasive procedures that disrupt the host’s natural barriers to infection, and the extensive empirical use of broad-spectrum antimicrobial agents that increases colonization with Candida species by changing the normal microbiota [6]. Aspergillus and Cryptococcus species are additional opportunistic fungi of great concern to clinicians [1, 2, 7]. Cryptococcal meningitis is one of the most important human immunodeficiency virus (HIV)-related opportunistic infections, especially in the developing world [8].
Systemic antifungal agents have been available since the 1950s; however, the challenges in the discovery and development of safe and efficient agents, stemming mainly from the molecular similarities of the eukaryotic fungi to mammalian cells [9, 10], have resulted in a slower antifungal discovery pipeline compared to those of antibacterial or antiviral agents. Amphotericin B deoxycholate (AmBD) has been the “gold standard” in antifungal chemotherapy for over 50 years, despite its frequent associated toxicities (reviewed in [11]). Ostrosky-Zeichner et al. [12] provide a comprehensive review of the evolution of our rather limited antifungal armamentarium from the highly toxic nystatin and amphotericin B to the reformulated for improved pharmacokinetics, lipid-based amphotericin B and the most commonly used triazoles, to the echinocandins recently; the latter lipopeptides represent the first fungus-specific class of antimycotics that inhibit the synthesis of a key component of many fungal cell walls, (1,3)-β-D-glucan (reviewed in [11, 13, 14]). The three marketed echinocandins (caspofungin, anidulafungin, and micafungin) are currently in late Phase III/IV clinical trials, while only one candidate, aminocandin (IP9601/HMR3270) is in early preclinical development [12]. An overview of the research into biologically active natural products with antifungal activity, which regained popularity among synthetic chemists in the late 1990s with the advent of the concept of diversity-oriented synthesis, is presented elsewhere [15].
Progress has been made in the development of therapeutics in the field of medical mycology during recent years; however, its pace must be increased to meet the growing demand for more efficient agents with improved pharmacological properties in the facet of drug resistance (reviewed in [16]), although antifungal drug resistance may actually be less of a problem than for bacterial infections [17]. Here, we review and discuss current developments of a recently introduced approach that can be undertaken to identify novel antifungal agents using the model nematode Caenorhabditis elegans as a biological probe in in vivo bioassays. Insights into the usage of invertebrate model hosts in antifungal drug discovery have also been presented by Pukkila-Worley et al. [18]. An overview of the molecular approaches that may be undertaken for the identification of new antifungal drug targets has been presented elsewhere [19].
C. ELEGANS: AN ALTERNATIVE MODEL HOST OF FUNGAL PATHOGENESIS
The study of the interaction of fungi with their natural environmental predators, such as insects, free-living amoebae, and nematodes, provides clues to the identification of novel antifungal therapies, especially those that could exert their activity by promoting host immune responses [20-25]. The hypothesis behind the usage of heterologous invertebrate model hosts to study fungal pathogenesis is that host innate immune responses were selected for over evolutionary time (which also explains their broad conservation across many phyla), while fungal pathogens developed virulence factors to adapt to, and potentially counteract, these host defense mechanisms; these same virulence traits would enable the evolution of fungi to successful opportunistic pathogens in mammalian hosts under the appropriate conditions (reviewed in [26-28]).
In 2002, Mylonakis and colleagues found that feeding C. elegans solely with non-pathogenic cryptococcal strains, such as Cryptococcus laurentii and Cryptococcus kuetzingii, did not affect progeny production or lifespan compared with growth on the usual laboratory food source, Escherichia coli OP50; in contrast, C. elegans were killed when fed the human pathogenic yeast Cryptococcus neoformans, and the killing of the nematodes depended on genes previously shown to be involved in mammalian virulence [25]. These observations support the hypothesis that the mammalian pathogenesis of C. neoformans may be a consequence of adaptations that have evolved during the interaction of the fungus with such environmental predators as free-living nematodes, and suggest that C. elegans can be used as a simple model host in which C. neoformans pathogenesis can be readily studied.
The C. elegans infection model also allows for the study of complex prokaryote-eukaryote interactions when the same niche is occupied within a living host. For example, Acinetobatcer baumannii was found to inhibit the filamentation process of C. albicans when the two pathogens co-infected the nematode [29]. These findings suggest that C. elegans provides a whole-animal model system useful for the study of the complex dynamics of bacterial-fungal interactions in vivo, possibly also providing clues to novel drug targets based on targets identified by the dominant pathogen that enable it to inhibit the other.
The C. elegans killing assay, which was used in these early studies of both Gram-negative [30-32] and Gram-positive [33] bacteria as well as of the human pathogenic fungus C. neoformans [25] as mentioned above, entailed observing a synchronized population of nematodes for kinetics of death on a lawn of the microbe under study. Living and dead worms were counted at approximately 24-hour intervals, and the time for 50% of the nematodes to die, was calculated. The development of two separate tools in the C. elegans-c. neoformans system, a method for screening a pool of random insertion mutants of C. neoformans and a progeny-based screen, led to the identification of novel cryptococcal virulence genes important for pathogenesis in mammalian models [34, 35].
On the opposite end of the spectrum, several conserved innate immune responses, possibly reflecting the selection pressure imposed by early environmental interactions with natural predators, were uncovered in C. elegans (reviewed in [36, 37]). A Toll/IL-l receptor domain adaptor protein that functions upstream of a conserved p38 mitogen activated protein (MAP) kinase pathway is one such conserved signal transduction component that is required for pathogen resistance in C. elegans, as shown by genetic and functional genomic studies [36, 38]. This ancestral innate immune signaling pathway present in the common ancestor of nematodes, arthropods and vertebrates, has been suggested to predate the canonical Toll signaling pathways in innate immunity [36]. A recent report furthermore showed that host defense against two prototypic fungal pathogens, C. neoformans and C. albicans, is mediated by two evolutionarily conserved beta-glucan binding receptors: the C. elegans CED-1 and C03F11.3 and their mammalian orthologues, the scavenger receptors SCARF1 and CD36, respectively [39]. The activation of CED-1 or C03Fl1.3 through C. neoformans exposure leads to the induction of such antimicrobial peptides as antibacterial factors abf-1, and abf-2 that could be explored further as potential antifungal compounds [40, 41].
Key pathogenesis features of fungal infections are thus shared between certain invertebrate and mammalian hosts and the study of the interactions between invertebrate model hosts and pathogenic fungi provides insights into the mechanisms underlying both host immunity and pathogen virulence (reviewed in [26-28, 36, 42]). Moreover, invertebrate model hosts complement the use of mammalian models by enabling whole-animal high-throughput infection assays for the in vivo screening of antifungal compounds as discussed below.
C. ELEGANS AS A PLATFORM FOR THE IDENTIFICATION OF NOVEL ANTIMYCOTICS
Mammalian models of infection have been traditionally used to discern the molecular mechanisms by which pathogens interact with the human host. The genetic complexity, long reproductive cycles and small progeny sizes of mammals and possibly the length of time post-infection required for the development of symptoms, render, nonetheless, pathogenesis studies in such hosts challenging; cost and ethical challenges constitute further shortcomings of mammalian experiments [43]. Certain heterologous hosts provide alternative model systems of direct relevance to higher order species for the examination of host-pathogen interactions and the identification of novel antimycotics (reviewed in [26-28, 42]). Invertebrate model hosts that have been used to study fungal pathogenesis include the fruit fly Drosophila melanogaster and the greater wax moth Galleria mellonella [26-28, 42]. Although many of these organisms provide excellent infection models and some (e.g. Drosophila) have been used successfully as alternative platforms for drug efficacy testing of antifungals [22, 44], several technical limitations currently limit their practical utility in high-throughput screens. In contrast, C. elegans, the small (about I mm long), temperate-region soil nematode that Sydney Brenner began using in the 1960’s to study the genetics of development and neurobiology, can also be used as a tractable model mainly due to its amenability for processing in liquid assays, which means that automation can be incorporated into the work flow as discussed in detail below.
Several aspects render the microscopic C. elegans a promising model host for the study of microbial pathogenesis and the discovery of novel drug targets and bioactive compounds. Its genome has been sequenced and genetic studies have shed light into the functions of many of its genes [45]. RNA interference (RNAi), in particular, proved to be a powerful molecular technique to silence gene expression through the formation of double-stranded RNA and systematically analyze the connection between gene sequence, chromosomal location and gene function in C. elegans [46]. The short reproductive cycle of the nematode worms and their hermaphroditism that lead to genetically identical populations in brief periods of time, are additional general advantages of this model organism. However, progeny production by wild-type C. elegans confounds killing assays because of an asynchronous population which can be affected differently by the pathogen at different life stages and because larvae often hatch inside the nematode, leading to the death of the worm by a mechanism not directly related to pathogen exposure (so-called matricidal killing). These problems can be solved by substituting wild-type nematodes with a C. elegans glp-4 mutant strain in screening assays. The glp-4 mutation renders the strain incapable of producing gonads or progeny at 25°C, without affecting the normal morphology or brood sizes at 15°C, the temperature that is typically used for the propagation and maintenance of the nematodes [47]. Auxotrophic strains of E. coli are the usual laboratory food source for the nematodes that can be easily infected with both a number of pathogenic bacteria and fungi by substituting this food source with the pathogen of interest. The development of C. elegans as an infection model is hindered only by the consumability and retention of the pathogen.
C. albicans, like C. neoformans, establishes a lethal infection in the digestive tract of the worms by employing several virulence traits that are also required for mammalian pathogenesis. In particular, C. albicans cells undergo morphological transitions from planktonic to filamenting forms, as shown in Fig. 1. Adhesion to host cells, biofilm formation, and direct invasion through host tissues are facilitated by these filaments that appear to be required for the full virulence potential of Candida albicans in both mammals and C. elegans [48, 49]. It should be noted that filamentation alone is not sufficient to kill other invertebrate model hosts, such as G. mellonella (Lepidoptera: the greater wax moth), as suggested by a recent report [50]. During C. elegans infection, however, a network of true hyphae is created by wild-type C. albicans resulting in the physical piercing of the worm cuticle and its death [51].
Fig. (1). Progression of Candida albicans infection in the C. elegans intestine.
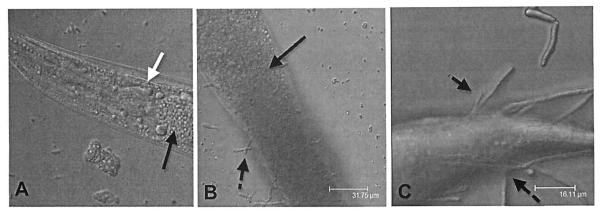
(A). The nematode becomes infected with fungi by consuming the yeast cells as a food source. The white arrow points to the C. elegans pharyngeal grinder. Black arrows point to the location of the fungal cells in the nematode intestinal lumen. (B). Once the C. albicans cells are in the nematode, they form filaments (indicated by the broken black arrows) (C) that penetrate through the worm cuticle.
The presence of filaments breaking through the worm and its killing served as specific end-points in pilot experiments in the C. elegans-C. albicans model that was used to screen a library of chemical compounds with established biological activities [49]. The assay was performed in liquid media using standard 96-well plate technology. A C. elegans glp4;sek-1 double mutant was used in the screening. As mentioned above, the glp-4 mutation renders the strain incapable of producing progeny at 25°C and is, thus, suited for these studies, although sterile animals are long-lived compared to wild-type animals [47, 52]. C. elegans SEK-I encodes a conserved mitogen-activated protein (MAP) kinase kinase involved in the innate immune response, and sek-I animals are relatively immunocompromised [53].
The constitutive expression of green fluorescent protein (GFP) by the C. albicans strain that was ingested by the nematode worms allowed for the detection of antifungal activity. C. albicans strain MLR62 expresses GFP (linked to the constitutively active TEF1 promoter) and exhibits similar killing kinetics to the parent strain DAY 185 in the C. elegans assay [49, 54]. Compounds with significant antifungal activity prevented the development of green fluorescence in the intestinal tract of infected worms that continued to move normally at the endpoint of the assay; in contrast, in wells containing inefficient antimycotic compounds, nematodes actively infected with C. albicans, as manifested by the high levels of intestinal fluorescence, were rod-shaped, non-mobile and engulfed in filament networks.
This reporter method was used in a screen of 1,266 compounds with known pharmaceutical activities, which identified 15 (approximately 1.2%) that prolonged the survival of C. albicans-infected nematodes and inhibited the in vivo filamentation of the fungus [49]. The compounds that were identified by this and subsequent screens in the Candida-mediated C. elegans assay arc listed in Table 1. Two identified compounds, caffeic acid phenethyl ester (CAPE), a major active component of honeybee propolis, and the fluoroquinolone agent enoxacin exhibited antifungal activity and prolonged survival in a murine model of candidiasis as well; however, of these two compounds only CAPE inhibited C. albicans growth in vitro. CAPE prolonged the survival of nematodes in the in vivo C. elegans-C. albicans assay at a concentration of 4 μg/mL [49], while its minimal inhibitory concentration (MIC) was 16-fold higher, suggesting that its beneficial actions for survival lay beyond the inhibition of fungal growth, possibly in the immunomodulation of the host, but this hypothesis warrants further studies. A similar screen for novel antibacterial agents nonetheless identified several compounds with immunomodulatory activity in a C. elegans live-animal infection model [55].
Table 1.
Compounds Exhibiting Antifungal Activity Identified in the C. elegans-C. albicans Infection Assay to Date
| Category | Compound | Action/Use | Reference |
|---|---|---|---|
| Established antifungals | |||
| Amphotericin B | Interaction with ergosterol to form transmembrane channels [81, 82] | [59] | |
| Bifonazole | Interference with ergosterol synthesis [83] | [59] | |
| Butoconazole nitrate | Interference with ergosterol synthesis [84, 85] | [59] | |
| Ketoconazole | Interference with ergosterol synthesis [83] | [59] | |
| Nystatin | Binding to ergosterol to induce permeability changes [86] | [59] | |
| Oxiconazole nitrate | Interference with ergosterol synthesis [84, 87] | [59] | |
| Terconazole | Interference with ergosterol synthesis [88] | [59] | |
| Immunosuppressant agents | |||
| Ascomycin (Ethyl analogue of FK-506) |
Binding to immunophilins and inhibition of Th1 & Th2 cytokine production [89] | [59] | |
| Cyclosporin A | Complex formation with cyclophilin to inhibit calcineurin [90] | [59] | |
| FK-506 (Tacrolimus) | Complex formation with FKBP-12 to inhibit calcineurin [91, 92] | [59] | |
| Plant-derived saponin natural products | |||
| A2 Sakurasosaponin | Molluscicidal and antifungal triterpenoid saponin [93] | [62] | |
| A6 Arvensoside B | No previous reports on antifungal activity | [62] | |
| A7 | No previous reports on antifungal activity | [62] | |
| A8 (Analogue of A16) | Glycosylated derivative of Aginoside [94, 95] | [62] | |
| All | No previous reports on antifungal activity | [62] | |
| A16 Aginoside | Known antifungal natural product [94, 95] | [62] | |
| A17 | Pentasaccharide that incorporates the distal furanose residue, related to maesabalide family; no reports on antifungal activity [96] |
[62] | |
| A19 | Related to barrigenol family, unique polyglycosylation [97,98] | [62] | |
| A20 | No previous reports on antifungal activity | [62] | |
| A21 | Pentasaccharide that incorporates the distal furanose residue, related to maesabalide family; no reports on antifungal activity [96] |
[62] | |
| A24 (Analogue of A 16) | Glycosylated derivative of Aginoside [94, 95] | [62] | |
| A25 | Related to barrigenol family, unique polyglycosylation; excellent activity against C. albicans in vitro [97] |
[62] | |
| Other compounds | |||
| Caffeic acid phenethyl ester (CAPE) |
Active component of propolis, a natural antimicrobial resinous plant product collected & used by honeybees for protection of hives [99] |
[49] | |
| Concanamycin A | Potent & specific inhibition of vacuolar-ATPase [100] | [59] | |
| Dequalinium chloride | Potent anti-tumor activity and protein kinase C alpha (PKC) inhibition [101] | [59] | |
| Ellipticine | Potent anti-tumor agent; DNA intercalation and/or topoisomerase II inhibition [102] | [59] | |
| Enoxacin | Fluoroquinolone antibiotic; enhancement of in vitro antifungal activity of amphotericin B & mepartricin; no inhibition of filament formation in C. elegans-killing assay [103] |
[49] | |
| Lapachol | Naphthoquinone found in tropical plant seeds & heartwood; anti-leishmanicidal activity [104, 105] |
[49] | |
| Malachite green carbinol base |
Metabolism to carbinol form, then to toxic leucomalachite green form; used for parasitic, fungal & bacterial infections in fish & fish eggs [106, 107] |
[59] | |
| Mepartricin | Used to treat chronic pelvic pain syndrome; reduction of estrogen plasma levels & their con- centration in the prostrate; antifungal activity against Candida [108] |
[59] | |
| Phenylmercuric acetate | Metabolism to diphenylmercury; used as fungicide, herbicide, slimicide & bacteriocide [109] | [59] | |
| Suloctidil | Used as a peripheral vasodilator, formerly in treating peripheral and cerebral vascular disor- ders; hepatotoxic [110] |
[59] | |
| Triadimefon | Gibberellin & sterol biosynthesis [111] | [59] | |
| Valinomycin | Apoptosis inducer by disruption of mitochondrial membrane potential; affects C. albicans morphology [112, 113] |
[59] | |
In particular, Moy et al. adapted a previously developed agarbased C. elegans-Enterococcus faecalis infection assay so that it could be carried out in liquid medium in standard 96-well microtiter plates and used this simple infection system to screen 6,000 synthetic compounds and 1,136 natural product extracts. The screen identified 16 compounds and 9 extracts that promoted the survival of E. faecalis-infected nematodes [55]. Some of these compounds and extracts also inhibited the growth of E. faecalis in vitro, but at significantly lower MICs compared to traditional antibiotics. Thus, the C. elegans infection model can be used not only for the identification of traditional antimicrobials, but also as a means to identify immunostimulatory compounds that could perhaps be effective against multi-drug resistant (MDR) pathogens. A new class of agents for countering efflux-mediated bacterial drug resistance may be hybrid antimicrobials containing an antibacterial linked to an MDR pump inhibitor; this strategy has been successfully evaluated using a C. elegans infection model [56]. The discovery that many bacteria use quorum-sensing (QS) systems to coordinate virulence and biofilm development may also lead to new, promising targets for antimicrobial drugs that can be assessed on the C. elegans model [57].
The feasibility of conducting the screen as a liquid assay process has transformed the initially tedious screening assay protocol and optimized the work flow by using robotic systems for the labor-intensive and time-consuming steps of precise dispensing of worms and compounds into assay wells and co-inoculation of nematodes with C. albicans [58, 59]. The microscopy/imaging and software scoring components of the C. elegans-based screening system also continues to evolve. One of the available imaging systems for scoring and analysis is the Molecular Devices Discovery-1 microscope and MetaExpress software. This system is capable of capturing light images and scoring C. albicans-infected worms 10 detect reduction in fungal growth or wells containing chemical compounds with antifungal activity; however, visual scoring is still required to evaluate nematode life or death based on the sinusoidal- or rod-shaped appearance of the worms, correspondingly [59].
The ability to increase the screening process to greater high-throughput capacity can be potentially aided by an increase in automation and the addition of florescence. Gosai et al. [60] showed that they could advance preclinical drug discovery campaigns by screening live C. elegans modeling α1-antitrypsin deficiency and other complex disease phenotypes on high-content imaging platforms. Using transgenic animals expressing fluorescently-tagged proteins, they first developed a high-quality, high-throughput work-flow utilizing an automated fluorescence microscopy platform with integrated image acquisition and data analysis modules to qualitatively assess different biological processes including growth, tissue development, cell viability and autophagy. Gosai el al. [60] subsequently adapted this technology to conduct a small molecule screen and identified compounds that altered the intracellular accumulation of the human aggregation prone mutant that causes liver disease in α1-antitrypsin deficiency.
Another means of scoring C. elegans in high-throughput screens is SYTOX Orange combined with the CellProfiler software, a method that has been used in the search of antibiotics when bacteria served as the infecting agents [61]. This method relies on the property of the fluorescent marker SYTOX Orange to target dead cells and worms. The extrapolation of this method to the fungal infection model has not been feasible yet due to the fact that C. albicans retain the stain, thus preventing the acquisition of accurate readings with the microscopy analysis software.
The assay protocol for the study of Candida cells in non-planktonic form and the concurrent evaluation of toxicity and antifungal activity of compounds is described in detail by Tampakakis el al. [58]. This improved, facile and relatively inexpensive high-throughput method was utilized to screen a library of a total of 3,228 compounds that consisted of 1,948 bioactive compounds and 1,280 small molecules derived via diversity-oriented synthesis [59]. The efficacy of most known antifungal compounds within the chemical library was confirmed, while twelve other compounds that are not primarily used as antifungal agents, including three immunosuppressive drugs, were also identified (Table 1).
A compound screen of natural products was also undertaken using the C. elegans assay system; twelve plant-derived saponins were found to have antifungal properties (Table 1), two of which (a representative one from each group), A16 (aginoside) and A19 (a saponin that shares a similar aglycone core which is related to the barrigenol family of natural products), were selected for further analysis [62]. C. albicans isolates, including strains resistant to clinically used antifungal agents, were inhibited by these compounds that also disrupted C. albicans hyphae and biofilm formation. Saponins increase the permeability of fungal cells, rendering them more susceptible to salt-induced osmotic stress. When used in combination with photosensitizer compounds, the fungus displayed increased susceptibility to photodynamic inactivation due to this property of saponins that facilitated the cell penetration of the photosensitizers [62]. Photodynamic therapy is a rapidly developing antimicrobial treatment strategy that employs a non-toxic dye, termed a photosensitizer, and low intensity visible light, which, in the presence of oxygen, combine to produce reactive oxygen species that can be targeted to specific destination cells or tissues [63, 64]. C. neoformans has been shown to be susceptible to photodynamic inactivation and this susceptibility of the fungal pathogen was associated with cell wall integrity [65]. Hence, compounds containing saponin structural features may be proven to be suitable chemical scaffolds for a new generation of antifungal compounds, either by themselves or in conjunction with currently used antimycotics.
ADVANTAGES AND LIMITATIONS OF THE C. ELEGANS SCREENING SYSTEM
Several attributes of C. elegans whole-animal bioassays render them particularly attractive for the discovery of novel antifungal therapeutics (reviewed in [18]). Thousands of compounds can be screened simultaneously in a high-throughput fashion since the experiments can be performed in liquid media using standard 96- or 384-well microtiter plates, thus providing an advantage over the use of other host options that lack a liquid assay aspect and, therefore, are not amenable to high-throughput automation techniques; automated robotic systems can now be used to fill the assay plates, sort the worms in the wells, and pin transfer compounds directly from dimethyl sulfoxide (DMSO) stocks to the assay plates. No additional dosing formulation development (e.g. for oral, intravenous, or subcutaneous use) is required as in the in vitro models, and the compounds are therefore administered directly from the DMSO stocks to the screening plates.
The Z’-factor, a dimensionless, simple statistical feature for each high-throughput screening assay, provides a useful tool for the evaluation of the quality of assays by comparing the assay performance of test compounds to those with known antifungal activity, such as amphotericin or ketoconazole [66]. Increasing the number of worms used to screen each compound obviously improves the statistical power of the assay. In contrast to traditional drug discovery models where individual properties of compounds have to be evaluated and optimized sequentially, several desirable compound characteristics (e.g. with respect to potency, solubility, or permeability) can be assessed simultaneously in this whole-animal in vivo assay. The efficiency of primary screens in the identification of potentially quality lead compounds is thus increased.
The C. elegans model may also be suitable for laboratory toxicity testing. For example, in the C. albicans infection model, a concentration of 32 μg/mL of fluconazole effectively prolonged the survival of nematodes exposed to a fluconazole-susceptible Candida strain, whereas higher drug concentrations (up to 100 μg/mL) negatively affected nematode survival, even in comparison to an untreated control group of worms [49]. The survival-promoting effects of the drug at lower concentrations may mask any existing toxic effects (reviewed in [18]).
Several lines of evidence support the use of C. elegans in early rounds of chemical toxicity screening. The model nematode has been used as a toxicity biomarker caused by soil pollutants such as heavy metals and organic solvents; the sensitivity of these tests in the worm is comparable to tests with other soil species (e.g. enchytraeids, earthworms and springtails), while they are less demanding to space and time [67]. C. elegans was also found to be a predictor of mammalian acute lethality to metallic salts, generating toxicity values that correlated well with rat and mouse values at about 10% of the cost in these conventionally used animals [68]. Similarly, the order of toxicity of the nematodes following organophosphate exposure was significantly correlated to those of both rats and mice, supporting the use of C. elegans as a mammalian neurological model as well [69].
Thus, the whole-animal C. elegans assay may help not only to monitor the toxicity of a candidate compound that would be then eliminated from further study, but it can also identify antifungal compounds with novel mechanisms of action that most likely would be missed by in vitro screens that target fungal growth. Compounds exerting their activity through the immunomodulation of the host, or by altering the expression patterns of fungal virulence factors that are induced only during infection can be identified by the C. elegans bioassay. The availability of RNAi in the model nematode could aid in this direction. Compounds identified in such screens can potentially be used as “probe compounds” that may well have antifungal activity against other fungi [49]. Compounds may have synergistic effects when used in combination, as described recently for anthelmintic combinations [70]; however, the properties (pharmacological and other) of any such combined compounds should be evaluated very carefully before such a conclusion is drawn, especially since results are to be extrapolated from the C. elegans nematode to higher-order host models.
Naturally, the C. elegans-C. albicans antifungal discovery assay has several limitations. A potential antifungal agent may be overlooked in this assay system in the following scenarios that were actually encountered recently in the cases of the clinically relevant albendazole, and the avermectin and abamectin, correspondingly [59]: 1) if the assayed compound is characterized by limited solubility in water; 2) if it has both antifungal and nematicidal activity; 3) if the compound is active against fungi other than C. albicans, the fungus used in the screen. The number and types of immunomodulatory compounds that can be identified in the C. elegans bioassays may be restricted due to the simplicity of the nematode innate immune system. Important pharmacodynamic properties that influence the systemic absorption of compounds, protein binding, and volume of distribution, may not be assessed accurately in the tiny nematodes. It is also of note that it is not possible to control the precise number of fungal cells (inoculum concentration) that are consumed by each nematode [26], although the use of a standardized fungal inoculum has been introduced to the assay system lately [59]. Toxic compounds from a compound library may not be all excluded by these bioassays.
It should be made clear that the liquid-based C. elegans-bioassay applies only to C. albicans in terms of fungal pathogens at present; C. elegans has been used as a model organism in the study of other fungi, such as C. neoformans, on solid media as previously mentioned [25, 34, 35]. The only limiting factors, in this case, are that the fungal cells need to be (a) of a consumable size for C. elegans and, (b) once consumed and after passing through the nematode grinder organ, they must be retained, intact, in the gut for the infection to be adequately established. In theory, the use of C. elegans could be extended to the study of other opportunistic fungal pathogens, possibly with different modes of transmission, and this is a possibility that should be explored in the near future.
Therefore, a reasonable overall strategic approach in drug discovery for novel antifungal agents could entail high-throughput compound screens in C. elegans and further validation of the compounds identified by these bioassays using traditional mammalian models. As such the C. elegans bioassay can complement, rather than substitute, the in vivo study of novel compounds in higher order hosts.
BIOAVAILABILITY SCREENING ADDRESSES THE CHALLENGE OF PERMEATION IN C. ELEGANS
The resistance of C. elegans to perturbation by pharmacologically active molecules is a significant problem that could limit its utility as a screening tool in drug discovery. The low in vivo permeability of the model nematode is attributable to such physical barriers as its four-layer cuticle [71] and its rapidly pumping intestine [72], as well as to its impressive enzymatic xenobiotic defenses that are controlled by over 140 genes [73]. As a result, only 2% of small-molecule pharmaceuticals induce a robust phenotypic response in C. elegans, even when they are applied in doses far in excess of those applied to mammalian cells [74, 75]. Burns et al. have recently presented an interesting approach to circumvent this problem by screening for bioaccumulation rather than bioactivity in C. elegans, in order to map a priori the chemical space that can permeate the worm’s formidable defenses [76].
More specifically, Burns and colleagues conducted a survey of the accumulation and metabolism of over 1,000 commercially available drug-like small molecules in C. elegans. Fewer than 10% of these molecules were found to accumulate to concentrations greater than 50% of that present in the external environment of the worm. Using the dataset of the chemical structures of the accumulated compounds, Burns et al. developed and validated a Bayesian model that identifies compounds with an increased likelihood of bioavailability and bioactivity in C. elegans [76]. Pre-selecting for molecules with chemical structures predicted by this model to confer accumulation to effective concentrations in the nematodes could increase the success of future small molecule screens in C. elegans.
The idea of the probabilistic relationship between the bioavailability of a compound and its chemical structure and physicochemical properties is not new (reviewed in [77]). Lipinski was the first to set the stage in this line of pharmacological research with his seminal work on the “Rule of Five” which describes how defined physicochemical parameters of drugs correlate with the likelihood of their oral bioavailability in humans [78]. The “Rule of Five” does not, however, apply to many antibacterial or antifungal drugs. According to Hopkins and Bickerton, the novelty of the Burns et al. work stems from the idea that screening for bioavailability to define the chemical space for bioactive discovery, a priori, should be a goal in its own right [79]. In a similar fashion, screening rules have been proposed recently in fungicide, herbicide, and insecticide design to increase the efficiency of finding leads [80]. This strategy could potentially address some fundamental challenges in drug discovery and improve the chances of discovering novel, more efficacious antimycotics in the near future.
ACKNOWLEDGEMENTS
This work was supported by NIH grants P01 AI 083214, R01 AI075286 and R21 AI079569 to E.M.
Footnotes
CONFLICTS OF INTEREST E.M. has served in an Advisory Board and received research support from Astellas Pharma Inc. C.G.A. and B.B.F. report no potential conflicts of interest.
REFERENCES
- [1].Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- [2].Lass-Florl C. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses. 2009;52:197–205. doi: 10.1111/j.1439-0507.2009.01691.x. [DOI] [PubMed] [Google Scholar]
- [3].Macphail GL, Taylor GD, Buchanan-Chell M, Ross C, Wilson S, Kureishi A. Epidemiology, treatment and outcome of candidemia: a five-year review at three Canadian hospitals. Mycoses. 2002;45:141–5. doi: 10.1046/j.1439-0507.2002.00741.x. [DOI] [PubMed] [Google Scholar]
- [4].Wenzel RP, Gennings C. Bloodstream infections due to Candida species in the intensive care unit: identifying especially high-risk patients to detennine prevention strategies. Clin Infect Dis. 2005;41(Suppl 6):S389–93. doi: 10.1086/430923. [DOI] [PubMed] [Google Scholar]
- [5].Rentz AM, Halpern MT, Bowden R. The impact of candidemia on length of hospital stay, outcome, and overall cost of illness. Clin Infect Dis. 1998;27:781–8. doi: 10.1086/514955. [DOI] [PubMed] [Google Scholar]
- [6].Smith JA, Kauffman CA. Recognition and prevention of nosocomial invasive fungal infections in the intensive care unit. Crit Care Med. 2010;38:S380–7. doi: 10.1097/CCM.0b013e3181e6cf25. [DOI] [PubMed] [Google Scholar]
- [7].Richardson M, Lass-Florl C. Changing epidemiology of systemic fungal infections. Clin Microbiol Infect. 2008;14(Suppl 4):5–24. doi: 10.1111/j.1469-0691.2008.01978.x. [DOI] [PubMed] [Google Scholar]
- [8].Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–30. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- [9].Baldauf SL, Palmer JD. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci USA. 1993;90:11558–62. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–2. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- [11].Petrikkos G, Skiada A. Recent advances in antifungal chemotherapy. Int J Antimicrob Agents. 2007;30:108–17. doi: 10.1016/j.ijantimicag.2007.03.009. [DOI] [PubMed] [Google Scholar]
- [12].Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov. 2010;9:719–27. doi: 10.1038/nrd3074. [DOI] [PubMed] [Google Scholar]
- [13].Denning DW. Echinocandin antifungal drugs. Lancet. 2003;362:1142–51. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- [14].Odds FC, Brown AJ, Gow NA. Antifungal agents: mechanisms of action. Trends Microbiol. 2003;11:272–9. doi: 10.1016/s0966-842x(03)00117-3. [DOI] [PubMed] [Google Scholar]
- [15].Di Santo R. Natural products as antifungal agents against clinically relevant pathogens. Nat Prod Rep. 2010;27:1084–98. doi: 10.1039/b914961a. [DOI] [PubMed] [Google Scholar]
- [16].Spanakis EK, Aperis G, Mylonakis E. New agents for the treatment of fungal infections: clinical efficacy and gaps in coverage. Clin Infect Dis. 2006;43:1060–8. doi: 10.1086/507891. [DOI] [PubMed] [Google Scholar]
- [17].Denning DW, Hope WW. Therapy for fungal diseases: opportunities and priorities. Trends Microbiol. 2010;18:195–204. doi: 10.1016/j.tim.2010.02.004. [DOI] [PubMed] [Google Scholar]
- [18].Pukkila-Worley R, Holson E, Wagner F, Mylonakis E. Antifungal drug discovery through the study of invertebrate model hosts. Curr Med Chem. 2009;16:1588–95. doi: 10.2174/092986709788186237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tournu H, Serneels J, Van Dijck P. Fungal pathogens research: novel and improved molecular approaches for the discovery of antifungal drug targets. Curr Drug Targets. 2005;6:909–22. doi: 10.2174/138945005774912690. [DOI] [PubMed] [Google Scholar]
- [20].Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci USA. 2001;98:15245–50. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Apidianakis Y, Rahme LG, Heitman J, Ausubel FM, Calderwood SB, Mylonakis E. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot Cell. 2004;3:413–9. doi: 10.1128/EC.3.2.413-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lionakis MS, Lewis RE, May GS, et al. Toll-deficient Drosophila flies as a fast, high-throughput model for the study of antifungal drug efficacy against invasive aspergillosis and Aspergillus virulence. J Infect Dis. 2005;191:1188–95. doi: 10.1086/428587. [DOI] [PubMed] [Google Scholar]
- [23].Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect Immun. 2003;71:4862–72. doi: 10.1128/IAI.71.9.4862-4872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Navas A, Cobas G, Talavera M, Ayala JA, Lopez JA, Martinez JL. Experimental validation of Haldane’s hypothesis on the role of infection as an evolutionary force for Metazoans. Proc Natl Acad Sci USA. 2007;104:13728–31. doi: 10.1073/pnas.0704497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci USA. 2002;99:15675–80. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fuchs BB, Mylonakis E. Using non-mammalian hosts to study fungal virulence and host defense. Curr Opin Microbiol. 2006;9:346–51. doi: 10.1016/j.mib.2006.06.004. [DOI] [PubMed] [Google Scholar]
- [27].Mylonakis E, Aballay A. Worms and flies as genetically tractable animal models to study host-pathogen interactions. Infect Immun. 2005;73:3833–41. doi: 10.1128/IAI.73.7.3833-3841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mylonakis E, Casadevall A, Ausubel FM. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 2007;3:e101. doi: 10.1371/journal.ppat.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC, Jr., Mylonakis E. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc Natl Acad Sci USA. 2008;105:14585–90. doi: 10.1073/pnas.0805048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aballay A, Ausubel FM. Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr Opin Microbiol. 2002;5:97–101. doi: 10.1016/s1369-5274(02)00293-x. [DOI] [PubMed] [Google Scholar]
- [31].Kurz CL, Chauvet S, Andres E, et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 2003;22:1451–60. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96:715–20. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Garsin DA, Sifri CD, Mylonakis E, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci USA. 2001;98:10892–7. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mylonakis E, Idnurm A, Moreno R, et al. Cryptococcus neoformans Kin1 protein kinase homologue, identified through a Caenorhabditis elegans screen, promotes virulence in mammals. Mol Microbiol. 2004;54:407–19. doi: 10.1111/j.1365-2958.2004.04310.x. [DOI] [PubMed] [Google Scholar]
- [35].Tang RJ, Breger J, Idnurm A, et al. Cryptococcus neoformans gene involved in mammalian pathogenesis identified by a Caenorhabditis elegans progeny-based approach. Infect Immun. 2005;73:8219–25. doi: 10.1128/IAI.73.12.8219-8225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim DH, Ausubel FM. Evolutionary perspectives on innate immunity from the study of Caenorhabditis elegans. Curr Opin Immunol. 2005;17:4–10. doi: 10.1016/j.coi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- [37].Pukkila-Worley R, Mylonakis E. From the outside in and the inside out: Antifungal immune responses in Caenorhabditis elegans. Virulence. 2010;1:111–2. doi: 10.4161/viru.1.3.11746. [DOI] [PubMed] [Google Scholar]
- [38].Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Means TK, Mylonakis E, Tampakakis E, et al. Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARFI and CD36. J Exp Med. 2009;206:637–53. doi: 10.1084/jem.20082109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mallo GV, Kurz CL, Couillault C, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–14. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- [41].Kato Y, Aizawa T, Hoshino H, Kawano K, Nitta K, Zhang H. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem J. 2002;361:221–30. doi: 10.1042/0264-6021:3610221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].London R, Orozco BS, Mylonakis E. The pursuit of cryptococcal pathogenesis: heterologous hosts and the study of cryptococcal host-pathogen interactions. FEMS Yeast Res. 2006;6:567–73. doi: 10.1111/j.1567-1364.2006.00056.x. [DOI] [PubMed] [Google Scholar]
- [43].Mylonakis E, Ausubel FM, Tang RJ, Calderwood SB. The art of serendipity: killing of Caenorhabditis elegans by human pathogens as a model of bacterial and fungal pathogenesis. Expert Rev Anti Infect Ther. 2003;1:167–73. doi: 10.1586/14787210.1.1.167. [DOI] [PubMed] [Google Scholar]
- [44].Chamilos G, Lionakis MS, Lewis RE, et al. Drosophila melanogasler as a facile model for large-scale studies of virulence mechanisms and antifungal drug efficacy in Candida species. J Infect Dis. 2006;193:1014–22. doi: 10.1086/500950. [DOI] [PubMed] [Google Scholar]
- [45].C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- [46].Kamath RS, Fraser AG, Dong Y, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- [47].Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–66. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- [48].Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–49. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- [49].Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 2007;3:e18. doi: 10.1371/journal.ppat.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fuchs BB, Eby J, Nobile CJ, El Khoury JB, Mitchell AP, Mylonakis E. Role of filamentation in Galleria me lionella killing by Candida albicans. Microbes Infect. 2010;12:488–96. doi: 10.1016/j.micinf.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E. Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell. 2009;8:1750–8. doi: 10.1128/EC.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Miyata S, Begun J, Troemel ER, Ausubel FM. DAF-16-dependent suppression of immunity during reproduction in Caenorhabditis elegans. Genetics. 2008;178:903–18. doi: 10.1534/genetics.107.083923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kim DH, Feinbaum R, Alloing G, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–6. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- [54].Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcrlp. Curr Biol. 2005;15:1150–5. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- [55].Moy TI, Ball AR, Anklesaria Z, Casadei G, Lewis K, Ausubel FM. Identification of novel antimicrobials using a live-animal infection model. Proc Natl Acad Sci USA. 2006;103:10414–9. doi: 10.1073/pnas.0604055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tomkiewicz D, Casadei G, Larkins-Ford J, et al. Berberine-INF55 (5-nitro-2-phenylindole) hybrid antimicrobials: effects of varying the relative orientation of the berberine and INF55 components. Antimicrob Agents Chemother. 2010;54:3219–24. doi: 10.1128/AAC.01715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Rasmussen TB, Bjarnsholt T, Skindersoe ME, et al. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol. 2005;187:1799–814. doi: 10.1128/JB.187.5.1799-1814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tampakakis E, Okoli I, Mylonakis E. A C. elegans-based, whole animal, in vivo screen for the identification of antifungal compounds. Nat Protoc. 2008;3:1925–31. doi: 10.1038/nprot.2008.193. [DOI] [PubMed] [Google Scholar]
- [59].Okoli I, Coleman JJ, Tampakakis E, et al. Identification of antifungal compounds active against Candida albicans using an improved high-throughput Caenorhabditis elegans assay. PLoS One. 2009;4:e7025. doi: 10.1371/journal.pone.0007025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gosai SJ, Kwak JH, Luke CJ, et al. Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin alphal-antitrypsin Z. PLoS One. 2010;5:e15460. doi: 10.1371/journal.pone.0015460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Moy TI, Conery AL, Larkins-Ford J, et al. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem Biol. 2009;4:527–33. doi: 10.1021/cb900084v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Coleman JJ, Okoli I, Tegos GP, et al. Characterization of plant-derived saponin natural products against Candida albicans. ACS Chem Bioi. 2010;5:321–32. doi: 10.1021/cb900243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Demidova TN, Hamblin MR. Photodynamic therapy targeted to pathogens. Int J Immunopathol Pharmacol. 2004;17:245–54. doi: 10.1177/039463200401700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hamblin MR, Hasan T. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem Photobiol Sci. 2004;3:436–50. doi: 10.1039/b311900a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fuchs BB, Tegos GP, Hamblin MR, Mylonakis E. Susceptibility of Cryptococcus neoformans to photodynamic inactivation is associated with cell wall integrity. Antimicrob Agents Chemother. 2007;51:2929–36. doi: 10.1128/AAC.00121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- [67].Sochova I, Hofman J, Holoubek I. Using nematodes in soil ecotoxicology. Environ Int. 2006;32:374–83. doi: 10.1016/j.envint.2005.08.031. [DOI] [PubMed] [Google Scholar]
- [68].Williams PL, Dusenbery DB. Using the nematode Caenorhabditis elegans to predict mammalian acute lethality to metallic salts. Toxicol Ind Health. 1988;4:469–78. doi: 10.1177/074823378800400406. [DOI] [PubMed] [Google Scholar]
- [69].Cole RD, Anderson GL, Williams PL. The nematode Caenorhabditis elegans as a model of organophosphate-induced mammalian neurotoxicity. Toxicol App1 Pharmacol. 2004;194:248–56. doi: 10.1016/j.taap.2003.09.013. [DOI] [PubMed] [Google Scholar]
- [70].Hu Y, Platzer EG, Bellier A, Aroian RV. Discovery of a highly synergistic anthelmintic combination that shows mutual hypersusceptibility. Proc Natl Acad Sci USA. 2010;107:5955–60. doi: 10.1073/pnas.0912327107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cox GN, Kusch M, Edgar RS. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J Cell Biol. 1981;90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Avery L, Shtonda BB. Food transport in the C. elegans pharynx. J Exp Biol. 2003;206:2441–57. doi: 10.1242/jeb.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lindblom TH, Dodd AK. Xenobiotic detoxification in the nematode Caenorhabditis elegans. J Exp Zool A Comp Exp Biol. 2006;305:720–30. doi: 10.1002/jez.a.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kwok TC, Ricker N, Fraser R, et al. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature. 2006;441:91–5. doi: 10.1038/nature04657. [DOI] [PubMed] [Google Scholar]
- [75].Burns AR, Kwok TC, Howard A, et al. High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nat Protoc. 2006;1:1906–14. doi: 10.1038/nprot.2006.283. [DOI] [PubMed] [Google Scholar]
- [76].Burns AR, Wallace IM, Wildenhain J, et al. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chern Biol. 2010;6:549–57. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- [77].Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–61. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- [78].Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- [79].Hopkins AL, Bickerton GR. Drug discovery: Know your chemical space. Nat Chern Biol. 2010;6:482–3. doi: 10.1038/nchembio.395. [DOI] [PubMed] [Google Scholar]
- [80].Liu B, Zhu F, Huang Y, et al. Screening rules for leads of fungicides, herbicides, and insecticides. J Agric Food Chern. 2010;58:2673–84. doi: 10.1021/jf902639x. [DOI] [PubMed] [Google Scholar]
- [81].Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–17. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Akins RA. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med Mycol. 2005;43:285–318. doi: 10.1080/13693780500138971. [DOI] [PubMed] [Google Scholar]
- [83].Carrilo-Munoz AJ, Tur C, Torres J. In-vitro antifungal activity of sertaconazole, bifonazole, ketoconazole, and miconazole against yeasts of the Candida genus. J Antimicrob Chemother. 1996;37:815–9. doi: 10.1093/jac/37.4.815. [DOI] [PubMed] [Google Scholar]
- [84].Beggs WH. Influence of growth phase on the susceptibility of Candida albicans to butoconazole, oxiconazole, and sulconazole. J Antimicrob Chemother. 1985;16:397–9. doi: 10.1093/jac/16.3.397. [DOI] [PubMed] [Google Scholar]
- [85].Hernandez Molina JM, Llosa J, Martinez Brocal A, Ventosa A. In vitro activity of cloconazole, sulconazole, butoconazole, isoconazole, fenticonazole, and five other antifungal agents against clinical isolates of Candida albicans and Candida spp. Mycopathologia. 1992;118:15–21. doi: 10.1007/BF00472565. [DOI] [PubMed] [Google Scholar]
- [86].Schafer-Korting M, Blechschmidt J, Korting HC. Clinical use of oral nystatin in the prevention of systemic candidosis in patients at particular risk. Mycoses. 1996;39:329–39. doi: 10.1111/j.1439-0507.1996.tb00149.x. [DOI] [PubMed] [Google Scholar]
- [87].Pariser DM, Pariser RJ. Oxiconazole nitrate lotion, 1 percent: an effective treatment for tinea pedis. Cutis. 1994;54:43–4. [PubMed] [Google Scholar]
- [88].Pfaller MA, Gerarden T. Susceptibility of clinical isolates of Candida spp. to terconazole and other azole antifungal agents. Diagn Microbiol Infect Dis. 1989;12:467–71. doi: 10.1016/0732-8893(89)90079-5. [DOI] [PubMed] [Google Scholar]
- [89].Arai T, Kouama Y, Suenaga T, Honda H. Ascomycin, an antifungal antibiotic. J Antibiot (Tokyo) 1962;15:231–2. [PubMed] [Google Scholar]
- [90].High KP, Washburn RG. Invasive aspergillosis in mice immunosuppressed with cyclosporin A, tacrolimus (FK506), or sirolimus (rapamycin) J Infect Dis. 1997;175:222–5. doi: 10.1093/infdis/175.1.222. [DOI] [PubMed] [Google Scholar]
- [91].Kino T, Hatanaka H, Hashimoto M, et al. FK-506, a novel immunosuppressant isolated from a Streptomyces. I. Fermentation, isolation, and physico-chemical and biological characteristics. J Antibiot (Tokyo) 1987;40:1249–55. doi: 10.7164/antibiotics.40.1249. [DOI] [PubMed] [Google Scholar]
- [92].Kino T, Hatanaka H, Miyata S, et al. FK-506, a novel immunosuppressant isolated from a Streptomyces. II. Immunosuppressive effect of FK-506 in vitro. J Antibiot (Tokyo) 1987;40:1256–65. doi: 10.7164/antibiotics.40.1256. [DOI] [PubMed] [Google Scholar]
- [93].Ohtani K, Mavi S, Hostettmann K. Molluscicidal and antifungal triterpenoid saponins from Rapanea melanophloeos leaves. Phytochemistry. 1993;33:83–86. [Google Scholar]
- [94].Sata N, Matsunaga S, Fusetani N, Nishikawa H, Takamura S, Saito T. New antifungal and cytotoxic steroidal saponins from the bulbs of an elephant garlic mutant. Biosci Biotechnol Biochem. 1998;62:1904–11. doi: 10.1271/bbb.62.1904. [DOI] [PubMed] [Google Scholar]
- [95].Carotenuto A, Fattorusso E, Lanzotti V, Magno S. Spirostanol saponins of Allium porrum L. Phytochemistry. 1999;51:1077–82. doi: 10.1016/s0031-9422(98)00712-2. [DOI] [PubMed] [Google Scholar]
- [96].Germonprez N, Maes L, Van Puyvelde L, Van Tri M, Tuan DA, De Kimpe N. In vitro and in vivo anti-leishmanial activity of triterpenoid saponins isolated from Maesa balansae and some chemical derivatives. J Med Chem. 2005;48:32–7. doi: 10.1021/jm031150y. [DOI] [PubMed] [Google Scholar]
- [97].Herlt AJ, Mander LN, Pongoh E, Rumampuk RJ, Tarigan P. Two major saponins from seeds of Barringtonia asiatica: putative antifeedants toward Epilachna sp. larvae. J Nat Prod. 2002;65:115–20. doi: 10.1021/np000600b. [DOI] [PubMed] [Google Scholar]
- [98].Liu HW, Yao XS, Wang NL, Cai GP. A New Triterpenoid Spaonin Isolated from the Seeds of Aesculus assamica Griff. Chinese Chemical Letters. 2006;17:211–214. [Google Scholar]
- [99].Banskota AH, Tezuka Y, Kadota S. Recent progress in pharmacological research of propolis. Phytother Res. 2001;15:561–71. doi: 10.1002/ptr.1029. [DOI] [PubMed] [Google Scholar]
- [100].Kinashi H, Someno K, Sakaguchi K. Isolation and characterization of concanamycins A, Band C. J Antibiot (Tokyo) 1984;37:1333–43. doi: 10.7164/antibiotics.37.1333. [DOI] [PubMed] [Google Scholar]
- [101].Della Casa V, Noll H, Gonser S, Grob P, Graf F, Pohlig G. Antimicrobial activity of dequalinium chloride against leading germs of vaginal infections. Arzneimittelforschung. 2002;52:699–705. doi: 10.1055/s-0031-1299954. [DOI] [PubMed] [Google Scholar]
- [102].Auclair C. Multimodal action of antitumor agents on DNA: the ellipticine series. Arch Biochem Biophys. 1987;259:1–14. doi: 10.1016/0003-9861(87)90463-2. [DOI] [PubMed] [Google Scholar]
- [103].Petrou MA, Rogers TR. In-vitro activity of antifungal agents in combination with four quinolones. Drugs Exp Clin Res. 1988;14:9–18. [PubMed] [Google Scholar]
- [104].Teixeira MJ, de Almeida YM, Viana JR, et al. In vitro and in vivo Leishmanicidal activity of 2-hydroxy-3-(3-methyl-2-butenyl)-1,4-naphthoquinone (lapachol) Phytother Res. 2001;15:44–8. doi: 10.1002/1099-1573(200102)15:1<44::aid-ptr685>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- [105].Lima NM, Correia CS, Leon LL, et al. Antileishmanial activity of lapachol analogues. Mem Inst Oswaldo Cruz. 2004;99:757–61. doi: 10.1590/s0074-02762004000700017. [DOI] [PubMed] [Google Scholar]
- [106].Cho BP, Yang T, Blankenship LR, et al. Synthesis and characterization of N-demethylated metabolites of malachite green and leucomalachite green. Chem Res Toxicol. 2003;16:285–94. doi: 10.1021/tx0256679. [DOI] [PubMed] [Google Scholar]
- [107].Allen JL, Gofus JE, Meinertz JR. Determination of malachite green residues in the eggs, fry, and adult muscle tissue of rainbow trout (Oncorhynchus mykiss) J AOAC Int. 1994;77:553–7. [PubMed] [Google Scholar]
- [108].Bacigalupo A, Van Lint MT, Frassoni F, Podesta M, Marmont A, Colombo L. Mepartricin: a new antifungal agent for the treatment of disseminated Candida infections in the immunocompromised host. Acta Haematol. 1983;69:409–13. doi: 10.1159/000206930. [DOI] [PubMed] [Google Scholar]
- [109].Matsumura F, Gotoh Y, Boush GM. Phenylmercuric acetate: metabolic conversion by microorganisms. Science. 1971;173:49–51. doi: 10.1126/science.173.3991.49. [DOI] [PubMed] [Google Scholar]
- [110].Roba J, Claeys M, Lambelin G. Antiplatelet and antithrombogenic effects of suloctidil. Eur J Pharmacol. 1976;37:265–74. doi: 10.1016/0014-2999(76)90034-0. [DOI] [PubMed] [Google Scholar]
- [111].Berg D, Draber W, von Hugo H, Hummel W, Mayer D. The effect of clotrimazole and triadimefon on 3-hydroxy-3-methyl-glutaryl-CoA-reductase-[EC 1.1.1.34]-activity in Saccharomyces cerevisiae. Z Naturforsch C. 1981;36:798–803. [PubMed] [Google Scholar]
- [112].Cockrell RS, Harris EJ, Pressman BC. Energetics of potassium transport in mitochondria induced by valinomycin. Biochemistry. 1966;5:2326–35. doi: 10.1021/bi00871a022. [DOI] [PubMed] [Google Scholar]
- [113].Watanabe H, Azuma M, Igarashi K, Ooshima H. Valinomycin affects the morphology of Candida albicans. J Antibiot (Tokyo) 2005;58:753–8. doi: 10.1038/ja.2005.102. [DOI] [PubMed] [Google Scholar]


