Abstract
Objective. Nevirapine is metabolized by cytochrome P450 (CYP) 2B6 and CYP3A4. We characterized relationships between clinical parameters, human genetics, pharmacokinetics, and human immunodeficiency virus type 1 (HIV-1) drug resistance mutations in pregnant women following single-dose intrapartum nevirapine.
Methods. In AIDS Clinical Trials Group study A5207, women received nevirapine at onset of labor and were randomly assigned to receive lamivudine/zidovudine, emtricitabine/tenofovir, or lopinavir/ritonavir for 7 or 21 days. Plasma nevirapine level was quantified on postpartum day 1 and on weeks 1, 3, and 5. We assayed 214 polymorphisms in CYP2B6 and other genes and evaluated associations with pharmacokinetic parameters, including elimination constant, time to protein-adjusted 50% inhibitory concentration (IC50), and week 5 nevirapine level below the quantification limit.
Results. Among 301 women with evaluable pharmacokinetic and genotype data, lower body mass index and random assignment to receive lopinavir/ritonavir were associated with more rapid nevirapine elimination. Among those of African ancestry, longer time to IC50 was associated with CYP2B6 983T→C (P = .004) but not with CYP2B6 516G→T (P = .8). Among Indians, slower nevirapine elimination was associated with CYP2B6 516G→T (P = .04). Emergent resistance was infrequent and not associated with pharmacokinetics or CYP2B6 genotype.
Conclusions. The effects on plasma drug exposure following single-dose nevirapine may be greater for CYP2B6 983T→C than for 516G→T and are less pronounced than at steady state.
Keywords: nevirapine, pharmacokinetics, CYP2B6, pharmacogenetics, pregnancy
Nevirapine has been a cornerstone of efforts to prevent mother-to-child transmission of human immunodeficiency virus type 1 (HIV-1) for more than a decade. In a study of HIV-infected pregnant women in Uganda, a single intrapartum dose of nevirapine 200 mg to the mother, followed by a single 2 mg/kg dose to the newborn, reduced mother-to-child transmission during the first several months of life by nearly 50% [1]. Due to its low genetic barrier to viral resistance and long plasma half-life, single-dose nevirapine without concomitant antiretrovirals selects for nevirapine-resistant HIV-1 in a substantial proportion of women [2], with increased risk possibly associated with higher plasma nevirapine concentrations [3]. Coadministering other antiretrovirals to cover the nevirapine “tail” decreases selection for drug-resistant HIV [4–8].
The median plasma half-life of single-dose nevirapine (200 mg) ranges from approximately 45 to 75 hours, with considerable interindividual variability [9–12]. Nevirapine is metabolized primarily by cytochrome P450 (CYP) 2B6 and CYP3A4, with secondary metabolism by other CYPs [13–15]. Nevirapine may also be a weak substrate for the efflux transporter P-glycoprotein (encoded by ABCB1) [16]. Repeated dosing of nevirapine increases plasma clearance by inducing pregnane X receptor (encoded by NR1I2) and perhaps other nuclear receptors [17], which upregulate drug metabolism and transport genes, including CYP2B6, CYP3A4, CYP3A5, and ABCB1 [18].
Retrospective analyses using data from AIDS Clinical Trials Group (ACTG) A5097s first showed that CYP2B6 516G→T (rs3745274) predicts increased steady-state plasma exposure of efavirenz [19], also a CYP2B6 substrate [20]. Subsequent studies associated higher steady-state plasma nevirapine exposure with CYP2B6 516G→T in HIV-infected patients in Europe [21–23], Africa [24], North America [25], India [26], and Southeast Asia [27, 28]. Following single-dose nevirapine, however, pharmacokinetic associations with CYP2B6 516G→T have not been apparent among nonpregnant, HIV-uninfected African Americans [12] nor among pregnant, HIV-infected women in Thailand [29].
A less frequent CYP2B6 polymorphism, 983T→C (rs28399499), also predicts steady-state increased plasma efavirenz exposure [22, 30, 31]. Data regarding 983T→C and nevirapine are lacking. Recently, a third CYP2B6 polymorphism, 15582C→T (rs4803419), has been shown to be associated with steady-state pharmacokinetics of both nevirapine and efavirenz [32, 33]. Both CYP2B6 516G→T and 15582C→T are much more frequent than 983T→C. In addition, CYP2B6 983T→C has been reported almost exclusively with African ancestry, CYP2B6 516G→T is generally more frequent with African ancestry than with European or Asian ancestry, and 15582C→T is generally more frequent with Asian or European ancestry than with African ancestry [34].
The present study characterizes clinical and genetic determinants of interindividual variability in plasma nevirapine exposure following a single intrapartum dose among pregnant women who were randomly assigned to receive lamivudine/zidovudine, emtricitabine/tenofovir, or lopinavir/ritonavir for 7 or 21 days. Genetic analyses focus primarily on CYP2B6 516G→T, 983T→C, 15582C→T, and rs7251950 and explore additional polymorphisms in ABCB1, CYP2B6, CYP2C19, CYP3A4, CYP3A5, and NR1I2. We hypothesized that drug metabolism and/or transport gene variants would be associated with prolonged plasma nevirapine exposure and, consequently, with increased likelihood of the emergence of nevirapine-resistant HIV-1.
METHODS
Study Participants
ACTG A5207 evaluated strategies to prevent emergent nevirapine resistance following intrapartum single-dose nevirapine. Women were enrolled in 8 sites in Haiti, India, Malawi, South Africa, Tanzania, and Uganda. At onset of labor, women received single-dose nevirapine (200 mg) and were randomized to receive either 7- or 21-day courses of lamivudine/zidovudine (150 mg/300 mg) twice daily, emtricitabine/tenofovir (200 mg/300 mg) once daily, or lopinavir/ritonavir (400 mg/100 mg) twice daily. The first dose of the latter antiretrovirals was administered together with the single dose of nevirapine. Women enrolled in Africa or Haiti are hereafter referred to as being of African ancestry. This study complied with the Helsinki Declaration and was approved by institutional review boards for each site, and women gave written informed consent.
Nevirapine Assays
Plasma specimens for the nevirapine assay were obtained on postpartum day 1 and on weeks 1, 3, and 5. Whole blood was collected into ethylenediaminetetraacetic acid tubes and centrifuged at 800 × g for 10 minutes, and the supernatant plasma was transferred to a fresh tube and centrifuged again for 10 minutes. Plasma was maintained at −70°C and shipped on dry ice until assayed. Nevirapine was quantified using a validated high-performance liquid chromatography (HPLC) method. Following liquid-liquid extraction (LLE) in 25:75 (by volume) methylene chloride (MeCl):tert-butyl methyl ether (tBME), 200-μL samples were separated via reversed-phase liquid chromatography on a Microsorb MV C8 analytical column under isocratic conditions (70% 50 mM KH2PO4, pH 3.0; 30% acetonitrile), with a 10-minute run time. UV detection at 284 nm provided adequate sensitivity with minimal interference from endogenous matrix components. The assay was validated from 50–10 000 ng/mL. Intraday and interday precision and accuracy are within 15% of nominal values. Approximately 18% of samples were analyzed using a separate HPLC method in a different laboratory. In this method, analyte was extracted from a 250-µL sample using a bonded silica solid-phase extraction column and analyzed chromatographically on a reversed-phase, 150 × 4.6 mm (length × internal diameter), 5-µm particle, Supelco LC-8 analytical column. Separation was achieved with an isocratic mobile phase of 63% phosphate buffer (0.025 M, pH 6.0) with 1-butanesulfonic acid as anion-pair reagent (21.5% methanol:15.5% acetonitrile). The ion-pair reagent was used to separate nevirapine from endogenous plasma components. Peaks were detected at a flow rate of 1.0 mL/minute at a wavelength of 280 nm, with an 8-minute run time. The assay is linear from 25–10 000 ng/mL. Specimens with results below the limit of assay quantification (either 50 or 25 ng/mL) were secondarily assayed by a validated liquid chromatography with tandem mass spectrometry (LC/MS/MS) method. After addition of internal standard (BIRH-414), a LLE procedure by means of 25:75 (by volume) MeCl:tBME was used for analyte extraction. Reversed-phase chromatographic separation was performed on a Waters Atlantis dC18 (2.1 × 100 mm, 3 μm particle size) analytical column under isocratic conditions. A binary mobile phase comprising 0.1% formic acid in water and methanol (45:55) provided adequate separation. Detection and quantitation were achieved by multiple reactions monitoring, and nevirapine and the internal standard were detected using the following transitions for protonated molecular products [M + H]+: m/z NVP 267.2 → 226.1; BIRH-414 255.2 → 227.1. The dynamic range is 0.25–100 ng/mL, using 200 μL of human plasma. All assay methods have been validated internally and by twice-yearly external proficiency testing through the DAIDS Clinical Pharmacology Quality Assurance and Quality Control Program.
Pharmacokinetic Parameters
To estimate nevirapine elimination constants (assumed to be linear on the log10 scale) for each woman, we used nevirapine concentration data from day 1, week 1, and week 3. Nevirapine concentrations below the lower limit of quantification were imputed to one-half of the limit of quantification threshold. Nevirapine elimination constants (ie, slopes of nevirapine decay) were estimated by linear mixed-effects models. Estimates were obtained separately for each race/ethnicity group. Nevirapine elimination constants based on linear mixed-effects models were used for genetic association analyses.
For week 5 nevirapine below the limit of quantification status, data from each of the 301 women were dichotomized as being either above or below limit of quantification. Lower limit of quantification specific to the LC/MS/MS method was used to determine whether the concentration was above or below the limit of quantification. Based on the individual-specific random slopes estimated from a linear mixed-effects model, we estimated, for each woman, the time when the concentration reached 46 ng/mL (nevirapine protein-adjusted IC50 [35]). This derived pharmacokinetic parameter, time to reach the IC50, was used as an additional dependent variable in association analyses.
Viral Resistance Testing by Allele-Specific Polymerase Chain Reaction (PCR)
In A5207, the presence of nevirapine-resistant virus in plasma at baseline and 6 weeks after completing study treatment was assessed using a highly sensitive allele-specific PCR assay, using methods previously described [36]. Plasma samples with an HIV-1 RNA load of ≥5000 copies/mL were tested for the following nevirapine resistance mutations: K103N (either ATT or AAC, allele-specific PCR detection limit 0.1%) and Y181C (allele-specific PCR detection limit 0.3%).
Characterization of Human Genetic Variants
Whole blood was stored at −80°C until DNA was extracted using Autopure systems (Minneapolis, MN). A total of 214 polymorphisms (68 in ABCB1, 55 in CYP2B6, 1 in CYP2C19, 38 in CYP3A4, 1 in CYP3A5, and 51 in NR1I2) were successfully assayed in the Vanderbilt DNA Resources Core, using MassARRAY iPLEX Gold (Sequenom). Our strategy for genotyping was as follows. For CYP2B6, CYP3A4, ABCB1, and NR1I2, we tagged each entire gene by use of SeattleSNPs [37], using a cosmopolitan strategy across populations (Yoruba, Asian, African American, European American, and Hispanic) with a 5% allelic frequency cutoff, a 0.80 threshold for r2, 85% data convergence for tagging polymorphisms, and 70% data convergence for clustering. For CYP2B6, we included 5 kB in each 5′ and 3′ untranslated region (UTR), and for ABCB1 and NR1I2, we included 20 kB in each UTR. The arbitrary inclusion of 5 kB versus 20 kB reflects that these assays were designed at different times. For CYP2B6, additional polymorphisms that were of interest (but were not extremely infrequent, based on a <1% minor allele frequency) were added on the basis of a previous report [38], as were polymorphisms with an allelic frequency of at least 5% in 20 kB of the 5′ UTR identified using Ensembl Genome Browser [39] and upstream polymorphisms possibly associated with CYP2B6 expression [40]. The CYP2B6 assay included 2 polymorphisms in nearby CYP2A6 (rs28399454 and rs28399433). We also included ABCB1 3435C→T (rs1045642) and 2677G/T/A (rs2032582) and CYP3A5 6986A→G (rs776746). The final Sequenom assay design is available on request. Genotypes were confirmed by visual inspection of plots. Laboratory personnel with no knowledge of clinical data performed genotyping. Ample duplicate and blank assays were included to ensure validity.
Statistical Methods
Quantitative analysis of genotype associations with derived pharmacokinetic parameters was conducted using R, version 2.10.1, and PLINK, version 1.07 (available at: http://pngu.mgh.harvard.edu/~purcell/plink/). Associations with single polymorphisms were based on parametric regression (linear or logistic) models, using nevirapine elimination constant, week 5 nevirapine below limit of quantification status, or time to reach protein-adjusted IC50 as the dependent variable and genotypes (coded as 0, 1, and 2 for the number of minor alleles), body mass index (BMI; defined as the weight in kilograms divided by the height in meters squared), and treatment arm (lopinavir/ritonavir vs nucleoside reverse-transcriptase inhibitors [NRTIs]) as independent variables. We grouped by lopinavir/ritonavir versus NRTIs because of known interactions between lopinavir/ritonavir and CYP3A4. P values from linear or logistic regression models were based on the Wald test. All analyses were conducted within each race/ethnicity group and were adjusted for BMI and treatment arm (except for analyses within treatment arm, which were only adjusted for BMI). Relationships between development of HIV-1 nevirapine resistance mutations and BMI, treatment arm, pharmacokinetic parameters, and host genotypes were assessed by the Fisher exact test (for discrete covariates) or the Wilcoxon rank sum test (for continuous covariates). For clinical variables and for genetic variants of primary interest, P values of < .05 were deemed statistically significant. For exploratory genetics associations, Bonferroni thresholds were calculated by dividing the nominal significance threshold by the number of polymorphisms tested.
RESULTS
Participant Characteristics
In ACTG A5207, a total of 422 women received single-dose nevirapine at the time of active labor onset, of whom 416 had at least 1 subsequent nevirapine plasma concentration determination. Of these 416 women, 315 had evaluable human genotype data based on 214 polymorphisms. The primary reason for lack of evaluable genotype data was declined consent for this optional genetic analysis, much of which reflected 1 site that, because of miscommunication, did not receive consent from any participants for genetic analysis. Nevirapine plasma concentration data were available at multiple time points (836 total specimens) from 301 of these 315 women, including 217 (72%) of African ancestry and 84 (28%) who were Indian. All analyses hereafter are based on these 301 women unless otherwise stated. Baseline characteristics of study participants, stratified by lopinavir/ritonavir randomization, are presented in Table 1. These 301 women did not differ significantly from the other 121 women with regard to baseline age, CD4+ T-cell count, and plasma HIV-1 RNA level.
Table 1.
Baseline Characteristics of 301 Women Included in Genetic Analyses
| Characteristic | Total (n = 301) | Lopinavir/r (n = 100) | Non-lopinavir/r (n = 201) |
|---|---|---|---|
| Age, y | 26 (23–30) | 26 (23–30) | 26 (23–30) |
| Weight, kg | 64 (57–74) | 65 (57–77) | 64 (57–73) |
| BMIa | 25.8 (23.5–30.0) | 26.0 (22.8–30.1) | 25.6 (23.7–29.5) |
| African ancestry | 217 (72) | 74 (74) | 143 (71) |
| Indian | 84 (28) | 26 (26) | 58 (29) |
| CD4+ T-cell count, cells/mm3 | 490 (384–668) | 493 (378–727) | 490 (393–661) |
| HIV-1 RNA load, log10 copies/mL | 3.5 (2.8–4.2) | 3.5 (2.7–4.2) | 3.5 (2.8–4.2) |
Data are median (interquartile range) or no. (%) of subjects.
Abbreviations: BMI, body mass index (defined as the weight in kilograms divided by the height in meters squared); HIV-1, human immunodeficiency virus type 1; lopinavir/r, lopinavir/ritonavir.
a Defined as the weight in kilograms divided by the height in meters squared.
Clinical Associations With Nevirapine Pharmacokinetics
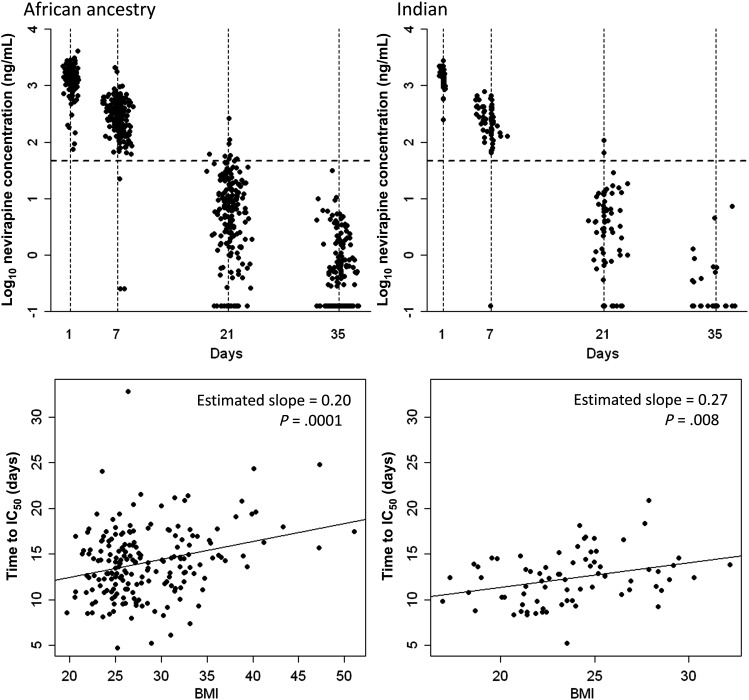
Nevirapine concentrations were obtained from women on postpartum day 1 and on weeks 1, 3, and 5. Scatter plots of measured plasma nevirapine concentrations over time are shown separately for African ancestry and Indian groups in Figure 1. Relationships between BMI, lopinavir/ritonavir status, and pharmacokinetic parameters are presented in Table 2. In both African ancestry and Indian groups, greater baseline BMI was associated with greater elimination constants and longer time to reach IC50. Relationships between BMI and time to IC50 are shown in Figure 1. There was also a modest association between randomization to a lopinavir/ritonavir-containing regimen (as compared to NRTI-containing regimens) and faster elimination, but not time to reach IC50.
Figure 1.
Plasma nevirapine concentration-time profiles following receipt of a single intrapartum 200-mg dose of nevirapine and relationship with body mass index (BMI). Top panels are scatter plots of measured plasma nevirapine concentrations around each time point at 1 day (±1 day), 7 days (±2 days), 21 days (±3 days), and 35 days (±3 days) after dose receipt. The x-axes represent days after dose. The y-axes represent plasma nevirapine concentrations (log10 scale). Horizontal dotted lines indicate the 50% inhibitory concentration (IC50) for nevirapine. Bottom panels represent relationships between BMI and time to reach nevirapine plasma IC50. Left panels denote data for 217 women of African ancestry, and right panels denote data for 84 Indian women.
Table 2.
Univariate Relationships Between Baseline Body Mass Index (BMI), Lopinavir/Ritonavir (LPV/r) Randomization, and Nevirapine Pharmacokinetics
| kel (h−1) |
Time to IC50 (days) |
BLQ at Week 5 |
||||
|---|---|---|---|---|---|---|
| Characteristic | Estimated Mean Difference | Pa | Estimated Mean Difference | Pa | Odds Ratio | Pa |
| African ancestry | ||||||
| Treatment arms | 0.0004 | .047 | 0.40 | .46 | 0.63 | .14 |
| BMIb(per 5-unit increase) | 0.0004 | <.0001 | 0.97 | .0001 | 0.75 | .054 |
| Indian | ||||||
| Treatment arms | 0.0006 | .030 | 1.01 | .14 | 0.48 | .15 |
| BMIb(per 5-unit increase) | 0.0007 | .001 | 1.33 | .008 | 0.30 | .006 |
Abbreviations: BLQ, below lower limit of quantification; IC50, protein-adjusted 50% inhibitory concentration; kel, nevirapine elimination rate constant.
The estimated mean difference represents the average difference in elimination constant (or time to IC50) in the non-LPV/r arm with respect to the LPV/r arm, and per 5-unit increase in BMI for the treatment arm and BMI effects, respectively. The odds ratio represents decreased odds of observing BLQ in the non-LPV arm with respect to the LPV/r arm, and per 5-unit increase in BMI for the treatment arm and BMI effects, respectively.
aBy the Wald test.
bDefined as the weight in kilograms divided by the height in meters squared.
Genetic Variants
A total of 214 genetic variants were successfully assayed. Of the 301 evaluable women, the average genotyping success was 99.7%, with a value of ≥95.8% for each women. Of the 214 polymorphisms, average genotyping success was 99.7%, with a value of ≥97.0% for each polymorphism. Among the 214 polymorphisms assayed, 18 and 61 had minor allele frequencies <0.01 in the African ancestry and Indian groups, respectively, and were excluded from analyses within each population. All polymorphisms passed Hardy-Weinberg equilibrium testing within each population, based on an adjusted P value threshold of ≥ .001, consistent with random assortment of alleles.
Genetic Associations With Nevirapine Pharmacokinetic Parameters
Associations between CYP2B6 polymorphisms of primary interest (516G→T, 983T→C, 15582C→T, and rs7251950) and nevirapine pharmacokinetic parameters are shown in Table 3. CYP2B6 983T→C was associated with longer time for plasma concentrations to fall to the protein-adjusted IC50 in women with African ancestry (P = .004). This corresponds to, on average, increases in time to reach protein adjusted IC50 by 47 hours with each additional CYP2B6 983 C allele (Figure 2). Results for CYP2B6 983 should be interpreted with caution because only 1 woman was homozygous for CYP2B6 983 CC. In Indians, CYP2B6 516G→T was associated with slower nevirapine elimination (P = .039).
Table 3.
Relationships Between Selected Genotypes and Nevirapine (NVP) Pharmacokinetic Parameters
| African Ancestry (n = 217) |
Indian (n = 84) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Polymorphism | No. of Subjects | kel (h−1) | Time to IC50 (d) | NVP Not BLQ at Week 5 | No. of Subjects | kel (h−1) | Time to IC50 (d) | NVP Not BLQ at Week 5 |
| CYP2B6 983T→C | ||||||||
| TT | 181 | 0.0051 ± 0.001 | 13.8 ± 3.2 | 54 | 84 | 0.0057 ± 0.001 | 12.5 ± 2.9 | 50 |
| TC | 35 | 0.0048 ± 0.001 | 15.3 ± 5.4 | 53 | 0 | … | … | … |
| CC | 1 | 0.0031 ± 0.000 | 24.0 ± 0.0 | 100 | 0 | … | … | … |
| Pa | .07 | .004 | .80 | … | … | … | ||
| CYP2B6 516G→T | ||||||||
| GG | 88 | 0.0051 ± 0.001 | 14.0 ± 4.1 | 56 | 29 | 0.0058 ± 0.001 | 12.1 ± 2.4 | 48 |
| GT | 101 | 0.0051 ± 0.001 | 14.0 ± 3.6 | 52 | 39 | 0.0058 ± 0.001 | 12.3 ± 3.0 | 56 |
| TT | 28 | 0.0048 ± 0.001 | 14.2 ± 2.9 | 58 | 16 | 0.0053 ± 0.001 | 13.5 ± 3.2 | 40 |
| Pa | .39 | .80 | .95 | .039 | .07 | .15 | ||
| CYP2B6 15582C→T | ||||||||
| CC | 188 | 0.0051 ± 0.001 | 14.1 ± 3.7 | 52 | 40 | 0.0058 ± 0.001 | 12.3 ± 3.0 | 41 |
| CT | 28 | 0.0050 ± 0.001 | 14.3 ± 3.7 | 64 | 35 | 0.0056 ± 0.001 | 12.7 ± 2.7 | 56 |
| TT | 1 | 0.0070 ± 0.000 | 9.8 ± 0.0 | 100 | 9 | 0.0058 ± 0.001 | 12.3 ± 3.0 | 67 |
| Pa | .75 | .79 | .13 | .96 | .63 | .36 | ||
| CYP2B6 rs7251950 | ||||||||
| CC | 168 | 0.0051 ± 0.001 | 14.1 ± 4.0 | 55 | 43 | 0.0058 ± 0.001 | 12.1 ± 2.9 | 42 |
| CT | 45 | 0.0051 ± 0.001 | 13.6 ± 2.6 | 51 | 33 | 0.0055 ± 0.001 | 13.0 ± 2.8 | 56 |
| TT | 3 | 0.0043 ± 0.001 | 16.5 ± 4.2 | 67 | 8 | 0.0058 ± 0.001 | 12.2 ± 3.2 | 62 |
| Pa | .95 | .68 | .98 | .73 | .40 | .69 | ||
Data are mean ± SD or %, unless otherwise indicated. For kel and time to IC50, mean and SDs are shown for each genotype. NVP not BLQ at week 5 represents the percentage of genotypes that had NVP concentration above BLQ.
Abbreviations: BLQ, below lower limit of quantification; IC50, protein-adjusted 50% inhibitory concentration; kel, NVP elimination rate constant.
a By the Wald test.
Figure 2.
Plasma nevirapine concentration-time profiles stratified by genotype following receipt of a single intrapartum 200-mg dose of nevirapine. Top panels denote data for 217 women of African ancestry, and bottom panels denote data for 84 Indian women. Left panels contain data stratified by CYP2B6 516G→T genotype, and right panels contain data stratified by CYP2B6 983T→C genotype. Mean nevirapine concentrations over time are connected by lines, and error bars represent SDs. The horizontal dotted line indicates the protein-adjusted 50% inhibitory concentration for nevirapine (46 ng/mL).
Among all 214 polymorphisms, association results with nevirapine pharmacokinetics and minor allele frequencies for the 10 with the lowest P values (based on association with time to reach protein-adjusted IC50) are provided for African descent and Indian groups separately in Table 4. Association results and minor allele frequencies for all polymorphisms with minor allele frequencies of >0.01 in each population are provided in the Supplementary Materials. Among all polymorphisms (196 and 153 polymorphisms in African ancestry and Indian groups, respectively), 22 were associated with nevirapine clearance at P < .05 (12 and 10, respectively), 16 were associated with time to reach protein-adjusted IC50 at P < .05 (10 and 6, respectively), and 16 were associated with week 5 below limit of quantification status (7 and 9, respectively). No polymorphism was consistently associated with nevirapine pharmacokinetics in both populations. Polymorphisms with a P value of < .05 were found in ABCB1, CYP2B6 (not including the above target polymorphisms), CYP2C19, CYP3A4, and NR1I2. No polymorphism in any analysis was significant after multiple testing adjustments based on the Bonferroni threshold.
Table 4.
Minor Allele Frequencies and Associations With Nevirapine (NVP) Pharmacokinetics
| Time to IC50 |
kel |
Not BLQ at Week 5 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Polymorphism | Minor Allele | Major Allele | MAF | Statistic | Pa | Statistic | Pa | Statistic | Pa |
| African ancestry | ||||||||||
| CYP2B6 | rs28399499 | C | T | 0.085 | 2.926 | .004 | 1.823 | .070 | 0.256 | .80 |
| CYP2B6 | rs1552222 | A | T | 0.245 | 2.896 | .004 | 2.092 | .038 | −1.097 | .27 |
| NR1I2 | rs1523127 | T | G | 0.106 | 2.693 | .008 | 2.596 | .010 | −0.687 | .49 |
| CYP2B6 | rs12721649 | A | G | 0.200 | 2.472 | .014 | 1.458 | .15 | −0.505 | .61 |
| CYP2B6 | rs28399433 | G | T | 0.070 | 2.409 | .017 | 2.014 | .045 | −0.602 | .55 |
| NR1I2 | rs9821892 | A | T | 0.180 | 2.276 | .024 | 2.451 | .015 | −0.773 | .44 |
| ABCB1 | rs3842 | G | A | 0.221 | 2.261 | .025 | 2.024 | .044 | −1.390 | .16 |
| CYP2C19 | rs4244285 | A | G | 0.153 | 2.066 | .040 | 1.816 | .071 | −2.009 | .044 |
| CYP2B6 | rs10411962 | A | G | 0.142 | 2.062 | .041 | 1.556 | .12 | −0.068 | .95 |
| CYP2B6 | rs36118214 | A | G | 0.224 | 2.010 | .046 | 0.937 | .35 | −0.298 | .77 |
| Indian | ||||||||||
| CYP3A4 | rs28988579 | G | T | 0.036 | −3.581 | .0006 | −3.559 | .0007 | 1.407 | .16 |
| CYP2B6 | rs4802101 | T | C | 0.235 | −2.853 | .006 | −2.607 | .011 | 1.774 | .076 |
| CYP2B6 | rs4803417 | C | A | 0.244 | −2.714 | .008 | −2.476 | .016 | 1.657 | .098 |
| CYP2B6 | rs10418990 | G | C | 0.241 | −2.658 | .010 | −2.424 | .018 | 1.570 | .12 |
| CYP2B6 | rs1987236 | G | A | 0.262 | −2.462 | .016 | −2.024 | .047 | 2.509 | .012 |
| CYP2B6 | rs2099361 | G | T | 0.214 | −2.373 | .020 | −2.224 | .029 | 1.323 | .19 |
| CYP3A4 | rs7801671 | A | C | 0.024 | 1.981 | .052 | 1.327 | .19 | −0.002 | .99 |
| CYP3A4 | rs28988568 | A | C | 0.024 | 1.981 | .052 | 1.327 | .19 | −0.002 | .99 |
| CYP3A4 | rs12114000 | A | G | 0.024 | 1.973 | .052 | 1.318 | .19 | −0.002 | .99 |
| CYP2B6 | rs7246465 | T | C | 0.185 | −1.953 | .055 | −1.431 | .16 | 2.258 | .023 |
Data are for polymorphisms with the 10 lowest P values for association with each pharmacokinetic parameter. For data regarding all 214 polymorphisms, refer to the Supplementary Materials.
Abbreviations: BLQ, below lower limit of quantification; IC50, protein-adjusted 50% inhibitory concentration; kel, NVP elimination rate constant; MAF, minor allele frequency.
a By the Wald test.
Associations With Emergent HIV-1 Nevirapine Resistance Mutations
In A5207, among 140 women evaluable by allele-specific PCR at both baseline and 6 weeks (after study treatment) and with no baseline nevirapine mutations detected, 16 (11%) had emergent nevirapine resistance mutations documented 6 weeks after study treatment. Of these 140 women, 104 were represented in the present pharmacogenetic data set. Among 76 women of African ancestry, 8 with emergent resistance mutations had a median time to nevirapine IC50 of 412 hours (interquartile range, 303–466 hours), while in 68 without emergent resistance mutations, the value was 320 hours (interquartile range, 288–366 hours). Among 28 Indian women, 2 with emergent resistance mutations had a median time to nevirapine IC50 of 388 hours, while in 26 without emergent resistance mutations, the value was 296 hours. No significant associations were detected between emergent resistance mutations and either BMI, lopinavir/ritonavir arm, nevirapine elimination constant, or week 5 below the limit of quantification status. No significant associations were detected with CYP2B6 983T→C in women of African ancestry (P = .33) or with CYP2B6 516G→T in women of African ancestry (P = .79) or Indians (P = .99).
DISCUSSION
Nevirapine, a cornerstone of efforts to prevent mother-to-child transmission of HIV, is metabolized primarily by CYP2B6, with secondary metabolism by CYP3A4 and other CYPs [13–15]. Previous studies have shown strong and independent associations between 3 CYP2B6 polymorphisms (CYP2B6 516G→T, 983T→C, and 15582C→T) and increased steady-state plasma nevirapine and/or efavirenz concentrations [19, 21–28, 30–33]. A provocative finding from the present study is that, among women of African ancestry who received a single 200-mg dose of nevirapine at the onset of labor, CYP2B6 983T→C was significantly associated with a longer time to reach the protein-adjusted nevirapine IC50, while CYP2B6 516G→T and 15582C→T were not. There was an 11% (36-hour) increase in time to IC50 in heterozygous subjects, while on the basis of the additive model, time to IC50 increased by 47 hours (on average) for each additional CYP2B6 983 C allele, adjusting for BMI and treatment. Among Indian women CYP2B6 516G→T was significantly associated with slower nevirapine elimination and tended to be associated with time to reach the protein-adjusted nevirapine IC50, while there was no apparent association with 15582C→T. The CYP2B6 983T→C variant was absent in Indians.
The finding of little or no association between CYP2B6 516G→T and plasma exposure following single-dose nevirapine with African ancestry is consistent with 2 previous reports, one involving 34 nonpregnant, HIV-uninfected African Americans [12] and the other involving 330 pregnant, HIV-infected women in Thailand [29]. We cannot, however, explain the apparent association between CYP2B6 516G→T and plasma exposure among Indian women in the present study.
With regard to the discrepant associations between CYP2B6 516G→T and nevirapine pharmacokinetics under single-dose versus steady-state conditions, it has been speculated that autoinduction may play a role. Autoinduction of CYP2B6 and CYP3A4 requires repeated dosing with nevirapine [29]. Under basal conditions (ie, following a single dose), the difference in levels of hepatic CYP2B6 between 516GG and 516TT homozygotes may be relatively modest. Following autoinduction (ie, at steady state), hepatic CYP2B6 activity may increase considerably more in 516GG homozygotes than in 516TT homozygotes, thus making the differential genetic effect on pharmacokinetics more apparent.
The association between CYP2B6 983T→C and single-dose nevirapine pharmacokinetics despite little or no association with CYP2B6 516G→T suggests a fundamental difference between these polymorphisms. It may be that 983T→C reduces hepatic CYP2B6 expression and/or activity to a much greater extent than does 516G→T, such that its effect is apparent even following a single dose of nevirapine. This is supported by recent evidence that CYP2B6 983T→C has a greater effect on steady-state plasma efavirenz concentrations than does CYP2B6 516G→T [33]. Alternatively, CYP2B6 516G→T may largely affect enzyme inducibility, whereas CYP2B6 983T→C may largely affect enzymatic activity. The lack of association between 15582C→T and single-dose nevirapine pharmacokinetics may reflect this polymorphism's relatively modest effect on CYP2B6 expression and/or activity. The effect of 15582C→T on steady-state plasma efavirenz concentrations was approximately one-half that of CYP2B6 516G→T [33].
In exploratory analysis of 210 additional polymorphisms, associations beyond CYP2B6 516G→T and 983T→C at a P value of < .05 are likely spurious, as none were significant after adjustment for multiple testing, none were consistent across populations, and the polymorphisms were present in almost every gene studied.
There are several implications of these findings. In general, the human genetic variants studied here do not profoundly affect plasma exposure following single-dose nevirapine. For example, although a strong association with the CYP2B6 983 was observed, this polymorphism is rare; in the HapMap Yoruban population, CC genotype is observed in <2% of individuals and is absent from white and Asian populations. Similarly, the effect of BMI was modest. In a multivariable model including the effect of CYP2B6 983T→C and BMI, a 5-unit increase in BMI was associated with, on average, a 22-hour increase in time to IC50. A uniform 200-mg dose of nevirapine is therefore appropriate without the need to individualize the dose on the basis of host genotype or BMI. Women who are heterozygous for CYP2B6 983 T/C or homozygous for CYP2B6 516 T/T may perhaps have a somewhat longer plasma nevirapine exposure, but it is not long enough to warrant regimen modification. We expect that homozygosity for CYP2B6 983 C/C, which, fortunately, is infrequent, will more greatly prolong nevirapine exposure following single-dose nevirapine, but data are limited.
The present study had limitations. Because concomitant antiretroviral were prescribed to cover the nevirapine “tail,” we are pleased that very few women experienced emergent nevirapine resistance mutations. This, however, limited our power to detect associations between viral resistance, pharmacokinetic parameters, and genetic variants. We only studied women of African ancestry and Indians, and results may differ in other populations. A candidate gene approach was used on the basis of a priori knowledge. More extensive, high-throughput genotyping might identify novel associations.
Mother-to-child transmission of HIV-1 is most effectively prevented by initiating combination antiretroviral therapy (cART) as early as week 14 of gestation and continuing cART until cessation of breast-feeding [41, 42]. This approach is not always feasible, particularly in resource-limited countries. On the basis of available data, the 2010 World Health Organization consensus guidelines for preventing mother-to-child transmission recommend 2 options: (1) administration of cART as described above or (2) administration of twice-daily zidovudine during pregnancy; single-dose nevirapine, zidovudine, and lamivudine during labor and delivery; and twice-daily zidovudine and lamivudine for 7 days postpartum [43].
In summary, among women who received a single 200-mg dose of nevirapine at the onset of labor, CYP2B6 983T→C may more greatly affect nevirapine exposure than CYP2B6 516G→T. These data suggest that single-dose nevirapine regimens do not require individual dose adjustment on the basis of host genotype.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the AIDS Clinical Trials Group (ACTG) funded by the National Institute of Allergy and Infectious Diseases, National Institute of Health (ACTG Network Leadership Grant: (U01 AI068636); ACTG Statistical and Data Analysis Center Grant: [U01 AI068634]). This work was also supported by the National Institute of Allergy and Infectious Diseases (University of Pittsburgh clinical trials unit Grant [U01 AI069494]), by Virology Support Laboratory Subcontract (204VC009) of the ACTG Central Group, (U01 68636), Vanderbilt University clinical trials unit Grant (U01 AI069439), (RO1 AI077505) to D. W. H., and the Vanderbilt Meharry Center for AIDS Research (P30 AI54999). In addition, the following clinical research sites (CRS) enrolled women to the study: Joint Clinical Research Center CRS, Kampala, Uganda (U01 AI069501); Blantyre, Malawi (U01 AI069518); YRG CARE Medical Centre, VHS Chennai CRS, Chennai, India (U01 AI069432); Les Centres GHESKIO CRS, Port-au-Prince, Haiti (U01 AI069421); Durban Adult HIV CRS, Durban, South Africa (U01 AI069426); WITS HIV CRS, Johannesburg, South Africa (U01 AI38858, U01 AI69463); Kilimanjaro Christian Medical Centre, Moshi, Tanzania (U01 AI069484); and BJ Medical College CRS, Pune, India (U01 AI069417). Study drugs were provided by Abbott Laboratories, Boehringer-Ingelheim, Gilead Sciences, and GlaxoSmithKline.
Potential conflicts of interest. U. L. serves on the advisory boards of Boehringer-Ingelheim and GSK for asthma and chronic obstructive pulmonary disease, has received honoraria from Boehringer-Ingelheim and GSK for lectures, and has received support to attend respiratory scientific meetings. D. M. has been supported by research grants to the University of Pittsburgh from Gilead Sciences. D. W. H. has been supported by research grants to Vanderbilt University from Boehringer-Ingelheim, Gilead Sciences, and Merck. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Guay LA, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 2.Jackson JB, Becker-Pergola G, Guay LA, et al. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS. 2000;14:F111–5. doi: 10.1097/00002030-200007280-00001. [DOI] [PubMed] [Google Scholar]
- 3.Aizire J, McConnell MS, Mudiope P, et al. Kinetics of nevirapine and its impact on HIV-1 RNA levels in maternal plasma and breast milk over time after perinatal single dose nevirapine. JAIDS. 2012;60:483–8. doi: 10.1097/QAI.0b013e318246bf9e. [DOI] [PubMed] [Google Scholar]
- 4.McIntyre JA, Hopley M, Moodley D, et al. Efficacy of short-course AZT plus 3TC to reduce nevirapine resistance in the prevention of mother-to-child HIV transmission: a randomized clinical trial. PLoS Med. 2009;6:e1000172. doi: 10.1371/journal.pmed.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi BH, Ellis GM, Chintu N, et al. Intrapartum tenofovir and emtricitabine reduces low-concentration drug resistance selected by single-dose nevirapine for perinatal HIV prevention. AIDS Res Hum Retroviruses. 2009;25:1099–106. doi: 10.1089/aid.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrive E, Chaix ML, Nerrienet E, et al. Tolerance and viral resistance after single-dose nevirapine with tenofovir and emtricitabine to prevent vertical transmission of HIV-1. AIDS. 2009;23:825–33. doi: 10.1097/QAD.0b013e32832949d5. [DOI] [PubMed] [Google Scholar]
- 7.Chi BH, Sinkala M, Mbewe F, et al. Single-dose tenofovir and emtricitabine for reduction of viral resistance to non-nucleoside reverse transcriptase inhibitor drugs in women given intrapartum nevirapine for perinatal HIV prevention: an open-label randomised trial. Lancet. 2007;370:1698–705. doi: 10.1016/S0140-6736(07)61605-5. [DOI] [PubMed] [Google Scholar]
- 8.Lallemant M, Ngo-Giang-Huong N, Jourdain G, et al. Efficacy and safety of 1-month postpartum zidovudine-didanosine to prevent HIV-resistance mutations after intrapartum single-dose nevirapine. Clin Infect Dis. 2010;50:898–908. doi: 10.1086/650745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunz A, Frank M, Mugenyi K, et al. Persistence of nevirapine in breast milk and plasma of mothers and their children after single-dose administration. J Antimicrob Chemother. 2009;63:170–7. doi: 10.1093/jac/dkn441. [DOI] [PubMed] [Google Scholar]
- 10.Muro E, Droste JA, Hofstede HT, Bosch M, Dolmans W, Burger DM. Nevirapine plasma concentrations are still detectable after more than 2 weeks in the majority of women receiving single-dose nevirapine: implications for intervention studies. JAIDS. 2005;39:419–21. doi: 10.1097/01.qai.0000167154.37357.f9. [DOI] [PubMed] [Google Scholar]
- 11.Mirochnick M, Siminski S, Fenton T, Lugo M, Sullivan JL. Nevirapine pharmacokinetics in pregnant women and in their infants after in utero exposure. Pediat Infect Dis J. 2001;20:803–5. doi: 10.1097/00006454-200108000-00017. [DOI] [PubMed] [Google Scholar]
- 12.Haas DW, Gebretsadik T, Mayo G, et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J Infect Dis. 2009;199:872–80. doi: 10.1086/597125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson DA, Mather G, Trager WF, Levy RH, Keirns JJ. Characterization of the in vitro biotransformation of the HIV-1 reverse transcriptase inhibitor nevirapine by human hepatic cytochromes P-450. Drug Metab Dispos. 1999;27:1488–95. [PubMed] [Google Scholar]
- 14.Wang H, Tompkins LM. CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Current Drug Metab. 2008;9:598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen B, Chen Y, Fitch WL. Metabolic activation of nevirapine in human liver microsomes: dehydrogenation and inactivation of cytochrome P450 3A4. Drug Metab Dispos. 2009;37:1557–62. doi: 10.1124/dmd.108.024851. [DOI] [PubMed] [Google Scholar]
- 16.Almond LM, Edirisinghe D, Dalton M, Bonington A, Back DJ, Khoo SH. Intracellular and plasma pharmacokinetics of nevirapine in human immunodeficiency virus-infected individuals. Clin Pharm Therap. 2005;78:132–42. doi: 10.1016/j.clpt.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Riska P, Lamson M, MacGregor T, et al. Disposition and biotransformation of the antiretroviral drug nevirapine in humans. Drug Metab Dispos. 1999;27:895–901. [PubMed] [Google Scholar]
- 18.Kojima K, Nagata K, Matsubara T, Yamazoe Y. Broad but distinct role of pregnane x receptor on the expression of individual cytochrome p450s in human hepatocytes. Drug Metab Pharmacokinet. 2007;22:276–86. doi: 10.2133/dmpk.22.276. [DOI] [PubMed] [Google Scholar]
- 19.Haas DW, Ribaudo HJ, Kim RB, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–400. [PubMed] [Google Scholar]
- 20.Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharm Exp Therap. 2003;306:287–300. doi: 10.1124/jpet.103.049601. [DOI] [PubMed] [Google Scholar]
- 21.Rotger M, Colombo S, Furrer H, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genom. 2005;15:1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Wyen C, Hendra H, Vogel M, et al. Impact of CYP2B6 983T→C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother. 2008;61:914–8. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahungu T, Smith C, Turner F, et al. Cytochrome P450 2B6 516G→T is associated with plasma concentrations of nevirapine at both 200 mg twice daily and 400 mg once daily in an ethnically diverse population. HIV Med. 2009;10:310–7. doi: 10.1111/j.1468-1293.2008.00689.x. [DOI] [PubMed] [Google Scholar]
- 24.Penzak SR, Kabuye G, Mugyenyi P, et al. Cytochrome P450 2B6 (CYP2B6) G516T influences nevirapine plasma concentrations in HIV-infected patients in Uganda. HIV Med. 2007;8:86–91. doi: 10.1111/j.1468-1293.2007.00432.x. [DOI] [PubMed] [Google Scholar]
- 25.Saitoh A, Sarles E, Capparelli E, et al. CYP2B6 genetic variants are associated with nevirapine pharmacokinetics and clinical response in HIV-1-infected children. AIDS. 2007;21:2191–9. doi: 10.1097/QAD.0b013e3282ef9695. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran G, Ramesh K, Hemanth Kumar AK, et al. Association of high T allele frequency of CYP2B6 G516T polymorphism among ethnic south Indian HIV-infected patients with elevated plasma efavirenz and nevirapine. J Antimicrob Chemother. 2009;63:841–3. doi: 10.1093/jac/dkp033. [DOI] [PubMed] [Google Scholar]
- 27.Uttayamakul S, Likanonsakul S, Manosuthi W, et al. Effects of CYP2B6 G516T polymorphisms on plasma efavirenz and nevirapine levels when co-administered with rifampicin in HIV/TB co-infected Thai adults. AIDS Res Ther. 2010;7:8. doi: 10.1186/1742-6405-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou M, Bertrand J, Segeral O, et al. Population pharmacokinetic-pharmacogenetic study of nevirapine in HIV-infected Cambodian patients. Antimicrob Ag Chemother. 2010;54:4432–9. doi: 10.1128/AAC.00512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chantarangsu S, Cressey TR, Mahasirimongkol S, et al. Influence of CYP2B6 polymorphisms on the persistence of plasma nevirapine concentrations following a single intra-partum dose for the prevention of mother to child transmission in HIV-infected Thai women. J Antimicrob Chemother. 2009;64:1265–73. doi: 10.1093/jac/dkp351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Sonnerborg A, Rane A, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet Genom. 2006;16:191–8. doi: 10.1097/01.fpc.0000189797.03845.90. [DOI] [PubMed] [Google Scholar]
- 31.Ribaudo HJ, Liu H, Schwab M, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis. 2010;202:717–22. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertrand J, Chou M, Richardson DM, et al. Multiple genetic variants predict steady-state nevirapine clearance in HIV-infected Cambodians. Pharmacogenet Genom. 2012;22:868–76. doi: 10.1097/FPC.0b013e32835a5af2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holzinger ER, Grady B, Ritchie MD, et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols. Pharmacogenet Genom. 2012;22:858–67. doi: 10.1097/FPC.0b013e32835a450b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.dbSNP - short genetic variations. http://www.ncbi.nlm.nih.gov/projects/SNP/ Accessed 6 November 2012. [Google Scholar]
- 35.Acosta EP, Limoli KL, Trinh L, et al. Novel method to assess antiretroviral target trough concentrations using in vitro susceptibility data. Antimicrob Ag Chemother. 2012;56:5938–45. doi: 10.1128/AAC.00691-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boltz VF, Zheng Y, Lockman S, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci U S A. 2011;108:9202–7. doi: 10.1073/pnas.1105688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SeattleSNPs variation discovery resource. http://pga.gs.washington.edu/ Accessed 6 November 2012. [Google Scholar]
- 38.Rotger M, Tegude H, Colombo S, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Therap. 2007;81:557–66. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 39.Ensembl. http://www.ensembl.org/index.html. Accessed 6 November 2012. [Google Scholar]
- 40.Lamba V, Lamba J, Yasuda K, et al. Hepatic CYP2B6 Expression: Gender and Ethnic Differences and Relationship to CYP2B6 Genotype and CAR Expression. J Pharmacol Exp Therap. 2003;307:906–22. doi: 10.1124/jpet.103.054866. [DOI] [PubMed] [Google Scholar]
- 41.de Vincenzi I. Triple antiretroviral compared with zidovudine and single-dose nevirapine prophylaxis during pregnancy and breastfeeding for prevention of mother-to-child transmission of HIV-1 (Kesho Bora study): a randomised controlled trial. Lancet Infect Dis. 2011;11:171–80. doi: 10.1016/S1473-3099(10)70288-7. [DOI] [PubMed] [Google Scholar]
- 42.Lehman DA, Chung MH, Mabuka JM, et al. Lower risk of resistance after short-course HAART compared with zidovudine/single-dose nevirapine used for prevention of HIV-1 mother-to-child transmission. JAIDS. 2009;51:522–9. doi: 10.1097/QAI.0b013e3181aa8a22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.HIV/AIDS Programme, World Health Organization (WHO) Antiretroviral drugs for treating pregnant women and preventing HIV infection in infants: recommendations for a public health approach, 2010 version. Geneva: WHO; 2010. http://whqlibdoc.who.int/publications/2010/9789241599818_eng.pdf . Accessed 6 June 2013. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.