Abstract
Background. Factor H (fH) binding protein (fHbp) is part of vaccines developed for prevention of meningococcal serogroup B disease. More than 610 fHbp amino acid sequence variants have been identified, which can be classified into 2 subfamilies. The extent of cross-protection within a subfamily has been difficult to assess because of strain variation in fHbp expression.
Methods. Using isogenic mutant strains, we compared cross-protective serum antibody responses of mice immunized with 7 divergent fHbp variants in subfamily B, including identification numbers (ID) 1 and 55, which were chosen for vaccine development.
Results and Conclusions. In the presence of the human complement downregulator fH, the ability of the anti-fHbp antibodies to deposit sufficient complement C3b on the bacterial surface to elicit bactericidal activity required inhibition of binding of fH by the anti-fHbp antibodies. With less bound fH, the bacteria became more susceptible to complement-mediated bactericidal activity. Among the different fHbp sequence variants, those more central in a phylogenic network than ID 1 or 55 elicited anti-fHbp antibodies with broader inhibition of fH binding and broader bactericidal activity. Thus, the more central variants show promise of extending protection to strains with divergent fHbp sequences that are covered poorly by fHbp variants in clinical development.
Keywords: Neisseria meningitidis, bactericidal antibody, phylogeny, fHbp, LP2086, vaccine, complement activation
Meningococcal factor H (fH) binding protein (fHbp) is an important virulence determinant. This protein recruits human fH to the surface of the bacteria, which downregulates complement activation and enhances bacterial survival in human serum. Recombinant fHbp is a component of 2 vaccines [1, 2], one of which recently was licensed in Europe [3]. These vaccines elicit complement-mediated serum bactericidal activity, which is the serologic hallmark of protection against meningococcal disease in humans [4].
For vaccine evaluation, fHbp sequence variants are assigned specific identification numbers (ID; available at: http://pubmlst.org/neisseria/fHbp/) and are classified into 2 subfamilies [5], or 3 variant groups [6]. One of the vaccines in clinical development contains recombinant lipidated fHbp ID 45 from subfamily A and ID 55 from subfamily B [7–9]. The other vaccine, which was licensed in Europe, contains recombinant fHbp ID 1 from subfamily B [10, 11], which is expressed as a fusion protein with genome-derived antigen 2091 [10]. There is general agreement that antibodies to fHbp confer protection primarily against strains with fHbp from the same subfamily or variant group as the fHbp antigen in the vaccine [5, 7]. The vaccine with fHbp ID 1 uses other recombinant protein antigens such as NadA [12] and NHBA [13] to elicit protective antibodies against strains with fHbp from subfamily A [1, 11, 14]. The licensed vaccine also includes detergent-extracted outer-membrane vesicles, which elicit protective antibodies against a major porin protein, PorA [15, 16].
To date, >618 fHbp sequence variants have been identified (available at: http://pubmlst.org/neisseria/fHbp/). The underlying premise of both fHbp vaccines is that one fHbp sequence variant is sufficient for broad protection against strains within a subfamily. The extent of cross-protection within an fHbp subfamily is controversial, however [2, 14]. When strains with low fHbp expression were tested, the breadth of coverage was considerably less than with high-fHbp-expressing strains, especially when the fHbp variant in the strain did not exactly match that of the vaccine [2, 7, 17–19]. Collectively, the data suggest that sequence divergence between the fHbp variant in the vaccine and the test strain, as well as low fHbp expression in strains, can decrease bactericidal activity.
In the present study, we immunized mice with 7 different recombinant fHbp vaccines representing diverse sequence variants from subfamily B and evaluated cross-protective human complement-mediated serum bactericidal antibody responses. To control for differences in fHbp expression, we tested bactericidal activity against isogenic mutant strains with similar respective levels of expression of each of the 7 fHbp variants chosen for immunization.
MATERIALS AND METHODS
Ethics Statement
The studies in mice were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The protocol was approved by the Institutional Animal Care and Use Committee at Children's Hospital Oakland Research Institute. Human serum was obtained with written informed consent from a healthy adult who participated in a protocol approved by the Institutional Review Board at Children's Hospital Oakland Research Institute.
Purification of Recombinant fHbp
Recombinant fHbp was expressed in Escherichia coli and purified by Ni2+ affinity chromatography as previously described [20]. A second purification step was performed using cation exchange chromatography (HiTrap SP HP; GE Life Sciences).
Immunogenicity Study
The recombinant fHbp vaccines were adsorbed with aluminum hydroxide (Alhydrogel, Brenntag-Biosector; 600 µg per dose). Groups of female CD-1 mice (12 per group) were immunized beginning at 4 weeks of age with each of the recombinant fHbp vaccines (20 µg per dose). A control group of 8 mice received aluminum hydroxide without a vaccine antigen. Three injections were given intraperitoneally at 3-week intervals, and blood was collected 3 weeks after the third dose.
Serum Antibody Responses
Serum bactericidal activity was measured against a panel of 7 isogenic mutant strains in which each of the fHbp sequences matched one of the fHbp variants used for immunization (see “Construction of Isogenic Mutant Strains,” below). The strains were grown to mid-exponential phase in Mueller-Hinton broth supplemented with 0.25% glucose and 0.02 mmol/L cytidine monophosphate N-acetylneuraminic acid. Mouse serum pools were heated for 30 minutes at 56°C to inactivate intrinsic complement activity. The exogenous human complement source was serum from an adult who lacked bactericidal activity [21]. This serum was depleted of immunoglobulin G (IgG) with a protein G column as previously described [22]. This depletion step eliminated naturally acquired nonbactericidal IgG antibodies, which can augment bactericidal activity of the test antisera [23]. After accounting for approximately 10% dilution of serum during the procedure, the hemolytic complement activity (EZ Complement CH50; Diamedix) was not significantly decreased, compared with the original nondepleted serum. The bactericidal titer was the reciprocal serum dilution that yielded a 50% decrease in colony-forming units after 60 minutes of incubation relative to that of control wells at 60 minutes.
Construction of Isogenic Mutant Strains
The parent serogroup B strain, NZ98/254, was from an epidemic in New Zealand [15]. The PorA variable region type was 7–2, 4 and the sequence type was ST-42. The strain was transformed with the plasmid pBSΔ741::erm [6], which had been modified to include a hybrid PorA/NadA promoter [21] and 1 of 7 fHbp genes. Single erythromycin-resistant colonies were selected. Complete replacement of the native fHbp gene by the desired fHbp gene was confirmed by DNA sequencing. The parental wild-type and mutant strains showed similar respective growth characteristics and similar binding to the control monoclonal antibodies to the capsule (SEAM 12, which was produced in the authors’ laboratory [24]) or to PorA (P1.4, obtained from the National Institute of Biological Standards and Control, Potters Bar, UK).
Flow Cytometry
Anti-fHbp monoclonal antibody JAR 41 was produced in the authors’ laboratory [25]. Binding of anti-fHbp antibodies or the anti-capsular monoclonal antibody to live bacteria was performed as described elsewhere [26], using a BD Fortessa flow cytometer. Inhibition of fH binding to the mutant strains by the anti-fHbp antisera and the ability of the anti-fHbp antibodies to deposit C4 and C3 fragments on the bacterial surface were measured using fluorescein isothiocyanate–conjugated rabbit anti-human C3c or C4c antibodies (Abcam) [26].
RESULTS
Rationale for Selection of fHbp Sequence Variants
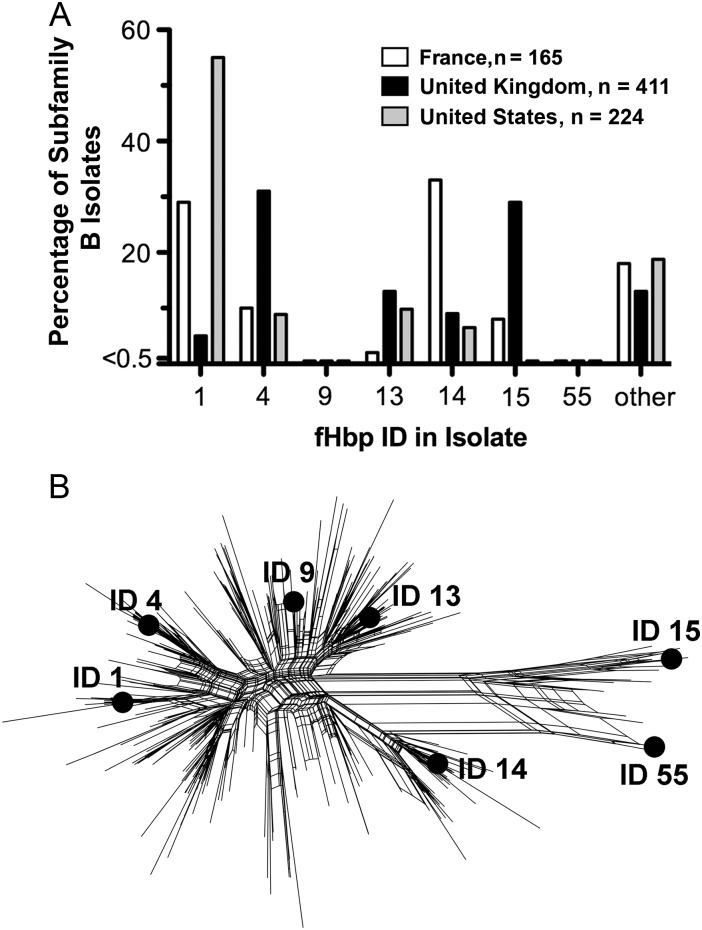
We selected 7 subfamily B sequence variants for preparing recombinant fHbp vaccines on the basis of their presence in meningococcal vaccines under development or licensed in Europe [1, 2, 7–9, 14] and/or their high prevalence among invasive isolates with fHbp in subfamily B (Figure 1A). In the United States ID 1 was the most prevalent sequence variant, whereas in the United Kingdom ID 4 and 13 were the most prevalent. In France, ID 14 was most prevalent. Although ID 9 was rare among serogroup B strains, we included this variant because of its high prevalence among serogroup W strains from sub-Saharan Africa [18]. The 7 fHbp subfamily B sequence variants selected were diverse, based on a phylogenic analysis of 325 subfamily B sequences (available at: http://pubmlst.org/neisseria/fHbp; Figure 1B). Interestingly, the fHbp subfamily B sequence variants ID 1 and 55, which had been selected by the manufacturers for clinical vaccine development, were among the most divergent and, overall, had 87% sequence identity to each other.
Figure 1.
Relative prevalence of factor H binding protein (fHbp) sequence variants in different geographic regions and phylogenic relationships among sequence variants. A, Percentage of serogroup B isolates with different fHbp sequence variants (ID) in subfamily B, adapted from published data [34, 35]. fHbp ID 9 was identified in a subset of serogroup W isolates from sub-Saharan Africa [18]. B, SplitsTree [36] network analysis of 325 known fHbp sequences in subfamily B (as of 4 May 2012). The sequence variants tested as recombinant fHbp vaccines in the present study are labeled.
Construction of Isogenic Mutant Strains
As described in Materials and Methods, we constructed a panel of isogenic mutant strains, each expressing one of the fHbp sequence variants used for immunization. The mutant strains were tested for fHbp expression by flow cytometry with a broadly cross-reactive anti-fHbp monoclonal antibody, JAR 41 [25]. The wild-type NZ98/254 strain was a relatively low expresser of fHbp ID 14, compared with strain H44/76, which is naturally a relatively higher expresser of fHbp ID 1 (Supplementary Figure 1A) [17]. Each of the NZ98/254 mutants with a different fHbp sequence variant expressed a similar amount of fHbp as that of the control H44/76 strain (Supplementary Figure 1B–H), and a similar amount of group B capsule (Supplementary Figure 1I).
Cross-protective Antibody Responses Elicited by Recombinant fHbp Sequence Variants ID 1 and 55
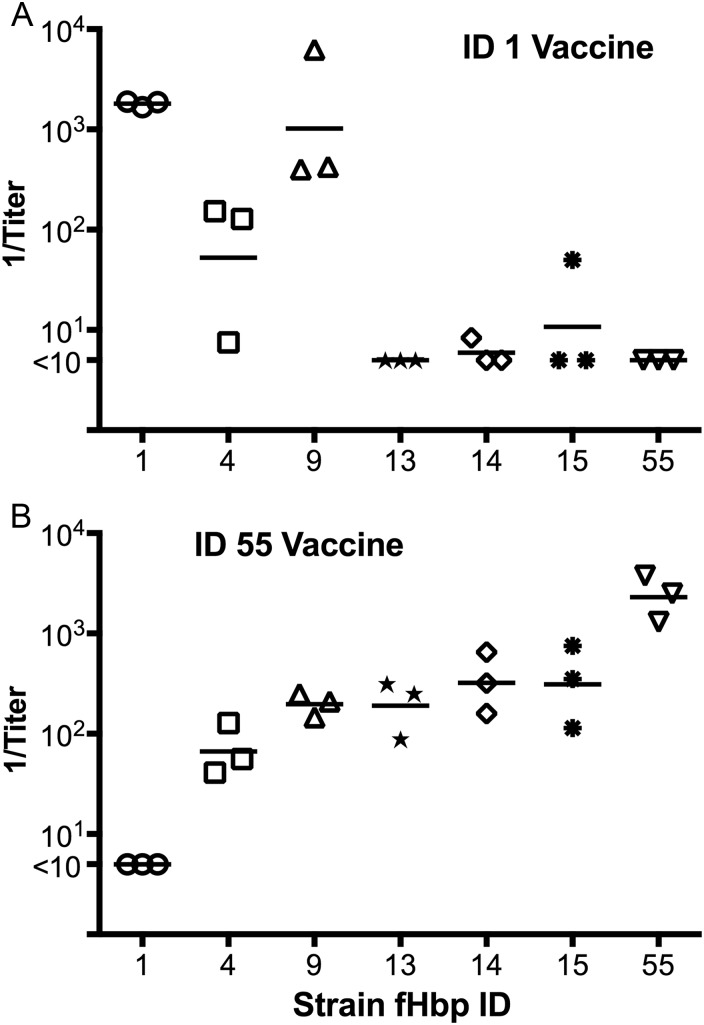
As described above, the 2 vaccines licensed or in clinical development contain fHbp ID 1 or 55 as subfamily B sequence variants. Mice immunized with the recombinant fHbp ID 1 vaccine developed the highest serum bactericidal titers when tested against the mutant strain with fHbp ID 1 that matched the vaccine; the titers were lower against the mutants with ID 4 or 9 and were even lower or not detectable against the mutants with ID 13, 14, 15, or 55 (Figure 2A). Mice immunized with the recombinant fHbp ID 55 vaccine also developed the highest serum bactericidal responses against the mutant strain that matched the vaccine. The titers were lower against the mutants with ID 15, 14, 13, 9, or 4 and were not detectable against the mutant with ID 1 (Figure 2B). In comparing the respective responses to the 2 vaccines, there was a trend for the ID 55 vaccine to elicit broader cross-protection than the ID 1 vaccine (geometric mean titer, >1:50 for 6 of 7 strains tested for the ID 55 vaccine, versus 3 of 7 strains for the ID 1 vaccine; P = .26, by the Fisher exact test).
Figure 2.
Complement-mediated serum bactericidal antibody responses of mice immunized with recombinant factor H binding protein (fHbp) vaccines ID 1 (A) or ID 55 (B) measured against each the 7 isogenic mutant strains. Each symbol represents the reciprocal titer of a serum pool (3 per vaccine group; 4 mice per pool); the horizontal lines represent the reciprocal geometric mean titers.
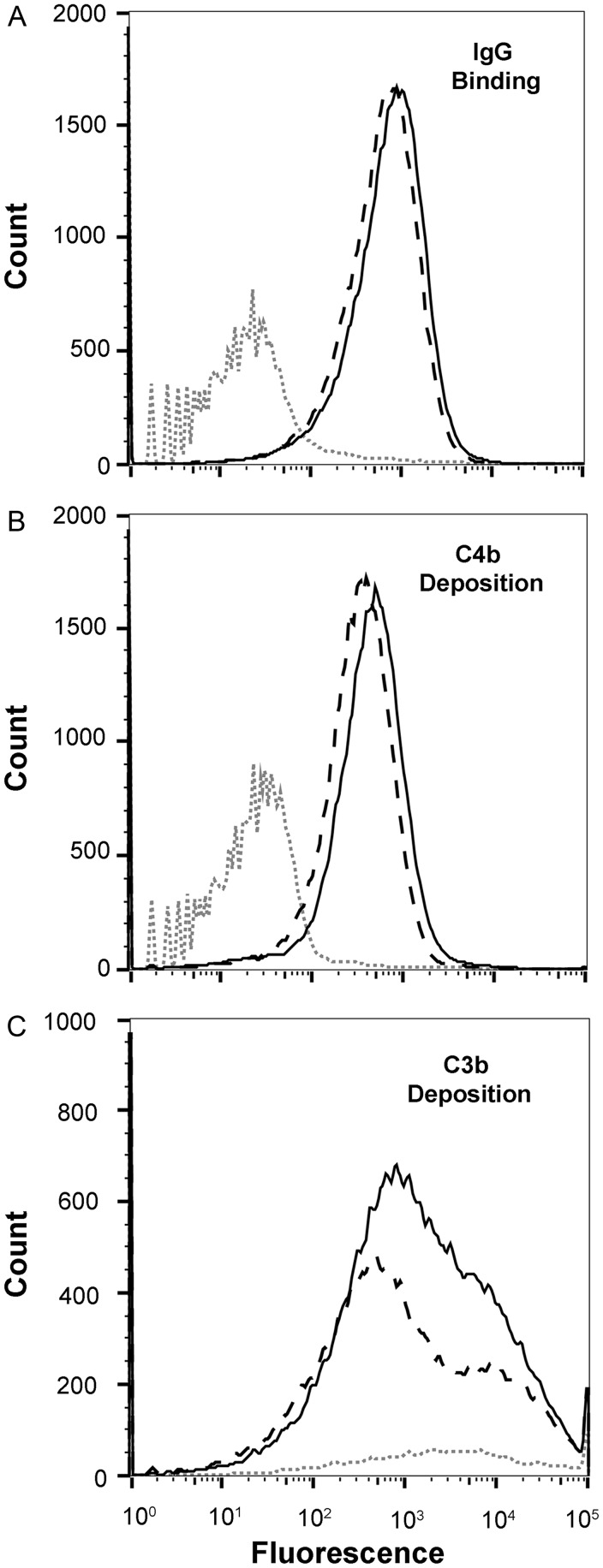
Activation of the Classical and Alternative Complement Pathways by Cross-reactive Anti-fHbp Antibodies
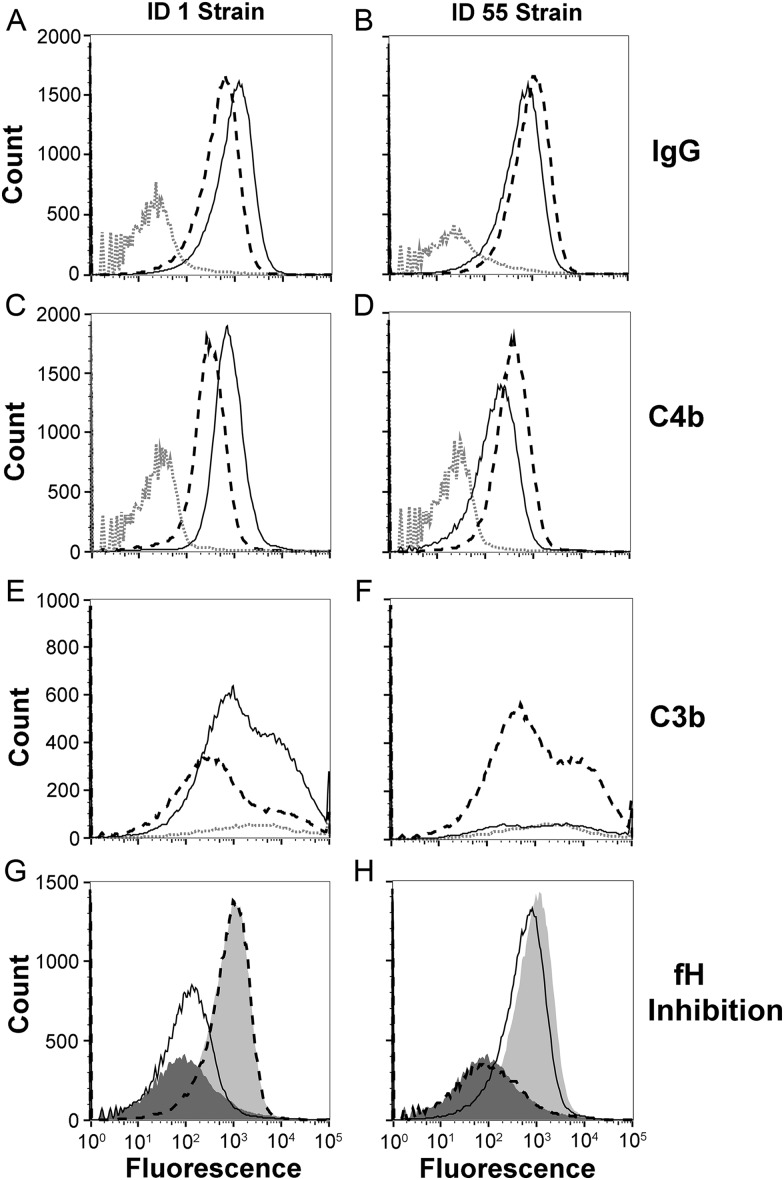
By flow cytometry, IgG antibodies in the antiserum to fHbp ID 1 bound slightly more to the isogenic mutant strain with ID 1 than did antibodies in the antiserum to ID 55 (Figure 3A). The antiserum to fHbp ID 55 bound slightly more to the mutant strain with ID 55 than did the antiserum to ID 1 (Figure 3B). Both antisera also deposited human C4b (a marker of classical complement pathway activation) on both mutant strains, which for each strain was slightly more for the respective matched than mismatched antiserum (Figure 3C and 3D). For measurement of C3 deposition (a marker of activation of both the alternative and classical pathways), we used sheep anti-human C3c, which reacted with both C3b and iC3b. Thus, the C3b deposition data reflected a combination of C3 activation and C3b inactivation. The anti-fHbp ID 1 antiserum deposited more C3b on the mutant with fHbp ID 1 than did the anti-fHbp ID 55 antiserum (Figure 3E). The anti-fHbp ID 55 antiserum also deposited more C3b on the mutant with fHbp ID 55 then did the anti-fHbp ID 1 antiserum (Figure 3F). Note that the anti-ID 55 antiserum elicited more C3b deposition on the mutant strain with fHbp ID 1 than did the anti-fHbp ID 1 antiserum on the mutant with fHbp ID 55. With both antisera, however, the amount of C3b deposition by the mismatched antisera was insufficient to elicit bactericidal activity (Figure 2).
Figure 3.
Binding of immunoglobulin G (IgG) anti–factor H binding protein (fHbp) antibodies, complement deposition, and inhibition of fH binding to live bacteria of isogenic mutant strains with fHbp ID 1 or ID 55, as measured by flow cytometry. A and B, Binding of IgG antibodies (1:50 serum dilution). Anti-fHbp ID 1 antiserum is denoted by the solid black line, anti-fHbp ID 55 antiserum is denoted by the dashed black line, and negative control serum (aluminum hydroxide without a vaccine antigen) is denoted by the dotted grey line. C and D, Complement C4b deposition on the mutant strains with ID 1 or ID 55, respectively, elicited by the mouse antisera (1:50). E and F, C3b deposition elicited by the mouse immune sera (1:200). Experiments in C–F used 5% IgG-depleted human serum from a single human donor as the source of complement. G and H, Inhibition of binding of fH to mutant strains with ID 1 (left) or 55 (right) by mouse antisera (1:50 dilution). The fH concentration was 90 µg/mL. The fHbp knockout strain, when incubated with fH and no antiserum, is denoted by dark grey filled histograms; isogenic mutant strains expressing fHbp when incubated with fH and no antiserum are denoted by light grey filled histograms; mutant strains expressing fHbp when incubated with fH and anti-fHbp ID 1 antiserum are denoted by solid black lines; and mutant strains expressing fHbp, when incubated with fH and anti-fHbp ID 55 antiserum, are denoted by dashed black lines. The light and dark shaded histograms correspond to no and complete fH inhibition, respectively. Data were visualized with FlowJo vX software (Tree Star).
Since binding of fH to the bacteria previously was reported to downregulate alternative pathway amplification [26, 27], we measured the ability of serum anti-fHbp antibodies to inhibit binding of fH to the mutant strains with matched or mismatched fHbp variants. The anti-fHbp antibodies elicited by the ID 1 vaccine strongly inhibited binding of fH to the mutant strain with fHbp ID 1 (Figure 3G). In contrast, the anti-fHbp ID 55 antibodies did not inhibit fH binding to the mutant strain with ID 1 (similar to fH binding in the absence of anti-fHbp antibodies). The converse was observed with the fHbp ID 55 mutant (strong inhibition of fH binding by the anti-fHbp ID 55 antibodies; no inhibition of fH binding by anti-fHbp ID 1 antibodies; Figure 3H).
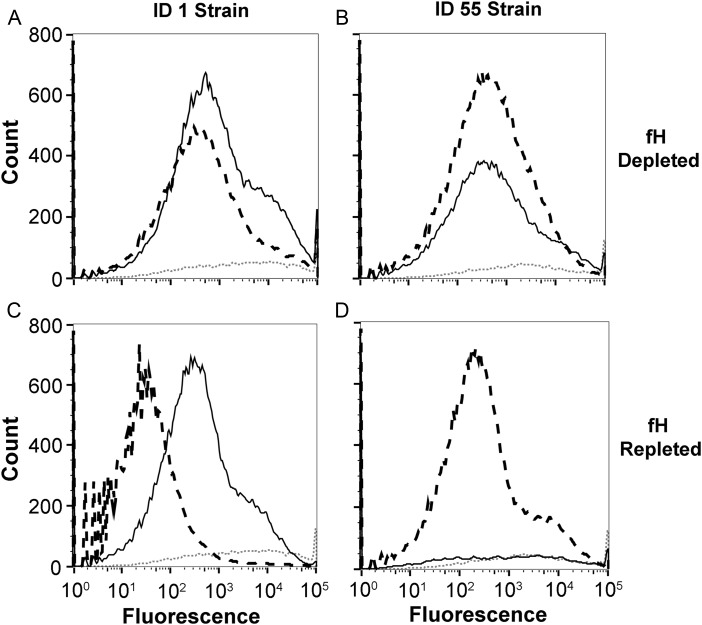
The results described above suggested that the ability of anti-fHbp antibodies to deposit sufficient C3b to elicit bactericidal activity against the mutant strains with the matched fHbp variants, but not the mutants with the mismatched variants, resulted from the ability of the matched antiserum to inhibit binding of fH to the bacteria. To test this hypothesis, we used fH-depleted human serum as a complement source to measure the ability of each of the antisera to deposit C3b on the 2 strains (Figure 4). As predicted, in the absence of fH, both the matched and mismatched antisera deposited C3b on both mutants (fHbp ID 1 mutant, Figure 4A; ID 55 mutant, Figure 4B). The addition of purified human fH to the reaction mixture markedly decreased C3b deposition on bacterial cells that had been incubated with the mismatched antisera, which did not inhibit fH binding. In contrast, the addition of human fH had much less effect on downregulating C3b deposition when the bacterial cells were incubated with the matched antisera, which inhibited fH binding (Figure 4C and 4D).
Figure 4.
C3b deposition by antiserum to factor H binding protein (fHbp) ID 1 or 55 with fH-depleted or fH-repleted human complement. A, fHbp ID 1 mutant strain incubated with mouse antisera and fH-depleted human serum. Antiserum to fHbp ID 1 is denoted by the solid black line, antiserum to fHbp ID 55 is denoted by the dashed black line, and negative control antiserum (adjuvant alone) is denoted by the dotted grey line. B, ID 55 mutant strain incubated with mouse antisera and fH-depleted complement. Line patterns are the same as in A. C, ID 1 mutant strain incubated with mouse antisera and fH-repleted complement (addition of 50 µg/mL purified human fH). D, ID 55 mutant strain incubated with mouse antisera and fH-repleted complement. Experiments used 5% fH-depleted, pooled human serum that also had been depleted of immunoglobulin G as the source of complement (see Materials and Methods).
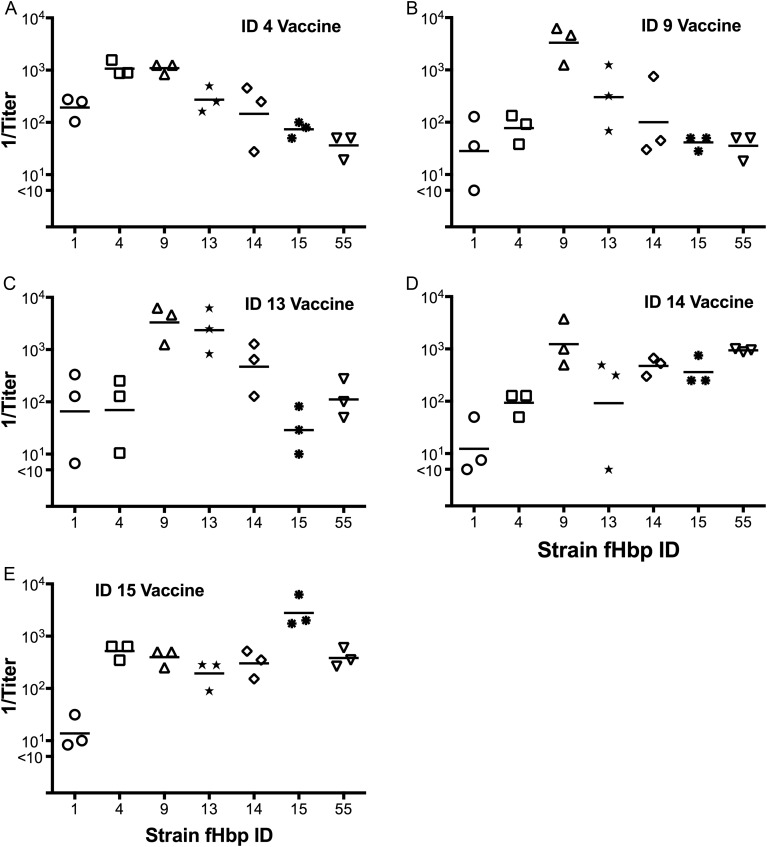
Cross-protective Responses to fHbp Vaccines With Prevalent Sequence Variants
Mice immunized with each of the 5 other recombinant fHbp vaccines tested developed high serum bactericidal antibody responses when measured against the respective mutant with the fHbp variant that matched the vaccine. The fHbp ID 14 and 15 vaccines (Figure 5D and 5E, respectively) elicited cross-reactive bactericidal responses similar to those of the ID 55 vaccine, which elicited high responses against most of the mutants but low or nondetectable titers against the mutant strain with fHbp ID 1 (Figure 2B). In contrast, the ID 4, 9, and 13 vaccines, which were the most centrally located in the phylogram (Figure 1B), elicited the broadest bactericidal antibody responses, which included positive titers against both of the mutants with outlying sequence variants, ID 1 or 55 (Figure 5A–C).
Figure 5.
Complement-mediated serum bactericidal antibody responses of mice immunized with 5 recombinant factor H binding protein (fHbp) subfamily B vaccines. Each panel is labeled with the specific fHbp sequence variant of the vaccine tested. Each symbol represents the reciprocal titer of an individual serum pool (4 mice per pool), and the horizontal lines represent the reciprocal geometric mean titers.
Since inhibition of binding of fH to the bacteria by the antisera to fHbp ID 1 or 55 corresponded with bactericidal activity, we tested the ability of antisera from mice immunized with each of the 3 broadly protective fHbp variants, ID 4, 9, or 13, to inhibit binding of fH to the mutant strains with fHbp ID 1 or 55. Against the mutant strain with ID 1, each of the antisera to fHbp ID 4, 9 or 13 showed partial inhibition of fH binding (Figure 6A). Against the mutant strain with ID 55, each of the 3 antisera showed even more inhibition of fH binding, which was similar to binding of fH to the fHbp knockout control strain in the absence of anti-fHbp antibodies (Figure 6B).
Figure 6.
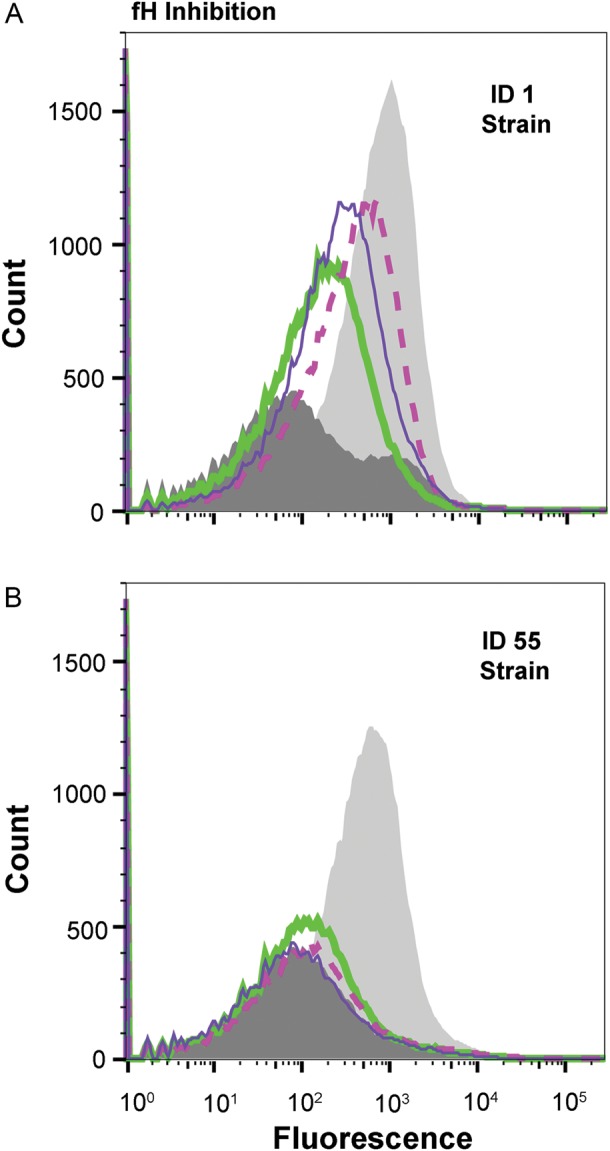
Inhibition of binding of factor H (fH) to mutant strains with ID 1 (A) or ID 55 (B) by mouse antisera (1:50). Negative control fH binding protein (fHbp) knockout strain with 90 µg/mL fH is denoted by the dark grey filled histogram, a mutant strain expressing fHbp incubated with fH alone (no antiserum) is denoted by the light grey filled histogram, a mutant strain expressing fHbp incubated with fH and anti-fHbp ID 4 antiserum is denoted by a thick green line, anti-fHbp ID 9 antiserum is denoted by a dashed pink line, and anti-fHbp ID 13 antiserum is denoted by a thin purple line.
Since all 3 anti-fHbp antisera raised to the “central” fHbp variants in the phylogram gave partial or complete inhibition of fH binding to the mutants with fHbp ID 1 or ID 55, respectively, we tested the ability of the anti-ID 9 antibodies to bind to each of these mutants and deposit C4b and C3b. The IgG antibodies in the antiserum to fHbp ID 9 showed similar respective binding to both mutant strains (Figure 7A) and similar respective deposition of C4b (classical complement pathway; Figure 7B). The anti-fHbp ID 9 antiserum also deposited C3b (the sum of activation of all pathways) on both strains, although slightly more C3b was deposited on the strain with fHbp ID 1 than the strain with ID 55 (Figure 7C). Collectively, these studies showed that the ability of anti-fHbp antibodies to inhibit binding of fH to the bacteria resulted in more amplification of the alternative complement pathway, which was important for eliciting C3b deposition and cross-protective bactericidal activity by antibodies to the recombinant fHbp vaccines.
Figure 7.
Binding of serum immunoglobulin G (IgG) anti–factor H binding protein (fHbp) antibodies from mice immunized with the recombinant fHbp ID 9 vaccine and complement deposition on strains with fHbp ID 1 (solid black line) or ID 55 (dashed black line). A, Binding of IgG anti-fHbp ID 9 antibodies (1:50 serum dilution) or negative control antiserum (1:50) from mice immunized with aluminum hydroxide alone (dotted grey line). B, C4b deposition by anti-fHbp ID 9 antiserum (1:50). C, C3b deposition by the anti-ID 9 antiserum (1:200). Experiments shown in B and C used 5% IgG-depleted human serum as the source of complement.
DISCUSSION
Despite use of mutants with relatively high fHbp expression, which maximized the extent of possible cross-protective anti-fHbp activity, the fHbp ID 55 vaccine did not elicit serum bactericidal activity against the mutant with fHbp ID 1, which is the most prevalent fHbp sequence variant among serogroup B stains in the United States (Figure 1A). The ID 1 vaccine elicited low bactericidal antibody responses against the mutants with ID 13, 14, or 15, which are prevalent among serogroup B strains from the United Kingdom and/or France (Figure 1A). Thus, neither fHbp sequence variant chosen for clinical development appears to be optimal for prevention of disease in all geographic regions. This result contrasted with previous studies that suggested that vaccine coverage was broad when the fHbp variant in the strain was from the same subfamily or variant group as the vaccine antigen [5, 7, 28, 29].
One possible explanation for the lack of broad coverage in the present study is that we used different recombinant fHbp antigen preparations than in the commercial vaccines. The Pfizer vaccine uses fHbp antigens with a lipid modification [2, 7], which was reported to elicit higher serum bactericidal antibody responses in mice than the corresponding nonlipidated antigens [5]. The Novartis vaccine uses a nonlipidated fHbp in a fusion protein with GNA 2091, which also may improve fHbp immunogenicity [10]. In infants and/or toddlers, however, the results of clinical studies with both vaccines were consistent with the present results in mice showing limited breadth of vaccine coverage against strains with fHbp variants that did not closely match the respective antigens in the vaccine. For example, toddlers immunized with the Pfizer lipidated fHbp vaccine developed high serum bactericidal antibody responses against a test strain with fHbp ID 87 [2], which had 93% amino acid sequence identity with ID 55 in the vaccine (Figure 1B). In contrast, the serum bactericidal titers were <1:4 against 2 other test strains with fHbp ID 1 or 13, which had greater amino acid sequence divergence than ID 87 from the subfamily B vaccine antigen. Similarly, infants and toddlers immunized with a prototype Novartis recombinant protein vaccine that did not contain outer membrane vesicles developed high serum bactericidal antibody responses against a test strain with fHbp ID 1 that matched the vaccine antigen and low responses against 3 strains with other subfamily B variants that did not match the vaccine antigen [1, 14]. Since these studies used wild-type test strains with different levels of fHbp expression, it is possible that some of the low bactericidal activity resulted from low fHbp strain expression [1]. Nevertheless, collectively the results were consistent with the present data in mice, which indicated limited breadth of coverage when measured with a panel of isogenic mutant test strains with uniformly high fHbp expression.
Because of the limited breadth of strain coverage, the manufacturer of the vaccine containing fHbp ID 1 combined the recombinant proteins with detergent-treated outer membrane vesicles, which enhanced the breadth of serum bactericidal antibody responses [1, 14]. The manufacturer of the vaccine containing the lipidated fHbp ID 55 antigen is targeting adolescents as the main group for immunization [8, 9]. Adolescents, because of greater previous natural exposure to N. meningitidis or related commensal organisms, have broader serum bactericidal antibody responses than immunized infants or toddlers [15, 16].
The lack of cross-protective activity between mouse anti-fHbp antibodies elicited by the ID 1 or 55 vaccines is of interest since the 2 proteins are 87% identical and most of the differences in amino acid sequence are located between residues 32 and 49 (Supplementary Figure 2A), which in a previous study were thought not to play a major role in eliciting bactericidal antibodies [30]. However, when a 3-dimensional structural model of fHbp was examined, many of the amino acid differences between ID 1 and 55 were residues previously identified as fH contact residues (Supplementary Figure 2B and C) [31]. Indeed, a key functional difference between antibodies elicited by the 2 vaccines was their ability to inhibit binding of fH to the bacterial surface of the mutant with the matched fHbp variant, but not the mismatched variant. This disparity appears to result from sequence differences in the epitopes involving fH contact residues in the 2 variants.
Among many wild-type meningococcal isolates, fHbp is expressed sparsely on the bacterial surface [32], and data from previous studies suggested that anti-fHbp bactericidal activity required activation of both the classical pathway and alternative complement pathway amplification for sufficient C3b deposition to elicit bactericidal activity [26, 33]. The results of the present study provided further evidence for this hypothesis since IgG antibodies in the anti-ID 1 and anti-ID 55 antisera showed similar binding and C4b deposition on both of the mutants with the respective matched or mismatched fHbp. Only the matched antisera, which inhibited fH binding, elicited sufficient C3b deposition for bactericidal activity. In previous studies, human IgG1-murine anti-fHbp chimeric monoclonal antibodies were bactericidal when tested with fH-depleted complement, but only the monoclonal antibodies that inhibited fH binding to fHbp had bactericidal activity in the presence of fH [26]. Collectively, these results suggested that inhibition of fH binding was important for bactericidal activity elicited by anti-fHbp antibodies by permitting sufficient C3b deposition by a combination of the classical and alternative complement pathways.
Another important finding of the present study was that fHbp sequence variants such as ID 4, 9, and 13, which are more central in a phylogenic network analysis than ID 1 or 55 (Figure 1B), elicited broader cross-protective bactericidal antibody responses than the 2 outlying sequence variants chosen for clinical vaccine development. Antisera to each of these more central sequence variants gave partial or nearly complete inhibition of fH binding against both of the mutants with fHbp ID 1 or 55. Although further studies are needed, the results are consistent with our hypothesis that fHbp vaccines chosen from phylogenically more central sequence variants than the ID 1 or 55 variants might elicit broader protection in humans.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Emily Braga and Mike Cheng, for expert technical assistance; Rolando Pajon, for initial phylogenic network analyses and for suggesting that a central fHbp variant might elicit broader protective antibody responses; Sanjay Ram (University of Massachusetts Medical School), Gregory Moe, and Rolando Pajon, for critical review of the manuscript; and Amanda Cohn (Centers for Disease Control), for providing fHbp frequency data for US isolates.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grants R01 AI 70955 and AI 99125 to P. T. B. and grants AI 46464 and AI 82263 to D. M. G) and the National Center for Research Resources (C06 RR 16226), National Institutes of Health.
Potential conflicts of interest. P. T. B. and D. M. G. are inventors on patents or patent applications in the area of meningococcal B vaccines. D. M. G. is principal investigator of laboratory research conducted on behalf of Children's Hospital Oakland Research Institute that is funded by grants from Novartis Vaccines and Diagnostics. He also holds a paid consultancy from Novartis. M. K. certifies no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Findlow J, Borrow R, Snape MD, et al. Multicenter, open-label, randomized phase II controlled trial of an investigational recombinant meningococcal serogroup B vaccine with and without outer membrane vesicles, administered in infancy. Clin Infect Dis. 2010;51:1127–37. doi: 10.1086/656741. [DOI] [PubMed] [Google Scholar]
- 2.Marshall HS, Richmond PC, Nissen MD, et al. Safety and immunogenicity of a meningococcal B bivalent rLP2086 vaccine in healthy toddlers aged 18–36 months: a phase 1 randomized-controlled clinical trial. Pediatr Infect Dis J. 2012;31:1061–8. doi: 10.1097/INF.0b013e31826327e4. [DOI] [PubMed] [Google Scholar]
- 3.Granoff DM. European medicines agency recommends approval of a broadly protective vaccine against Serogroup B meningococcal disease. Pediatr Infect Dis J. 2012;32:372–3. doi: 10.1097/INF.0b013e318282942f. [DOI] [PubMed] [Google Scholar]
- 4.Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med. 1969;129:1307–26. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fletcher LD, Bernfield L, Barniak V, et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun. 2004;72:2088–100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masignani V, Comanducci M, Giuliani MM, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med. 2003;197:789–99. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang HQ, Hoiseth SK, Harris SL, et al. Broad vaccine coverage predicted for a bivalent recombinant factor H binding protein based vaccine to prevent serogroup B meningococcal disease. Vaccine. 2010;28:6086–93. doi: 10.1016/j.vaccine.2010.06.083. [DOI] [PubMed] [Google Scholar]
- 8.Richmond PC, Nissen MD, Marshall HS, et al. A bivalent Neisseria meningitidis recombinant lipidated factor H binding protein vaccine in young adults: Results of a randomised, controlled, dose-escalation phase 1 trial. Vaccine. 2012;30:6163–74. doi: 10.1016/j.vaccine.2012.07.065. [DOI] [PubMed] [Google Scholar]
- 9.Richmond PC, Marshall HS, Nissen MD, et al. Safety, immunogenicity, and tolerability of meningococcal serogroup B bivalent recombinant lipoprotein 2086 vaccine in healthy adolescents: a randomised, single-blind, placebo-controlled, phase 2 trial. Lancet Infect Dis. 2012;12:597–607. doi: 10.1016/S1473-3099(12)70087-7. [DOI] [PubMed] [Google Scholar]
- 10.Giuliani MM, Adu-Bobie J, Comanducci M, et al. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A. 2006;103:10834–9. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toneatto D, Ismaili S, Ypma E, Vienken K, Oster P, Dull P. The first use of an investigational multicomponent meningococcal serogroup B vaccine (4CMenB) in humans. Hum Vaccin. 2011;7:646–53. doi: 10.4161/hv.7.6.15482. [DOI] [PubMed] [Google Scholar]
- 12.Comanducci M, Bambini S, Brunelli B, et al. NadA, a novel vaccine candidate of Neisseria meningitidis. J Exp Med. 2002;195:1445–54. doi: 10.1084/jem.20020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serruto D, Spadafina T, Ciucchi L, et al. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci U S A. 2010;107:3770–5. doi: 10.1073/pnas.0915162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snape MD, Dawson T, Oster P, et al. Immunogenicity of two investigational serogroup B meningococcal vaccines in the first year of life: a randomized comparative trial. Pediatr Infect Dis J. 2010;29:e71–9. doi: 10.1097/INF.0b013e3181f59f6d. [DOI] [PubMed] [Google Scholar]
- 15.O'Hallahan J, Lennon D, Oster P, et al. From secondary prevention to primary prevention: a unique strategy that gives hope to a country ravaged by meningococcal disease. Vaccine. 2005;23:2197–201. doi: 10.1016/j.vaccine.2005.01.061. [DOI] [PubMed] [Google Scholar]
- 16.Tappero JW, Lagos R, Ballesteros AM, et al. Immunogenicity of 2 serogroup B outer-membrane protein meningococcal vaccines: a randomized controlled trial in Chile. JAMA. 1999;281:1520–7. doi: 10.1001/jama.281.16.1520. [DOI] [PubMed] [Google Scholar]
- 17.Pajon R, Beernink PT, Harrison LH, Granoff DM. Frequency of factor H-binding protein modular groups and susceptibility to cross-reactive bactericidal activity in invasive meningococcal isolates. Vaccine. 2010;28:2122–9. doi: 10.1016/j.vaccine.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pajon R, Fergus AM, Koeberling O, Caugant DA, Granoff DM. Meningococcal factor H binding proteins in epidemic strains from Africa: implications for vaccine development. PLoS Negl Trop Dis. 2011;5:e1302. doi: 10.1371/journal.pntd.0001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donnelly J, Medini D, Boccadifuoco G, et al. Qualitative and quantitative assessment of meningococcal antigens to evaluate the potential strain coverage of protein-based vaccines. Proc Natl Acad Sci U S A. 2010;107:19490–5. doi: 10.1073/pnas.1013758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beernink PT, Granoff DM. Bactericidal antibody responses induced by meningococcal recombinant chimeric factor H-binding protein vaccines. Infect Immun. 2008;76:2568–75. doi: 10.1128/IAI.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beernink PT, Shaughnessy J, Pajon R, Braga EM, Ram S, Granoff DM. The effect of human factor H on immunogenicity of meningococcal native outer membrane vesicle vaccines with over-expressed factor H binding protein. PLoS Pathog. 2012;8:e1002688. doi: 10.1371/journal.ppat.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beernink PT, Shaughnessy J, Braga EM, et al. A meningococcal factor H binding protein mutant that eliminates factor H binding enhances protective antibody responses to vaccination. J Immunol. 2011;186:3606–14. doi: 10.4049/jimmunol.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vu DM, Wong TT, Granoff DM. Cooperative serum bactericidal activity between human antibodies to meningococcal factor H binding protein and Neisserial heparin binding antigen. Vaccine. 2011;29:1968–73. doi: 10.1016/j.vaccine.2010.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granoff DM, Bartoloni A, Ricci S, et al. Bactericidal monoclonal antibodies that define unique meningococcal B polysaccharide epitopes that do not cross-react with human polysialic acid. J Immunol. 1998;160:5028–36. [PubMed] [Google Scholar]
- 25.Vu DM, Pajon R, Reason DC, Granoff DM. A broadly cross-reactive monoclonal antibody against an epitope on the N-terminus of meningococcal fHbp. Scientific Reports. 2012;2:341. doi: 10.1038/srep00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giuntini S, Reason DC, Granoff DM. Combined roles of human IgG subclass, alternative complement pathway activation, and epitope density in the bactericidal activity of antibodies to meningococcal factor H binding protein. Infect Immun. 2012;80:187–94. doi: 10.1128/IAI.05956-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madico G, Welsch JA, Lewis LA, et al. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol. 2006;177:501–10. doi: 10.4049/jimmunol.177.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seib KL, Brunelli B, Brogioni B, et al. Characterization of diverse subvariants of the meningococcal factor H (fH) binding protein for their ability to bind fH, to mediate serum resistance, and to induce bactericidal antibodies. Infect Immun. 2011;79:970–81. doi: 10.1128/IAI.00891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunelli B, Del Tordello E, Palumbo E, et al. Influence of sequence variability on bactericidal activity sera induced by Factor H binding protein variant 1.1. Vaccine. 2011;29:1072–81. doi: 10.1016/j.vaccine.2010.11.064. [DOI] [PubMed] [Google Scholar]
- 30.Giuliani MM, Santini L, Brunelli B, et al. The region comprising amino acids 100 to 255 of Neisseria meningitidis lipoprotein GNA 1870 elicits bactericidal antibodies. Infect Immun. 2005;73:1151–60. doi: 10.1128/IAI.73.2.1151-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider MC, Prosser BE, Caesar JJ, et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature. 2009;458:890–3. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welsch JA, Ram S, Koeberling O, Granoff DM. Complement-dependent synergistic bactericidal activity of antibodies against factor H-binding protein, a sparsely distributed meningococcal vaccine antigen. J Infect Dis. 2008;197:1053–61. doi: 10.1086/528994. [DOI] [PubMed] [Google Scholar]
- 33.Giuntini S, Reason DC, Granoff DM. Complement-mediated bactericidal activity of anti-factor H binding protein monoclonal antibodies against the meningococcus relies upon blocking factor H binding. Infect Immun. 2011;79:3751–9. doi: 10.1128/IAI.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Cohn A, Comanducci M, et al. Prevalence and genetic diversity of candidate vaccine antigens among invasive Neisseria meningitidis isolates in the United States. Vaccine. 2011;29:4739–44. doi: 10.1016/j.vaccine.2011.04.092. [DOI] [PubMed] [Google Scholar]
- 35.Murphy E, Andrew L, Lee KL, et al. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J Infect Dis. 2009;200:379–89. doi: 10.1086/600141. [DOI] [PubMed] [Google Scholar]
- 36.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.