Abstract
Influenza A/California/4/2009 (H1N1/09) is a recently emerged influenza virus capable of causing serious illness or death in otherwise healthy individuals. Serious outcomes were most common in young adults and children, suggesting that pre-existing heterologous immunity may influence the severity of infection. Using tetramers, we identified CD4+ T-cell epitopes within H1N1/09 hemagglutinin (HA) that share extensive homology with seasonal influenza and epitopes that are unique to H1N1/09 HA. Ex vivo tetramer staining revealed that T cells specific for conserved epitopes were detectable within the memory compartment, whereas T cells specific for unique epitopes were naive and infrequent prior to infection or vaccination. Following infection, the frequencies of T cells specific for unique epitopes were 11-fold higher, reaching levels comparable to those of T cells specific for immunodominant epitopes. In contrast, the frequencies of T cells specific for conserved epitopes were only 2- to 3-fold higher following infection. In general, H1HA-reactive T cells exhibited a memory phenotype, expressed CXCR3 and secreted IFN-γ, indicating a predominantly Th1-polarized response. A similar Th1 response was seen in vaccinated subjects, but the expansion of T cells specific for HA epitopes was comparatively modest after vaccination. Our findings indicate that CD4+ T cells recognize both strain-specific and conserved epitopes within the influenza HA protein and suggest that naive T cells specific for HA epitopes undergo significant expansion, whereas memory T cells specific for the conserved epitopes undergo more restrained expansion.
Keywords: ex vivo, HLA, naive, peptide, tetramer
Introduction
The critical role of both humoral and cellular immunity in dictating the course of an influenza infection is most clearly highlighted by studying newly emerging influenza viruses. Epidemiologic studies have suggested that young adults and children are more vulnerable to ‘novel’ viral pathogens such as H1N1/1918 (1–3) and H5N1 (4–6). Likewise, it was reported that a disproportionate number of critically ill patients infected with the newly emerged influenza A/California/4/2009 (H1N1/09) virus were young adults, children and pregnant women (7–9). Detrimental pathogenic changes in these critically ill patients were frequently associated with elevated serum levels of pro-inflammatory cytokines and chemokines, including some known to influence the intensity of innate responses and others that mediate the development of Th1 or Th17 responses (10–16). These observations in patients lacking previous exposure to the virus could indicate exaggerated T-cell responses, perhaps originating from the naive compartment. A recent influenza challenge study demonstrated that CD4+ T cells appear to play multiple roles in dictating the course of influenza infection (17). Although T-cell frequency post-challenge was positively correlated with influenza viral shedding and symptoms, pre-existing CD4+ T-cell immunity against internal proteins was inversely correlated with viral shedding and symptoms. However, much of that study focused on cross-reactivity between epitopes from the conserved matrix protein (M1) and nucleoproteins (NP). As such, a targeted study of CD4+ immunity directed against the more poorly conserved influenza hemagglutinin (HA) protein is warranted.
Cellular immunity to influenza viruses in adults is typically a memory response. There is diversity among influenza strains and most individuals have been exposed to multiple strains via natural infection and vaccination. Seasonal antigenic drift introduces some amino acid mutations, but these are not necessarily significant enough to generate novel T-cell epitopes. As a result, HLA class II-restricted CD4+ T-cell epitopes identified in previous influenza viruses are often present as identical or highly homologous sequences in newly emerging influenza strains. The recent influenza H1N1/09 virus is a re-assortment of swine, human and avian influenza strains. In particular, its re-assorted HA is of both swine and human origin (18, 19). Despite having considerable sequence homology to previously circulating strains (e.g. 78% homology with influenza A/Brisbane/59/2007), the HA1 domain of the H1N1/09 HA protein contains several discrete regions of lower homology. We hypothesized that these low-homology regions contain T-cell epitopes unique to H1N1/09 and that the dynamics of CD4+ T-cell responses against these unique HA epitopes following vaccination or influenza infection would differ from those of memory T cells recognizing conserved epitopes present within this newly circulating strain.
In this study, we utilized the Tetramer Guided Epitope Mapping (TGEM) (20) approach to identify CD4+ T-cell epitopes within the HA protein of the H1N1/09 virus restricted by a few common HLA-DR types. As expected, we observed novel HA epitopes that were unique to the H1N1/09 virus and conserved epitopes present within multiple strains. Using our recently described ex vivo HLA class II tetramer enrichment approach, we directly measured the frequencies of HA-specific T cells in subjects who had no exposure to this novel influenza strain, in subjects who had recently recovered from infection by the H1N1/09 virus and in subjects who received the influenza vaccine. These tools enabled us to compare the frequencies and phenotypes of T cells that recognized conserved HA epitopes with those that recognize unique HA epitopes.
Methods
Patient and vaccination subject recruitment and HLA-DR typing
For epitope mapping studies, a group of 27 subjects with no influenza symptoms was recruited. In addition, we studied eight subjects who had received one dose of either the injected Influenza A (H1N1) 2009 Monovalent Vaccine (Sanofi Pasteur) or the Influenza A (H1N1) 2009 Monovalent Vaccine (MedImmune) live attenuated vaccine and seven more subjects who had received the trivalent seasonal influenza vaccine (Fluzone, Sanofi Pasteur). These subjects were 25–56 years of age (average age = 39±10 years). In addition, we studied seven patients with H1N1/09 infection (diagnosis based on clinical symptoms and confirmed by detection of H1N1/09 viral RNA in nasopharyngeal aspirates by real-time reverse transcription–PCR with protocols, probes, primers and reagents approved by Centers for Disease Control and Prevention) recruited from the Virginia Mason Medical Center, Seattle, WA, USA. These naturally infected patients were 21–54 years of age (average age = 41±11 years). All subjects were recruited with informed consent under a study that had been approved by the Benaroya Research Institute Institutional Review Board. For each subject, HLA-DR was typed by PCR amplification with sequence-specific primers followed by second round high-resolution typing using Dynal Unitray SSP Kits according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). HLA information for all subjects is summarized in Supplementary Table 1, available at International Immunology Online.
Fluorescent antibodies, tetramer reagents and peptides
The fluorescent antibodies used in this study were obtained from eBioscience (San Diego, CA, USA), BD Biosciences (San Jose, CA, USA) and BioLegend (San Diego, CA, USA). MHC class II tetramer reagents were generated as previously described (21). Briefly, ‘empty’ HLA-DR proteins were expressed and purified from insect cell culture supernatants. Following in vitro biotinylation, class II monomers were loaded with either peptide pools or individual peptides by incubating for 48h at 37°C with 25-fold molar excess of peptide (total) in phosphate buffer, pH 6.0 in the presence of 0.2% n-octyl-d-glucopyranoside. Tetramers were formed by incubating class II molecules with PE-labeled streptavidin for 6–18h at room temperature at a molar ratio of 8:1. A peptide library was generated based on the HA sequence of H1N1/09-like virus (accession: ACQ76318), a strain used by manufacturers for vaccine development. The library consisted of 70 peptides, each 20 amino acids long (except the last peptide, which was 14 amino acids long), with a 12 amino acid overlap. The peptide library was purchased from Mimotopes (Clayton, Australia).
HLA-DR-restricted HA epitope identification and ex vivo epitope-specific T-cell analysis
HLA-DR-restricted epitopes were identified by stimulating CD4+ T cells with 14 pools of overlapping peptides (one peptide pool per well) representing the H1N1/09 HA protein and staining with the corresponding class II tetramers, using our previously described methodology (20, 21). Briefly, after 14 days of in vitro stimulation, 100 µl of cell suspension from each well (100 000–250 000 cells) was stained using 2 µl of peptide pool-loaded tetramer (10 µg ml-1 final) at 37°C for 1–2h and then stained with antibodies (5 µl of anti-CD3-FITC, anti-CD4-PerCP and anti-CD25-APC) at room temperature for 10min. Cells were washed once in 1ml of PBS and analyzed using a FACS Calibur (BD Biosciences). Pools that gave positive staining were decoded by staining another 100 µl of cell suspension using tetramers loaded with the corresponding individual peptides.
To analyze epitope-specific T cells ex vivo, 30–50 million peripheral blood mononuclear cells (PBMCs) per epitope in 1ml T-cell culture medium were treated with Dasatinib (22) for 10min at 37°C followed by staining with 20 µl of tetramer at room temperature for 2h. After tetramer staining, a 1/100th fraction of the cells was removed and reserved for counting to estimate the total number of cells used in the tetramer enrichment assay. The remaining cells were labeled with anti-PE magnetic beads and enriched using a magnetic column according to the manufacturer’s instructions (Miltenyi Biotec, Auburn, CA, USA). Magnetically enriched cells and the previously reserved 1/100th cell fraction were stained with anti-CD4 APC-H7, anti-CD3 V500 and anti-CD14 PerCP, anti-CD19 PerCP and Via-Probe (dumping antibody mixture). Gated CD4+/CD3+/CD14– CD19–Via-Probe–live CD4+ T cells were analyzed for tetramer (PE) staining and their expression of one or more of the following phenotypic markers: CD38 FITC, CXCR3 APC, CD45RA V450, CCR7 PE-Cy7, Ki-67 FITC, CD62L APC. Samples were analyzed on a 17-color BD LSR II instrument. Each sample was diluted and collected to completion. The frequency (F) of epitope-specific T cells per million CD4+ T cells was calculated as follows: F = (1 000 000 × tetramer positive events from enriched tube)/(100 × number of CD4+ T cells from pre-enriched fraction).
Cytokine profile analysis of antigen-specific CD4+ T cells
Antigen-specific CD4+ T-cell lines were generated by stimulating PBMCs with selected HA peptides in vitro for 14 days. After verifying that at least 2% of the CD4+ T cells were tetramer positive, the cells were collected, re-suspended in T-cell culture medium containing 5 µg ml1 anti-CD28 and 1 µg ml1 anti-CD49d and seeded into a flat-bottom 96-well plate (0.25 × 106 cells in 100 µl per well) coated with MHC class II tetramer loaded with the stimulatory peptide or an irrelevant peptide (negative control well). Twenty-four hours later, supernatants from each well were collected and frozen at −80°C. After a series of samples had been collected, cytokines were assayed using an MSD Th1/Th2 7-plex plate and read on a Sector Imager-2400 (Meso Scale Discovery, Gaithersburg, MD, USA).
Results
Identification of epitopes unique to H1N1/09 HA
The first objective of this study was to identify epitopes within the H1N1/09 HA proteins that are recognized by CD4+ T cells. Among all influenza antigens, HA is the most diversified. Sequence alignment analysis [NCBI, specialized BLAST multiple sequence analysis using BLAST (bl2seq)] indicated that the H1N1/09 HA protein (accession: ACQ76318) has a maximum of 79% sequence identity to the HA proteins of previously circulated H1N1 influenza viruses (influenza A/Brisbane/59/2007, accession: ADE28750) and that the low-homology regions mainly lie within the HA1 subunit. H1N1/09 HA epitopes restricted by multiple HLA-DR alleles (HLA-DRB1*01:01, *04:01, *04:04, *07:01, *11:01, *15:01 and HLA-DRB5*01:01—abbreviated as DR0101, DR0401, DR0404, DR0701, DR1101, DR1501 and DRB5) were investigated using the TGEM approach. For each HLA allele, 2–7 subjects were recruited between May 2009 and September 2009 for epitope identification. As summarized in Table 1, a total of 27 antigenic peptides were identified, several of which consisted of two consecutive overlapping peptides that, based on in silico prediction, appear to share the same minimal binding register (23).
Table 1.
Sequences of the 2009 H1N1 HA protein that contain CD4+ T-cell epitopes
| Amino acid region | Peptide sequencesa,b,c | HLA restriction | Response frequency |
|---|---|---|---|
| 1–20 | MKAILVVLLYTFATANADTL | DR0404 | 1 of 4 |
| 33–52 | VDTVLEKNVTVTHSVNLLED | DR0701 | 7 of 7 |
| 57–76 | KLCKLRGVAPLHLGKCNIAG | DR0701 | 1 of 7 |
| 113–132 | IDYEELREQLSSVSSFERFE | DRB5 | 4 of 4 |
| 121–140 | QLSSVSSFERFEIFPKTSSW | DR1101 | 2 of 3 |
| DRB5 | 4 of 4 | ||
| 129–148 | ERFEIFPKTSSWPNHDSNKG | DR1101 | 2 of 3 |
| 161–180 | FYKNLIWLVKKGNSYPKLSK | DR1101 | 3 of 3 |
| DRB5 | 3 of 4 | ||
| 209–228 | YQNADAYVFVGSSRYSKKFK | DR1101 | 2 of 3 |
| 217–236 | FVGSSRYSKKFKPEIAIRPK | DRB5 | 1 of 4 |
| 225–244 | KKFKPEIAIRPKVRDQEGRM | DRB5 | 1 of 4 |
| 241–260 | EGRMNYYWTLVEPGDKITFE | DRB5 | 1 of 4 |
| 249–268 | TLVEPGDKITFEATGNLVVP | DR0401 | 2 of 7 |
| DR0701 | 6 of 7 | ||
| 257–276 | ITFEATGNLVVPRYAFAMER | DR0401 | 1 of 7 |
| DR0701 | 7 of 7 | ||
| 265–284 | LVVPRYAFAMERNAGSGIII | DR0401 | 7 of 7 |
| DR0404 | 4 of 4 | ||
| 305–324 | TSLPFQNIHPITIGKCPKYV | DR0701 | 6 of 7 |
| 313–332 | HPITIGKCPKYVKSTKLRLA | DR1101 | 2 of 3 |
| 321–340 | PKYVKSTKLRLATGLRNIPS | DR1101 | 2 of 3 |
| 329–348 | LRLATGLRNIPSIQSRGLFG | DR0401 | 4 of 7 |
| DR0404 | 2 of 4 | ||
| 393–412 | TNKVNSVIEKMNTQFTAVGK | DR0101 | 2 of 4 |
| DR0401 | 4 of 7 | ||
| DR0404 | 2 of 4 | ||
| 401–420 | EKMNTQFTAVGKEFNHLEKR | DR0401 | 1 of 7 |
| DR1101 | 2 of 3 | ||
| 409–428 | AVGKEFNHLEKRIENLNKKV | DR1101 | 2 of 3 |
| 433–452 | LDIWTYNAELLVLLENERTL | DR0101 | 2 of 4 |
| DR1501 | 2 of 2 | ||
| 441–460 | ELLVLLENERTLDYHDSNVK | DR0101 | 2 of 4 |
| DR0401 | 5 of 7 | ||
| DR0404 | 2 of 4 | ||
| 457–476 | SNVKNLYEKVRSQLKNNAKE | DRB5 | 3 of 4 |
| 465–484 | KVRSQLKNNAKEIGNGCFEF | DR1101 | 2 of 3 |
| 513–532 | NREEIDGVKLESTRIYQILA | DR0701 | 1 of 7 |
| 521–540 | KLESTRIYQILAIYSTVASS | DR0101 | 2 of 4 |
| DR0404 | 2 of 4 |
aSequences based on H1N1/09 strain (accession: ACQ76318). bPeptides chosen for T-cell frequency analysis are shown in bold. c2009 H1N1 HA unique epitopes are in italics.
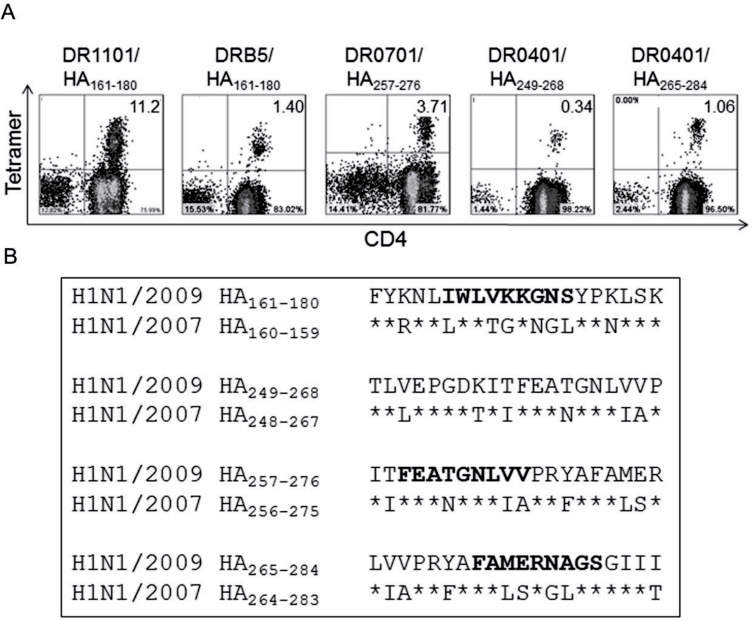
As expected, the sequences for many of these epitope-containing peptides were either identical or highly homologous to sequences within the HA proteins of previously circulating viruses. However, antigenic HA peptides that were unique to the H1N1/09 HA protein were also identified. Four unique H1N1/09 HA epitopes: DR0401-HA/09265–284, DR0701-HA/09257–276, DR1101-HA/09161–180 and DRB5-HA/09161–180, and five conserved epitopes, DR0401-HA393–412, DR0701-HA33–52, DR0701-HA305–324, DR1101-HA121–140 and DRB5-HA113–132 were selected for further analysis based on their ability to elicit an in vitro response in nearly every subject (Table 1). Figure 1A shows tetramer staining of in vitro expanded T cells with HLA-DR loaded with unique HA peptides. Detailed sequence alignments for four of these H1N1/09 HA peptides with the corresponding regions of influenza A/Brisbane/59/2007 are shown in Fig. 1B. Bold residues indicate the predicted minimal HLA-DR-binding registers: HA/09164–176 for DR1101, HA/09257–269 for DR0701 and HA/09270–282 for DR0401. These minimal epitopes were verified experimentally by stimulating PBMCs with 20-mer peptides for 2 weeks and then staining with tetramers loaded with either the 20-mer peptide or 13-mer peptide. In each case, tetramers loaded with the 20-mer or 13-mer versions of the peptide gave equivalent staining, indicating that each truncated peptide contained the minimal epitope (data not shown).
Fig. 1.
Identification of H1N1/09 HA-specific T epitopes. (A) Tetramer staining of in vitro expanded PBMCs with tetramers loaded with peptides containing H1N1/09 HA-specific epitopes (accession: ACQ76318). The percentages of tetramer+ cells are indicated in the upper corners of the FACS plots. The two DR0401 epitopes were analyzed in the same subject; the others are from different individuals. (B) Sequence alignment of peptides containing unique H1N1/09 epitopes with the corresponding regions of influenza A/Brisbane/59/2007 (accession: ADE28750). Asterisks denote identical residue in the two strains. Bold indicates HLA class II-binding registers, predicted in silico (25) and verified using truncated 13-mer peptides designed based on the predicted binding register.
T cells that recognize unique H1N1/09 HA epitopes are naive before exposure
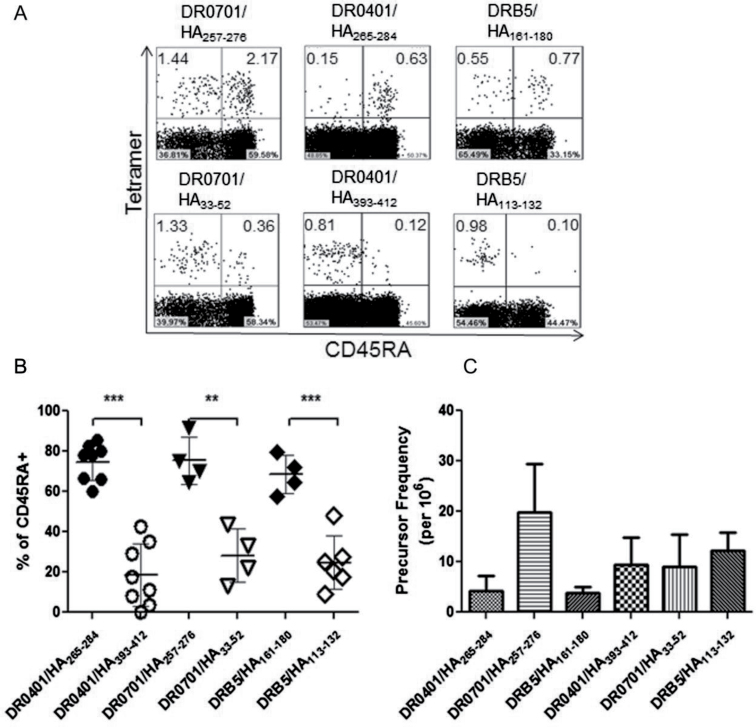
We first measured the frequency and phenotype of H1N1/09 HA-specific T-cell responses in recruited subjects who were self-reported as having no H1N1/09 exposure or symptoms of influenza infection. Because of the lack of circulation of the H1N1/09 virus in the Pacific Northwest during this phase of our study (widespread exposure did not occur until October 2009), most individuals had no exposure to the virus. Therefore, it might be expected that T cells that recognize epitopes that are unique to H1N1/09 would have a naive phenotype and occur at low frequencies in these subjects. As we recently reported for anthrax-specific T cells, the frequencies of epitope-specific T cells in the naive repertoire can be directly examined using ex vivo tetramer staining (24). Using this approach, we investigated the frequency and phenotype of CD4+ T cells specific for the following epitopes: HA/09265–284 (DR0401 restricted), HA/09257–276 (DR0701 restricted) and HA/09161–180 (DRB5 restricted). Frequencies were calculated as detailed in Methods. Naive versus memory phenotype was determined primarily by surface CD45RA staining and confirmed using additional markers, including CCR7 and CD27. For comparative purposes, the frequency and phenotype of CD4+ T cells specific for the following conserved HA epitopes were also investigated in the same subjects: HA393–412 (DR0401 restricted), HA33–52 (DR0701 restricted) and HA113–132 (DRB5 restricted). As shown in Fig. 2A and B, T cells recognizing unique H1N1/09 epitopes were predominantly CD45RA+, suggesting that this population of T cells was naive. These CD45RA+ T cells typically expressed CCR7 and CD27 (Supplementary Figure 1, available at International Immunology Online), supporting the conclusion that these cells have a naive phenotype. In contrast, T cells recognizing conserved HA epitopes were predominantly CD45RA–, suggesting a memory phenotype, most likely generated as a result of previous vaccination and/or infection. These data suggest that the non-homologous H1N1/09 peptides in our panel represent unique epitopes that are not present in widely circulated influenza A viruses. Figure 2C summarizes the frequencies of HA-specific T cells that recognize unique or conserved epitopes. Surprisingly, the frequency of T cells specific for one of the unique epitopes (HA/09257–276, 20±10 per million) was similar to the frequency of T cells specific for conserved epitopes (10±5 per million for HA393–412, 8±16 per million for HA33–52 and 12±4 per million for HA113–132). The frequencies of T cells specific for the other unique epitopes were lower (4±3 per million for HA/09265–284 and 4±1 per million for HA/09161–180).
Fig. 2.
Naive/memory phenotypes and frequencies of CD4+ T cells recognizing unique H1N1/09 HA epitopes or conserved influenza A HA epitopes. (A) Ex vivo tetramer staining, enrichment and naive/memory phenotype analysis for representative non-exposed subjects. Top panels: unique H1N1/09 HA epitopes (DR0701-HA257–276, DR0401-HA/09265–284 and DRB5-HA161–180). Bottom panels: conserved influenza A HA epitopes (DR0701-HA33–52, DR0401-HA393–412 and DRB5-HA113–132). The plots for each given HLA-DR allele are from a single subject. The percentages of tetramer+ cells are indicated in the upper corners of the FACS plots. (B) Summary of CD45RA expression by T cells specific for unique and conserved H1N1 HA epitopes for multiple non-exposed individuals. (C) Summary of the frequencies of T cells specific for unique and conserved H1N1 HA epitopes for multiple non-exposed individuals. ** denotes P < 0.01 and *** denotes P < 0.001.
CD4+ T-cell responses to unique and conserved H1N1/09 HA epitopes in H1N1/09-infected subjects
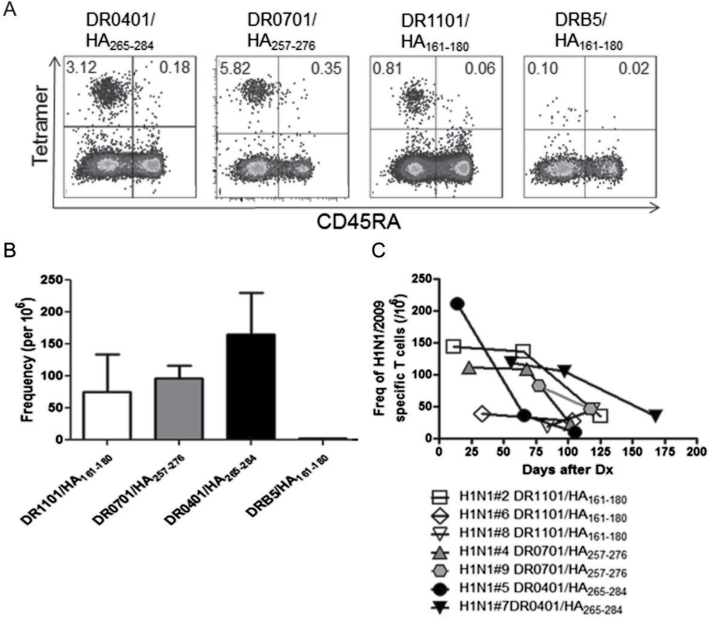
If CD4+ T cells that recognize unique H1N1/09 epitopes are immunologically relevant, the frequency and phenotype of these cells would be expected to change following exposure to virus. To investigate this, 17 subjects with laboratory confirmed H1N1/09 infection were recruited between November 2009 and January 2010; 7 of these had at least one HLA allele of interest. CD4+ T cells isolated from these seven patients were analyzed by direct ex vivo staining using tetramers loaded with four unique epitopes: DR0401-HA/09265–284, DR0701-HA/09257–276, DR1101-HA/09161–180 and DRB5/HA161–180. Again, the frequency and phenotype of H1N1/09 HA-specific T-cell responses were examined using ex vivo tetramer staining (Fig. 3A). As shown in Fig. 3B, three of these tetramers (DR0401-HA/09265–284, DR0701-HA/09257–276, DR1101-HA/09161–180) detected high frequencies of H1N1/09-specific T cells (165±66, 97±20 and 76±59 per million, respectively) in each of the individuals tested, whereas the fourth (DRB5-HA/09161–180) detected low frequencies (<5 tetramer+ T cells per million). As expected, tetramer+ CD4+ T cells were CD45RA– for each of these specificities, indicating that in contrast to unexposed donors, the HA-specific T cells in H1N1/09-infected subjects were antigen-experienced memory cells (Fig. 3A).
Fig. 3.
Naive/memory phenotype and frequency of H1N1/09 HA-specific CD4+ T cells in naturally infected subjects. (A) Ex vivo analysis of the naive/memory (CD45RA) phenotype of H1N1/09 HA-specific T cells after staining and enrichment using DR0401-HA/09265–284, DR0701-HA257–276, DR1101-HA161–180 and DRB5-HA161–180 tetramers. The percentages of tetramer+ cells are indicated in the upper corners of the FACS plots. (B) The peak frequencies (the first time point of blood draw after infection) of H1N1/09 HA-specific T cells specific for epitopes restricted by HLA-DR1101 (three subjects), DR0701 (two subjects), DR0401 (two subjects) and DRB5 (two subjects). (C) Dynamics of frequency changes of T cells specific for unique H1N1/09 HA epitopes in clinically diagnosed subjects 2–24 weeks after infection.
To study the in vivo dynamics of T cells specific for unique H1N1/09 epitopes following infection, T-cell frequencies were measured in multiple subjects by direct ex vivo tetramer staining at multiple time points from 2 to 24 weeks following infection. As shown in Fig. 3C, most individuals had moderate or high early T-cell frequencies (~50–200 per million). These frequencies were stable for the first 2 months and then gradually declined (Fig. 3C), except in Subject 5 for whom T-cell frequency declined sooner. At the end of observation, the frequency in most of the subjects was <50/106 (average of 32±12/106) (Fig. 3C). These frequencies were markedly higher than those of the naive T cells observed in subjects with no virus exposure (Fig. 2C), especially at early time points after infection.
We also monitored the frequencies of T cells specific for several conserved epitopes following infection, including HLA-DR0401-restricted HA393–412, DR0701-restricted HA33–52 and HA309–424 and DR1101-restricted HA121–140. The overall dynamics were similar to those of T cells specific for unique epitopes. However, the peak frequencies of T cells specific for conserved epitopes were generally much lower (typically ~15 cells per million CD4+ T cells). Taken together with our observation that T cells specific for unique epitopes were present at frequencies in the periphery that were typically lower than conserved epitopes prior to exposure to virus, it appears that T cells specific for unique epitopes may expand more extensively than T cells specific for conserved epitopes in response to infection with H1N1/09.
Expansion of influenza-specific naive and memory T cells after vaccination
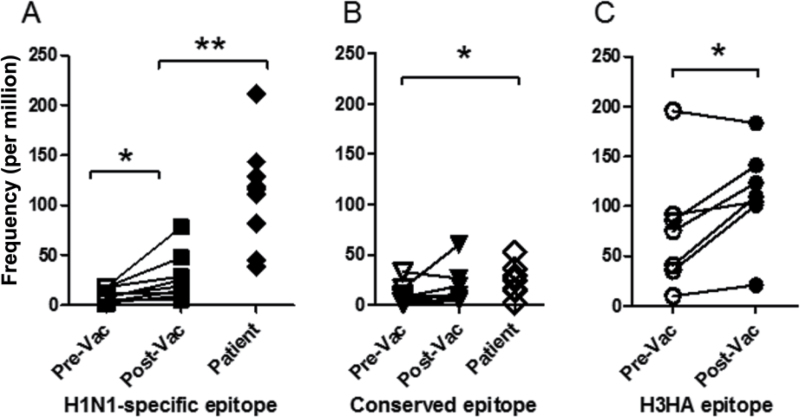
In naturally infected patients, we observed elevated frequencies of T cells specific for unique H1N1/09 epitopes and more modest frequencies of memory T cells specific for conserved epitopes. We next examined the frequency and phenotype of T cells specific for these unique and conserved H1N1/09 HA epitopes in healthy subjects before and <30 days after vaccination. Eight non-exposed healthy volunteers (six individuals with DR0401, DR0701 or DR1101 haplotypes and two subjects that were heterozygous for these alleles) were recruited and vaccinated with a single intramuscular dose of the monovalent H1N1/09 vaccine. We compared the frequencies of T cells specific for unique HA epitopes (DR0401-HA/09265–284, DR0701-HA/09257–276, DR1101-HA/09161–180) with those for T cells specific for conserved HA epitopes (DR0401-HA393–412, DR0701-HA305–324, DR1101-HA121–140) before and after vaccination, measuring the frequencies of these epitope-specific T cells simultaneously. The average frequencies of T cells specific for unique and conserved epitopes were low prior to vaccination (Fig. 4A and B). After vaccination there was a modest but significant increase in the frequency of T cells specific for unique epitopes (10±6 and 25±23 per million for pre- and post-vaccination, respectively, a 2.5-fold increase, paired t-test, P < 0.05), but no significant increase (10±9 and 15±18 per million for pre- and post-vaccination, respectively, 1.5-fold increase, paired t-test, P = 0.27) in the frequency of CD4+ T cells specific for conserved epitopes. As depicted in Fig. 4A, the frequencies of T cells specific for unique epitopes observed in infected subjects were 11-fold higher than the naive frequencies seen in unexposed subjects (107±60 per million, a highly significant increase over the baseline, Student’s t-test, P < 0.01). As depicted in Fig. 4B, T cells specific for conserved epitopes in infected subjects were 2.6-fold higher than the baseline memory frequencies (25±16 per million, a significant increase over the pre-vaccination baseline, Student’s t-test, P = 0.027). These differences suggest that the expansion of HA-specific T cells after vaccination may be modest compared with the expansion of HA-specific T cells after natural infection for unique epitopes (11-fold higher than baseline after infection versus 2.5-fold expansion after vaccination) but fairly similar for conserved epitopes (2.6-fold higher than baseline after infection versus 1.5-fold expansion after vaccination). However, because T-cell frequencies vary with time after vaccination or natural infection, these apparent differences in T-cell expansion may be confounded by variations in the timing of the samples.
Fig. 4.
T-cell frequencies in pre-vaccination and post-vaccination subjects, and in naturally infected patients. (A) Frequency of CD4+ T cells specific for unique H1N1/09 epitopes, including DR1101-HA/09161–180, DR0701-HA/09257–276 and DR0401-HA/09265–284, in unexposed subjects, vaccinated subjects and naturally infected subjects. (B) Frequency of CD4+ T cells specific for conserved H1N1 influenza epitopes, including DR1101/HA121–140, DR0701/HA33–52, DR0701/HA305–324 and DR0401/HA393–412, in unexposed subjects, vaccinated subjects and naturally infected subjects. (C) Frequency of CD4+ T cells specific for H3HA epitopes, including DR0401/H3HA306–318, DR0404/ H3HA306–318 and DR0101/ H3HA306–318, in unexposed subjects and vaccinated subjects. * denotes P ≤ 0.05 and ** denotes P ≤ 0.01.
A lack expansion of T cells specific for conserved epitopes after vaccination could indicate that these epitopes are not immunodominant. To investigate this possibility, we next examined the frequency and phenotype of T cells specific for an immunodominant H3HA epitope in seven subjects with DR0401 and DR0101 haplotypes. These subjects were recruited for a pre-vaccination blood draw and then vaccinated intramuscularly with Fluzone (which contains A/Uruguay/716/2007, an H3N2 strain). The frequencies of DR0101-H3HA306–318- or DR0401-H3HA306–318-specific T cells were determined using ex vivo tetramer analysis. As expected, the frequencies of H3HA306–318-specific T cells before vaccination were relatively high in these subjects (76±60 per million CD4+ T cells, Fig. 4C), presumably due to repeated vaccination and/or natural infection. After vaccination, there was a modest increase in the frequency of these T cells (1.9-fold, from 76±60 to 110±49 per million, a significant increase over the pre-vaccination baseline, paired t-test, P < 0.05). These results indicate that T cells specific for immunodominant epitopes occur at high frequencies but expand modestly in the periphery after vaccination. Therefore, modest expansion in response to vaccination may be a trait that is exhibited by HA-specific memory T cells regardless of their initial frequency or relative immunodominance.
Surface phenotype and cytokine secretion of H1N1/09 HA-specific T cells
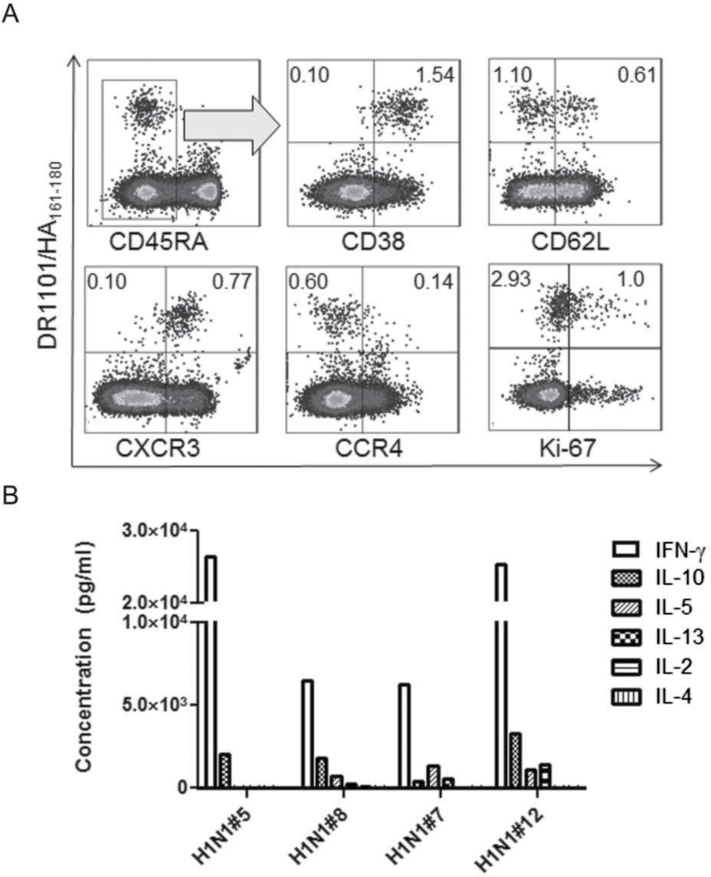
Although protective immunity to influenza A virus is primarily mediated by neutralizing antibodies, CD4+ T-cell responses play an important role in the generation of humoral immunity and direct cellular antiviral immunity is correlated with protection. It can be expected that these important effects would be influenced by the effector functions of the responding T cells (25, 26). To directly analyze the polarization of H1N1/09 HA-specific T cells, tetramer+ T cells were enriched and analyzed ex vivo for expression of surface markers that correlate with Th1, Th2 and Th17 phenotypes. Figure 5A depicts representative results for an infected subject sampled 11 days after diagnosis. At this time point, this subject’s H1N1/09 HA-specific CD4+ T cells had a distinct memory (CD45RA–) phenotype, with a large proportion expressing high levels of CD38, an activation marker for memory CD4+ T cells. Approximately 30% of these cells had also down-regulated CD62L. These markers suggest a population of recently activated memory cells. In addition, most of these cells expressed CXCR3 (CD183), but were CCR4–, suggesting a predominantly Th1-polarized population. Intracellular Ki-67 staining was measured in the same subject 17 days after diagnosis. Approximately 25% of the tetramer+ cells expressed Ki-67, indicating active cell division. Using the same approach, we examined the activation of H1N1/09 HA-specific T cells in additional subjects for several months (Supplementary Figure 2, available at International Immunology Online). Over time, the level of activation for these memory T cells declined steadily, reaching 50% after 2 months and slowly decreasing to 30% after 5 months. HA-reactive T cells in multiple subjects were found to be CCR6– (data not shown) suggesting that responses were not Th17 polarized, at least in the periphery.
Fig. 5.
Phenotype analysis for H1N1/09 HA-specific T cells. (A) Phenotype analysis of H1N1/09-specific T cells. The ex vivo surface marker expression of DR1101/HA161–180-specific CD4+ T cells (left panels) for a representative naturally infected subject sampled 11 days after diagnosis was analyzed by multicolor flow cytometry. The intracellular proliferation marker Ki-67 (last panels) was analyzed 17 days after diagnosis. The percentages of tetramer+ cells are indicated in the upper corners of the FACS plots. (B) Cytokine profiles of H1N1/09-specific T cells. Antigen-specific T cells were expanded from PBMCs in vitro, verified by tetramer staining, and restimulated overnight in plates coated with the corresponding tetramers (or a negative control). Antigen-specific cytokine secretion into the supernatant was measured using a 7-plex Th1/Th2 MSD plate. The data shown are from four representative subjects.
To correlate the implied polarization of HA-reactive T cells (based on their cell surface phenotype) with actual effector function, H1N1/09 HA-specific T cells were expanded in vitro, re-activated in tetramer coated plates and analyzed for cytokine secretion. In agreement with the observed expression of CXCR3, these cells predominantly secreted IFN-γ with virtually undetectable levels of IL-4, IL-2 and IL-13 (Fig. 5B). Occasionally, low levels of IL-10 and IL-5 secretion were observed. A similar Th1 cytokine profile was also observed for vaccinated subjects (data not shown).
Discussion
H1N1/09 is a recently emerged influenza strain with an HA protein sequence that differs significantly from that of strains that had previously circulated (18, 19). The arrival of this strain in our region provided an opportunity to investigate CD4+ T-cell responses to this novel strain prior to and following widespread exposure in our study population. Hoping to differentiate responses to H1N1/09 from pre-existing memory responses to previously circulating influenza strains, we first sought to identify strain-specific epitopes within the HA protein of H1N1/09. As expected, we identified a number of CD4+ T-cell epitopes, including multiple unique epitopes from the low-homology regions of the HA protein and conserved epitopes that were present in various influenza strains that had circulated in previous years (Table 1). We then utilized recently developed ex vivo tetramer methodology to directly examine the frequency and phenotype of T cells specific for HA epitopes. As reported in our recent anthrax study, this approach can reliably measure the frequencies of epitope-specific T cells in the naive repertoire of unexposed subjects by labeling and enriching epitope-specific cells in a highly specific fashion (24). Our subjects were self-reported as having no H1N1/09 exposure or symptoms of influenza infection, but our experimental design did not permit serological testing to confirm lack of exposure to the pandemic H1N1 strain. Therefore, in principle, it is plausible that some of these subjects could have been infected but asymptomatic. However, based on the timing of our experiments (the work was conducted early in the first phase of the pandemic) and the non-activated phenotype of the strain-specific T cells, we expect that our results accurately reflect immune status prior to exposure to the pandemic H1N1 virus.
In our population of unexposed subjects, we observed low but measurable frequencies of H1N1/09-specific T cells. These HA-specific T cells were CD45RA+ and typically expressed CCR7 and CD27, strongly suggesting a naive phenotype. In contrast, T cells that recognized conserved epitopes were CD45RA–, suggesting a memory phenotype. The naive frequencies of T cells specific for two of the unique epitopes (HA/09265–284 and HA/09161–180) were low (~4 cells per million), whereas the third (HA/09257–276) was similar to that of T cells specific for conserved epitopes (~20 cells per million). In subjects with H1N1/09 infection, the frequency of these H1N1/09 HA-reactive T cells expanded to ~100 cells per million. Although measured in different subjects, the absolute frequency of H3HA306–318-specific CD4+ T cells was comparable to the frequencies of H1N1/09 HA-specific T cells found in infected patients (110±49 cells per million versus 107±60 cells per million, respectively). This similarity suggests that these unique HA epitopes may also be immunodominant epitopes.
In non-exposed subjects, the frequencies of T cells specific for unique epitopes were generally lower than those of T cells specific for conserved epitopes. However, T cells that recognized the unique epitopes were present at significantly higher frequencies than T cells specific for conserved epitopes in subjects that were exposed to H1N1/09 by natural infection (or vaccination, but to a lesser extent). Thus, our results suggest that naive T cells that recognize unique epitopes expand to a much greater extent than memory T cells that recognize conserved epitopes. These observations agree with those of Martin et al. (27), who observed that naive CD8+ T cells undergo greater expansion and generate higher numbers of long-lived memory T cells than memory T cells co-transferred into the same recipient. It has been observed that repeated antigen stimulation affects both the gene expression and function of memory T cells. Key differences in the expression profiles of naive and memory T cells may account for the reduced expansion of memory T cells, as compared with naive T cells (28). It has also been demonstrated that memory CD4+ T cells can exhibit anomalous signaling pathways (lack of the T cell-specific ZAP-70 kinase phosphorylation and alterations in CD3ζ-associated proteins) when compared with naive and effector CD4+ T cells (29, 30). As such, it could be argued that these T cells have a reduced intrinsic proliferative capacity. At the same time, the lack of expansion of conserved HA epitope-specific memory T cells could simply indicate that these epitopes are sub-dominant or cryptic epitopes, as T cells specific for these epitopes are present at low frequencies in the memory repertoire at baseline and these frequencies do not increase after vaccination. This lack of expansion is in contrast to influenza memory T cells specific for conserved M1 and NP epitopes, which are present at high frequencies that increase significantly after vaccination (data not shown).
Previous studies, including an influenza challenge study, have demonstrated that the frequency of pre-existing CD4+ memory T cells that recognize conserved M1 and NP epitopes is correlated with disease protection (17). In contrast, after heterologous challenge, higher overall T-cell frequencies were correlated with increased viral shedding and symptoms. Taken in this context, our results raise the possibility that existing HA-specific memory CD4+ T cells may provide only minimal protection from heterologous influenza infection based on their modest frequencies and lack of expansion. The observed increase in the number of HA-specific T cells following vaccination and natural infection is apparently due primarily to the expansion of naive T cells that recognize unique epitopes within the heterologous virus. It could be speculated that the degree of expansion may correspond to viral load, as T-cell frequencies were much higher in naturally infected subjects (reaching levels of 100 cells per million, roughly a 10-fold increase) than in vaccinated subjects (reaching levels of 25 cells per million, roughly a 3-fold increase). However, it was not feasible to assess viral load in our study. In the setting of both natural infection and vaccination, T cells that recognize unique H1N1/09 HA epitopes expanded to a greater extent compared with T cells that recognized the cross-reactive conserved epitopes. This would agree with a recent study (31), which documented higher increases in influenza-specific CD4+ T cells in response to H1N1/09 vaccination (4.6-fold increase) than in response to the seasonal vaccine (2.7-fold increase). These observed changes in frequency (both in our study and that of Schmidt et al.) must be interpreted with the caveat that all observations were obtained using peripheral blood samples. T-cell numbers could very well be higher in certain tissues, as antigen-specific T cells are often retained in primary-infected tissues and their draining lymph nodes (32). However, we expect that the relative proportions of T cells that recognize unique and conserved epitopes would probably be similar.
The naive frequencies of H1N1/09-reactive T cells ranged from 4 to 20 cells per million total CD4+ T cells. In a previous study, we observed that T cells specific for the protective antigen of Bacillus anthracis had naive frequencies ranging from 0.1 to 10 per million total CD4+ T cells (24). The frequencies observed for these HA-specific T cells, although somewhat higher, are similar in magnitude. In our anthrax study, we observed that anthrax-reactive T cells could expand >60-fold after vaccination. In contrast, the average increase was <3-fold after H1N1/09 vaccination and only 11-fold after infection. These differences could suggest that the H1N1/09 virus is less immunogenic than B. anthracis or that the current influenza vaccine is less effective than the anthrax vaccine. However, the vaccination schedule for anthrax includes five doses (plus recommended annual booster doses), whereas the vaccinated subjects in the present study received only a single dose of the H1N1/09 vaccine. Therefore, it could be expected that repeated administration of the H1N1/09 vaccine over time might increase the frequency of these influenza-specific T cells to a higher magnitude.
Longitudinal analysis in naturally infected subjects indicated that T-cell frequencies declined modestly during the first 2 months after diagnosis and then had a more pronounced decline in subsequent months (Fig. 3C). However, for logistical reasons, our earliest samples were typically at least 2 weeks after initial virus exposure. It is possible that the peak of the T-cell response could have occurred during the first 2 weeks since previous studies indicate that viral shedding typically lasts for only 1 week (33, 34). Due to similar technical constraints, the timing of our analysis of vaccinated subjects also varied. As such, the observed differences in T-cell expansion may be confounded by variations in timing. It is also possible that our observations were influenced by the important limitation that only peripheral blood could be interrogated. T-cell frequency and phenotypes could differ significantly between the periphery and relevant tissues such as the lung and lymph nodes. Influenza-specific T cells can be maintained by residual antigen in lymph nodes and infected tissues, driving proliferation even after viral clearance. Indeed, it has been demonstrated in animal studies of influenza infection that residual antigen presentation is sufficient to prime and expand naive influenza-specific CD4+ and CD8+ T cells (32, 35, 36). More recently, it was demonstrated that secondary effectors accumulate in significant numbers in the infected lung following influenza A infection (37). In spite of these caveats, we observed that H1N1/09-specific CD4+ T cells were almost exclusively CXCR3+ regardless of the time elapsed after vaccination or infection (Fig. 4A) indicating a Th1 lineage. This is consistent with observations from animal studies (38, 39) and one human study (7) indicating that effective anti-viral immunity is correlated with Th1 responses and suggesting that cytokines from Th1 cells are sufficient for providing help in differentiation of B cells into IgG-producing cells. As expected, H1N1/09 HA-specific T cells that expressed CXCR3 predominantly secreted IFN-γ in an in vitro re-stimulation assay (Fig. 4B). Recent studies have noted early Th17-type responses in critical H1N1/09 cases (13), some suggesting that Th17 effector functions enhance the persistence and immunopathology of viral infection (36, 40). We did not observe any evidence of H1N1/09 HA-specific Th17 cells. However, cells with a Th17 phenotype could have been present at earlier time points than those that were sampled in this study or could be confined to primary-infected tissues and lymph nodes.
In summary, in this study, we characterize CD4+ T-cell responses to H1N1/09 influenza epitopes in human subjects. The emergence of the H1N1/09 virus allowed us to independently assay for the first time the expansion of CD4+ T cells that recognized either unique or conserved HA epitopes in both infected and vaccinated human subjects. The relatively modest expansion of T cells directed against the conserved HA epitopes either after vaccination or infection may indicate that these HA-reactive memory T cells offer minimal protection from heterologous infection. Naive T cells specific for H1N1/09 unique HA epitopes expanded more robustly than memory T cells specific for conserved epitopes, both after natural infection and vaccination. Although our study offers no direct evidence that these robust naive responses were actually protective, these HA-specific T cells were consistently CXCR3+ and secreted IFN-γ. Therefore, it is plausible that these cells may provide some benefit through secretion of Th1 cytokines. On the basis of our findings and other recent reports, we suggest that heterologous protection against influenza infection is most likely to be mediated by T cells specific for conserved internal proteins such as M1 and NP.
Supplementary data
Supplementary data are available at International Immunology Online.
Funding
National Institutes of Health and National Institute of Allergy and Infectious Diseases (HHSN272200900043C; 1U01A089859 to W.W.K.).
Supplementary Material
Acknowledgements
We wish to thank our study participants for making this work possible. The administrative support of Diana Sorus is greatly appreciated.
References
- 1. Kash J. C., Basler C. F., Garcia-Sastre A., et al. 2004. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J. Virol. 78:9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simonsen L., Clarke M. J., Schonberger L. B., Arden N. H., Cox N. J., Fukuda K. 1998. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J. Infect. Dis. 178:53. [DOI] [PubMed] [Google Scholar]
- 3. Taubenberger J. K., Morens D. M. 2006. 1918 influenza: the mother of all pandemics. Emerg. Infect. Dis. 12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schroedl A. 2010. Age-based human influenza A virus (H5N1) infection patterns, Egypt. Emerg. Infect. Dis. 16:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smallman-Raynor M., Cliff A. D. 2007. Avian influenza A (H5N1) age distribution in humans. Emerg. Infect. Dis. 13:510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu H., Gao Z., Feng Z., et al. 2008. Clinical characteristics of 26 human cases of highly pathogenic avian influenza A (H5N1) virus infection in China. PLoS ONE. 3:e2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balter S., Gupta L. S., Lim S., Fu J., Perlman S. E. 2010. Pandemic (H1N1) 2009 surveillance for severe illness and response, New York, New York, USA, April-July 2009. Emerg. Infect. Dis. 16:1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Echevarría-Zuno S., Mejia-Arangure J. M., Mar-Obeso A. J., et al. 2009. Infection and death from influenza A H1N1 virus in Mexico: a retrospective analysis. Lancet. 374:2072. [DOI] [PubMed] [Google Scholar]
- 9. Louie J. K., Acosta M., Jamieson D. J., Honein M. A. 2010. Severe 2009 H1N1 influenza in pregnant and postpartum women in California. N. Engl. J. Med. 362:27. [DOI] [PubMed] [Google Scholar]
- 10. Bermejo-Martin J. F., Ortiz de Lejarazu R., Pumarola T., et al. 2009. Th1 and Th17 hypercytokinemia as early host response signature in severe pandemic influenza. Crit. Care. 13:R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan M. C., Cheung C. Y., Chui W. H., et al. 2005. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 6:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crowe C. R., Chen K., Pociask D. A., et al. 2009. Critical role of IL-17RA in immunopathology of influenza infection. J. Immunol. 183:5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang K. J., Su I. J., Theron M., et al. 2005. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 75:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Headley A. S., Tolley E., Meduri G. U. 1997. Infections and the inflammatory response in acute respiratory distress syndrome. Chest. 111:1306. [DOI] [PubMed] [Google Scholar]
- 15. de Jong M. D., Simmons C. P., Thanh T. T., et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. To K. K., Hung I. F., Li I. W., et al. 2010. Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin. Infect. Dis. 50:850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilkinson T. M., Li C. K., Chui C. S., et al. 2012. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 18:274. [DOI] [PubMed] [Google Scholar]
- 18. Garten R. J., Davis C. T., Russell C. A., et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 325:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith G. J., Vijaykrishna D., Bahl J., et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 459:1122. [DOI] [PubMed] [Google Scholar]
- 20. Novak E. J., Liu A. W., Gebe J. A., et al. 2001. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J. Immunol. 166:6665. [DOI] [PubMed] [Google Scholar]
- 21. Roti M., Yang J., Berger D., Huston L., James E. A., Kwok W. W. 2008. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J. Immunol. 180:1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lissina A., Ladell K., Skowera A., et al. 2009. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J. Immunol. Methods. 340:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh H., Raghava G. P. 2001. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 17:1236. [DOI] [PubMed] [Google Scholar]
- 24. Kwok W. W., Tan V., Gillette L., et al. 2012. Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire. J. Immunol. 188:2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abbas A. K., Murphy K. M., Sher A. 1996. Functional diversity of helper T lymphocytes. Nature. 383:787. [DOI] [PubMed] [Google Scholar]
- 26. Mosmann T. R., Coffman R. L. 1989. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 7:145. [DOI] [PubMed] [Google Scholar]
- 27. Martin M. D., Condotta S. A., Harty J. T., Badovinac V. P. 2012. Population dynamics of naive and memory CD8 T cell responses after antigen stimulations in vivo. J. Immunol. 188:1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wirth T. C., Xue H. H., Rai D., et al. 2010. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 33:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farber D. L., Luqman M., Acuto O., Bottomly K. 1995. Control of memory CD4 T cell activation: MHC class II molecules on APCs and CD4 ligation inhibit memory but not naive CD4 T cells. Immunity. 2:249. [DOI] [PubMed] [Google Scholar]
- 30. Farber D. L., Acuto O., Bottomly K. 1997. Differential T cell receptor-mediated signaling in naive and memory CD4 T cells. Eur. J. Immunol. 27:2094. [DOI] [PubMed] [Google Scholar]
- 31. Schmidt T., Dirks J., Enders M., et al. 2012. CD4+ T-cell immunity after pandemic influenza vaccination cross-reacts with seasonal antigens and functionally differs from active influenza infection. Eur. J. Immunol. 42:1755. [DOI] [PubMed] [Google Scholar]
- 32. Jelley-Gibbs D. M., Brown D. M., Dibble J. P., Haynes L., Eaton S. M., Swain S. L. 2005. Unexpected prolonged presentation of influenza antigens promotes CD4 T cell memory generation. J. Exp. Med. 202:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suess T., Buchholz U., Dupke S., et al. 2010. Shedding and transmission of novel influenza virus A/H1N1 infection in households–Germany, 2009. Am. J. Epidemiol. 171:1157. [DOI] [PubMed] [Google Scholar]
- 34. Witkop C. T., Duffy M. R., Macias E. A., et al. 2010. Novel Influenza A (H1N1) outbreak at the U.S. Air Force Academy: epidemiology and viral shedding duration. Am. J. Prev. Med. 38:121. [DOI] [PubMed] [Google Scholar]
- 35. Turner D. L., Cauley L. S., Khanna K. M., Lefrançois L. 2007. Persistent antigen presentation after acute vesicular stomatitis virus infection. J. Virol. 81:2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zammit D. J., Turner D. L., Klonowski K. D., Lefrançois L., Cauley L. S. 2006. Residual antigen presentation after influenza virus infection affects CD8 T cell activation and migration. Immunity. 24:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strutt T. M., McKinstry K. K., Kuang Y., Bradley L. M., Swain S. L. 2012. Memory CD4+ T-cell-mediated protection depends on secondary effectors that are distinct from and superior to primary effectors. Proc. Natl Acad. Sci. USA. 109:E2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baumgarth N., Kelso A. 1996. In vivo blockade of gamma interferon affects the influenza virus-induced humoral and the local cellular immune response in lung tissue. J. Virol. 70:4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Graham M. B., Dalton D. K., Giltinan D., Braciale V. L., Stewart T. A., Braciale T. J. 1993. Response to influenza infection in mice with a targeted disruption in the interferon gamma gene. J. Exp. Med. 178:1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hou W., Kang H. S., Kim B. S. 2009. Th17 cells enhance viral persistence and inhibit T cell cytotoxicity in a model of chronic virus infection. J. Exp. Med. 206:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.