Abstract
Central corneal thickness (CCT) is associated with eye conditions including keratoconus and glaucoma. We performed a meta-analysis on >20,000 individuals in European and Asian populations that identified 16 new loci associated with CCT at genome-wide significance (P < 5 × 10−8). We further showed that 2 CCT-associated loci, FOXO1 and FNDC3B, conferred relatively large risks for keratoconus in 2 cohorts with 874 cases and 6,085 controls (rs2721051 near FOXO1 had odds ratio (OR) = 1.62, 95% confidence interval (CI) = 1.4–1.88, P = 2.7 × 10−10, and rs4894535 in FNDC3B had OR = 1.47, 95% CI = 1.29–1.68, P = 4.9 × 10−9). FNDC3B was also associated with primary open-angle glaucoma (P = 5.6 × 10−4; tested in 3 cohorts with 2,979 cases and 7,399 controls). Further analyses implicate the collagen and extracellular matrix pathways in the regulation of CCT.
Human ocular biometric parameters comprise a set of highly heritable and often correlated quantitative traits. One notable example is CCT, which has an estimated heritability of up to 95% (ref. 1). Whereas extreme corneal thinning is a dramatic clinical feature for rare congenital connective tissue disorders, including brittle cornea syndrome (BCS) and several types of osteogenesis imperfecta2,3, mildly reduced CCT is involved in more common and late-onset eye diseases. It is a hallmark of keratoconus and a risk factor for primary open-angle glaucoma (POAG) in individuals with ocular hypertension4,5. Previous genome-wide association studies (GWAS) conducted on both European and Asian populations have identified 11 CCT-associated loci6–9. Among these loci, mutations in ZNF469 (refs. 10–12), COL5A1 (ref. 13) and COL8A2 (refs. 14,15) are known to cause rare disorders of BCS, Ehlers-Danlos syndrome (EDS) and corneal dystrophy, respectively. However, none was found to be associated with common eye diseases.
Keratoconus is a common corneal ectasia, affecting 1 in 2,000 in the general population16. It is a progressive eye disease characterized by thinning and asymmetrical conical protrusion of the cornea, which causes variable and severe visual impairment. Owing to the limited availability of medical treatments, keratoconus is one of the leading causes of corneal transplantation worldwide17. Two GWAS have been conducted on susceptibility for keratoconus, and these studies suggested some new genetic associations, but neither study reported genome-wide significant loci18,19. POAG is the most common form of glaucoma, which is the second leading cause of blindness worldwide20. Several risk loci for POAG have been identified through early linkage and candidate gene studies21,22 and recent GWAS23–27. In both diseases, affected individuals have reduced CCT relative to the general population.
Dissecting the genetics of key risk factors may provide insights into associated disease etiology. Therefore, we conducted a large meta-analysis of GWAS on CCT from over 20,000 individuals, including individuals of European and Asian descent who were affected or unaffected with glaucoma to identify new CCT-associated loci. To evaluate potential clinical relevance, we tested the identified CCT-associated loci in 2 keratoconus susceptibility studies (totaling 874 keratoconus cases and 6,085 controls) and 3 studies on POAG risk (totaling 2,979 POAG cases and 7,399 controls). The overall study design is shown in Supplementary Figure 1.
RESULTS
Meta-analysis of CCT from >20,000 samples
We collected 13 GWAS on CCT, totaling over 20,000 individuals (Supplementary Table 1). Because of differences in the sample attributes, we performed meta-analyses of CCT within each subset, including 13,057 individuals with European ancestry who were unaffected with eye disease in set 1, 6,963 individuals with Asian ancestry who were unaffected with eye disease in set 2, 1,936 POAG cases with European ancestry in set 3 and 198 normal tension glaucoma (NTG, a common form of open-angle glaucoma) cases with Asian ancestry in set 4. All samples were genotyped on commercially available genotyping arrays. The majority of studies in the first two sets were imputed according to the phased haplotypes of HapMap reference samples, whereas few studies in the other two sets had imputation data available by the time of this study.
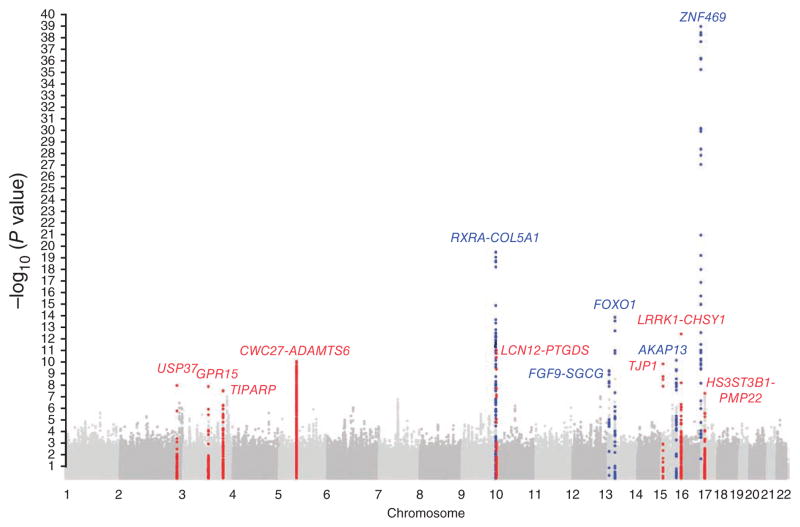
We separated the twin studies (Australian BATS and TEST twin studies and UK twin studies) from the first set of samples with European ancestry. Because family studies are less affected by potential population stratification, these twin studies were analyzed as a direct replication of association results from the discovery set consisting of all the remaining set 1 samples. All loci from the discovery set were found in the twin studies with similar effect sizes and with at least nominal replication (Table 1). This result, together with a low genomic control parameter (λ = 1.05; Supplementary Fig. 2), suggested that population stratification had little effect on the meta-analysis. We then performed meta-analysis on the results from the discovery set and the twin studies, reported as combined set 1. This combined set identified 13 loci that were associated with CCT at genome-wide significance in the populations of European descent (Table 1, Supplementary Fig. 3a–m and Supplementary Table 2). Those included five known loci, COL5A1, AVGR8 (reported here as FGF9-SGCG), FOXO1, AKAP13 and ZNF469, in European populations7,8, one locus, LRRK1-CHSY1, previously identified in Asian populations6 but unknown in European populations and seven new loci (Fig. 1 and Table 1).
Table 1.
CCT-associated loci from meta-analysis of set 1 samples (13,057 individuals of European descendant unaffected with eye disease)
| Locusa,b | Chr. | Lead SNP | Base paire | A1/A2 | AF1 | Discovery set (n = 9,584)
|
Australian and UK twins (n = 3,473 from 1,905 families)
|
Combined set 1 (n = 13,057 individuals)
|
Conditional and joint analysis
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (s.e.)d | P | β (s.e.)d | P | β (s.e.)d | P | I2 | Phet | β (s.e.)d | P | ||||||
| USP37 | 2 | rs10189064 | 219035744 | A/G | 0.04 | −0.19 (0.05) | 5.9 × 10−5 | −0.30 (0.07) | 1.8 × 10−5 | −0.23 (0.04) | 1.0 × 10−8 | 30.2 | 0.15 | −0.23 (0.04) | 1.1 × 10−8 |
| GPR15 | 3 | rs3749260 | 99733552 | A/C | 0.13 | −0.12 (0.02) | 9.5 × 10−7 | −0.11 (0.04) | 3.8 × 10−3 | −0.12 (0.02) | 1.3 × 10−8 | 0 | 0.94 | −0.12 (0.02) | 1.2 × 10−8 |
| TIPARPc | 3 | rs9822953 | 157954765 | T/C | 0.67 | 0.08 (0.02) | 7.5 × 10−7 | 0.07 (0.03) | 1.1 × 10−2 | 0.08 (0.01) | 2.7 × 10−8 | 0 | 0.69 | 0.08 (0.01) | 3.0 × 10−8 |
| CWC27- ADAMTS6 | 5 | rs1117707 | 64425421 | A/G | 0.70 | −0.11 (0.02) | 2.8 × 10−10 | −0.06 (0.03) | 4.0 × 10−2 | −0.09 (0.01) | 8.4 × 10−11 | 0 | 0.75 | −0.09 (0.02) | 9.0 × 10−11 |
| RXRA- COL5A1b | 9 | rs3118520 | 136581416 | A/G | 0.63 | 0.14 (0.02) | 1.3 × 10−18 | 0.09 (0.03) | 1.5 × 10−3 | 0.13 (0.01) | 3.4 × 10−20 | 12.1 | 0.33 | 0.13 (0.01) | 5.2 × 10−21 |
| COL5A1b | 9 | rs7044529 | 136707872 | T/C | 0.15 | −0.12 (0.02) | 1.5 × 10−−8 | −0.14 (0.04) | 1.5 × 10−4 | −0.13 (0.02) | 1.1 × 10−11 | 0 | 0.61 | −0.13 (0.02) | 9.4 × 10−12 |
| LCN12- PTGDS | 9 | rs11145951 | 138980085 | T/C | 0.49 | 0.09 (0.01) | 1.4 × 10−9 | 0.08 (0.03) | 1.7 × 10−3 | 0.09 (0.01) | 9.2 × 10−12 | 0 | 0.6 | 0.09 (0.01) | 1.1 × 10−11 |
| FGF9-SGCG | 13 | rs1034200 | 22126691 | A/C | 0.23 | 0.09 (0.02) | 6.4 × 10−7 | 0.12 (0.03) | 1.8 × 10−4 | 0.10 (0.02) | 6.1 × 10−10 | 31.6 | 0.12 | 0.09 (0.01) | 5.4 × 10−10 |
| Near FOXO1 (3′) | 13 | rs2721051 | 40008884 | T/C | 0.11 | −0.15 (0.02) | 1.4 × 10−9 | −0.22 (0.04) | 6.8 × 10−7 | −0.17 (0.02) | 1.3 × 10−14 | 10.9 | 0.33 | −0.17 (0.02) | 1.6 × 10−14 |
| Near TJP1 (5′) | 15 | rs785422 | 27961177 | T/C | 0.11 | −0.14 (0.02) | 1.8 × 10−8 | −0.13 (0.04) | 2.1 × 10−3 | −0.14 (0.02) | 1.4 × 10−10 | 33.1 | 0.1 | −0.14 (0.02) | 1.5 × 10−10 |
| Near AKAP13 (5′) | 15 | rs6496932 | 83626571 | A/C | 0.20 | −0.09 (0.02) | 4.1 × 10−7 | −0.15 (0.03) | 1.2 × 10−5 | −0.11 (0.02) | 6.8 × 10−11 | 0 | 0.97 | −0.11 (0.02) | 6.7 × 10−11 |
| LRRK1b | 15 | rs2034809 | 99372922 | A/G | 0.50 | −0.05 (0.02) | 5.2 × 10−4 | −0.05 (0.03) | 5.7 × 10−2 | −0.05 (0.01) | 7.5 × 10−5 | 0 | 0.89 | −0.08 (0.01) | 2.4 × 10−8 |
| LRRK1b | 15 | rs930847 | 99376085 | T/G | 0.77 | −0.12 (0.02) | 3.2 × 10−11 | −0.10 (0.03) | 2.8 × 10−3 | −0.11 (0.02) | 3.7 × 10−13 | 0 | 0.9 | −0.13 (0.02) | 4.0 × 10−16 |
| CHSY1b | 15 | rs752092 | 99599457 | A/G | 0.66 | −0.08 (0.02) | 4.7 × 10−8 | −0.06 (0.03) | 3.4 × 10−2 | −0.08 (0.01) | 5.9 × 10−9 | 44.3 | 0.03 | −0.09 (0.01) | 5.7 × 10−10 |
| BANP- ZNF469 | 16 | rs6540223 | 86878937 | T/C | 0.64 | 0.19 (0.02) | 3.0 × 10−31 | 0.18 (0.03) | 5.8 × 10−10 | 0.18 (0.01) | 1.3 × 10−39 | 22.5 | 0.22 | 0.18 (0.01) | 5.9 × 10−39 |
| HS3ST3B1- PMP22 | 17 | rs2323457 | 14494915 | A/C | 0.29 | −0.07 (0.02) | 6.0 × 10−6 | −0.09 (0.03) | 1.9 × 10−3 | −0.08 (0.01) | 4.7 × 10−8 | 0 | 0.48 | −0.08 (0.01) | 5.4 × 10−08 |
Chr., chromosome; A1/A2, reference allele/other allele; AF, allele frequency; s.e., standard error.
Locus assigned to the RefSeq protein-coding gene within or near the association signal interval (defined by LD plot using the measure r2 > 0.9 with the lead SNP implemented in SNAP using the CEU reference). The locus was assigned to the interval defined by the two flanking RefSeq protein-coding genes if clearly intergenic. New loci for European populations are shown in bold. The FGF9-SGCG locus was previously reported as AVGR8.
These loci contain multiple independent associations, as suggested from the results of the conditional and joint analysis.
The lead SNP is within a validated non-coding mRNA, LOC730091.
Effect and s.e. are in standardized units.
Genomic positions are based on NCBI Build 36/hg18.
Figure 1.
Manhattan plot of set 1 meta-analysis results. Association results (–log10 P values) are plotted for each chromosome. A blue dot and the corresponding gene name indicate that the locus is known in European populations. The red dot and the corresponding gene name indicate that the locus has not previously been reported as associated with CCT in European populations.
Using a recent approach of approximate conditional and joint multiple-SNP analysis28, we found that two loci harbored multiple independent variants that were associated with CCT (Table 1). One example was the COL5A1 region. Previous studies based on European and Asian samples both suggested the presence of two independent signals in this region8,9. In the current meta-analysis, we observed that the two variants rs3118520 and rs7044529, which were 126 kb apart with linkage disequilibrium (LD) r2 < 0.01 in our reference samples from the Queensland Institute of Medical Research (QIMR) cohort28,29, represented two independent LD blocks. The first LD block was located upstream of COL5A1 (RXRA-COL5A1), and the second was in an intron of COL5A1. Similarly, the locus at LRRK1-CHSY1 harbored three independent associations that could be captured by rs2034809, rs930847 and rs752092. Because the trait-increasing alleles of rs2034809 and rs930847 were negatively correlated (r = −0.17), the association at rs2034809 was not found to reach genome-wide significance in the single-SNP analysis (P = 7.5 × 10−5), and the marginal effects of both SNPs were underestimated (Table 1). The five known loci (including two independent signals at COL5A1) explained 4% of additive variance in CCT in European populations. The eight loci not known in European populations (including multiple signals at the LRRK1-CHSY1 locus) explained an additional 3.5% of additive variance, adding up to 7.5% of the total variance explained in European populations.
The two published GWAS of CCT in three Asian ancestry groups–Indian, Malay and Chinese—successfully replicated associations at COL5A1, AKAP13 and ZNF469 (refs. 6,9). To investigate whether CCT-associated loci are important across populations on a larger scale, we tested the loci identified from European samples in set 1 in these three Asian populations. All loci had effect directions consistent with the ones found in European samples except for rs10189064 at the USP37 locus, which was very rare in the Asian samples. Furthermore, 11 of 15 lead SNPs (or their proxies) were at least nominally significant at P < 0.05 (Supplementary Table 3). These observations suggest that most of the CCT-associated loci identified from populations of European descent are shared in Asian populations. Therefore, we performed another meta-analysis using Fisher’s method to combine association P values from GWAS of European samples and Asian samples. Fisher’s method makes no assumptions regarding trait distribution or allele frequencies across studies. This meta-analysis showed that the known CCT-associated loci in the Asian populations, including COL8A2, FAM46A-IBTK, C7orf42 and 9p23 (reported here as MPDZ-NF1B), replicated in European samples and further identified ten new loci associated with CCT, including COL4A3, which had been suggested in a set of Croatian samples but had not reached genome-wide significance8 (Fig. 2a,b and Table 2).
Figure 2.
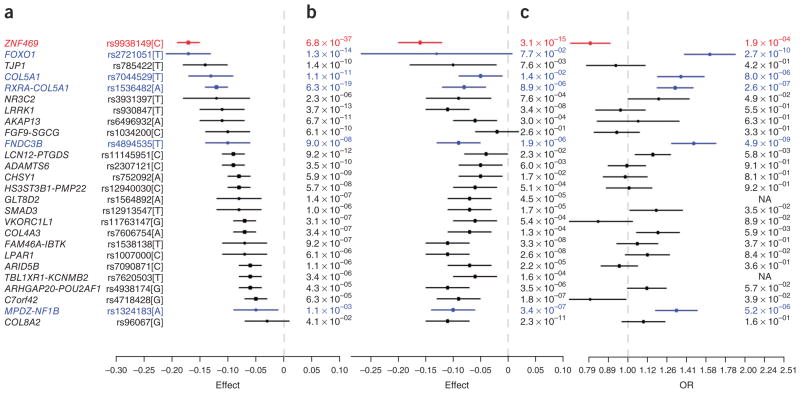
Association with CCT in European and Asian populations, and with keratoconus risk in European populations. (a) Association with CCT in European populations (set 1). (b) Association with CCT in Asian populations (set 2). (c) Association with keratoconus risk in European populations. This plot is a standard forest plot, but, instead of showing study-specific effects, it shows the association effects in the three panels for each locus. The allele is chosen as the CCT-reducing allele (boxes that represent the point estimates in a and b are on the left side of the dashed line, effect on CCT < 0). Effect on CCT is in standardized units. Given the association between reduced CCT and elevated keratoconus risk, for the effect directions to be consistent, the CCT-reducing allele would be the keratoconus risk allele (boxes that represent the OR of keratoconus in c on the right side of the dashed line, OR > 1). Five loci in blue, FOXO1, two SNPs at COL5A1, FNDC3B and MPDZ-NF1B, are the CCT-associated loci that are significantly associated with keratoconus risk after correction for multiple testing and also show consistent effect directions. The ZNF469 locus is marked in red because the CCT-reducing allele is significantly associated with a lower risk of keratoconus. rs1564892 and rs7620503 were not genotyped in the keratoconus case-control studies and are therefore presented as missing values (NA).
Table 2.
CCT-associated loci from meta-analysis of set 1 and set 2 samples
| Locusa | Chr. | Lead SNP | A1/A2 | Base pairg | Set 1–European samples (n = 13,057)
|
Set 2–Asian samples (n = 6,963)
|
Fisher’s method P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF1 | βc | s.e.c | P | AF1 | βc | s.e.c | P | ||||||
| COL8A2 | 1 | rs96067 | A/G | 36344507 | 0.80 | 0.03 | 0.02 | 4.1 × 10−2 | 0.58 | 0.11 | 0.02 | 2.3 × 10−11 | 2.7 × 10−11 |
| COL4A3 | 2 | rs7606754 | A/G | 227843424 | 0.35 | −0.07 | 0.01 | 3.4 × 10−7 | 0.36 | −0.07 | 0.02 | 1.3 × 10−4 | 1.1 × 10−9 |
| FNDC3B | 3 | rs4894535 | T/C | 173478299 | 0.17 | −0.10 | 0.02 | 9.0 × 10−8 | 0.27 | −0.09 | 0.02 | 1.9 × 10−6 | 5.1 × 10−12 |
| TBL1XR1-KCNMB2b | 3 | rs7620503 | T/C | 178786992 | 0.39 | −0.06 | 0.01 | 3.4 × 10−6 | 0.51 | −0.06 | 0.02 | 1.6 × 10−4 | 1.2 × 10−8 |
| NR3C2 | 4 | rs3931397 | T/G | 149298947 | 0.07 | −0.12 | 0.03 | 2.3 × 10−6 | 0.12 | −0.09 | 0.03 | 7.6 × 10−4 | 3.7 × 10−8 |
| ADAMTS6 | 5 | rs2307121 | T/C | 64661268 | 0.34 | 0.09 | 0.01 | 3.5 × 10−10 | 0.30 | 0.05 | 0.02 | 6.0 × 10−3 | 5.9 × 10−11 |
| FAM46A-IBTK | 6 | rs1538138 | T/C | 82851313 | 0.25 | −0.07 | 0.02 | 9.2 × 10−7 | 0.23 | −0.11 | 0.02 | 3.3 × 10−8 | 9.8 × 10−13 |
| VKORC1L1 | 7 | rs11763147 | A/G | 64964256 | 0.45 | 0.07 | 0.01 | 3.1 × 10−7 | 0.32 | 0.06 | 0.02 | 5.4 × 10−4 | 4.0 × 10−9 |
| C7orf42 | 7 | rs4718428 | T/G | 66058881 | 0.65 | 0.05 | 0.01 | 6.3 × 10−5 | 0.44 | 0.09 | 0.02 | 1.8 × 10−7 | 3.0 × 10−10 |
| MPDZ-NF1B | 9 | rs1324183 | A/C | 13547491 | 0.20 | −0.05 | 0.02 | 1.1 × 10−3 | 0.25 | −0.10 | 0.02 | 3.4 × 10−7 | 8.1 × 10−9 |
| LPAR1 | 9 | rs1007000 | T/C | 112702502 | 0.22 | 0.07 | 0.02 | 6.1 × 10−6 | 0.21 | 0.11 | 0.02 | 2.6 × 10−8 | 4.8 × 10−12 |
| RXRA-COL5A1d | 9 | rs1536482 | A/G | 136580349 | 0.34 | −0.12 | 0.01 | 6.3 × 10−19 | 0.34 | −0.08 | 0.02 | 8.9 × 10−6 | 3.0 × 10−22 |
| COL5A1 | 9 | rs7044529 | T/C | 136707872 | 0.15 | −0.13 | 0.02 | 1.1 × 10−11 | 0.20 | −0.05 | 0.02 | 1.4 × 10−2 | 4.8 × 10−12 |
| LCN12-PTGDS | 9 | rs11145951 | T/C | 138980085 | 0.49 | 0.09 | 0.01 | 9.2 × 10−12 | 0.69 | 0.04 | 0.02 | 2.3 × 10−2 | 6.4 × 10−12 |
| ARID5B | 10 | rs7090871 | T/C | 63500292 | 0.59 | 0.06 | 0.01 | 1.1 × 10−6 | 0.64 | 0.07 | 0.02 | 2.2 × 10−5 | 6.3 × 10−10 |
| ARHGAP20-POU2AF1 | 11 | rs4938174 | A/G | 110418450 | 0.31 | 0.06 | 0.01 | 4.3 × 10−5 | 0.15 | 0.11 | 0.02 | 3.5 × 10−6 | 3.6 × 10−9 |
| Near GLT8D2 (5′) | 12 | rs1564892 | A/G | 102969872 | 0.76 | −0.08 | 0.02 | 1.4 × 10−7 | 0.43 | −0.07 | 0.02 | 4.5 × 10−5 | 1.7 × 10−10 |
| FGF9-SGCGe | 13 | rs1034200 | A/C | 22126691 | 0.23 | 0.10 | 0.02 | 6.1 × 10−10 | 0.27 | 0.02 | 0.02 | 2.6 × 10−1 | 3.7 × 10−9 |
| Near FOXO1 (3′) | 13 | rs2721051 | T/C | 40008884 | 0.11 | −0.17 | 0.02 | 1.3 × 10−14 | 0.03 | −0.13 | 0.07 | 7.7 × 10−2 | 3.6 × 10−14 |
| Near TJP1 (5′) | 15 | rs785422 | T/C | 27961177 | 0.11 | −0.14 | 0.02 | 1.4 × 10−10 | 0.08 | −0.10 | 0.04 | 7.6 × 10−3 | 3.1 × 10−11 |
| SMAD3 | 15 | rs12913547 | T/C | 65254561 | 0.77 | −0.08 | 0.02 | 1.0 × 10−6 | 0.64 | −0.07 | 0.02 | 1.7 × 10−5 | 4.6 × 10−10 |
| Near AKAP13 (5′)f | 15 | rs6496932 | A/C | 83626571 | 0.20 | −0.11 | 0.02 | 6.8 × 10−11 | 0.36 | −0.06 | 0.02 | 3.1 × 10−4 | 6.7 × 10−13 |
| LRRK1 | 15 | rs930847 | T/G | 99376085 | 0.77 | −0.11 | 0.02 | 3.7 × 10−13 | 0.73 | −0.11 | 0.02 | 3.4 × 10−8 | 5.9 × 10−19 |
| CHSY1 | 15 | rs752092 | A/G | 99599457 | 0.66 | −0.08 | 0.01 | 5.9 × 10−9 | 0.78 | −0.05 | 0.02 | 1.7 × 10−2 | 2.4 × 10−9 |
| BANP-ZNF469 | 16 | rs9938149 | A/C | 86889141 | 0.62 | 0.17 | 0.01 | 6.8 × 10−37 | 0.74 | 0.16 | 0.02 | 3.1 × 10−15 | 2.4 × 10−49 |
| HS3ST3B1-PMP22 | 17 | rs12940030 | T/C | 14501741 | 0.71 | 0.08 | 0.01 | 5.7 × 10−8 | 0.54 | 0.06 | 0.02 | 5.1 × 10−4 | 7.4 × 10−10 |
Locus assigned to the RefSeq protein-coding gene within or near the association signal interval (defined by LD plot using the measure r2 > 0.9 with the lead SNP implemented in SNAP using the CEU reference). The locus was assigned to the interval defined by the two flanking RefSeq protein-coding genes if clearly intergenic. Additional new loci for both European and Asian populations are shown in bold. The MPDZ-NF1B locus was previously reported as 9p23.
The lead SNP is within a validated non-coding mRNA, LINC00578.
Effect and s.e. are in standardized units.
An imputed SNP, rs3118516, in high LD with rs1536482 (complete LD in HapMap CEU set, r2 = 0.97 in the HapMap CHB and JPT set), has a lower association P value, with Fisher’s method P = 3.5 × 10−23.
An imputed SNP, rs9552680, in high LD with rs1034200 (r2 = 0.57 and 0.95 in HapMap CEU and CHB and JPT sets, respectively), has a lower association P value, with Fisher’s method P = 3.2 × 10−9.
An imputed SNP, rs10163187, in high LD with rs6496932 (r2 = 0.73 and 0.68 in HapMap CEU and CHB and JPT sets, respectively), has a lower association P value, with Fisher’s method P = 1.7 × 10−13.
Genomic positions are based on NCBI Build 36/hg18.
Many studies observed that individuals with glaucoma, especially those with the advanced form, have thinner corneas than the general population30. Owing to phenotypic differences and potential confounding factors, for example, the fact that individuals with glaucoma take intraocular pressure–lowering medication, which has been shown to be associated with corneal thinning31, we conducted separate meta-analyses of CCT in the clinical cohorts. We collected 5 cohorts comprising a total of 1,936 POAG cases with European ancestry (set 3) and an additional cohort from Hong Kong comprising 198 NTG cases with Han Chinese ancestry (set 4), all of whom had CCT measured. Neither of these two sample sets had adequate statistical power to identify genetic loci significantly associated with CCT (in the lower range observed in individuals with glaucoma). We tested the CCT-associated loci identified from the general population in these two sets. The results from meta-analysis in set 3 showed that all genome-wide significant loci except one had the same direction of effect in open-angle glaucoma (OAG) samples compared to those in the general population, and 12 loci were at least nominally significant (Supplementary Table 4). In set 4, most of the loci were found to have effect directions consistent with those from the other sets, but, owing to the very limited power in this study, only one locus showed nominal association (Supplementary Table 4). We applied polygenic modeling in a subset of POAG cases from set 3 and found that polygenic overlap existed beyond the loci detected in healthy individuals and in POAG cases (Supplementary Fig. 4a). Despite the slight phenotypic differences, we have shown that there is substantial overlap between the genetic variants that account for CCT variation in the general population and the variants found in individuals with glaucoma. This suggests that similar pathway(s) regulate CCT regardless of eye disease status.
Pathway analysis of CCT-associated loci
To investigate whether the CCT-associated loci were enriched in functional units in terms of genes and pathways, we performed a gene-based analysis using the Versatile Gene–based Association Study (VEGAS)32 and a further pathway analysis using a new tool extended from VEGAS (VEGAS-Pathway). These analyses were first run on the meta-analysis results from set 1; therefore, HapMap phase 2 Utah residents of Northern and Western European ancestry (CEU) samples were used as the reference to estimate patterns of LD. We defined the gene regulatory region as 50 kb both upstream and downstream of a gene. This definition includes most but not all regulatory effects. For example, the lead SNPs at the ZNF469 locus were >100 kb upstream of the gene and were thus not accounted for in the gene-based test (the gene-based P value for ZNF469 was 0.18). The most significant gene was the CWC27-ADAMTS6 locus (the gene-based P values for CWC27 and ADAMTS6 were <1 × 10−6), followed by COL5A1 (P value of 3 × 10−6) (Supplementary Table 5).
On the basis of the VEGAS results, we tested whether the genes associated with CCT clustered into certain pathways, using the 4,628 pathways defined by the Gene Ontology (GO) database. We found that the top-ranking pathways were collagen (GO 0005581, empirical pathway P = 1 × 10−5, P = 0.046 after Bonferroni correction for multiple testing), extracellular matrix (ECM; GO 0031012, empirical pathway P = 3.2 × 10−5, P = 0.146 after multiple-testing correction) and their related pathways, including proteinaceous ECM, ECM part, fibrillar collagen and collagen fibril organization, as well as multicellular organismal metabolic process and myosin binding (Supplementary Table 6). A number of collagen genes were common in these pathways, for example, COL2A1, COL5A1, COL5A2, COL5A3, COL11A1 and COL11A2. Removing all of the genome-wide significant genes and repeating the pathway analysis reduced the significance of the collagen pathway, as expected, but the pathway remained nominally significant (empirical P = 0.005), suggesting that more of the remaining collagen pathway genes also underlie variation in CCT. The three pathways involving ECM contain various components, for example, connective tissue components, such as HAPLN1, and molecules regulating cell migration and adhesion, for example, EDIL3 and TGFB2. We ran another gene set enrichment analysis using the MAGENTA tool33. The only pathway that reached significance (false discovery rate (FDR) < 0.05) was myosin binding; however, the proteinaceous ECM and collagen pathways remained among the significant pathways (empirical P values = 5.2 × 10−5 and 0.01, respectively) (Supplementary Table 7).
The same framework of VEGAS-Pathway analysis was applied to the meta-analysis of the three Asian populations, with the HapMap 2 Japanese in Tokyo, Japan (JPT) and Han Chinese in Beijing, China (CHB) populations set as the reference for LD patterns. None of the GO pathways was significant after Bonferroni correction for multiple testing. But the top-ranking pathways overlapped well with the ones from the European populations, which included collagen fibril organization, ECM organization and ECM part (Supplementary Table 8). Notably, two pathways that were among the top—face morphogenesis and head morphogenesis—although not significant after correction for multiple testing, contained the gene PDGFRA, which has been reported to associate with human corneal curvature in Asian populations34. In a meta-analysis of pathways obtained from both European and Asian samples using Fisher’s method, four pathways were significant after correction for multiple testing: collagen fibril organization (Fisher’s P = 3.2 × 10−7, 1.5 × 10−3 after correction), collagen (Fisher’s P = 3.4 × 10−6, 0.016 after correction), fibrillar collagen (Fisher’s P = 4.8 × 10−6, 0.022 after correction) and ECM part (Fisher’s P = 5.2 × 10−6, 0.024 after correction), and ECM was borderline significant (Fisher’s P = 1.3 × 10−5, 0.06 after correction) (Supplementary Table 9).
Finally, we used the GRAIL tool35 to explore the functional connectivity between the CCT-associated loci using the GO database as the knowledge base. The top two pathways suggested from this gene set enrichment approach were ECM structural constituents conferring tensile strength (GO 0030020) and the collagen pathway (Supplementary Table 10). The results were thus consistent using different pathway approaches. Using PubMed Text as a knowledge base, the top key words showing functional connection between the 27 loci included collagen, syndrome, cornea, mutation and mitochondrial. We also considered each locus singly, but none was significantly connected with the remaining loci (GRAIL Ptext > 0.05 for all genes).
These results together highlight the importance of the collagen and ECM pathways in the regulation of CCT.
Clinical relevance to rare and common eye diseases
Corneal thinning is reported as one of the clinical features in a number of rare disorders that affect a variety of connective tissues. Rare mutations in ZNF469 (refs. 10–12) and a range of collagen genes2,3,13–15 have been mapped to these disorders. We and others have identified associations of the common variants in these genes, for example, ZNF469, COL5A1 and COL8A2, with normal corneal thickness7–9. In a more recent study, mutations in PRDM5 (4q27) were identified in two affected families with BCS who were known to be negative for mutations in the ZNF469 gene36. Although none of the CCT-associated loci were mapped to PRDM5, the common SNP rs10518367 70 kb upstream of the gene was associated with CCT in the European samples at the significance level of P = 8.9 × 10−5.
Keratoconus and POAG are common conditions with milder corneal thinning but more complex genetic etiologies. To investigate whether the CCT-associated loci influence genetic susceptibility to these conditions, we tested association of these loci in case-control studies with a prespecified P-value threshold for testing 26 independent SNPs (Table 2) in 2 diseases of 0.001 (P < 0.05/52). As reduced CCT is associated with POAG5 and progressive corneal thinning is observed in keratoconus17, we hypothesized that, for the effect directions to be consistent in the epidemiological sense, the CCT-reducing allele would also be the keratoconus or POAG risk allele.
We genotyped the CCT-associated loci from Table 2 in 652 keratoconus cases and 2,761 controls and also evaluated these loci in a published GWAS of keratoconus susceptibility with 222 cases and 3,324 controls19 (Supplementary Table 11). The meta-analysis of these 2 sets (874 keratoconus cases and 6,085 controls in total) identified 6 loci that were strongly associated with keratoconus risk: FOXO1 (OR = 1.62, 95% CI = 1.4–1.88, P = 2.7 × 10−10 for rs2721051), FNDC3B (OR = 1.47, 95% CI = 1.29–1.68, P = 4.9 × 10−9 for rs4894535), RXRA-COL5A1 (OR = 1.32, 95% CI = 1.19–1.47, P = 2.6 × 10−7 for rs1536482), MPDZ-NF1B (OR = 1.33, 95% CI = 1.18–1.51, P = 5.2 × 10−6 for rs1324183), COL5A1 (OR = 1.37, 95% CI = 1.19–1.57, P = 8 × 10−6 for rs7044529) and ZNF469 (OR = 1.25, 95% CI = 1.11–1.40, P = 1.9 × 10−4 for rs9938149) (Fig. 2c and Table 3). However, among the six significant loci, the rs9938149 SNP in ZNF469 showed an unexpected effect direction, with the CCT-increasing allele leading to increased risk for keratoconus (Fig. 2c). Applying polygenic modeling to the 222 keratoconus cases and 3,324 controls, we found that the aggregate effects of loci with more modest effect on CCT did not clearly enhance the prediction of keratoconus risk over and above the risk conferred by the six significantly associated loci (Supplementary Fig. 4b). We note, however, that this lack of increased prediction might have been caused by the polygenic modeling only being applicable to a subset of our available keratoconus cases (only 222 of the total 874 cases had GWAS genotyping data available, meaning that most keratoconus samples did not contribute to the polygenic modeling result).
Table 3.
Testing CCT-associated loci for susceptibility to keratoconus and POAG
| Locusb | Chr. | Lead SNP | A1/A2 | OR (95% CI)a | Pc | Effect direction as expected?d | OR (95% CI)a | Pc | Effect direction as expected?d |
|---|---|---|---|---|---|---|---|---|---|
| COL8A2 | 1 | rs96067 | A/G | 0.91 (0.80–1.04) | 0.16 | + | 0.98 (0.89–1.08) | 0.64 | + |
| COL4A3 | 2 | rs7606754 | A/G | 1.19 (1.05–1.35) | 6.0 × 10−3 | + | 0.96 (0.89–1.05) | 0.38 | − |
| FNDC3B | 3 | rs4894535 | T/C | 1.47 (1.29–1.68) | 4.9 × 10−9 | + | 0.83 (0.74–0.92) | 5.6 × 10−4 | − |
| TBL1XR1-KCNMB2 | 3 | rs7620503 | T/C | NAe | NAe | NAe | 0.91 (0.84–0.98) | 0.02 | − |
| NR3C2 | 4 | rs3931397 | T/G | 1.20 (1.00–1.44) | 0.05 | + | 0.90 (0.78–1.05) | 0.17 | − |
| ADAMTS6 | 5 | rs2307121 | T/C | 1.01 (0.90–1.12) | 0.91 | − | 0.94 (0.87–1.02) | 0.12 | + |
| FAM46A-IBTK | 6 | rs1538138 | T/C | 1.06 (0.94–1.19) | 0.37 | + | 0.99 (0.90–1.07) | 0.74 | − |
| VKORC1L1 | 7 | rs11763147 | A/G | 1.19 (0.97–1.46) | 0.09 | − | 1.07 (0.97–1.18) | 0.16 | − |
| C7orf42 | 7 | rs4718428 | T/G | 1.25 (1.01–1.54) | 0.04 | − | 1.01 (0.93–1.10) | 0.80 | − |
| MPDZ-NF1B | 9 | rs1324183 | A/C | 1.33 (1.18–1.51) | 5.2 × 10−6 | + | 1.07 (0.97–1.18) | 0.16 | + |
| LPAR1 | 9 | rs1007000 | T/C | 0.89 (0.78–1.02) | 0.08 | + | 0.99 (0.90–1.09) | 0.87 | + |
| RXRA-COL5A1 | 9 | rs1536482 | A/G | 1.32 (1.19–1.47) | 2.6 × 10−7 | + | 1.00 (0.92–1.09) | 0.94 | + |
| COL5A1 | 9 | rs7044529 | T/C | 1.37 (1.19–1.57) | 8.0 × 10−6 | + | 0.98 (0.88–1.10) | 0.77 | − |
| LCN12-PTGDS | 9 | rs11145951 | T/C | 0.86 (0.78–0.96) | 5.8 × 10−3 | + | 1.02 (0.94–1.10) | 0.72 | − |
| ARID5B | 10 | rs7090871 | T/C | 1.05 (0.94–1.17) | 0.36 | − | 1.00 (0.92–1.09) | 0.96 | − |
| ARHGAP20-POU2AF1 | 11 | rs4938174 | A/G | 0.89 (0.80–1.00) | 0.06 | + | 0.92 (0.85–1.00) | 0.06 | + |
| Near GLT8D2 (5′) | 12 | rs1564892 | A/G | NAe | NAe | NAe | 1.24 (1.06–1.45) | 6.7 × 10−3 | + |
| FGF9-SGCG | 13 | rs1034200 | A/C | 1.07 (0.93–1.22) | 0.33 | − | 0.96 (0.88–1.04) | 0.34 | + |
| Near FOXO1 (3′) | 13 | rs2721051 | T/C | 1.62 (1.40–1.88) | 2.7 × 10−10 | + | 1.08 (0.95–1.22) | 0.24 | + |
| Near TJP1 (5′) | 15 | rs785422 | T/C | 0.93 (0.78–1.11) | 0.42 | − | 1.05 (0.93–1.18) | 0.46 | + |
| SMAD3 | 15 | rs12913547 | T/C | 1.18 (1.01–1.38) | 0.03 | + | 1.00 (0.92–1.10) | 0.95 | + |
| Near AKAP13 (5′) | 15 | rs6496932 | A/C | 1.06 (0.83–1.35) | 0.63 | + | 1.00 (0.91–1.11) | 0.94 | + |
| LRRK1 | 15 | rs930847 | T/G | 0.96 (0.83–1.11) | 0.55 | − | 0.99 (0.91–1.09) | 0.89 | − |
| CHSY1 | 15 | rs752092 | A/G | 0.98 (0.86–1.12) | 0.81 | − | 1.03 (0.95–1.11) | 0.53 | + |
| BANP-ZNF469 | 16 | rs9938149 | A/C | 1.25 (1.11–1.40) | 1.9 × 10−4 | − | 1.02 (0.93–1.13) | 0.63 | − |
| HS3ST3B1-PMP22 | 17 | rs12940030 | T/C | 0.99 (0.87–1.14) | 0.92 | + | 1.04 | 0.39 | − |
Study-specific results are given in supplementary tables 11 and 12.
Locus assigned to the RefSeq protein-coding gene within or near the association signal interval (defined by LD plot using the measure r2 > 0.9 with the lead SNP implemented in SNAP using the CEU reference). The locus was assigned to the interval defined by the two flanking RefSeq protein-coding genes if clearly intergenic.
P values that are significant after correction for multiple testing (P < 0.001 for testing 26 independent SNPs in 2 diseases) are shown in bold.
Given the association between reduced CCT and elevated keratoconus or glaucoma risk, it is expected that the CCT-reducing allele is the keratoconus or POAG risk allele.
+, the effect direction is as expected; −, the effect direction is not as expected.
SNPs were not genotyped in the two keratoconus case-control studies and are therefore presented as missing values (NA).
Similarly, we examined the CCT-associated loci for a role in POAG risk in 3 clinical cohorts (2,979 cases and 7,399 controls in total): advanced POAG cases and controls from the Australian and New Zealand Registry of Advanced Glaucoma (ANZRAG)24,37 and POAG cases and controls from GLAUGEN and NEIGHBOR25,38,39 (Supplementary Table 12). Performing meta-analysis of these results, we found that FNDC3B was also significantly associated with POAG risk (P = 5.6 × 10−4) (Table 3). However, unlike its association with keratoconus, the CCT-reducing allele of rs4894535 was associated with reduced risk of POAG (OR = 0.83, 95% CI = 0.74–0.92). Furthermore, polygenic modeling on these three sets showed limited evidence for CCT aggregate effects predicting POAG risk (Supplementary Fig. 4c).
Taken together, variants identified through GWAS of CCT were not only relevant to rare disorders but to more common and complex eye diseases. Other CCT-associated loci might also be valuable candidate genes when studying the genetics of keratoconus.
Gene expression in human and inbred mouse cornea
To further establish genes from the identified loci as candidates for influencing CCT, we examined publicly available microarray gene expression data from the corneas of two human donor samples and three strains of inbred mice40,41 (Supplementary Table 13). For lead SNPs with a single associated nearby gene, the vast majority were expressed in the corneas of both humans and mice. For lead SNPs with two nearby genes, presence-absence of corneal expression suggested prioritization among the two candidates in two loci (expressed in CWC27, not expressed in ADAMTS6; expressed in PTGDS, not expressed in LCN12). To our knowledge, except for collagen loci, none of the lead genes are known to influence corneal thickness in mice, although some strains with mutations influencing collagen or ECM have previously been described42–45.
We also queried the publicly available expression quantitative trait locus (eQTL) database GHS Express46 to determine potential SNP function in terms of transcript regulation. We found that rs930847 and rs752092 were cis eQTLs for LRRK1 (P = 1.04 × 10−11, R2 = 3.4%) and CHSY1 (P = 1.2 × 10−6, R2 = 1.8%), respectively, both in monocytes. There is limited evidence of functional roles for the other CCT-associated loci identified in the meta-analysis.
DISCUSSION
Previous GWAS of CCT have identified 11 loci, 5 of which were found in studies of individuals with European ancestry7,8 and 6 of which came from studies of 3 Asian populations6,9. We performed CCT meta-analysis on an overall sample size of >20,000 individuals and brought the number of CCT-associated loci to 27. Using a recent approach of approximate conditional and joint analysis28, we found that two loci harbored multiple independent variants in association with CCT. In the subset of normal samples with European ancestry, we estimated that the five known loci explained 4% of additive variance in CCT and that eight new loci in European populations explained another 3.5%. The loci associated with CCT in European and Asian populations (Table 2) together explained 8.3% of additive variance in Europeans and 7% in Asians.
The differences in the sample attributes enabled various comparisons between the subsets of meta-analysis results. We observed similar CCT distributions in European and Asian populations and found that the underlying genetic effects were largely shared between the two ancestry groups. Despite a slightly lower CCT range in the group of individuals with glaucoma, there was significant overlap in CCT loci from samples affected and unaffected with glaucoma, suggesting similar pathways regulating CCT regardless of disease status. Our results also showed that genes harboring rare variants causing Mendelian disorders with clinical feature of extreme corneal thinning (for example, BCS and osteogenesis imperfecta) also harbor common variants that influence CCT in the general population.
We evaluated the relevance of CCT-associated loci for two major eye diseases, keratoconus and glaucoma, using independent case-control sets. We found that, despite the modest effect on CCT, 11 SNPs showed nominal association with keratoconus, with 6 significant after correction for multiple testing. The effect of the CCT-associated loci on glaucoma was less pronounced, although the FNDC3B locus was significantly associated with POAG. Our results showed that part of the genetic predisposition to these diseases was mediated through the genes underlying CCT, with the remaining predisposition attributable to alternative mechanisms.
We expected CCT-reducing alleles to be associated with increased risk of keratoconus and POAG, given the known association between thin corneas and these two diseases. In general, this was true for keratoconus, with most CCT-reducing alleles associated with increased risk of keratoconus. However, there were exceptions. The CCT-reducing allele near ZNF469 (rs9938149) led to decreased keratoconus risk. At FNDC3B, the CCT-reducing allele resulted in elevated keratoconus risk (OR = 1.47, 95% CI = 1.29–1.68), but it lowered POAG risk (OR = 0.83, 95% CI = 0.74–0.92). The pattern of complex associations seen at ZNF469 for keratoconus and at FNDC3B for POAG, where the effect directions did not always agree with the epidemiological prediction, might either reflect a false positive association or a genuine pleiotropic action of these genes in disease. An example was shown in which a body fat percentage–decreasing allele near IRS1 also associated with adverse metabolic profile, and the authors showed that it could be elucidated by a complex mechanism of pleiotropic actions47.
The six SNPs that were significantly associated with keratoconus each had a relatively large OR (range of 1.25–1.62). Unlike the inflated ORs obtained from testing thousands of variants for association between a trait and marker in GWAS48, our targeted typing of SNPs on the basis of the CCT findings, led to unbiased estimates for the ORs. Because all six SNPs had moderately high risk allele frequencies, a risk profile based on even just these SNPs yielded reasonable risk prediction. For example, ~1% of the population carry 1 or 2 risk-associated loci at each of the 6 loci, and these individuals are at 7.2-fold increased risk (assuming a multiplicative model) of keratoconus relative to the ~1% of the population who are homozygous for the protective allele at each SNP. Although these six loci combined only explained ~2% of the variation in CCT, the effect of these SNPs on keratoconus risk is large, and further evaluation of the clinical relevance of these SNPs is merited.
Our data clearly show that the endophenotype approach yields disease-relevant loci. Notably, the new CCT-associated locus from our meta-analysis, FNDC3B, was also significantly associated with both keratoconus and POAG risk. This locus did not reach genome-wide significant association with CCT in either set 1 (European ancestry) or set 2 (Asian ancestry), and, owing to the small effect on CCT, meta-analysis across groups was required to detect genome-wide significance. This raises the possibility that, among the hundreds of SNPs approaching genome-wide significance, there remain additional loci that may prove clinically relevant in determining keratoconus and glaucoma risk. The steady increase in the number of significant loci as sample size has increased for the CCT GWAS agrees with the findings that, for most quantitative traits, in a rough approximation, doubling the sample size leads to doubling of the number of significant loci49. Therefore, performing genome-wide meta-analysis of CCT and of other ocular biometric parameters on an ever-larger sample set would likely lead to further insights into associated eye diseases, such as keratoconus and glaucoma.
Finally, using three different methods of pathway analysis, we showed that CCT-associated loci converge on collagen and ECM pathways (and their related pathways). We also showed that similar pathways regulate cornea thickness in both European and Asian populations. The identified collagen pathway allows rationalization of the existing findings on the associated collagen genes and suggests more collagen genes may be important for CCT. We showed that the causal genes for BCS, ZNF469 (ref. 10) and PRDM5 (ref. 36), had a role in CCT. Although neither of the genes was included in any of the GO pathway definitions, recent microarray expression analysis showed that both genes participate in a pathway regulating ECM development and maintenance36. Therefore, the ECM pathway probably has a significant role in the regulation of CCT.
Taken together, the results from this meta-analysis further illuminate the genetic architecture of CCT and reveal that CCT-associated loci convey eye disease risk.
ONLINE METHODS
Samples
We collected 13 GWAS on CCT (n > 20,000) and an additional 2 keratoconus and 3 glaucoma case-control studies (874 cases and 6,085 controls for keratoconus; 2,979 cases and 7,399 controls for glaucoma). Each cohort was approved by a research ethics committee, and all participants gave informed consent.
Meta-analysis of set 1 (13,057 individuals with European ancestry unaffected by eye disease)
Details of the specific studies are provided in the Supplementary Note. The majority of the sites controlled for demographic variables, such as age and sex. Additional covariates that were controlled in the analysis, for example, principal components, study site or CCT measurement, were variable across individual studies. Most of the study sites have imputed data available (with the HapMap phase 2 CEU population set as the reference). Before the meta-analysis, additional checks were imposed on the SNPs, including for MAF (MAF > 1%) and imputation quality (R2 from MACH > 0.3 or proper_info from IMPUTE/SNPTEST > 0.3), and we required that SNPs be autosomal and not strand ambiguous if strand information was not available. We also compared the sample allele frequencies with HapMap allele frequencies to ensure that the allele frequencies were concordant to the ones from the reference. The quantile-quantile plot and the genomic control parameters from individual studies were also examined to assure no obvious inflation in association P values owing to residual population stratification or other confounding factors at each site.
We applied inverse variance–based meta-analysis, using METAL50 to combine individual association results in each of the four sample sets. Even though the genomic control parameter from each study was close to 1, we applied genomic control correction within each study before inclusion in the meta-analysis. A second genomic control correction on the meta-analysis statistics was suggested to be overly conservative51; therefore, we did not apply it in this study. The overall λ value was 1.048 (Supplementary Fig. 2). The heterogeneity test based on Cochran’s Q test was also used to examine whether observed effect sizes were homogeneous across samples. We also searched for the proxies of the most associated variants, that is, the genotyped SNPs that were in high LD with the lead SNPs (Supplementary Table 2). This process was carried out to determine whether there was any spurious association due to genotyping or imputation artifacts.
We separated the Australian and UK twin studies from set 1, so that the remaining set 1 samples were analyzed as a discovery set, and Australian and UK twin studies were analyzed as an independent target set. Because all associated variants from the discovery set had the same effect direction and nominal significance in the target set, we conducted a meta-analysis on the whole set referred to as combined set 1 (Table 1). Regional association plots are shown in Supplementary Figure 3a–m.
We also applied the recent approach of approximate conditional and joint multiple-SNP analysis28 to the set 1 meta-analysis results. This approach uses the genotypes from a large reference set to approximate LD, instead of requiring the actual genotypes from the study samples as in conventional conditional analysis. Here, we used the QIMR cohort as the reference, which included 3,924 unrelated individuals with European ancestry and with genotypes available for 2,410,957 SNPs28,29.
Testing CCT-associated loci in set 2 (6,963 Asian samples unaffected by eye disease)
To investigate whether the associated variants identified in European populations had similar effects on corneal thickness in Asian individuals, we collected samples from three Asian populations (Indian, Malay and Chinese) and tested the CCT-associated loci identified from set 1 in these samples. These Asian samples were imputed using HapMap reference panels: the phase 2 JPT and CHB reference was used for the Chinese samples, and the phase 3 cosmopolitan panel (1,011 individuals from Africa, Asia, Europe and North America) were used for the Indian and Malay samples. We used HapMap JPT and CHB data as an approximate reference to identify proxies for the variants that were not genotyped or imputed in any of the Asian populations. To increase power, we conducted a meta-analysis on the three Asian data sets using the same scheme as described for the meta-analysis of the set 1 samples (Supplementary Table 3).
Meta-analysis of set 1 and set 2
We performed a meta-analysis on the normal samples with European and Asian ancestry using Fisher’s method (Table 2). This method combines the P values from the two sets of results; therefore, it evaluates the strength of association instead of weighting on the basis of effect size. We also performed this meta-analysis using the inverse-variance weighting method—this method is potentially more powerful than Fisher’s method but is inappropriate if the trait distribution or allele frequencies vary across studies. We considered loci to be independent if LD was very low (r2 < 0.01) in both European and Asian samples.
Testing CCT-associated loci in the clinical cohorts (set 3: 1,936 POAG cases with European ancestry and set 4: 198 normal-tension glaucoma cases with Asian ancestry)
We analyzed the set of individuals with glaucoma for separate association testing to determine whether the CCT-associated loci identified in the general population also influenced CCT values in glaucoma cases. The diagnostic criteria for POAG are provided in the Supplementary Note. We performed meta-analysis on the set 3 samples, which included five sets of POAG cases with European ancestry, using a similar protocol to that described for the meta-analysis of set 1 (Supplementary Table 4). An additional 198 NTG cases, all of Chinese ancestry, formed set 4 and were tested for CCT-associated loci separately (Supplementary Table 4).
Testing CCT-associated loci in glaucoma case-control studies and kera-toconus case-control studies
To investigate whether the CCT-associated loci were relevant to disease, we directly tested the lead SNPs for association with risk of two common ophthalmological disorders, glaucoma and keratoconus. The sample included 652 keratoconus cases from Australia (n = 517) and Northern Ireland (n = 135), who were genotyped in 2 Sequenom multiplexes at the Australian Genotyping Research Facility; 2,761 samples typed on Illumina HumanHap610 arrays from the Blue Mountains Eye Study were used as controls. US keratoconus cases (n = 222) and controls (n = 3,324) were genotyped on Illumina HumanHap370 arrays19. The combined test of association was determined by a weighted fixed-effects meta-analysis of the two clinical cohorts (Supplementary Table 11). Similarly, we obtained three cohorts to evaluate POAG risk, 590 advanced POAG cases and 3,956 controls from South Australia24 and GLAUGEN and NEIGHBOR case- control collections25,38,39 including 1,669 and 720 high-tension and normal- tension glaucoma cases and 3443 controls (Supplementary Table 12). The diagnostic criteria for both diseases are provided in the Supplementary Note.
Pathway analysis
We used three pathway analysis approaches, an extension of the VEGAS32 gene-based test to pathway analysis (VEGAS-Pathway), MAGENTA33 and GRAIL35.
For VEGAS-Pathway analysis, we first performed gene-based tests using VEGAS software on the set 1 and set 2 meta-analysis results separately. VEGAS incorporates information from the full set of markers within a gene and accounts for LD between markers by using simulations from the multivariate normal distribution. Because the set 1 samples were all of European descent, we used the HapMap 2 CEU population as the reference to estimate patterns of LD. The set 2 samples were from three Asian ancestry groups; however, we used the combined HapMap 2 JPT and CHB populations as the reference to approximate LD patterns. The gene regulatory region was defined as 50 kb both upstream and downstream of a gene. The gene-based results for meta-analysis of European samples were presented for the genes of interest, including known CCT-associated loci, newly identified loci and their neighboring genes (Supplementary Table 5).
Pathway analysis was carried out using prespecified pathways from the GO database. Pathways with 10 to 1,000 genes were selected, yielding 4,628 pathways. Pathway analysis was based on combining gene-based test results from VEGAS32. Pathway P values were computed by summing χ2 test statistics derived from VEGAS P values. Empirical VEGAS-Pathway P values for each pathway were computed by comparing the summed χ2 test statistics from real data with those generated in 500,000 simulations where the relevant number (according to the size of the pathway) of randomly drawn χ2 test statistics was summed. To ensure that clusters of genes did not adversely affect results, within each pathway, gene sets were pruned such that each gene was >500 kb away from all other genes in the pathway. Where required, all but one of the clustered genes was dropped at random when genes were clustered. The top-ranking pathways regulating CCT in European and Asian populations are listed in Supplementary Tables 6 and 8, respectively. We also performed meta-analysis on the two sets of pathway P values using Fisher’s method (Supplementary Table 9).
We then used a gene set enrichment test implemented in MAGENTA software33. The same definition of gene regulatory region used for VEGAS was maintained here, that is, 50 kb both upstream and downstream of a gene. The first difference in the methods used in MAGENTA and in VEGAS-Pathway is that MAGENTA assigns the best-SNP P value within the gene regulatory region as the gene score and corrects this gene score for confounding factors, such as gene size, whereas VEGAS takes the full set of markers and accounts for LD between markers by simulation. The second difference is in how the gene-based results are used to form a pathway test statistic. VEGAS-Pathway uses the full set of VEGAS P values within a given pathway to form the test statistic. In contrast, MAGENTA uses a non-parametric framework to conduct the pathway enrichment test, only using genes that reach a prespecified enrichment cutoff to construct a test statistic. We used the 95th percentile of all gene scores as the enrichment cutoff. The GO and PANTHER (2010) databases available in MAGENTA were used to query the meta-analysis results from the set 1 samples of European ancestry (Supplementary Table 7).
GRAIL is an algorithm examining the functional connection between associated loci35. We queried the GO database (available in GRAIL) to identify the key pathways that show the connectivity between the CCT-associated loci listed in Table 2 (Supplementary Table 10).
Polygenic modeling
We applied the previously described polygenic approach52,53, using CCT meta-analysis results as the discovery set, to the following target sets: ANZRAG subjects (to test whether aggregation of CCT effects predicts CCT values in individuals with POAG), the US keratoconus case-control set (to test whether aggregation of CCT effects predicts keratoconus risk) and ANZRAG, GLAUGEN and NEIGBOR case-control sets (similarly, to test whether CCT effects predict POAG risk on a polygenic basis). We used genotyped data in the target sets, retaining only SNPs with clear, non-ambiguous strand coding. The polygenic profile score for each individual in the target sets was calculated as the summation of the SNP genotypes in different P-value bins defined by the CCT meta-analysis, weighted by the CCT effects. Finally, polygenic scores were used to predict trait values in the target sets. When the target set included keratoconus or POAG cases and controls, logistic regression was used to assess association between the disease trait and the polygenic profile score. Analogously, when testing CCT in POAG cases, linear regression was used.
Gene expression analyses
Previously published microarray data were used to initially characterize the expression of genes associated with lead SNPs40,41. To confirm the mouse microarray data, quantitative RT-PCR (qRT-PCR) analysis was performed using independent tissue samples. Corneas were dissected at the limbus from enucleated eyes in PBS, and the corneas from each mouse were pooled to form one sample; three samples (mice) were analyzed per strain. Corneal samples were homogenized, and RNA was extracted, treated with DNase I, purified (Aurum Total RNA Fatty and Fibrous Tissue Pack, Bio-Rad) and converted to cDNA (iScript cDNA Synthesis kit, Bio-Rad). Quantitative PCR was performed with a SYBR green master mix (iQ SYBR Green Supermix, Bio-Rad) in an RT-PCR detection system (CFX Connect, Bio-Rad). Each reaction contained 5.9 μl of water, 7.5 μl of 2× iQ SYBR green master mix, 0.3 μl of 5′-primer (5 μM), 0.3 μl of 3′ primer (5 μM) and 1 μl of cDNA (2 ng/μl). Sequences for the primer pairs used in the PCR reactions are available upon request. PCR conditions involved incubation at 95 °C for 3 min and 40 cycles of 95 °C for 10 s, 55 °C for 10 s and 72 °C for 30 s. PCR products were subjected to melting curve analysis to ensure that only a single product was amplified. Each experiment included three technical replicates of each RNA sample. Expression data were quantified on the basis of threshold cycle (CT) values. For each transcript, CT values for each sample were averaged and normalized to the values for β-actin (ACTB). Change analysis was based on ΔΔCT values and the amplification efficiency of the transcripts54.
Supplementary Material
Acknowledgments
A list of acknowledgments by study is provided in the Supplementary Note.
Footnotes
AUTHOR CONTRIBUTIONS
S.M., V.V., D.A.M., T.Y.W. and Y.L. conceived and designed the study, and liaised with the International Glaucoma Genetics Consortium for this project. Y.L. performed the primary analyses. S.M., J.Y., M.U., X.L., C.C.K., E.N.V., T.A., K.P.B., G.T., F.J., V.V., O.P., D.Y.L.L., L.J.C., C.C.Y.T., R.C., D.K., W.A., W.D.R., V.J.M.V., H.S., J.G., A.J.C., A. MacLeod, S.E., P.G.H., Y.B. and X.L. contributed to analysis. S.M. and Y.L. performed pathway analysis. J.E.C., P.M., U.T., A.F.W., N.P., C.P.P., M.G.A., J.L.W., M.A.H., L.R.P., C.E.W., N.G.M., D.A.M., C.M.v.D., T.Y.W., A.J.L., C.J.H. and Y.S.R. were the overseeing principal investigators of the individual studies. J.E.C., K.P.B., D.P.D., R.A.M., G.T., K.S., F.J., U.T., A.F.W., V.V., I.R., Z.V., C.H., O.P., H.C., J.F.W., B.F., N.P., A. Mirshahi, T.Z., R.H., F.G., R.C., K.J.L., C.P.P., D.Y.L.L., L.J.C., C.C.Y.T., M.G.A., D.K., J.L.W., L.R.P., M.U., J. Liu, B.L.Y., A.B.O., J.E.R., S.E.M., J.L.H., J.H.K., L.R.P., R.R.A., A.A.-K., J.L.W., M.A.H., N.G.M., Y.L., G.W.M., S.M., D.A.M., A.W.H., J.M., W.A., S.Y., C.P., T.L.Y., W.D.R., V.J.M.V., R.W., H.S., C.C.W.K., C.M.v.D., C.C.K., E.N.V., B.K.C., W.-T.T., E.S.T., C.-Y.C., J.-N.F., J. Li, S.M.S., T.A., T.Y.W., J.G., A.J.C., A. MacLeod, S.E., A.J.L., P.G.H., T.D.S., T.L.Y. and C.J.H. contributed reagents or methods to the genotyping, phenotyping and data analysis of corneal thickness data sets. J.E.C., K.P.B., D.P.D., R.A.M., C.P.P., D.Y.L.L., L.J.C., C.C.Y.T., J.L.W., L.R.P., M.U., J. Li, B.L.Y., A.B.O., J.E.R., S.E.M., J.L.H., J.H.K., L.R.P., R.R.A., A.A.-K., J.L.W., M.A.H., C.E.W., A.J.L., J.G., A.J.C., A. MacLeod, S.E., Y.S.R., Y.B., X.L., D.S., K.D.T., J.J.W., A.C.V. and J.I.R. contributed reagents or the genotyping, phenotyping and data analysis of the glaucoma, and keratoconus samples. Y.L. and S.M. wrote the first draft of this manuscript. K.P.B., V.V., C.C.K., Y.B., A. Mirshahi, A.W.H., D.K., P.G.H., W.D.R., J.L.W., C.M.v.D., Y.S.R., D.A.M., J.E.C. and T.Y.W. provided critical comments for manuscript revision. All authors reviewed the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
URLs. GHS Express, http://genecanvas.ecgene.net/uploads/ForReview/; Gene Ontology, http://www.geneontology.org/; SNAP, http://www.broadinstitute.org/mpg/snap/ldsearchpw.php; MAGENTA, http://www.broadinstitute.org/mpg/magenta/; GRAIL, http://www.broadinstitute.org/mpg/grail/.
Note: Supplementary information is available in the online version of the paper.
References
- 1.Dimasi DP, Burdon KP, Craig JE. The genetics of central corneal thickness. Br J Ophthalmol. 2010;94:971–976. doi: 10.1136/bjo.2009.162735. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen U, Bramsen T. Central corneal thickness in osteogenesis imperfecta and otosclerosis. ORL J Otorhinolaryngol Relat Spec. 1984;46:38–41. doi: 10.1159/000275682. [DOI] [PubMed] [Google Scholar]
- 3.Evereklioglu C, et al. Central corneal thickness is lower in osteogenesis imperfecta and negatively correlates-vith the presence of blue sclera. Ophthalmic Physiol Opt. 2002;22:511–515. doi: 10.1046/j.1475-1313.2002.00062.x. [DOI] [PubMed] [Google Scholar]
- 4.Cohen EJ. Keratoconus and normal-tension glaucoma: a study of the possible association with abnormal biomechanical properties as measured by corneal hysteresis (An AOS Thesis) Trans Am Ophthalmol Soc. 2009;107:282–99. [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon MO, et al. The Ocular Hypertension Treatment Study: baseline factors that predict the onset of primary open-angle glaucoma. Arch Ophthalmol. 2002;120:714–720. doi: 10.1001/archopht.120.6.714. discussion 829–830. [DOI] [PubMed] [Google Scholar]
- 6.Cornes BK, et al. Identification of four novel variants that influence central corneal thickness in multi-ethnic Asian populations. Hum Mol Genet. 2012;21:437–445. doi: 10.1093/hmg/ddr463. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, et al. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010;6:e1000947. doi: 10.1371/journal.pgen.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vitart V, et al. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum Mol Genet. 2010;19:4304–4311. doi: 10.1093/hmg/ddq349. [DOI] [PubMed] [Google Scholar]
- 9.Vithana EN, et al. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum Mol Genet. 2011;20:649–658. doi: 10.1093/hmg/ddq511. [DOI] [PubMed] [Google Scholar]
- 10.Abu A, et al. Deleterious mutations in the zinc-finger 469 gene cause brittle cornea syndrome. Am J Hum Genet. 2008;82:1217–1222. doi: 10.1016/j.ajhg.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen AE, et al. Brittle cornea syndrome associated with a missense mutation in the zinc-finger 469 gene. Invest Ophthalmol Vis Sci. 2010;51:47–52. doi: 10.1167/iovs.09-4251. [DOI] [PubMed] [Google Scholar]
- 12.Khan AO, Aldahmesh MA, Mohamed JN, Alkuraya FS. Blue sclera with and without corneal fragility (brittle cornea syndrome) in a consanguineous family harboring ZNF469 mutation (p E1392X) Arch Ophthalmol. 2010;128:1376–1379. doi: 10.1001/archophthalmol.2010.238. [DOI] [PubMed] [Google Scholar]
- 13.Segev F, et al. Structural abnormalities of the cornea and lid resulting from collagen V mutations. Invest Ophthalmol Vis Sci. 2006;47:565–573. doi: 10.1167/iovs.05-0771. [DOI] [PubMed] [Google Scholar]
- 14.Gottsch JD, et al. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of fuchs corneal dystrophy. Invest Ophthalmol Vis Sci. 2005;46:1934–1939. doi: 10.1167/iovs.04-0937. [DOI] [PubMed] [Google Scholar]
- 15.Biswas S, et al. Missense mutations in COL8A2, the gene encoding the α2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet. 2001;10:2415–2423. doi: 10.1093/hmg/10.21.2415. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy RH, Bourne WM, Dyer JA. A 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol. 1986;101:267–273. doi: 10.1016/0002-9394(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 17.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 18.Burdon KP, et al. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest Ophthalmol Vis Sci. 2011;52:8514–8519. doi: 10.1167/iovs.11-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2012;21:421–429. doi: 10.1093/hmg/ddr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone EM, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 22.Pasutto F, et al. Heterozygous NTF4 mutations impairing neurotrophin-4 signaling in patients with primary open-angle glaucoma. Am J Hum Genet. 2009;85:447–456. doi: 10.1016/j.ajhg.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorleifsson G, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42:906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdon KP, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011;43:574–578. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- 25.Wiggs JL, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramdas WD, et al. Common genetic variants associated with open-angle glaucoma. Hum Mol Genet. 2011;20:2464–2471. doi: 10.1093/hmg/ddr120. [DOI] [PubMed] [Google Scholar]
- 27.van Koolwijk LM, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012;8:e1002611. doi: 10.1371/journal.pgen.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medland SE, et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet. 2009;85:750–755. doi: 10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herndon LW, et al. Central corneal thickness in normal, glaucomatous, and ocular hypertensive eyes. Arch Ophthalmol. 1997;115:1137–1141. doi: 10.1001/archopht.1997.01100160307007. [DOI] [PubMed] [Google Scholar]
- 31.Harasymowycz PJ, Papamatheakis DG, Ennis M, Brady M, Gordon KD. Relationship between travoprost and central corneal thickness in ocular hypertension and open-angle glaucoma. Cornea. 2007;26:34–41. doi: 10.1097/ICO.0b013e31802e3ce4. [DOI] [PubMed] [Google Scholar]
- 32.Liu JZ, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segrè AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han S, et al. Association of variants in FRAP1 and PDGFRA with corneal curvature in Asian populations from Singapore. Hum Mol Genet. 2011;20:3693–3698. doi: 10.1093/hmg/ddr269. [DOI] [PubMed] [Google Scholar]
- 35.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burkitt Wright EM, et al. Mutations in PRDM5 in brittle cornea syndrome identify a pathway regulating extracellular matrix development and maintenance. Am J Hum Genet. 2011;88:767–777. doi: 10.1016/j.ajhg.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Souzeau E, et al. The Australian and New Zealand Registry of Advanced Glaucoma: methodology and recruitment. Clin Experiment Ophthalmol. 2012;40:569–575. doi: 10.1111/j.1442-9071.2011.02742.x. [DOI] [PubMed] [Google Scholar]
- 38.Wiggs JL, et al. The NEIGHBOR Consortium Primary Open-Angle Glaucoma Genome-wide Association Study: rationale, study design, and clinical variables. J Glaucoma. 2012 Jul 23; doi: 10.1097/IJG.0b013e31824d4fd8. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiggs JL, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011;20:4707–4713. doi: 10.1093/hmg/ddr382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lively GD, et al. Genetic dependence of central corneal thickness among inbred strains of mice. Invest Ophthalmol Vis Sci. 2010;51:160–171. doi: 10.1167/iovs.09-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez-Miranda A, Nakatsu MN, Zarei-Ghanavati S, Nguyen CV, Deng SX. Keratin 13 is a more specific marker of conjunctival epithelium than keratin 19. Mol Vis. 2011;17:1652–1661. [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper LJ, et al. The role of dermatopontin in the stromal organization of the cornea. Invest Ophthalmol Vis Sci. 2006;47:3303–3310. doi: 10.1167/iovs.05-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashida Y, et al. Matrix morphogenesis in cornea is mediated by the modification of keratan sulfate by GlcNAc 6-O-sulfotransferase. Proc Natl Acad Sci USA. 2006;103:13333–13338. doi: 10.1073/pnas.0605441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao M, Hedberg-Buenz A, Koehn D, John SW, Anderson MG. Anterior segment dysgenesis and early-onset glaucoma in nee mice with mutation of Sh3pxd2b. Invest Ophthalmol Vis Sci. 2011;52:2679–2688. doi: 10.1167/iovs.10-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weaving L, et al. Twist2: role in corneal stromal keratocyte proliferation and corneal thickness. Invest Ophthalmol Vis Sci. 2010;51:5561–5570. doi: 10.1167/iovs.09-5123. [DOI] [PubMed] [Google Scholar]
- 46.Zeller T, et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilpeläinen TO, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Göring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet. 2001;69:1357–1369. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Painter JN, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43:51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.