Abstract
Epigenetic dysregulation comprising DNA hypermethylation and hypomethylation, enhancer of zeste homologue 2 (EZH2) overexpression and altered patterns of histone modifications is associated with the progression of prostate cancer. DNA methylation, EZH2 and histone modifications also ensure the parental-specific monoallelic expression of at least 62 imprinted genes. Although it is therefore tempting to speculate that epigenetic dysregulation may extend to imprinted genes, expression changes in cancerous prostates are only well documented for insulin-like growth factor 2 (IGF2). A literature and database survey on imprinted genes in prostate cancer suggests that the expression of most imprinted genes remains unchanged despite global disturbances in epigenetic mechanisms. Instead, selective genetic and epigenetic changes appear to lead to the inactivation of a sub-network of imprinted genes, which might function in the prostate to limit cell growth induced via the PI3K/Akt pathway, modulate androgen responses and regulate differentiation. Whereas dysregulation of IGF2 may constitute an early change in prostate carcinogenesis, inactivation of this imprinted gene network is rather associated with cancer progression.
Keywords: androgen signalling, imprinted genes, loss of imprinting, PI3K signalling, prostate cancer
Epigenetic dysregulation in prostate cancer
Like other cancers of old age, prostate cancer develops over many years. Multiple pathogenic events adding on each other drive initially the development of prostatic intraepithelial neoplasia from which invasive carcinomas emerge that metastasize in more aggressive cases. Many genetic changes responsible for altered differentiation, sustained cell proliferation and invasive growth of prostate cancers have been elucidated in the last decade. Prominently among them figure chromosomal rearrangements generating novel fusion genes from which ETS transcription factors like ERG are overexpressed in an androgen-dependent manner.1 Another common change in prostate cancer is inappropriate activation of PI3K/Akt signalling, which is most efficiently achieved by homozygous deletions of the PTEN gene that encodes a crucial inhibitor of the pathway.2 Like oncogenic ETS transcription factors, PI3K signalling interacts with and modulates androgen action.3, 4, 5, 6 A major question in current prostate cancer research is how these and other genetic changes conspire to bring about the spectrum of prostate cancer phenotypes ranging from indolent, well-differentiated local nodules to highly aggressive and often poorly differentiated systemic cancers. Epigenetic abnormalities play a seminal role in shaping the varying phenotypes of prostate cancer.7, 8 Well-characterized abnormalities comprise DNA hypermethylation, DNA hypomethylation, overexpression of the histone methyltransferase enhancer of zeste homologue 2 (EZH2) and several distinctive changes in histone modification patterns. Typically, these epigenetic changes aggravate with tumour stage and Gleason grade and can be employed as prognostic biomarkers.9, 10 A prominent exception to this relation is constituted by consistent and distinctive DNA hypermethylation events associated with early development.11, 12 These events induce aberrant methylation in more than 80% of prostate cancers at the CpG islands of a dozen specific genes, which are normally free of DNA methylation. Additional hypermethylation events are observed in smaller subsets of the cases and many of these correlate with tumour progression. Accordingly, consistent hypermethylation events can be employed for detection of prostate cancers, whereas some additional hypermethylation events can serve as indicators of worse prognosis.9, 13 Hypermethylation of GSTP1 and PITX2, respectively, exemplifies the two categories.14, 15 The hypermethylation of CpG islands at the transcriptional start region of genes is often associated with silencing. More than 25 genes have so far been verified to be silenced by hypermethylation in prostate cancer.9, 13 While many hypermethylation events have been investigated for their potential as biomarkers, their functional relevance to the development of malignancy is not well understood.13 In prostate cancer cell lines, restoration of expression of several silenced genes, e.g., PDLIM4 and RASSF1A, arrested proliferation or induced apoptosis.16, 17 The contribution of these genes to the phenotype of prostate cancers in patients remains unclear.
Despite the aberrantly increased methylation at certain sites in the genome, its overall content of methylcytosine decreases during progression of prostate cancer. This decrease reflects hypomethylation of sequences that are methylated in normal tissues, i.e., intronic and intergenic sequences and prominently, interspersed and centromeric repeat sequences. Therefore, hypomethylation is primarily associated with regional or global changes in chromatin organisation that facilitate epigenetic reprogramming and genomic instability across the genome.18, 19 Locally, hypomethylation may also lead to reactivation of some genes that are normally repressed by DNA methylation. Although the number of such genes is limited, they comprise important regulators of pluripotency, lineage determination and development.18, 20
The global changes in epigenetic regulation are also reflected in altered patterns of histone modifications, some of which may also be prognostic.10 However, neither the mechanisms underlying these changes nor their consequences are very clear at this point. A large number of studies have addressed changes in the Polycomb factor EZH2 in prostate cancer (reviewed in Refs. 21–23). The Polycomb group proteins are normally involved in lineage determination during embryonic development and tissue maintenance, especially by suppressing alternative differentiation choices. They aid in gene silencing or in postponing gene activation to later differentiation stages. This function is achieved—among others—by specific histone modifications. Thus, trimethylation of histone H3 at lysine 27 (H3K27me3) is catalysed by the EZH2 subunit of the PRC2 Polycomb complex. Overexpression of EZH2 in prostate cancer is believed to contribute to altered patterns of histone modifications and to silence tumour suppressor genes in a complementary or cooperative fashion with DNA hypermethylation.24, 25 Increases in EZH2 are brought about by various mechanisms, including induction by E2F transcription factors in proliferating cells,26 gene amplification and loss of the regulatory microRNA miR-101.27 Interestingly, EZH2 expression is elevated as a consequence of ETS gene fusions in prostate cancer and may in turn conspire with these transcription factors to block cell differentiation, increase cell proliferation, facilitate invasiveness and alter androgen responses.6, 28 Accordingly, EZH2 expression levels have been linked to cancer recurrence.25, 29, 30, 31
EZH2 is by far not the only histone modifying enzyme upregulated in prostate cancer. In particular and somewhat curiously, certain histone demethylases of the JMJD2 family and KDM1/LSD1, which counteract the biochemical activity of EZH2, are likewise upregulated in prostate cancer.32, 33 They interact as cofactors with the androgen receptor and hypoxia-inducible factors and are implicated in the epigenetic changes during the development of castration resistance.34, 35, 36, 37
Genomic imprinting and its regulationa
DNA methylation, histone modifications and Polycomb factors, in particular EZH2, are also instrumental in the prototypic epigenetic phenomenon of ‘imprinting'. At least 62 genes in humans and twice as many in mice are known to be differentially expressed depending on their parental origin.38 The regulation of many imprinted genes depends on DNA methylation marks that differ between the alleles inherited from mother or father, designated differentially methylated regions (DMR). Most DMRs are established during gametogenesis and inherited and propagated by the zygote, whereas others, designated somatic DMRs, are established upon fertilisation, still in a parent-of-origin-specific fashion. Most of these methylation marks persist throughout life and are only erased in the embryonic primordial germ cells to be re-established once more in the new generation. Depending on the gene, imprinting is maintained in all cell types or in a tissue-specific manner. In addition or alternatively to DNA methylation, other epigenetic mechanisms like histone modifications and inter- or intrachromosomal interactions such as looping are employed to ensure the monoallelic expression of imprinted genes.39, 40, 41, 42 With respect to histone modifications, a striking characteristic of imprinted genes is a seemingly bivalent pattern, with modifications typical of actively transcribed genes such as trimethylation of H3K4 on one allele and H3K27 trimethylation on the other one.43, 44, 45
Imprinted genes are often situated in clusters, allowing coordinate regulation by common control regions consequently designated as imprinting control centres (ICRs). ICRs often contain DMRs. Many imprinted gene clusters encompass genes encoding non-translatable regulatory RNAs, which are termed long non-coding (lnc-RNA) to distinguish them from shorter species like microRNAs. Seemingly isolated imprinted genes are often actually gene pairs, of which one encodes an lnc-RNA acting on its partner gene. The regulatory RNAs may function by recruiting chromatin modifying proteins, including EZH2, to silence selected genes in the cluster,45, 46, 47 and by directing them into specific higher-order chromatin structures located in transcriptionally inactive subnuclear compartments.46, 48, 49, 50 In contrast, the expressed alleles of imprinted genes may be enabled to enter active subnuclear compartments to gain access to ‘transcriptional factories'.51
The extraordinarily complex multi-level mode of regulation of imprinted genes probably aims to tightly control the levels of gene products. Indeed, similar mechanisms are employed in the analogous process of X-chromosome inactivation to adjust the dosage of X-chromosomal genes.52, 53 In addition, imprinting may limit the influences of foetal or maternal factors on certain genes in the developing embryo.54 Although the biological rationale for imprinting is not firmly established, current thinking favours the ‘battle of the sexes' theory.55 This theory is based on the observation that many imprinted genes expressed from the maternal chromosome limit the acquisition of maternal resources by the foetus through the placenta, while paternally expressed genes function in the opposite direction to enhance foetal supply and growth. In another mirror fashion, maternally expressed genes regulate placental growth, while paternally expressed genes are crucial for foetal growth and development.54, 56
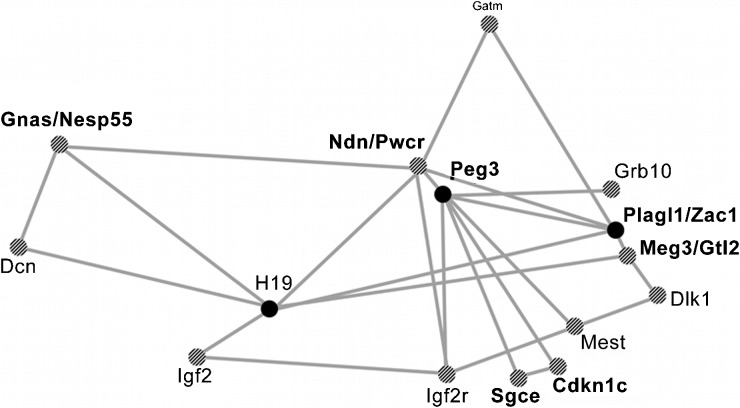
In addition, imprinted genes regulate the specification of certain lineages such as the musculoskeletal system and the brain, but also postnatal metabolism and social behaviour.56, 57, 58 In these functions, imprinted genes cooperate with each other as well as with many non-imprinted genes. In particular, a subnetwork containing the imprinted genes IGF2, H19, PLAGL1/(ZAC1), CDKN1C, DLK1 and PEG3 was characterized by remarkable co-expression across different organs and developmental stages and by functional association (Figure 1).59, 60, 61 The genes of the network are thought to control energy homeostasis at the levels of signal-sending (hypothalamus, pituitary and pancreas) and signal-receiving (liver, fat, muscle, cartilage and bone) organs to regulate body size, energy storage and expenditure during embryonic and postnatal development.56 The function of this imprinted gene network may thus consist in ‘programming' the growing embryo for optimal resource acquisition during foetal and adult life, with adjustments to the current environment.56 However, the increasing number of imprinted genes implicated in cancer and other diseases of adults underlines that their function is not restricted to the placenta and foetus.
Figure 1.
A network of imprinted genes. On the basis of 116 mouse microarray experiments, Varrault et al.60 discovered significant co-expression of selected imprinted genes. The figure shows a simplified version of the original Pajek network representation of coregulated imprinted genes disregarding the exact degree of co-expression and omitting non-imprinted genes. The enlarged circles represent the central imprinted genes of the network (modified from Ref. 60). The genes indicated in bold are deregulated in prostate cancer according to our analysis (Table 1). Note that IGF2R is not imprinted in humans. IGF2, insulin-like growth factor 2.
Defects in imprinting are responsible for a number of congenital paediatric diseases and syndromes. These often involve metabolic disturbances, neurological deficiencies and either extensive or scanty growth together with specific morphological abnormalities. Perturbations in imprinted gene expression occur in Beckwith–Wiedemann syndrome (BWS), Silver–Russell syndrome, intrauterine growth restriction, transient neonatal diabetes mellitus and congenital Wilms tumours.62, 63, 64, 65, 66, 67, 68 The lack of imprinting in such diseases originates from a failure in establishing epigenetic marks in germ cells or maintaining them in the zygote or early embryo.
Aberrant imprinting patterns may also represent a major factor in male infertility.69, 70, 71 In the gametes of some infertile men, imprinted gene loci displayed abnormal methylation states.70, 72, 73, 74 Moreover, gametes of some infertile men contain reduced amounts of normal H3K4me or H3K27me bivalent marks at certain imprinted genes and at genes encoding essential transcription factors for early development.75, 76 These changes are associated with excessive methylation of imprinted gene promoters and silencing.44, 75, 77, 78, 79
While failure to establish imprinting is a cause of paediatric diseases and infertility, secondary loss of imprinting (LOI) contributes to diseases of adults. In particular, it is a common epigenetic disturbance in cancers, but is also quite often observed in preneoplastic and ageing tissues.80, 81, 82, 83 Strictly spoken, LOI denotes the loss of parent-of-origin monoallelic expression of genes such that the two alleles may become either both transcriptionally active or both silenced. Since monoallelic expression is typically maintained by differential DNA methylation and allelic histone modifications, LOI is also reflected by a convergence of these epigenetic marks on both alleles. Experimentally, monoallelic expression can be most easily ascertained through single nucleotide or short repeat polymorphisms in gene transcripts. Loss of differential DNA methylation and histone modifications are sometimes considered as surrogate indicators of LOI. In prostate cancer, the most studied imprinted genes are IGF2 and H19.
Insulin-like growth factor 2 (IGF2)
IGF2 is the most widely studied imprinted gene in the context of tumourigenesis.81, 84 It belongs to a small imprinted gene cluster at 11p15.5 that also includes the H19 gene. The cluster is organized around a differentially methylated ICR separating IGF2 from H19 and common enhancer regions. Normally, IGF2 is expressed exclusively from the paternal allele and H19 from the maternal allele. Paternal-specific IGF2 expression is ensured by interactions between its promoter and distant enhancers. These interactions depend on the methylation status of a second DMR, viz. DMR0 upstream of the IGF2 promoters, and on the epigenetic status of the major ICR. On the maternal chromosome both regions are unmethylated, whereas on the paternal chromosome both are methylated. Expression of the gene cluster is ultimately regulated by differential formation of three-dimensional chromatin loops that separate actively transcribed and silenced genes and bring together different regulatory regions on the two alleles on both chromosomes.85, 86, 87 The correct looping is ensured by specific methylation patterns of the DMRs, and in particular by the insulator protein CTCF, which binds only unmethylated DNA in the central ICR.88 The intensely studied regulation of IGF2 and H19 serves as a paradigm in the field.
The product of IGF2 is a growth factor related to insulin that is essential for foetal development and growth. In adults, it acts as a cohormone together with follicle-stimulating hormone or luteinizing hormone. In general, IGF2 promotes cell growth, and also—like insulin—affects cell metabolism. Like its non-imprinted paralogue IGF1, IGF2 exerts its growth-promoting and anti-apoptotic effects mainly through the type 1 IGF receptor (IGFR1). Both factors act in a paracrine fashion on epithelial and stromal cells, but are more strongly expressed in stroma than in epithelium of normal prostate tissue.89 Their expression is enhanced in malignant epithelial cells, especially in tumours with high Gleason scores.90, 91, 92, 93, 94 IGFR1 activates PI3K/Akt signalling, a major determinant of prostate cancer development and progression.95
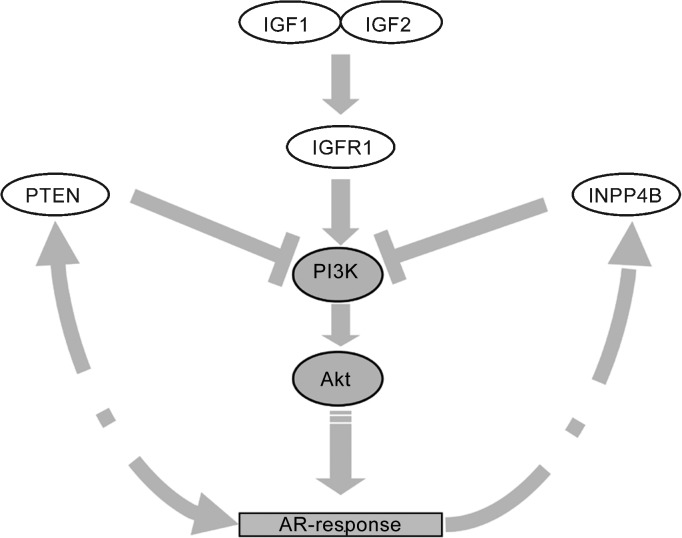
In prostate cancer, PI3K/Akt signalling is deregulated by different mechanisms, most severely by loss of the negative regulator PTEN, which reverses phosphorylation of phosphatidylinositol triphosphate by PI3K.96, 97 Particularly notable is downregulation of an inositol-triphosphate phosphatase, INPP4B, which restricts the substrate supply of PI3K.98, 99 In the absence of these negative regulators, activation of PI3K signalling by IGFs via IGFR1 is enhanced and sustained (Figure 2).
Figure 2.
A sketch of the relationship between IGF signalling, the PI3K pathway and androgen responses in the prostate. IGF, insulin-like growth factor.
In prostate cancer, PI3K and androgen receptor (AR) signalling are linked by a mutual crosstalk with inhibitory and stimulating interactions.5, 37, 100, 101 On the one hand, PI3K signalling enhances AR transcriptional activity by stabilizing active AR homodimers and sensitizing AR to low levels of androgen/DHT.95, 102 On the other hand, the androgen receptor interacts with or influences the expression of several pathway components, especially feedback inhibitors like PTEN and INPP4B. Via PI3K signalling, IGFs regulate the androgen response. Accordingly, the IGFR1 inhibitory antibody A12 blocked AR transactivation induced by IGFR1 in the absence of androgens.103
Interactions between IGF signalling and the androgen response occur at some further points. To name just a few, the pathway affects the activity of AR cofactors, e.g., via stabilisation of β-catenin by inhibition of its negative regulator GSK3. Furthermore, IGFR1 signalling stimulates AR cofactors like GRIP and IDE.104, 105 The AR coactivators SRC1 and TIF2 are substrates for MAP kinases activated by IGFs and EGF tyrosine kinase receptors.106 Consequently, endogenous IGF1 or constitutive PI3K/Akt signalling is required for androgen-induced prostate-specific antigen expression.95 IGF signalling relieves Foxo-corepressor mediated inhibition of AR ligand-mediated transactivation.107
Together, enhancement of PI3K/Akt signalling by increased IGFR1 activity and loss of negative and feedback regulators like PTEN promote AR transcriptional activity even at the low androgen concentrations achieved by androgen deprivation therapy. Ultimately, these mechanisms permit androgen-independent activation of the AR, termed illustratively ‘outlaw' AR activation.108 Apart from their effect on androgen signalling, IGFs through Akt create a potent survival signal by inactivating the pro-apoptotic tumour suppressor proteins FOXO1A and BAD, and activating the anti-apoptotic MDM2.95, 107, 109 This part of the signalling pathway may compensate for the lack of androgen pro-survival signals in castration-resistant prostate cancers. Therefore, pharmacological targeting of the IGFR1/PI3K/AKT signalling pathway by direct interference at the receptor or by decreasing circulating IGF1 levels is currently investigated in clinical trials.110
The deregulation of IGF2 in the cancerous prostate is likely caused by epigenetic mechanisms that resemble those observed in other cancers.80, 84, 111 IGF2 overexpression, often caused by loss of imprinting, is frequent across the range of paediatric tumours seen in BWS, in congenital and sporadic cases.112 LOI at the IGF2/H19 locus has been found in various cancers, including bladder and breast cancers, and in colorectal carcinoma.81, 84 Moreover, IGF2 LOI has a very promising diagnostic potential having been associated with an increased risk for developing several adult-type cancers.80, 83 For instance, LOI of IGF2 in blood cells has been proposed as a biomarker for colorectal cancer risk.113 A similar association has not yet been reported in prostate cancer patients.
A common mechanism leading to biallelic IGF2 expression is gain of methylation at the maternal ICR, which allows the interaction of the IGF2 promoter with the enhancers on both chromosomes. This mechanism has been observed in BWS patients.112 Alternatively, loss of methylation of the paternal DMR0 is associated with increased IGF2 expression. This mechanism is not completely understood, since the unmethylated maternal allele ought to function as a silencer, suppressing IGF2 expression independent of H19 imprinting. Instead, loss of methylation at the DMR0 is associated with increased IGF2 expression in breast, colorectal and Wilms tumours, perhaps because silencing of the maternal allele depends on additional repressor proteins that are no longer available in somatic cells.87, 112 In some cases, a switch to a different promoter may reactivate the normally silent maternal IGF2 allele independent of ICR methylation status.114
Importantly, in the prostate, as in some other organs, IGF2 LOI is also observed in the ageing tissue and especially in normal tissues adjacent to the cancer. Due to this ‘field effect' the highest expression of IGF2 and H19 in the prostate is actually found in benign tissues adjacent to cancer in comparison to fully normal tissues and cancer tissues. Two studies reported significant associations between overexpression and epigenetic aberrations at the locus in cancer adjacent tissues.82, 115 One study found >80% methylation of the ICR in benign prostatic hyperplasia samples, while 9/30 prostate carcinoma samples showed ∼40% methylation. Furthermore, the ICR was associated with the repressive H3K9me2 modification in benign prostatic hyperplasia but not in cancer tissues.115 In addition to ICR hypermethylation, the second study82 reported an association between increased IGF2 expression in tumour-adjacent tissue and decreased methylation of the DMR0, but no such changes in cancer tissues. Thus, DNA methylation aberrations and altered chromatin structures are likely to cause widespread IGF2 LOI in ageing prostates. This idea is supported by an investigation of cultured primary prostate epithelial cells demonstrating a conversion of the IGF2 imprinting status from monoallelic to biallelic, with a 10-fold induction of IGF2 expression, when these cells became senescent. Mechanistically, IGF2 LOI was traced to a diminished CTCF expression in the senescent cells.80, 116
H19
As compared to IGF2, its fellow gene H19 has hardly been studied in prostate cancer. While under negative regulation by p53, H19 is induced by c-myc, E2F1, FOXA and ZAC1.117 It is also upregulated by hypoxia, where it promotes cell survival.117 All these factors are also relevant in prostate cancer. H19 might potentiate IGF2 levels by inhibiting its suppressor IGFBP4 and has been implicated in epithelial–mesenchymal transition in a breast cancer metastasis model.118 Knockdown of H19 during hypoxia led to decreased expression of several genes which include AKT1, the prostate cancer biomarker AMACR and the imprinted genes CDKN1C and INPP5A.119 In transgenic mice, H19 activated the expression of imprinted genes including Dlk1, Rtl1, Gnas, Peg3, Slc38a4, Igf2R and Cdkn1c.59 Broader disturbances of imprinted genes are also observed following H19 deregulation in human congenital diseases.120, 121
The molecular mechanism by which the H19 RNA exerts its effects is not entirely clear.119 By a better established mechanism, during germ cell development, the H19 ICR interacts upon CTCF-binding with other imprinted genes in cis and in trans generating chromatin structures poised for transcription, establishing an ‘imprinted interactome', and regulating the replication timing of further interacting imprinted genes.85 H19 was shown to be regulated by steroid hormones, including androgens, and prolactin in androgen-sensitive LNCaP, but not in androgen-insensitive DU145 prostate cancer cells. If the imprinted interactome persists in adult prostatic cells, H19 and other imprinted genes might be influenced by androgen responses in prostate cancer.122
Taken together, these findings infer that relaxed IGF/H19 imprinting may occur early in prostate carcinogenesis where environmental factors or ageing may influence its epigenetic state. One could speculate that the resulting overexpression of IGF2 and H19 may promote the enlargement of an epigenetically unstable but not neoplastic progenitor cell pool and skew the androgen response, thereby changing the microenvironment and facilitating cancer formation.123
A systematic survey of imprinted gene expression in prostate cancer
In addition to IGF2, activation or inactivation of further imprinted genes may influence prostate cancer development and progression. One wonders, in particular, to which extent imprinted genes may manage to uphold their exquisitely regulated epigenetic patterns in the face of the severe and progressive disturbances of epigenetic regulation in prostate cancer. There are therefore many open questions. How many imprinted genes are deregulated in prostate cancer? Might their deregulation result straightforwardly from an ‘epigenetic chaos' in these carcinomas? How might imprinted genes contribute to the phenotype of prostate cancer?
For a systematic approach to these questions, we have used the Oncomine database (http://www.oncomine.org) in order to search for changes in the expression of imprinted genes in the results of 16 microarray studies comparing benign vs. cancerous tissues from prostatectomies. At the time of the search, the website ‘Geneimprint' (http://www.geneimprint.com/) listed 62 genes proven to be imprinted in humans. Of these, 52 had been investigated in at least two independent microarray studies. Of them only 12 genes showed consistent changes which at least approached statistical significance across multiple studies (Table 1).
Table 1. Changes of imprinted gene expression in prostate benign vs. cancerous tissues.
| Gene symbol | Gene product | Chromosomal localisation | Expression | Differential expression in PCa | Number of studies with | Total number of studies | ||
|---|---|---|---|---|---|---|---|---|
| Upregulating | Downregulating | |||||||
| HYMAI | Hydatidiform mole associated and imprinted (non-protein coding) | 6q24.2 | Paternal | ↓ | 0 | 3 | 4 | |
| PLAGL/ZAC1 | Zinc finger protein PLAGL1 | 6q24–q25 | Paternal | ↓ | 1 | 12 | 16 | |
| SGCE | Epsilon-sarcoglycan | 7q21–q22 | Paternal | ↓ | 1 | 11 | 15 | |
| PEG10 | Paternally expressed gene 10 protein | 7q21 | Paternal | ↓ | 1 | 6 | 14 | |
| PPP1R9A | Protein phosphatase 1, regulatory (inhibitor) subunit 9A | 7q21.3 | Maternal | ↓ | 5 | 0 | 6 | |
| INPP5F | Inositol polyphosphate-5-phosphatase F | 10q26.11 | Paternal | ↓ | 0 | 10 | 15 | |
| CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57,Kip2) | 11p15.5 | Maternal | ↓ | 0 | 12 | 12 | |
| MEG3/GTL2 | maternally expressed 3 (non-protein coding) | 14q32.2 | Maternal | ↓ | 4 | 13 | 15 | |
| NDN/PWCR | Necdin | 15q11.2–q12 | Paternal | ↓ | 0 | 10 | 14 | |
| SNRPN | Small nuclear ribonucleoprotein-associated protein N | 15q11.2 | Paternal | ↓ | ↑ | 6 | 7 | 16 |
| PEG3 | Paternally expressed gene 3 protein | 19q13.4 | Paternal | ↓ | 2 | 10 | 16 | |
| GNAS/NESP55 | Guanine nucleotide binding protein (G protein), alpha stimulating activity polypeptide 1 | 20q13.32 | Paternal | ↓ | 11 | 6 | 15 | |
Abbreviation: PCa, prostate cancer.
Imprinted genes found to be frequently differentially expressed in prostate benign vs. cancerous tissues, as found by in silico analysis of changes in 16 microarray studies available in Oncomine. The arrows indicate over- or underexpression. Note: in cases when some studies measured no difference in expression between benign and cancerous tissues, the total number of studies is bigger than the sum of studies with up- and downregulation. The sum of studies reporting up- or downregulation can be bigger than the total number of studies due to differences between identifiers in complex genes such as GNAS.
Most genes identified by this approach belong to the network of coordinately regulated imprinted genes that regulate growth and differentiation in the mouse embryo (Figure 1). Interestingly, IGF2 or H19 were not among them, in keeping with the idea that deregulation of these genes may occur across the entire organ. Instead, CDKN1C followed by MEG3 presented the most significant changes. Other significant expression changes concerned the 7q and 15q clusters and a pair of imprinted genes at 6q24, of which PLAGL1 encodes the ZAC1 transcription factor and HYMAI a non-coding RNA. With the exception of PPP1R9A and GNAS, all genes were expressed at reduced levels in the cancers compared to benign tissues. Of note, only the 7q and 10q regions and to a lesser extent 6q undergo significant copy number changes in prostate cancer.96 For SNRPN at 15q an almost equal number of studies described significant up- and downregulation. This may be due to the extremely complex transcription pattern of the gene with multiple splice variants confounding microarray analyses. It encodes a splicing factor acting mainly in the brain.124, 125 There is no information on SNRPN function in the prostate or in cancer.
Below, we will summarize the current knowledge on the function and regulation of the imprinted genes listed in Table 1 (except SNRPN), with an accent on their potential influence on prostate cancer progression mechanisms. Although not formally identified by our bio-informatic analysis, DLK1 and DCN will be treated, since they are part of the imprinted gene network and some evidence already links them to prostate cancer.
PLAGL1/ZAC1 and HYMAI
The chromosomal region 6q24 lost in various cancers, including a fraction of prostate carcinomas, is thought to contain a tumour suppressor gene. The most likely candidate is pleiomorphic adenoma gene-like 1 (PLAGL1), also known as ZAC1, which encodes a C2H2 seven-zinc-finger transcription factor and nuclear receptor coactivator. Two isoforms are generated via usage of alternative promoters. Transcription of the shorter isoform starts from a DMR downstream of the start of the longer isoform, which also includes the first exon of the non-coding RNA gene HYMAI. Both genes are ubiquitously paternally expressed. Defects in imprinting of the twin locus are implicated in transient neonatal diabetes mellitus, a rare disease characterized by intrauterine growth retardation, dehydration, and failure to thrive due to a lack of normal insulin secretion.68, 126
During embryonic development ZAC1 participates in molecular switches controlling progenitor cell fate decisions, proliferation and differentiation of bone, muscle and adipose tissue, secretory organs, including those of the gonadotropic axis (hypothalamus, pituitary and gonads) and the endocrine pancreas.127 Its function during development depends on its ability as a transcriptional activator. Monomeric or dimeric ZAC1 binds to GC-rich palindromic DNA elements128, 129 in the regulatory regions of the imprinted protein-coding genes IGF2, CDKN1C, DLK1 and MEST, the imprinted ncRNA genes H19, KCNQ1OT1 and MEG3; as well as in the non-imprinted genes PPARG, CK14 and PACAP1.129, 130, 131 Zac1 was suggested to represent a key regulator of the imprinted gene network in Figure 1.60
Apart from its function as a DNA-binding transcriptional activator, ZAC1 acts as a powerful coactivator for the hormone-dependent activity of nuclear receptors, including the androgen, estrogen, glucocorticoid, and thyroid hormone receptors. In this role, it functions as a scaffolding protein recruiting chromatin activators (p160 as well as histone acetyltransferases CBP, p300 and PCAF), but also corepressors (HDAC1 and mSin3a) to nuclear receptor target genes.132, 133, 134 Specifically, when interacting with the AR ZAC1 binds other coactivators (GRIP1 and SRC1) to enhance AR transcriptional activity in a hormone-dependent manner. The physiological role of ZAC1 may be the prevention of AR activation by other coactivators in the absence of hormone. Thus, ZAC1 synergized with GRIP1 in activating AR at lower androgen concentrations but suppressed the ability of GRIP1 to coactivate AR in a hormone-independent fashion.133 In effect, loss of ZAC1 may therefore promote castration-resistance in prostate cancer. However, this has not been investigated explicitly in human cancer tissues.
ZAC1 may exert antitumour effects by enhancing the transcriptional activity of p53 in the induction of certain antiproliferative proteins such as p21Cip1 or apoptosis protease-activating factor-1.134, 135, 136 Moreover, ZAC1-can inhibit proliferation by directly activating the PPARG gene, eliciting differentiation and growth arrest.130
Loss of ZAC1 expression has accordingly been observed in numerous tumour types, including breast and ovarian tumours, melanomas, astrocytomas, renal cell carcinomas and pituitary adenomas.137, 138, 139, 140, 141, 142 ZAC1 overexpression inhibited tumour cell growth through induction of apoptosis or cell cycle arrest.131, 143 PLAGL1/ZAC1 could therefore be relevant as a tumour suppressor in prostate cancer beyond its function in androgen action.
SGCE, PEG10 and the 7q21 imprinted gene cluster
Chromosome 7q is among the most frequently altered chromosomes in prostate cancer and subject to both gains and losses.96 These changes also involve the 7q21 region that contains an imprinted gene cluster. Several imprinted genes are clustered around two head-to-head oriented paternally expressed genes—paternally-expressed gene 10 (PEG10) and sarcoglycan epsilon (SGCE). PEG10 is derived from an ancient retrotransposon integration, which has been conserved in many mammalian genomes. A maternally methylated region in the first exon comprises an ICR which is marked by allele-specific histone modifications, i.e., H3K9me3 and H4K20me3 on the inactive maternal allele vs. H3K4me2 and acetylated H3K9 on the active paternal allele.144 Of note, PEG10 provides a striking example of how retrotransposon insertions contribute to evolution.145 Due to the sharing of a common promoter, PEG10 and SGCE are often coexpressed, but in some cases their expression diverges.146
The 7q21 cluster further contains the maternally expressed gene tissue factor pathway inhibitor 2 (TFPI2). It is considered as a tumour suppressor candidate, because it is silenced by genetic and epigenetic aberrations in several types of cancer, including prostate cancer.147, 148, 149, 150 TFPI2 and its non-imprinted homolog TFPI inhibit the coagulation-initiating protease tissue factor. Besides its well-established role in control of coagulation, it is a component of prostatic secretions contributing to stroma remodelling and angiogenesis. In a previous study, we investigated expression and methylation of TFPI2 and two further reciprocally imprinted genes in the locus—the maternally expressed paraoxonase 2 (PON2) and the paternally expressed SGCE.151 Whereas prostate cancer cell lines varied quite strongly in the expression of these genes, no significant differences were observed between prostate cancer and adjacent normal tissues, with the exception of PON2. Nevertheless, we observed a substantial variation of TFPI2 expression between different prostate cancer tissues. Moreover, the TFPI2 promoter was found to be partially methylated in several samples with low gene expression. Repressive chromatin modifications were more enriched in the weakly expressing LNCaP cell line with low TFPI2 expression in comparison to the PC3 cell line with high expression. Thus, TFPI2 might be prone to hypermethylation in prostate cancer. In contrast, the epigenetic status of the ICR did not correlate with TFPI2 expression in the prostate cancer cell lines. We concluded that epigenetic disturbances in the 7q21 cluster affect imprinted genes in a non-coordinated manner, suggesting an unstable epigenetic state prone to selection for specific expression changes.151
Our in silico analysis (Table 1) reveals that the expression of three imprinted genes from the 7q21 locus differs significantly between normal and cancer prostate tissues. Peculiarly, while the adjacent paternally expressed PEG10 and SGCE genes were downregulated in the majority of prostate cancer studies, PPP1R91A was upregulated. Since the epigenetic state of the ICR seemed unchanged in our former analysis,151 these expression changes might derive rather from gains and losses at 7q21, as observed in other cancer types. This explanation would also account for the lack of difference in SGCE expression in our sample series, which contains a relatively low fraction of cases with 7q alterations compared to other studies.152
PEG10 promotes embryonic growth and is overexpressed in several malignancies including B-cell ALL and CLL, hepatocarcinoma and Wilms tumour.153, 154, 155 In hepatocellular carcinoma (HCC), the 7q21 region was reported to be frequently amplified, which correlated with overexpression of both PEG10 and SGCE.156, 157 The oncogenic function of PEG10 might be mediated by several mechanisms. In HCC cells PEG10 inhibited SIAH1-mediated apoptosis.154 In androgen-dependent HCC cells, PEG10 expression was elevated upon treatment with androgen and necessary for induction of hTERT.158 Ablation of PEG10 by siRNA blocked the ability of DHT to enhance cell growth and apoptotic resistance in HCC cells.158 Another inducer of PEG10 is c-Myc.159 Furthermore, PEG10 can interfere with TGF-β signalling through interaction with two of its receptors—the activin receptor-like kinases ALK1 and ALK5. Moreover, the balance between these receptors is known to be crucial in determining proliferation and migration vs. quiescence of vascular endothelial cells during angiogenesis.160 Conceivably, this effect might also contribute to metastasis of prostate cancer cells.
The SGCE gene encodes the protein sarcoglycan epsilon which is part of a dystrophin—sarcoglycan complex in muscle and other tissues, thought to functionally link the cytoskeleton to the extracellular matrix.161 SGCE aberrations are implicated in intrauterine and postnatal growth retardation. The inherited disease Myoclonic dystonia characterized by irregular arrhythmic movements and behavioural disorders is caused by SGCE mutations.162 As in prostate cancer, SGCE is often downregulated in colorectal cancers, especially in cases with microsatellite instability and its loss may favour a less invasive phenotype.163
PPP1R9A encodes the protein phosphatase 1 regulatory (inhibitory) subunit 9a, also called neurabin I. It is an F-actin binding protein targeting protein phosphatase 1 to the actin cytoskeleton to limit the activity of the growth regulatory kinases PKA and p70S6K.164 Therefore, neurabin I was proposed to regulate adhesion-dependent signalling and function as a tumour suppressor.165
INPP5F
The INPP5F gene was recently shown to contain an imprinted transcript variant,166 coding for a phosphatidylinositol phosphatase which limits the substrate for PI3K and phospholipase C that are activated by tyrosine kinase receptors. To some extent, therefore, the phosphatidylinositol phosphatases function in a similar way to PTEN. For instance, INPP5F limits cardiomyocyte cell size and cardiac hypertrophy by inhibiting PI3K signalling during stress responses. PTEN loss likewise favours cardiac hypertrophy.167 A homolog of INPP5F, INPP4B, was recently found to be downregulated in prostate cancers. INPP4B is induced by AR in prostate cancer cells and diminishes phosphorylation of Akt.99 It is not known whether any of the INPP5F transcripts is regulated in a similar manner. However, phosphatidylinositol phosphatases are expected to dampen signalling from growth factor receptors like IGFR1 and EGFR in general. In particular, they may act as feedback inhibitors in the interaction of PI3K/Akt and AR signalling (Figure 2). Their potential as suppressors of development of prostate cancer and especially castration resistance is evident.
CDKN1C
The maternally expressed gene CDKN1C is located in a second imprinted gene cluster at 11p15.5 together with KCNQ, encoding an ion channel protein, and other genes. CDKN1C codes for the protein p57KIP2, a cyclin-dependent kinase inhibitor. It binds to various CDK-cyclin heterodimers and at low concentrations promotes their assembly, but at high concentrations abrogates the activity of CDKs 1, 2, 3, 4 and 6 leading to cell cycle arrest.168
Expression of p57KIP2 is high during embryogenesis, typically peaking at key stages of differentiation for each organ, after which it declines. In many developing tissues, particularly strong protein expression can be observed at the interface between mesenchymal and epithelial layers, perhaps corresponding to the sites where tissue-specific stem cells are suspected to reside in stem cell niches. These observations suggest a function of p57KIP2 in stem cell determination. Less speculatively, the protein is involved in regulating cell cycle exit during differentiation of many tissues.168, 169 In this function, p57KIP2 cooperates with RB family proteins to inactivate E2F proteins and induce G1 arrest and eventually a G0 state associated with terminal differentiation. Chromatin immunoprecipitation revealed p57KIP2 to be associated with E2F1-regulated promoters of genes crucial for DNA synthesis, where the protein inhibits RNA polymerase CTD phosphorylation and activity.170
Due to its growth-regulatory functions, CDKN1C has been studied extensively in various human cancers and is generally considered a tumour suppressor.171 Somewhat surprisingly, data on its role in prostate cancer are relatively limited. In cultured primary human normal prostate epithelial cells, p57KIP2 has been implicated in the acquisition of a senescent phenotype. Loss of the protein may be required for immortalisation of prostate cells.172 Accordingly, CDKN1C was shown to be a target of cancer-specific CpG hypermethylation in 56% of primary prostate cancer tissue samples. Promoter hypermethylation was also found in prostate cancer cell lines, where treatment with the DNA methylation inhibitor 5-aza-deoxycytidine and the histone deacetylase inhibitor trichostatin A significantly upregulated CDKN1C mRNA levels.173 In contrast to other cancers, such as urothelial carcinoma,174 allelic losses of CDKN1C at 11p15.5 are rare in prostate cancer. Very likely, p57KIP2 provides an important barrier to tumourigenesis in the prostate.
Aberrant methylation and histone modifications seem to dominate as mechanisms of CDKN1C inactivation in many malignancies. In breast cancer, EZH2 directly suppresses CDKN1C through H3K27 trimethylation.175 Likewise, restoration of the chromatin remodelling factor SMARCB1, which is frequently inactivated in cancer, led to CDKN1C upregulation via promoter histone H3 and H4 acetylation. Factors like SMARCB1 are crucial for pRB-mediated repression of E2F factors and their target genes.176 Another transcription factor regulating CDKN1C gene expression is ZAC1 (see below). Together, such findings suggest that multiple regulatory mechanisms converge at the CDKN1C gene to ensure adequate levels of p57KIP2 and that these may be disrupted by multiple mechanisms in different cancers.
In paediatric cancers associated with BWS, CDKN1C suppression involves loss of methylation at the differentially methylated ICR of the 11p15.5 imprinted gene cluster.65 This region contains the promoter of the ncRNA KCNQ1OT1 (formerly named LIT-1) which is methylated on the maternal allele. The ncRNA expressed from the paternal allele antagonizes the expression of CDKN1C on the same allele (i.e., in cis) by recruiting Polycomb complexes and histone modifying enzymes to the CDKN1C promoter and silencing its transcription. Upon LOI with loss of maternal methylation of the ICR, KCNQ1OT1 RNA is biallelically expressed, leading to silencing of CDKN1C on both chromosomes. In the paediatric tumours, a concomitant overactivation of the more distant IGF2 locus is also often observed. At least partial hypomethylation of the KCNQ1OT1 DMR has also been observed in cancers of adults including hepatocellular carcinoma and bladder cancer.174, 177 It is not known yet whether it also occurs in prostate cancer.
MEG3/Gtl2, DLK1 and the 14q32.2 imprinted gene cluster
The imprinted gene cluster on human chromosome 14q32.2 comprises protein coding genes like DLK1, RTL1 and DIO3, non-coding RNA genes (MEG3/GTL2, RIAN and anti-RTL1) as well as genes for micro- and small nucleolar RNAs.178 These genes have important roles in embryogenesis and post-natal behaviour. In particular, they have been implicated in regulation of stem-cell properties of embryonic stem cells as well as cancer stem cells,179, 180 especially the reciprocally imprinted gene pair consisting of the paternally expressed DLK1 gene and the maternally expressed non-coding RNA gene MEG3.181, 182
The epigenetic regulation of the 14q32.2 cluster is similar to that of the IGF2/H19 cluster on chromosome 11p15.5, where the promoter of an ncRNA (H19) contains a DMR whose methylation status influences the activity of a protein-coding gene (IGF2).183 In the DLK1/MEG3 cluster, the expression of MEG3 is dependent on the activity of the DMR in its promoter. In mice, the epigenetic patterning and transcriptional activity of the Meg3/Gtl2 promoter DMR together with a neighbouring intergenic DMR has been demonstrated to regulate the expression of paternally (Dlk1, Rtl1 and Dio3) and maternally expressed genes (Mirg, Rian, anti-Rtl1, miR-127 and miR-410) alike.184 Targeted deletion of the unmethylated maternally inherited Meg3/Gtl2 DMR or the intergenic DMR resulted in loss of imprinting across the entire cluster.184, 185 Accordingly, hypermethylation of the MEG3 DMR resulted in abnormal inactivation of DLK1 in human renal cell carcinoma.186 The MEG3 ncRNA regulates expression of the cluster in cis probably in a similar way as the lnc-RNA XIST mediates X-chromosomal silencing, i.e., through recruitment of heterochromatin to RNA-coated regions. Furthermore, the MEG3 gene hosts miRNAs that might also participate in silencing of transcripts from the imprinted locus.
Apart from its regulatory functions in the 14q32.2 gene cluster, MEG3 RNA may control cell proliferation by suppressing MDM2 and enhancing p16INK4A expression, thereby promoting activity of p53 and RB1 respectively.187 The MEG3-encoded miRNAs likewise have an established role in tumourigenesis. Furthermore, MEG3 functions bear on the Notch pathway.
Notch signalling is crucial for the development and homeostasis of many tissues including the neural system, skeletal muscle, pituitary, pancreas, gut and the prostate.188, 189 Typically, the Notch pathway regulates the size of the progenitor cell pool and its proliferative and differentiation capacity. Active Notch signalling results in translocation of the C-terminal part of the receptor protein to the nucleus to induce several target genes like HES1 and HEY1 capable of repressing lineage-specific genes and controlling choices between alternative differentiation pathways.
The best studied function of the paternally expressed DLK1 protein also concerns Notch signalling. As originally discovered in Drosophila, Dlk1 is a non-canonical Notch ligand capable of heterodimerizing with the Notch1 receptor proteins without eliciting their activation. Thereby, it limits Notch1 activation by canonical Jagged or delta ligands in a competitive manner.190
In the mature human prostate, DLK1 was reported to be expressed in some cells of the basal epithelial layer and in rarer neuroendocrine cells. It has been speculated that these DLK1-positive basal cells may represent a common prostate epithelial cell which has the potential to give rise to neuroendocrine, basal and luminal epithelial cells.191 NOTCH1 and its ligand JAGGED1 are predominantly expressed in the luminal cell layer, but since no NOTCH1 nuclear immunoreactivity can be observed, it is unclear whether Notch signalling is active. In cells at intermediate differentiation stages, DLK1 is lower and NOTCH1 expression rises.191 This suggests that the balance between NOTCH1 and DLK1 in prostate cells may be associated with epithelial differentiation.
The Notch target gene HEY1 is especially interesting in the context of prostate cancer, as it acts as an AR corepressor. Ceder et al.191 therefore proposed a dual role of Notch signalling in the human prostate. On the one hand, it may allow early luminal commitment by inhibition of neuroendocrine differentiation, by downregulating DLK1 and by upregulating HES1. On the other hand, Notch signalling may interfere with AR-regulated transcription through HEY1 to inhibit luminal differentiation.192 The authors suggest that Notch activity becomes downregulated as cells terminally differentiate.
Notch signalling contributes to the development of cancers in several tissues,193 but its role in prostate cancer is unclear. According to one study, prostate tumour foci exhibited fewer cleaved NOTCH1 as compared with surrounding benign tissue and lower HEY1.194 In other studies, NOTCH1 and JAGGED1 were reported to be overexpressed in metastatic cancers.195, 196, 197 It has therefore been suggested that not expression of NOTCH1 protein as such but its activation status may be essential for the onset and maintenance of prostate cancer.
The proposed downregulation of HEY1 in prostate cancers194 may relieve repression of the androgen receptor allowing increased cancer cell growth in the presence of androgen as well as under castration conditions.198 In contrast, the other Notch-induced transcriptional repressor HES1 might favour cancer development through binding and repression of the PTEN promoter.199 In this fashion Notch activity could enhance PI3K/Akt signalling. Therefore, the effects of active Notch signalling in prostate cancer, if it occurs, might be strongly context-dependent. In any case, the expression level and distribution of DLK1, which is expected to depend on MEG3, would be crucial.
NDN
Necdin is the product of the paternally expressed NDN gene from the melanoma antigen family that is lost in Prader–Willi syndrome, a hereditary imprinting disease characterized by mental retardation, hypogonadism and obesity.200, 201 Numerous studies suggest major functions of necdin in regulating cell cycle and differentiation. It contributes to myogenic and neuronal differentiation and survival, while inhibiting adipogenic programs. In order to promote myogenic differentiation, necdin acts as a transcriptional repressor silencing adipogenic differentiation genes like PPARG and DLK1.202 Similar to the pRB1 protein, necdin interacts with cell cycle factors like E2F1 and E2F4, resulting in suppression of proliferation and terminal differentiation.203, 204 A particular intriguing facet of necdin repressor activity is its interaction with hnRNP-U in a specific nuclear compartment, where it might be involved in maintaining a higher order chromatin structure, suppressing the transcriptional activity of colocalized cell cycle regulators.205 The pro-survival function of necdin is exerted at least in part through attenuation of p53. The protein interacts with SIRT1 and p53 to potentiate p53 deacetylation by SIRT1.206 In summary, necdin, on the one hand, has a potential to function as a tumour suppressor by inhibiting the cell cycle, but on the other hand, it might promote cell survival during stress by its attenuating effect on p53.207 The function of necdin in the prostate and in human cancers is poorly studied.
PEG3
Paternally expressed gene 3 (PEG3), also known as zinc finger imprinted domain 2 (ZIM2), is situated on chromosome 19q13.4 and is paternally expressed. A CpG island within the gene is methylated on the maternal chromosome and participates in its silencing in cis. This DMR contains tandem binding sites for the Polycomb-interacting protein YY1, which binds to the unmethylated paternal allele. This constellation suggests an important role of DNA methylation and Polycomb-dependent histone modifications in the regulation of PEG3 imprinting and expression.208 In glioma, ovarian and other gynaecological cancers, PEG3 was found to be silenced by DNA hypermethylation of the DMR, which could be relieved by demethylating drugs, especially in combination with a histone deacetylase inhibitor.209, 210, 211, 212, 213 Interestingly, the methylation of the intragenic PEG3 DMR correlated also with the expression of the neighbouring ITUP1 gene oriented head-to-head to PEG3.214
The PEG3 gene product is a Krüppel-type zinc finger protein. High levels of PEG3 are detected during embryo development and in the placenta. In the adult, it remains mainly expressed in the ovary, testis, muscle and brain, characterizing in particular adult stem and progenitor cell populations.215 Like other imprinted genes, PEG3 affects metabolic set-up. It regulates hypothalamic circuits that program energy homeostasis, and thereby growth and development of the entire body, evident from the phenotype of knockout mice.216 Various studies in mice and men show its role in neuronal development, parental and sexual behaviour, as well as in muscle development and homeostasis.
Although growth promoting in the embryo, in the adult PEG3 is suspected to have a tumour suppressor function that may be exerted by interactions with several pathways regulating cell proliferation and apoptosis. In cortical neurons Peg3 was upregulated after DNA damage in a p53-dependent manner, mediating between p53 and Bax in the induction of cell death.217 Overexpression of PEG3 and SIAH1 induced apoptosis independently of p53.218 PEG3 also promoted TNF/TNFR-dependent apoptosis by interaction with TRAF2.219
PEG3 moreover limits the activity of the canonical WNT pathway, in particular by decreasing β-catenin levels. Inhibition of PEG3 expression led to enhanced β-catenin expression, inhibited p53-dependent apoptosis and promoted proliferation in human glioma stem cells.220 This function of PEG3 might be particularly pertinent to prostate cancer, as deregulation of the WNT/β-catenin pathway is thought to contribute to prostate cancer progression, prominently by influencing AR signalling.221, 222, 223, 224, 225, 226
Through its involvement in major important pro-apoptotic pathways and its participation in the control and fine tuning of WNT signalling which impacts on androgen responses, PEG3 may serve to maintain the homeostasis in the normal prostate. Therefore, the strong tendency towards downregulated PEG3 expression in prostate cancer indicated by our in silico analysis might indicate a major function in suppressing cancer development. However, so far no direct evidence for its functional significance in prostate cancer has been reported. Moreover, compared to the tissues with the highest levels in the ovary, placenta, adrenal and pituitary glands PEG3 levels in human prostate tissues are relatively low. An early study reported downregulation of PEG3 mRNA by androgen treatment in LNCaP cells and expression could be detected neither in the androgen-responsive MDA-PCA-2 prostate cancer cell lines nor the poorly differentiated androgen-independent cell lines PC-3 and DU145. While the authors concluded that both androgens and loss of differentiation may affect PEG3 expression in the prostate,227 this remains to be ascertained in tissue samples and any consequences need to be elucidated further.
GNAS
The extremely complex GNAS locus is situated at chromosome 20q13.11 and codes for a variety of alternative transcripts with parental-of-origin-dependent expression. Best characterized are the protein-coding GNAS (Gsα) and its variants GNASXL (XL-Gsα) and NESP55, and the non-coding NESP55as, and Exon 1A. Although most of the transcripts share exons 2-13 of the Gsα transcript, their function, especially of the non-coding transcripts, may be to antagonize Gsα (XLsα) and to ensure its tissue-specific imprinted expression.228, 229, 230 Three DMRs control the monoallelic expression of the transcripts in the GNAS locus. Differential histone modifications, too, are involved in regulating its imprinting.231 In addition, remote cis-acting elements have been suggested to impact on imprinted expression. For a detailed review of GNAS locus regulation refer to ref.232.
Gsα is a guanine nucleotide binding heterotrimeric G-protein stimulatory subunit that activates adenylate cyclase in response to ligand binding to G-protein coupled receptors. It functions in signal transduction from hormone and growth factor receptors to induce cyclic AMP, protein kinase A activity and activation of cAMP-responsive genes through the transcription factor CREBBP. This signalling directs metabolic processes like glycogenolysis and lipolysis, but also secretion or cell survival. In particular, Gsα/cAMP signalling is involved in the stimulation of many endocrine glands by trophic hormones.230, 233, 234 Very likely, the complex regulation of the GNAS gene reflects the requirement to achieve a precise setting of energy metabolism.
This conclusion is underlined by the highly tissue-specific imprinting pattern of GNAS. Its expression is biallelic in most tissues, but silenced on the paternal allele in specific hormone-secretory or hormone-responsive tissues like the kidney, anterior pituitary, thyroid and ovary. In these tissues, cAMP acts as a growth factor for endocrine cells, and Gsα overactivation is often oncogenic.235 Accordingly, activating mutations of GNAS, which alter key residues required for its GTPase activity, are found in kidney, thyroid, pituitary, adenocortical and colorectal tumours, bone fibrous dysplasia and in patients with McCune-Albright syndrome.236 In these diseases, the activating mutations cause overactivity of Gsα, XLsα and adenylate cyclase, resulting in autonomous synthesis of cAMP and constitutive activation of PKA and its downstream pathways.235 GNAS gene amplification and overexpression have been found and may be relevant in ovarian and colorectal cancers.237, 238
Relaxed imprinting of GNAS locus and biallelic Gsα expression may constitute further mechanisms for oncogenic activation in secretory organs like the pituitary, thyroid and ovary, as observed, e.g., in pituitary tumours.239 Mutations in exon 1 inactivate Gsα but do not affect XLsα and NESP55 and may cause haploinsufficiency in tissues that express Gsα biallelically, and complete insufficiency in tissues with monoallelic expression. Maternally inherited mutations of this kind cause resistance to PTH, TSH and gonadotropins in their target organs (kidney, thyroid and ovaries) in addition to the bone growth defects of Albright hereditary osteodystrophy patients. Paternal transmission of the mutations, in contrast, does not cause hormonal resistance.233
Polymorphisms constitute a further factor to be considered in GNAS physiological and oncogenic functions. Homozygosity for the GNAS T393C polymorphism, which correlated with increased Gsα levels, was linked to a more favourable clinical course in bladder and colorectal cancer, as compared to patients with TC or CC genotypes.240, 241 Similarly, in clear cell renal cell carcinoma and gastric cancer,242 homozygous CC patients were found to be at highest risk for progression.242, 243 In prostate cancer, no effect of this polymorphism could be found.244
In view of the involvement of GNAS in endocrine regulation and many human cancers, it is surprising that the gene has hardly been studied yet in prostate cancer. The results of our bioinformatic analysis, with 11 studies reporting significant upregulation, clearly call for a study of GNAS expression, mutation and function in prostate cancer, even though this would have to detailed, given the complex regulation of the locus.
DCN
The product of the DCN gene, decorin (also known as PG-40), is a proteoglycan secreted by mesenchymal cells into the extracellular matrix, where it regulates growth factor signalling and receptor tyrosine kinase activity. In this fashion, it influences signalling by IGFs, EGF-like growth factors and TGF-β.
Altered expression of decorin has been linked to ageing and carcinogenesis in various organs. DCN was reported to be hypomethyated and overexpressed in the stroma of human colon carcinoma.245 In the Tiensin Albino 2 mouse, which develops spontaneous breast cancer, Dcn along with Igf2, Mest, Ndn and Peg3 is differentially expressed between normal mammary gland and cancer tissues. The decrease in decorin is supposed to contribute to carcinogenesis in the mammary gland via increased EGFR signalling.246 In human breast tissues, decorin accumulation was observed in areas of increased mammographic density and malignant-appearing microcalcifications, and was associated with in situ carcinomas.247 Decorin together with other small leucine-rich proteoglycans was proposed to be involved in the regulation of inflammatory and fibrotic renal disorders.248 Taken together, these studies suggest a major function of decorin in the remodelling and regeneration of inflamed and ageing tissues that might be highly pertinent to benign and malignant tumours in the prostate.
Indeed, decorin blocked AR nuclear translocation and decreased prostate-specific antigen levels in the androgen-dependent LNCaP cell line. Importantly, decorin inhibited the growth of both androgen-dependent and androgen–independent prostate tumour cells by suppressing signalling by EGFR, PI3K-Akt and the androgen receptor.249 In mouse prostates, decorin inhibited PI3K and Akt phosphorylation even after PTEN knockout. Downregulation of decorin has been reported during the transition from high-grade prostatic intraepithelial neoplasia to invasive carcinoma.250 Together, these properties suggest an ability of decorin to function as a tumour suppressor in the human prostate by influencing the crucial interacting PI3K/Akt and androgen response pathways.
Conclusions
In this final section, we will summarize the implications of our data mining and literature survey and draw some tentative conclusions as working hypotheses for further investigations.
The first conclusion from our analysis is evident. Despite the extensive epigenetic changes associated with the initiation and progression of prostate cancer, most imprinted genes remain stably regulated. In particular, there is surprisingly little evidence for LOI, apart from the IGF2/H19 locus. Notably, the changes altering the imprinted state at this locus are most likely early events in prostate carcinogenesis that precede deregulation of global DNA methylation patterns and histone modifications. In particular, IGF2 deregulation appears to be involved in the field effect evident in cancerous prostates that manifests in changes in gene expression and DNA methylation.251 There is so far little evidence that changes in the actual imprinting mechanism are responsible for altered expression of other imprinted genes in prostate cancer. At the 7q21 cluster and to a lesser extent at the 6q PLAGL1/HYMAI locus, chromosomal gains and losses may contribute to deregulation. At TFPI2 and CDKN1C, promoter hypermethylation has been observed,151, 173 but the state of the corresponding ICR is rather unchanged (in the former case) or has not been investigated yet (in the latter case).
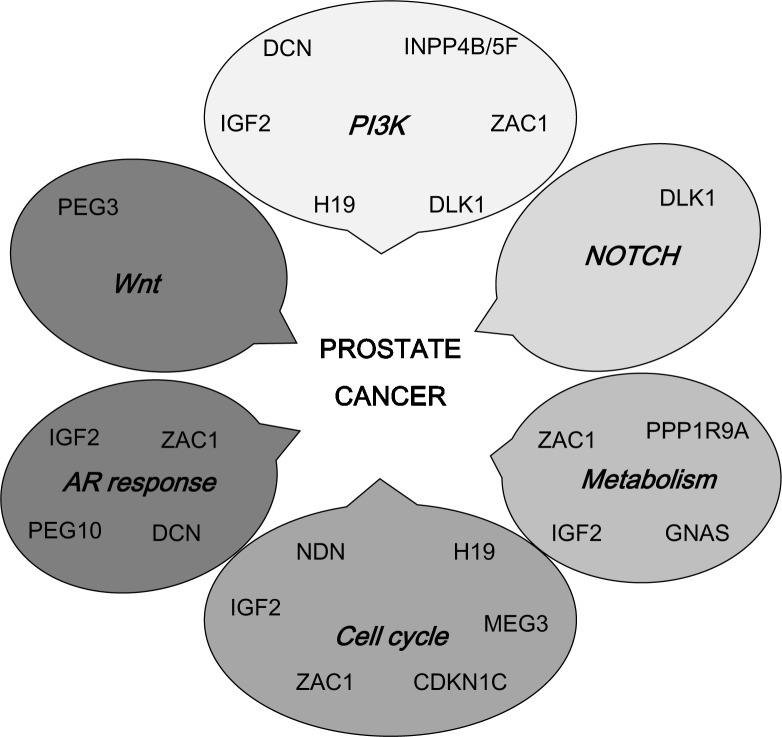
The second conclusion is that the changes in the expression of imprinted genes in prostate are strongly selected for by their function. A large fraction of those that are deregulated have functions related to PI3K/Akt signalling and androgen response, which interact in the development and progression of prostate cancer to CRPC. Additionally, there is preliminary evidence that other pathways, including WNT and Notch signalling, could be influenced by altered expression of individual imprinted genes (Figure 3). This relationship is not incidental, since the regulation of foetal growth and the establishment of set points for endocrine regulation are key functions of imprinted genes during development. In that context, they influence some of the same pathways that are relevant for prostate carcinogenesis and development. Regulation of the involved imprinted genes may be as critical in prostate carcinogenesis.
Figure 3.
An overview of signalling pathways and cellular regulatory systems influenced by and acting upon imprinted genes with altered expression in prostate cancer.
Thirdly, the imprinted genes controlling foetal growth in this fashion are linked with each other in a tight network (Figure 1). Thus, they do not only exert common functions, but are also subject to common regulatory mechanisms. In particular, some of the gene products of this network serve as hubs by regulating many other genes and the overall activity of the network. ZAC1 and MEG3 are prime candidates for such a hub function and H19 in some instances may play a similar role. This means that deregulation of hub genes in prostate cancer may spread through the network (Figure 1) and lead to coordinated changes as during embryonic development. Notably, such coordinated changes can also occur during regenerative processes in adult organs.155, 252, 253
In embryonic development and during tissue regeneration, the function of the network resides in controlling the extent of cell growth and proliferation as well as the expansion of progenitor cell compartments. In brief, the network is first activated to aid in the growth phase of tissues and ensures its transition to a stable and final size, whereupon the network genes are downregulated. In analogy, one might speculate that during prostate cancer development, activation of the network may occur at an early stage of carcinogenesis, but would be selected against once it begins to limit the tumour progression. This argument is supported by the observation that CDKN1C, which encodes one of the strongest antiproliferative proteins in the network, is most consistently downregulated in prostate cancer.
Of note, apart from coordinated regulation in the imprinted gene network, overexpression or downregulation of imprinted genes may occur in normal and neoplastic tissues by mechanisms unrelated to imprinting, e.g., by tissue-specific transcription factors or as a consequence of responses to hypoxia, stress or infection. To give a few examples, expression of CDKN1C is highly tissue-specific, although the factors involved are not well characterized.168 Overexpression of IGF2 in some cancers is associated with a switch to a different promoter.254 H19 is upregulated by hypoxic stress dependent on the p53 status.117 PLAGL1 is induced upon TLR3 activation during virus infection.255 Certain chemical mutagens change the expression level of the imprinted genes SNRPN, PEG3, NDN and ZAC1.256
Finally, several imprinted genes subject to altered expression in prostate cancer have been postulated to regulate stem cell and precursor population. Obviously, since expression analyses by microarrays or quantitative PCR measure mRNA levels across tissue samples, neither our in silico bio-informatic analysis nor the published data from the literature can reveal the expression status of such imprinted genes in subpopulations of epithelial cells in the normal prostate or in prostate cancer. Clearly, their expression and function needs to be considered in the context of cellular hierarchies. Unfortunately, the issue of cancer stem cells in normal and cancerous prostate is highly controversial (see Ref. 257 for a balanced account). However, a few points might be considered.
In the adult, imprinted genes are expressed at low levels and are thought to be mostly restricted to progenitor cell populations. Upregulation of imprinted genes with growth-promoting functions during organ regeneration might therefore be accounted for by the expansion of progenitor cells.258 Furthermore, a similar overexpression of imprinted genes is reported during immortalisation of cells and to contribute to their clonal expansion.259 Deregulation of imprinted genes in progenitor cells could therefore affect not only their number and their response to the microenvironment, but also their ability to give rise to correctly differentiated progeny. Such considerations underlie the theory of the epigenetically disrupted progenitor cells from which tumours may arise.258 According to this theory, epigenetic events perturb the regulation of precursor cells, creating a population of ‘neoplasia-ready cells' from which cancer originates. These cells would be present as well in morphologically normal tissue and explain the broader distribution of epigenetic changes in many neoplastic organs.258 In prostate carcinogenesis, changes at the IGF2/H19 locus may reflect this situation. Interestingly, it has been postulated that microenvironmental influences like cytokines and growth factors secreted during infection or altered hormone homeostasis provide the milieu that causes LOI in tissue-resident stem cells.260 Accordingly, increased levels of IGF2 have been postulated to cause cancer by increasing the progenitor cell population in tumours.261
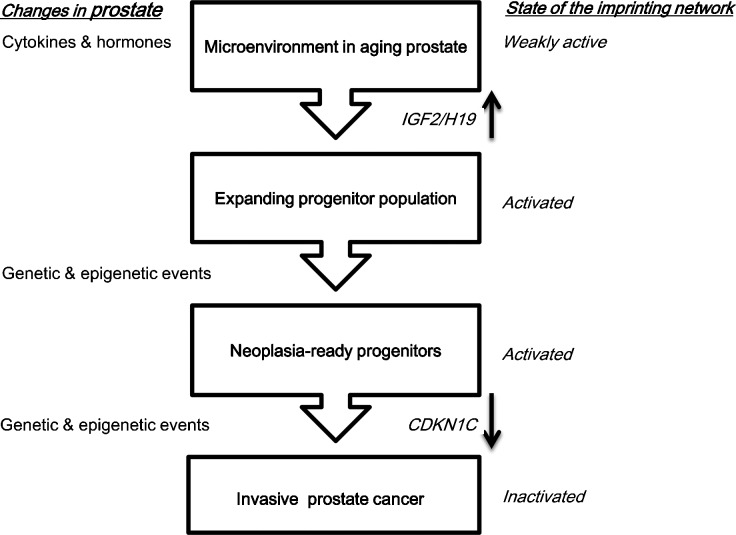
In summary, thus, we may propose the following model to account for the specificity of changes in imprinted gene expression in prostate cancer (Figure 4). Changes in IGF2 imprinting might be caused by micro-environmental changes in the ageing prostate and consequently contribute to the emergence of a tumourigenic precursor population. Genetic changes such as ERG translocations and certain chromosomal losses and gains may contribute to the establishment of that population8 or may follow. Certainly, they are required for the emergence of invasive cancers. These genetic changes along with epigenetic alterations elicit changes in the growth factor/PI3K/androgen response axis that are crucial for tumour growth and progression. In response to the expansion of the precursor cell pool or the activation of growth-stimulating signals, the imprinted gene network controlling tissue growth becomes activated and becomes subject to selection against its effects that limit proliferation and drive differentiation. This selection appears to result in the specific inactivation of individual genes that act as effectors or hubs within the network, eliciting broader changes in expression.
Figure 4.
A hypothesis on the role of imprinted genes in prostate cancer initiation and progression. IGF2, insulin-like growth factor 2.
Acknowledgments
This project was supported by the Deutsche Forschungsgemeinschaft and the Deutsche Krebshilfe.
The authors declare no competing commercial interests in relation to the submitted work.
Footnotes
There is another usage of ‘imprinting' that should not be confused with the phenomenon of ‘genomic imprinting' discussed in this article. Accumulating evidence suggests that environmental effects on endocrine regulation in utero can have lasting influences throughout life, including the development of diseases like diabetes and prostate cancer. These effects are likely to be mediated by epigenetic mechanisms too. The phenomenon should better be designated as “developmental reprogramming” to avoid confusion.
References
- Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assinder SJ, Dong Q, Kovacevic Z, Richardson DR. The TGF-beta, PI3K/Akt and PTEN pathways: established and proposed biochemical integration in prostate cancer. Biochem J. 2009;417:411–21. doi: 10.1042/BJ20081610. [DOI] [PubMed] [Google Scholar]
- Lin HK, Hu YC, Lee DK, Chang C. Regulation of androgen receptor signaling by PTEN (phosphatase and tensin homolog deleted on chromosome 10) tumor suppressor through distinct mechanisms in prostate cancer cells. Mol Endocrinol. 2004;18:2409–23. doi: 10.1210/me.2004-0117. [DOI] [PubMed] [Google Scholar]
- Kaarbo M, Mikkelsen OL, Malerod L, Qu S, Lobert VH, et al. PI3K–AKT–mTOR pathway is dominant over androgen receptor signaling in prostate cancer cells. Cell Oncol. 2010;32:11–27. doi: 10.3233/CLO-2009-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2–ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, de Marzo AM, Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology. 2009;150:3991–4002. doi: 10.1210/en.2009-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz WA, Hoffmann MJ. Epigenetic mechanisms in the biology of prostate cancer. Semin Cancer Biol. 2009;19:172–80. doi: 10.1016/j.semcancer.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Hoque MO. DNA methylation changes in prostate cancer: current developments and future clinical implementation. Expert Rev Mol Diagn. 2009;9:243–57. doi: 10.1586/erm.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Florl AR, Steinhoff C, Muller M, Seifert HH, Hader C, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer. 2004;91:985–94. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–86. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- Park JY. Promoter hypermethylation in prostate cancer. Cancer Control. 2010;17:245–55. doi: 10.1177/107327481001700405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci USA. 1994;91:11733–7. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G, Cottrell S, Distler J, Schatz P, Kristiansen G, et al. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. J Urol. 2009;181:1678–85. doi: 10.1016/j.juro.2008.11.120. [DOI] [PubMed] [Google Scholar]
- Vanaja DK, Grossmann ME, Cheville JC, Gazi MH, Gong A, et al. PDLIM4, an actin binding protein, suppresses prostate cancer cell growth. Cancer Invest. 2009;27:264–72. doi: 10.1080/07357900802406319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I, Gillespie JW, Protopopov A, Geil L, Dreijerink K, et al. The RASSF1A tumor suppressor gene is inactivated in prostate tumors and suppresses growth of prostate carcinoma cells. Cancer Res. 2002;62:3498–502. [PubMed] [Google Scholar]
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–62. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–74. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- de Smet C, Loriot A. DNA hypomethylation in cancer: epigenetic scars of a neoplastic journey. Epigenetics. 2010;5:206–13. doi: 10.4161/epi.5.3.11447. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–8. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- Cooper CS, Foster CS. Concepts of epigenetics in prostate cancer development. Br J Cancer. 2009;100:240–5. doi: 10.1038/sj.bjc.6604771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–63. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, et al. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunderfranco P, Mello-Grand M, Cangemi R, Pellini S, Mensah A, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS ONE. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen S, Martikainen PM, Tolonen T, Isola J, Tammela TL, et al. EZH2, Ki-67 and MCM7 are prognostic markers in prostatectomy treated patients. Int J Cancer. 2008;122:595–602. doi: 10.1002/ijc.23145. [DOI] [PubMed] [Google Scholar]
- Yu J, Rhodes DR, Tomlins SA, Cao X, Chen G, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–63. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- Hoffmann MJ, Engers R, Florl AR, Otte AP, Muller M, et al. Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer. Cancer Biol Ther. 2007;6:1403–12. doi: 10.4161/cbt.6.9.4542. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–11. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006;66:11341–7. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–53. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Schule R. Histone demethylation and androgen-dependent transcription. Curr Opin Genet Dev. 2006;16:513–7. doi: 10.1016/j.gde.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226–31. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabjeesh NJ, Willard MT, Frederickson CE, Zhong H, Simons JW. Androgens stimulate hypoxia-inducible factor 1 activation via autocrine loop of tyrosine kinase receptor/phosphatidylinositol 3′-kinase/protein kinase B in prostate cancer cells. Clin Cancer Res. 2003;9:2416–25. [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–65. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, et al. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum Mol Genet. 2009;18:3375–83. doi: 10.1093/hmg/ddp277. [DOI] [PubMed] [Google Scholar]
- Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell. 2007;27:353–66. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen KR, Ferguson-Smith AC. Distinguishing epigenetic marks of developmental and imprinting regulation. Epigenetics Chromatin. 2010;3:2. doi: 10.1186/1756-8935-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Singh P, Wu X, Lee DH, Li AX, Rauch TA, et al. Chromosome-wide analysis of parental allele-specific chromatin and DNA methylation. Mol Cell Biol. 2011;31:1757–70. doi: 10.1128/MCB.00961-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, et al. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 2007;26:720–9. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacem S, Feil R. Chromatin mechanisms in genomic imprinting. Mamm Genome. 2009;20:544–56. doi: 10.1007/s00335-009-9223-4. [DOI] [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP. Silencing by imprinted noncoding RNAs: is transcription the answer. Trends Genet. 2007;23:284–92. doi: 10.1016/j.tig.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Robson JE. Imprinted noncoding RNAs. Mamm Genome. 2008;19:493–502. doi: 10.1007/s00335-008-9139-4. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–37. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HA, Bernstein E. Partners in imprinting: noncoding RNA and polycomb group proteins. Dev Cell. 2008;15:637–8. doi: 10.1016/j.devcel.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Reik W, Murrell A, Lewis A, Mitsuya K, Umlauf D, et al. Chromosome loops, insulators, and histone methylation: new insights into regulation of imprinting in clusters. Cold Spring Harb Symp Quant Biol. 2004;69:29–37. doi: 10.1101/sqb.2004.69.29. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20:127–33. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–33. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiolini E, Fowden A, Coan P, Sandovici I, Smith P, et al. Regulation of placental efficiency for nutrient transport by imprinted genes. Placenta. 2006;27 Suppl A:S98–102. doi: 10.1016/j.placenta.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–9. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes. 2007;14:3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- Mann JR, Gadi I, Harbison ML, Abbondanzo SJ, Stewart CL. Androgenetic mouse embryonic stem cells are pluripotent and cause skeletal defects in chimeras: implications for genetic imprinting. Cell. 1990;62:251–60. doi: 10.1016/0092-8674(90)90363-j. [DOI] [PubMed] [Google Scholar]
- Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–43. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–21. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–22. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Berg JS, Lin KK, Sonnet C, Boles NC, Weksberg DC, et al. Imprinted genes that regulate early Mammalian growth are coexpressed in somatic stem cells. PLoS ONE. 2011;6:e26410. doi: 10.1371/journal.pone.0026410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann T. Epigenetic regulation of growth: lessons from Silver–Russell syndrome. Endocr Dev. 2009;14:10–9. doi: 10.1159/000207472. [DOI] [PubMed] [Google Scholar]
- Delaval K, Wagschal A, Feil R. Epigenetic deregulation of imprinting in congenital diseases of aberrant growth. Bioessays. 2006;28:453–9. doi: 10.1002/bies.20407. [DOI] [PubMed] [Google Scholar]
- Diaz-Meyer N, Yang Y, Sait SN, Maher ER, Higgins MJ. Alternative mechanisms associated with silencing of CDKN1C in Beckwith–Wiedemann syndrome. J Med Genet. 2005;42:648–55. doi: 10.1136/jmg.2004.030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliek J, Verde G, Callaway J, Maas SM, de Crescenzo A, et al. Hypomethylation at multiple maternally methylated imprinted regions including PLAGL1 and GNAS loci in Beckwith–Wiedemann syndrome. Eur J Hum Genet. 2009;17:611–9. doi: 10.1038/ejhg.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubertus J, Lacher M, Rottenkolber M, Muller-Hocker J, Berger M, et al. Altered expression of imprinted genes in Wilms tumors. Oncol Rep. 2011;25:817–23. doi: 10.3892/or.2010.1113. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Fujisawa D, Hashimoto K, Asano T, Maimaiti M, et al. Partial paternal uniparental disomy of chromosome 6 in monozygotic twins with transient neonatal diabetes mellitus and macroglossia. Clin Genet. 2010;78:580–4. doi: 10.1111/j.1399-0004.2010.01433.x. [DOI] [PubMed] [Google Scholar]
- Temple IK, Shield JP. Transient neonatal diabetes, a disorder of imprinting. J Med Genet. 2002;39:872–5. doi: 10.1136/jmg.39.12.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, et al. Specific epigenetic alterations of IGF2–H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–33. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–51. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- Filipponi D, Feil R. Perturbation of genomic imprinting in oligozoospermia. Epigenetics. 2009;4:27–30. doi: 10.4161/epi.4.1.7311. [DOI] [PubMed] [Google Scholar]
- Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res. 2011;727:62–71. doi: 10.1016/j.mrrev.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, et al. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod. 2011;26:2558–69. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Fu VX, Dobosy JR, Desotelle JA, Almassi N, Ewald JA, et al. Aging and cancer-related loss of insulin-like growth factor 2 imprinting in the mouse and human prostate. Cancer Res. 2008;68:6797–802. doi: 10.1158/0008-5472.CAN-08-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol. 2007;211:261–8. doi: 10.1002/path.2116. [DOI] [PubMed] [Google Scholar]
- Bhusari S, Yang B, Kueck J, Huang W, Jarrard DF. Insulin-like growth factor-2 (IGF2) loss of imprinting marks a field defect within human prostates containing cancer. Prostate. 2011;71:1621–30. doi: 10.1002/pros.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Correa M, Cui H, Giardiello FM, Powe NR, Hylind L, et al. Loss of imprinting of insulin growth factor II gene: a potential heritable biomarker for colon neoplasia predisposition. Gastroenterology. 2004;126:964–70. doi: 10.1053/j.gastro.2003.12.051. [DOI] [PubMed] [Google Scholar]
- Chao W, D'Amore PA. IGF2: epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev. 2008;19:111–20. doi: 10.1016/j.cytogfr.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Shi C, Sjolinder M, Zhao Z, Gondor A, et al. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 2009;23:2598–603. doi: 10.1101/gad.552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–93. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Vu TH, Nguyen AH, Hoffman AR. Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells. Hum Mol Genet. 2010;19:901–19. doi: 10.1093/hmg/ddp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, et al. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24:3497–504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada M, Inoue H, Masuda T, Ikeda D. Insulin-like growth factor I secreted from prostate stromal cells mediates tumor-stromal cell interactions of prostate cancer. Cancer Res. 2006;66:4419–25. doi: 10.1158/0008-5472.CAN-05-4239. [DOI] [PubMed] [Google Scholar]
- Cardillo MR, Monti S, Di Silverio F, Gentile V, Sciarra F, et al. Insulin-like growth factor (IGF)-I, IGF-II and IGF type I receptor (IGFR-I) expression in prostatic cancer. Anticancer Res. 2003;23:3825–35. [PubMed] [Google Scholar]
- Tennant MK, Thrasher JB, Twomey PA, Drivdahl RH, Birnbaum RS, et al. Protein and messenger ribonucleic acid (mRNA) for the type 1 insulin-like growth factor (IGF) receptor is decreased and IGF-II mRNA is increased in human prostate carcinoma compared to benign prostate epithelium. J Clin Endocrinol Metab. 1996;81:3774–82. doi: 10.1210/jcem.81.10.8855837. [DOI] [PubMed] [Google Scholar]
- Turney BW, Turner GD, Brewster SF, Macaulay VM. Serial analysis of resected prostate cancer suggests up-regulation of type 1 IGF receptor with disease progression. BJU Int. 2011;107:1488–99. doi: 10.1111/j.1464-410X.2010.09556.x. [DOI] [PubMed] [Google Scholar]
- Le H, Arnold JT, McFann KK, Blackman MR. DHT and testosterone, but not DHEA or E2, differentially modulate IGF-I, IGFBP-2, and IGFBP-3 in human prostatic stromal cells. Am J Physiol Endocrinol Metab. 2006;290:E952–60. doi: 10.1152/ajpendo.00451.2005. [DOI] [PubMed] [Google Scholar]
- Liu X, Allen JD, Arnold JT, Blackman MR. Lycopene inhibits IGF-I signal transduction and growth in normal prostate epithelial cells by decreasing DHT-modulated IGF-I production in co-cultured reactive stromal cells. Carcinogenesis. 2008;29:816–23. doi: 10.1093/carcin/bgn011. [DOI] [PubMed] [Google Scholar]
- Liu X, Choi RY, Jawad SM, Arnold JT. Androgen-induced PSA expression requires not only activation of AR but also endogenous IGF-I or IGF-I/PI3K/Akt signaling in human prostate cancer epithelial cells. Prostate. 2010;71:766–77. doi: 10.1002/pros.21293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sheikh SS, Romanska HM, Abel P, Domin J, et al. Predictive value of PTEN and AR coexpression of sustained responsiveness to hormonal therapy in prostate cancer—a pilot study. Neoplasia. 2008;10:949–53. doi: 10.1593/neo.08582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoulnik IU, Hodgson MC, Bowden WA, Ittmann MM. INPP4B: the new kid on the PI3K block. Oncotarget. 2011;2:321–8. doi: 10.18632/oncotarget.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson MC, Shao LJ, Frolov A, Li R, Peterson LE, et al. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res. 2011;71:572–82. doi: 10.1158/0008-5472.CAN-10-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker D, Reid AH, Yap TA, de Bono JS. Targeting the PI3K/AKT pathway for the treatment of prostate cancer. Clin Cancer Res. 2009;15:4799–805. doi: 10.1158/1078-0432.CCR-08-0125. [DOI] [PubMed] [Google Scholar]
- Plymate SR, Tennant MK, Culp SH, Woodke L, Marcelli M, et al. Androgen receptor (AR expression in AR-negative prostate cancer cells results in differential effects of DHT and IGF-I on proliferation and AR activity between localized and metastatic tumors. Prostate. 2004;61:276–90. doi: 10.1002/pros.20099. [DOI] [PubMed] [Google Scholar]
- Wu JD, Haugk K, Woodke L, Nelson P, Coleman I, et al. Interaction of IGF signaling and the androgen receptor in prostate cancer progression. J Cell Biochem. 2006;99:392–401. doi: 10.1002/jcb.20929. [DOI] [PubMed] [Google Scholar]
- Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, et al. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 2001;61:4315–9. [PubMed] [Google Scholar]
- Kupfer SR, Wilson EM, French FS. Androgen and glucocorticoid receptors interact with insulin degrading enzyme. J Biol Chem. 1994;269:20622–8. [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Yanase T, Morinaga H, Okabe T, Nomura M, et al. Insulin-like growth factor 1/insulin signaling activates androgen signaling through direct interactions of Foxo1 with androgen receptor. J Biol Chem. 2007;282:7329–38. doi: 10.1074/jbc.M610447200. [DOI] [PubMed] [Google Scholar]
- Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- McKenzie S, Kyprianou N. Apoptosis evasion: the role of survival pathways in prostate cancer progression and therapeutic resistance. J Cell Biochem. 2006;97:18–32. doi: 10.1002/jcb.20634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan EE. Plasma and tissue insulin-like growth factor-I receptor (IGF-IR) as a prognostic marker for prostate cancer and anti-IGF-IR agents as novel therapeutic strategy for refractory cases: a review. Mol Cell Endocrinol. 2011;344:1–24. doi: 10.1016/j.mce.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Jarrard DF, Bussemakers MJ, Bova GS, Isaacs WB. Regional loss of imprinting of the insulin-like growth factor II gene occurs in human prostate tissues. Clin Cancer Res. 1995;1:1471–8. [PubMed] [Google Scholar]
- Murrell A, Ito Y, Verde G, Huddleston J, Woodfine K, et al. Distinct methylation changes at the IGF2–H19 locus in congenital growth disorders and cancer. PLoS ONE. 2008;3:e1849. doi: 10.1371/journal.pone.0001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–5. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci USA. 1996;93:11757–62. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradowska A, Fenic I, Konrad L, Sturm K, Wagenlehner F, et al. Aberrant epigenetic modifications in the CTCF binding domain of the IGF2/H19 gene in prostate cancer compared with benign prostate hyperplasia. Int J Oncol. 2009;35:87–96. doi: 10.3892/ijo_00000316. [DOI] [PubMed] [Google Scholar]
- Fu VX, Schwarze SR, Kenowski ML, Leblanc S, Svaren J, et al. A loss of insulin-like growth factor-2 imprinting is modulated by CCCTC-binding factor down-regulation at senescence in human epithelial cells. J Biol Chem. 2004;279:52218–26. doi: 10.1074/jbc.M405015200. [DOI] [PubMed] [Google Scholar]
- Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–51. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliek J, Terhal P, van den Bogaard MJ, Maas S, Hamel B, et al. Hypomethylation of the H19 gene causes not only Silver–Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am J Hum Genet. 2006;78:604–14. doi: 10.1086/502981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliek J, Maas SM, Ruijter JM, Hennekam RC, Alders M, et al. Increased tumour risk for BWS patients correlates with aberrant H19 and not KCNQ1OT1 methylation: occurrence of KCNQ1OT1 hypomethylation in familial cases of BWS. Hum Mol Genet. 2001;10:467–76. doi: 10.1093/hmg/10.5.467. [DOI] [PubMed] [Google Scholar]
- Berteaux N, Lottin S, Adriaenssens E, van Coppenolle F, Leroy X, et al. Hormonal regulation of H19 gene expression in prostate epithelial cells. J Endocrinol. 2004;183:69–78. doi: 10.1677/joe.1.05696. [DOI] [PubMed] [Google Scholar]
- Cui H. Loss of imprinting of IGF2 as an epigenetic marker for the risk of human cancer. Dis Markers. 2007;23:105–12. doi: 10.1155/2007/363464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA. 1999;96:5616–21. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls RD. The impact of genomic imprinting for neurobehavioral and developmental disorders. J Clin Invest. 2000;105:413–8. doi: 10.1172/JCI9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya M, Judson H, Okazaki Y, Kusakabe M, Muramatsu M, et al. The cell cycle control gene ZAC/PLAGL1 is imprinted—a strong candidate gene for transient neonatal diabetes. Hum Mol Genet. 2000;9:453–60. doi: 10.1093/hmg/9.3.453. [DOI] [PubMed] [Google Scholar]
- Valente T, Junyent F, Auladell C. Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: differential phenotype of the Zac1-expressing cells during development. Dev Dyn. 2005;233:667–79. doi: 10.1002/dvdy.20373. [DOI] [PubMed] [Google Scholar]
- Bilanges B, Varrault A, Mazumdar A, Pantaloni C, Hoffmann A, et al. Alternative splicing of the imprinted candidate tumor suppressor gene ZAC regulates its antiproliferative and DNA binding activities. Oncogene. 2001;20:1246–53. doi: 10.1038/sj.onc.1204237. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Ciani E, Boeckardt J, Holsboer F, Journot L, et al. Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol Cell Biol. 2003;23:988–1003. doi: 10.1128/MCB.23.3.988-1003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barz T, Hoffmann A, Panhuysen M, Spengler D. Peroxisome proliferator-activated receptor gamma is a Zac target gene mediating Zac antiproliferation. Cancer Res. 2006;66:11975–82. doi: 10.1158/0008-5472.CAN-06-1529. [DOI] [PubMed] [Google Scholar]
- Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, et al. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 1997;16:2814–25. doi: 10.1093/emboj/16.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Spengler D. A new coactivator function for Zac1's C2H2 zinc finger DNA-binding domain in selectively controlling PCAF activity. Mol Cell Biol. 2008;28:6078–93. doi: 10.1128/MCB.00842-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Stallcup MR. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol Cell Biol. 2000;20:1855–67. doi: 10.1128/mcb.20.5.1855-1867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, Schonthal AH, Stallcup MR. Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1. Oncogene. 2001;20:2134–43. doi: 10.1038/sj.onc.1204298. [DOI] [PubMed] [Google Scholar]
- Rozenfeld-Granot G, Krishnamurthy J, Kannan K, Toren A, Amariglio N, et al. A positive feedback mechanism in the transcriptional activation of Apaf-1 by p53 and the coactivator Zac-1. Oncogene. 2002;21:1469–76. doi: 10.1038/sj.onc.1205218. [DOI] [PubMed] [Google Scholar]
- Liu PY, Chan JY, Lin HC, Wang SL, Liu ST, et al. Modulation of the cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by Zac1 through the antagonistic regulators p53 and histone deacetylase 1 in HeLa Cells. Mol Cancer Res. 2008;6:1204–14. doi: 10.1158/1541-7786.MCR-08-0123. [DOI] [PubMed] [Google Scholar]
- Abdollahi A, Godwin AK, Miller PD, Getts LA, Schultz DC, et al. Identification of a gene containing zinc-finger motifs based on lost expression in malignantly transformed rat ovarian surface epithelial cells. Cancer Res. 1997;57:2029–34. [PubMed] [Google Scholar]
- Abdollahi A, Roberts D, Godwin AK, Schultz DC, Sonoda G, et al. Identification of a zinc-finger gene at 6q25: a chromosomal region implicated in development of many solid tumors. Oncogene. 1997;14:1973–9. doi: 10.1038/sj.onc.1201034. [DOI] [PubMed] [Google Scholar]
- Basyuk E, Coulon V, Le Digarcher A, Coisy-Quivy M, Moles JP, et al. The candidate tumor suppressor gene ZAC is involved in keratinocyte differentiation and its expression is lost in basal cell carcinomas. Mol Cancer Res. 2005;3:483–92. doi: 10.1158/1541-7786.MCR-05-0019. [DOI] [PubMed] [Google Scholar]
- Cvetkovic D, Pisarcik D, Lee C, Hamilton TC, Abdollahi A. Altered expression and loss of heterozygosity of the LOT1 gene in ovarian cancer. Gynecol Oncol. 2004;95:449–55. doi: 10.1016/j.ygyno.2004.08.051. [DOI] [PubMed] [Google Scholar]
- Pagotto U, Arzberger T, Theodoropoulou M, Grubler Y, Pantaloni C, et al. The expression of the antiproliferative gene ZAC is lost or highly reduced in nonfunctioning pituitary adenomas. Cancer Res. 2000;60:6794–9. [PubMed] [Google Scholar]
- Koy S, Hauses M, Appelt H, Friedrich K, Schackert HK, et al. Loss of expression of ZAC/LOT1 in squamous cell carcinomas of head and neck. Head Neck. 2004;26:338–44. doi: 10.1002/hed.10386. [DOI] [PubMed] [Google Scholar]
- Varrault A, Ciani E, Apiou F, Bilanges B, Hoffmann A, et al. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc Natl Acad Sci USA. 1998;95:8835–40. doi: 10.1073/pnas.95.15.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D, Wagschal A, Arnaud P, Muller PS, Parker-Katiraee L, et al. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing. Genome Res. 2008;18:1270–81. doi: 10.1101/gr.077115.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux H, Flammann H, Hafner M, Lux A. Genetic and molecular analyses of PEG10 reveal new aspects of genomic organization, transcription and translation. PLoS ONE. 2010;5:e8686. doi: 10.1371/journal.pone.0008686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner SC, Dhir M, Yi JM, McGarvey KE, van Neste L, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–9. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Lin Y, Zhang H, Liu J, Zhang N, et al. Tissue factor pathway inhibitor-2 was repressed by CpG hypermethylation through inhibition of KLF6 binding in highly invasive breast cancer cells. BMC Mol Biol. 2007;8:110. doi: 10.1186/1471-2199-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konduri SD, Tasiou A, Chandrasekar N, Rao JS. Overexpression of tissue factor pathway inhibitor-2 (TFPI-2), decreases the invasiveness of prostate cancer cells in vitro. . Int J Oncol. 2001;18:127–31. [PubMed] [Google Scholar]
- Rao CN, Segawa T, Navari JR, Xu L, Srivastava S, et al. Methylation of TFPI-2 gene is not the sole cause of its silencing. Int J Oncol. 2003;22:843–8. [PubMed] [Google Scholar]
- Ribarska T, Ingenwerth M, Goering W, Engers R, Schulz WA. Epigenetic inactivation of the placentally imprinted tumor suppressor gene TFPI2 in prostate carcinoma. Cancer Genomics Proteomics. 2010;7:51–60. [PubMed] [Google Scholar]
- Kobayashi M, Ishida H, Shindo T, Niwa S, Kino M, et al. Molecular analysis of multifocal prostate cancer by comparative genomic hybridization. Prostate. 2008;68:1715–24. doi: 10.1002/pros.20832. [DOI] [PubMed] [Google Scholar]
- Kainz B, Shehata M, Bilban M, Kienle D, Heintel D, et al. Overexpression of the paternally expressed gene 10 (PEG10) from the imprinted locus on chromosome 7q21 in high-risk B-cell chronic lymphocytic leukemia. Int J Cancer. 2007;121:1984–93. doi: 10.1002/ijc.22929. [DOI] [PubMed] [Google Scholar]
- Okabe H, Satoh S, Furukawa Y, Kato T, Hasegawa S, et al. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. . Cancer Res. 2003;63:3043–8. [PubMed] [Google Scholar]
- Tsou AP, Chuang YC, Su JY, Yang CW, Liao YL, et al. Overexpression of a novel imprinted gene, PEG10, in human hepatocellular carcinoma and in regenerating mouse livers. J Biomed Sci. 2003;10:625–35. doi: 10.1159/000073528. [DOI] [PubMed] [Google Scholar]
- Dong H, Zhang H, Liang J, Yan H, Chen Y, et al. Digital karyotyping reveals probable target genes at 7q21.3 locus in hepatocellular carcinoma. BMC Med Genomics. 2011;4:60. doi: 10.1186/1755-8794-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji K, Yasui K, Gen Y, Endo M, Dohi O, et al. PEG10 is a probable target for the amplification at 7q21 detected in hepatocellular carcinoma. Cancer Genet Cytogenet. 2010;198:118–25. doi: 10.1016/j.cancergencyto.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Jie X, Lang C, Jian Q, Chaoqun L, Dehua Y, et al. Androgen activates PEG10 to promote carcinogenesis in hepatic cancer cells. Oncogene. 2007;26:5741–51. doi: 10.1038/sj.onc.1210362. [DOI] [PubMed] [Google Scholar]
- Li CM, Margolin AA, Salas M, Memeo L, Mansukhani M, et al. PEG10 is a c-MYC target gene in cancer cells. Cancer Res. 2006;66:665–72. doi: 10.1158/0008-5472.CAN-05-1553. [DOI] [PubMed] [Google Scholar]
- Lux A, Beil C, Majety M, Barron S, Gallione CJ, et al. Human retroviral gag- and gag-pol-like proteins interact with the transforming growth factor-beta receptor activin receptor-like kinase 1. J Biol Chem. 2005;280:8482–93. doi: 10.1074/jbc.M409197200. [DOI] [PubMed] [Google Scholar]
- Bonnemann CG, McNally EM, Kunkel LM. Beyond dystrophin: current progress in the muscular dystrophies. Curr Opin Pediatr. 1996;8:569–82. [PubMed] [Google Scholar]
- Ritz K, van Schaik BD, Jakobs ME, van Kampen AH, Aronica E, et al. SGCE isoform characterization and expression in human brain: implications for myoclonus-dystonia pathogenesis. Eur J Hum Genet. 2011;19:438–44. doi: 10.1038/ejhg.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega P, Moran A, Fernandez-Marcelo T, de Juan C, Frias C, et al. MMP-7 and SGCE as distinctive molecular factors in sporadic colorectal cancers from the mutator phenotype pathway. Int J Oncol. 2010;36:1209–15. doi: 10.3892/ijo_00000604. [DOI] [PubMed] [Google Scholar]
- Oliver CJ, Terry-Lorenzo RT, Elliott E, Bloomer WA, Li S, et al. Targeting protein phosphatase 1 (PP1) to the actin cytoskeleton: the neurabin I/PP1 complex regulates cell morphology. Mol Cell Biol. 2002;22:4690–701. doi: 10.1128/MCB.22.13.4690-4701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhadaway J, Rowe F, Elliott K, Mao NC, Prendergast GC. Bau, a splice form of Neurabin-I that interacts with the tumor suppressor Bin1, inhibits malignant cell transformation. Cell Adhes Commun. 1999;7:99–110. doi: 10.3109/15419069909034394. [DOI] [PubMed] [Google Scholar]
- Choi JD, Underkoffler LA, Wood AJ, Collins JN, Williams PT, et al. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol Cell Biol. 2005;25:5514–22. doi: 10.1128/MCB.25.13.5514-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Trivedi CM, Zhou D, Yuan L, Lu MM, et al. Inpp5f is a polyphosphoinositide phosphatase that regulates cardiac hypertrophic responsiveness. Circ Res. 2009;105:1240–7. doi: 10.1161/CIRCRESAHA.109.208785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateras IS, Apostolopoulou K, Niforou K, Kotsinas A, Gorgoulis VG. p57KIP2: “Kip”ing the cell under control. Mol Cancer Res. 2009;7:1902–19. doi: 10.1158/1541-7786.MCR-09-0317. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Wood MD, Tunster SJ, Barton SC, Surani MA, et al. Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev Biol. 2007;7:53. doi: 10.1186/1471-213X-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Chen L, Wright GM, Pillai SR, Chellappan SP, et al. CDKN1C negatively regulates RNA polymerase II C-terminal domain phosphorylation in an E2F1-dependent manner. J Biol Chem. 2010;285:9813–22. doi: 10.1074/jbc.M109.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh E, Joseph B. The hallmarks of CDKN1C (p57, KIP2) in cancer. Biochim Biophys Acta. 2011;1816:50–6. doi: 10.1016/j.bbcan.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Schwarze SR, Shi Y, Fu VX, Watson PA, Jarrard DF. Role of cyclin-dependent kinase inhibitors in the growth arrest at senescence in human prostate epithelial and uroepithelial cells. Oncogene. 2001;20:8184–92. doi: 10.1038/sj.onc.1205049. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218–27. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- Hoffmann MJ, Florl AR, Seifert HH, Schulz WA. Multiple mechanisms downregulate CDKN1C in human bladder cancer. Int J Cancer. 2005;114:406–13. doi: 10.1002/ijc.20749. [DOI] [PubMed] [Google Scholar]
- Yang X, Karuturi RK, Sun F, Aau M, Yu K, et al. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS ONE. 2009;4:e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algar EM, Muscat A, Dagar V, Rickert C, Chow CW, et al. Imprinted CDKN1C is a tumor suppressor in rhabdoid tumor and activated by restoration of SMARCB1 and histone deacetylase inhibitors. PLoS ONE. 2009;4:e4482. doi: 10.1371/journal.pone.0004482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienbacher C, Gramantieri L, Scelfo R, Veronese A, Calin GA, et al. Gain of imprinting at chromosome 11p15: A pathogenetic mechanism identified in human hepatocarcinomas. Proc Natl Acad Sci USA. 2000;97:5445–9. doi: 10.1073/pnas.090087497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1–Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Lujan E, Wernig M. An imprinted signature helps isolate ESC-equivalent iPSCs. Cell Res. 2010;20:974–6. doi: 10.1038/cr.2010.117. [DOI] [PubMed] [Google Scholar]
- Luk JM, Burchard J, Zhang C, Liu AM, Wong KF, et al. DLK1–DIO3 genomic imprinted microrna cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem. 2011;286:30706–13. doi: 10.1074/jbc.M111.229831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah BM, Jensen CH, Gutierrez G, Leslie RG, Jensen TG, et al. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res. 2004;19:841–52. doi: 10.1359/JBMR.040118. [DOI] [PubMed] [Google Scholar]
- Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, et al. Activation of the imprinted Dlk1–Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. 2010;285:19483–90. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M, O'Sullivan MJ, Green AJ, Watabe Y, Arisaka O, et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Cheunsuchon P, Nakayama Y, Lawlor MW, Zhong Y, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137:2643–52. doi: 10.1242/dev.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving MD, Buiting K, Kanber D, Donaghue C, Schulz R, et al. Segmental paternal uniparental disomy (patUPD) of 14q32 with abnormal methylation elicits the characteristic features of complete patUPD14. Am J Med Genet A. 2010;152A:1942–50. doi: 10.1002/ajmg.a.33449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Chano T, Minami K, Okabe H, Okada Y, et al. Imprinted DLK1 is a putative tumor suppressor gene and inactivated by epimutation at the region upstream of GTL2 in human renal cell carcinoma. Hum Mol Genet. 2006;15:821–30. doi: 10.1093/hmg/ddl001. [DOI] [PubMed] [Google Scholar]
- Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–9. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–63. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Wang XD, Shou J, Wong P, French DM, Gao WQ. Notch1-expressing cells are indispensable for prostatic branching morphogenesis during development and re-growth following castration and androgen replacement. J Biol Chem. 2004;279:24733–44. doi: 10.1074/jbc.M401602200. [DOI] [PubMed] [Google Scholar]
- Baladron V, Ruiz-Hidalgo MJ, Nueda ML, Diaz-Guerra MJ, Garcia-Ramirez JJ, et al. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res. 2005;303:343–59. doi: 10.1016/j.yexcr.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Ceder JA, Jansson L, Helczynski L, Abrahamsson PA. Delta-like 1 (Dlk-1), a novel marker of prostate basal and candidate epithelial stem cells, is downregulated by notch signalling in intermediate/transit amplifying cells of the human prostate. Eur Urol. 2008;54:1344–53. doi: 10.1016/j.eururo.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Belandia B, Parker MG. Nuclear receptor regulation gears up another Notch. Nucl Recept Signal. 2006;4:e001. doi: 10.1621/nrs.04001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746–62. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan JT, Kellogg A, Shewchuk BM, Hewan-Lowe K, Bertrand FE. Notch-1 signaling is lost in prostate adenocarcinoma and promotes PTEN gene expression. J Cell Biochem. 2009;107:992–1001. doi: 10.1002/jcb.22199. [DOI] [PubMed] [Google Scholar]
- Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–7. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- Christofori G. Metastatic colon cancer cells negotiate the intravasation Notch. Cancer Cell. 2011;19:6–8. doi: 10.1016/j.ccr.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–48. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaronga MA, Lavery DN, Bevan CL, Llanos S, Belandia B. HEY1 Leu94Met gene polymorphism dramatically modifies its biological functions. Oncogene. 2010;29:411–20. doi: 10.1038/onc.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A, Look AT. NOTCH and PI3K–AKT pathways intertwined. Cancer Cell. 2007;12:411–3. doi: 10.1016/j.ccr.2007.10.027. [DOI] [PubMed] [Google Scholar]
- Kuwajima T, Nishimura I, Yoshikawa K. Necdin promotes GABAergic neuron differentiation in cooperation with Dlx homeodomain proteins. J Neurosci. 2006;26:5383–92. doi: 10.1523/JNEUROSCI.1262-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliardini S, Ren J, Wevrick R, Greer JJ. Developmental abnormalities of neuronal structure and function in prenatal mice lacking the Prader–Willi syndrome gene necdin. Am J Pathol. 2005;167:175–91. doi: 10.1016/S0002-9440(10)62964-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu NL, Wang J, Tsukamoto H. The Necdin–Wnt pathway causes epigenetic peroxisome proliferator-activated receptor gamma repression in hepatic stellate cells. J Biol Chem. 2010;285:30463–71. doi: 10.1074/jbc.M110.156703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniura H, Taniguchi N, Hara M, Yoshikawa K. Necdin, a postmitotic neuron-specific growth suppressor, interacts with viral transforming proteins and cellular transcription factor E2F1. J Biol Chem. 1998;273:720–8. doi: 10.1074/jbc.273.2.720. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Osawa M, Jakt LM, Yoshikawa K, Nishikawa S. Necdin restricts proliferation of hematopoietic stem cells during hematopoietic regeneration. Blood. 2009;114:4383–92. doi: 10.1182/blood-2009-07-230292. [DOI] [PubMed] [Google Scholar]
- Taniura H, Yoshikawa K. Necdin interacts with the ribonucleoprotein hnRNP U in the nuclear matrix. J Cell Biochem. 2002;84:545–55. [PubMed] [Google Scholar]
- Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci. 2008;28:8772–84. doi: 10.1523/JNEUROSCI.3052-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Knowles MA. Necdin: a multi functional protein with potential tumor suppressor role. Mol Carcinog. 2009;48:975–81. doi: 10.1002/mc.20567. [DOI] [PubMed] [Google Scholar]
- Kim JD, Kang K, Kim J. YY1's role in DNA methylation of Peg3 and Xist. Nucleic Acids Res. 2009;37:5656–64. doi: 10.1093/nar/gkp613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MY, Liao WS, Lu Z, Bornmann WG, Hennessey V, et al. Decitabine and suberoylanilide hydroxamic acid (SAHA) inhibit growth of ovarian cancer cell lines and xenografts while inducing expression of imprinted tumor suppressor genes, apoptosis, G2/M arrest, and autophagy. Cancer. 2011;117:4424–38. doi: 10.1002/cncr.26073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy SC, Gostout BS, Shridhar V, Wu X, Smith DI, et al. Biallelic methylation and silencing of paternally expressed gene 3 (PEG3) in gynecologic cancer cell lines. Gynecol Oncol. 2005;99:126–34. doi: 10.1016/j.ygyno.2005.05.036. [DOI] [PubMed] [Google Scholar]
- Feng W, Marquez RT, Lu Z, Liu J, Lu KH, et al. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112:1489–502. doi: 10.1002/cncr.23323. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Yoshioka H, Itaba N, Kubota N, Nishihara S, et al. Epigenetic silencing of PEG3 gene expression in human glioma cell lines. Mol Carcinog. 2001;31:1–9. doi: 10.1002/mc.1034. [DOI] [PubMed] [Google Scholar]
- Otsuka S, Maegawa S, Takamura A, Kamitani H, Watanabe T, et al. Aberrant promoter methylation and expression of the imprinted PEG3 gene in glioma. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:157–65. doi: 10.2183/pjab.85.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Itaba N, Otsuka S, Kamitani H, Watanabe T, et al. Coordinate downregulation of a novel imprinted transcript ITUP1 with PEG3 in glioma cell lines. DNA Res. 2004;11:37–49. doi: 10.1093/dnares/11.1.37. [DOI] [PubMed] [Google Scholar]
- Besson V, Smeriglio P, Wegener A, Relaix F, Nait Oumesmar B, et al. PW1 gene/paternally expressed gene 3 (PW1/Peg3) identifies multiple adult stem and progenitor cell populations. Proc Natl Acad Sci USA. 2011;108:11470–5. doi: 10.1073/pnas.1103873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, et al. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–3. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Wu X, Aithmitti N, Morrison RS. Peg3/Pw1 is a mediator between p53 and Bax in DNA damage-induced neuronal death. J Biol Chem. 2002;277:23000–7. doi: 10.1074/jbc.M201907200. [DOI] [PubMed] [Google Scholar]
- Relaix F, Wei X, Li W, Pan J, Lin Y, et al. Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis. Proc Natl Acad Sci USA. 2000;97:2105–10. doi: 10.1073/pnas.040378897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Wei XJ, Wu X, Sassoon DA. Peg3/Pw1 is an imprinted gene involved in the TNF–NFkappaB signal transduction pathway. Nat Genet. 1998;18:287–91. doi: 10.1038/ng0398-287. [DOI] [PubMed] [Google Scholar]
- Jiang X, Yu Y, Yang HW, Agar NY, Frado L, et al. The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. J Biol Chem. 2010;285:8472–80. doi: 10.1074/jbc.M109.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronauer MV, Schulz WA, Ackermann R, Burchardt M. Effects of WNT/beta-catenin pathway activation on signaling through T-cell factor and androgen receptor in prostate cancer cell lines. Int J Oncol. 2005;26:1033–40. doi: 10.3892/ijo.26.4.1033. [DOI] [PubMed] [Google Scholar]
- Li H, Kim JH, Koh SS, Stallcup MR. Synergistic effects of coactivators GRIP1 and beta-catenin on gene activation: cross-talk between androgen receptor and Wnt signaling pathways. J Biol Chem. 2004;279:4212–20. doi: 10.1074/jbc.M311374200. [DOI] [PubMed] [Google Scholar]
- Song LN, Gelmann EP. Interaction of beta-catenin and TIF2/GRIP1 in transcriptional activation by the androgen receptor. J Biol Chem. 2005;280:37853–67. doi: 10.1074/jbc.M503850200. [DOI] [PubMed] [Google Scholar]
- Song LN, Herrell R, Byers S, Shah S, Wilson EM, et al. Beta-catenin binds to the activation function 2 region of the androgen receptor and modulates the effects of the N-terminal domain and TIF2 on ligand-dependent transcription. Mol Cell Biol. 2003;23:1674–87. doi: 10.1128/MCB.23.5.1674-1687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry S, Yang X, Chen MW, Vacherot F, Buttyan R. Multifaceted interaction between the androgen and Wnt signaling pathways and the implication for prostate cancer. J Cell Biochem. 2006;99:402–10. doi: 10.1002/jcb.20983. [DOI] [PubMed] [Google Scholar]
- Yang X, Chen MW, Terry S, Vacherot F, Bemis DL, et al. Complex regulation of human androgen receptor expression by Wnt signaling in prostate cancer cells. Oncogene. 2006;25:3436–44. doi: 10.1038/sj.onc.1209366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrix W, Swinnen JV, Heyns W, Verhoeven G. Androgens down-regulate the expression of the human homologue of paternally expressed gene-3 in the prostatic adenocarcinoma cell line LNCaP. Mol Cell Endocrinol. 1999;155:69–76. doi: 10.1016/s0303-7207(99)00113-6. [DOI] [PubMed] [Google Scholar]
- Plagge A, Gordon E, Dean W, Boiani R, Cinti S, et al. The imprinted signaling protein XL alpha s is required for postnatal adaptation to feeding. Nat Genet. 2004;36:818–26. doi: 10.1038/ng1397. [DOI] [PubMed] [Google Scholar]
- Plagge A, Kelsey G. Imprinting the Gnas locus. Cytogenet Genome Res. 2006;113:178–87. doi: 10.1159/000090830. [DOI] [PubMed] [Google Scholar]
- Plagge A, Kelsey G, Germain-Lee EL. Physiological functions of the imprinted Gnas locus and its protein variants Galpha(s) and XLalpha(s) in human and mouse. J Endocrinol. 2008;196:193–214. doi: 10.1677/JOE-07-0544. [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Liu J, Greene A, Chen M, Weinstein LS. Tissue-specific imprinting of the G protein Gsalpha is associated with tissue-specific differences in histone methylation. Hum Mol Genet. 2004;13:819–28. doi: 10.1093/hmg/ddh098. [DOI] [PubMed] [Google Scholar]
- Kelsey G. Imprinting on chromosome 20: tissue-specific imprinting and imprinting mutations in the GNAS locus. Am J Med Genet C Semin Med Genet. 2010;154C:377–86. doi: 10.1002/ajmg.c.30271. [DOI] [PubMed] [Google Scholar]
- Weinstein LS, Liu J, Sakamoto A, Xie T, Chen M. Minireview: GNAS: normal and abnormal functions. Endocrinology. 2004;145:5459–64. doi: 10.1210/en.2004-0865. [DOI] [PubMed] [Google Scholar]
- Mariot V, Wu JY, Aydin C, Mantovani G, Mahon MJ, et al. Potent constitutive cyclic AMP-generating activity of XLalphas implicates this imprinted GNAS product in the pathogenesis of McCune–Albright syndrome and fibrous dysplasia of bone. Bone. 2011;48:312–20. doi: 10.1016/j.bone.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CH, McIntyre RE, Arends MJ, Adams DJ. The activating mutation R201C in GNAS promotes intestinal tumourigenesis in Apc(Min/+) mice through activation of Wnt and ERK1/2 MAPK pathways. Oncogene. 2010;29:4567–75. doi: 10.1038/onc.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- Middeldorp A, van Eijk R, Oosting J, Forte GI, van Puijenbroek M, et al. Increased frequency of 20q gain and copy-neutral loss of heterozygosity in mismatch repair proficient familial colorectal carcinomas. Int J Cancer. 2012;130:837–46. doi: 10.1002/ijc.26093. [DOI] [PubMed] [Google Scholar]
- Tominaga E, Tsuda H, Arao T, Nishimura S, Takano M, et al. Amplification of GNAS may be an independent, qualitative, and reproducible biomarker to predict progression-free survival in epithelial ovarian cancer. Gynecol Oncol. 2010;118:160–6. doi: 10.1016/j.ygyno.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Mantovani G, Lania AG, Spada A. GNAS imprinting and pituitary tumors. Mol Cell Endocrinol. 2010;326:15–8. doi: 10.1016/j.mce.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Frey UH, Alakus H, Wohlschlaeger J, Schmitz KJ, Winde G, et al. GNAS1 T393C polymorphism and survival in patients with sporadic colorectal cancer. Clin Cancer Res. 2005;11:5071–7. doi: 10.1158/1078-0432.CCR-05-0472. [DOI] [PubMed] [Google Scholar]
- Frey UH, Eisenhardt A, Lummen G, Rubben H, Jockel KH, et al. The T393C polymorphism of the G alpha s gene (GNAS1) is a novel prognostic marker in bladder cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:871–7. doi: 10.1158/1055-9965.EPI-04-0720. [DOI] [PubMed] [Google Scholar]
- Alakus H, Monig SP, Warnecke-Eberz U, Alakus G, Winde G, et al. Association of the GNAS1 T393C polymorphism with tumor stage and survival in gastric cancer. World J Gastroenterol. 2009;15:6061–7. doi: 10.3748/wjg.15.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey UH, Lummen G, Jager T, Jockel KH, Schmid KW, et al. The GNAS1 T393C polymorphism predicts survival in patients with clear cell renal cell carcinoma. Clin Cancer Res. 2006;12:759–63. doi: 10.1158/1078-0432.CCR-05-1722. [DOI] [PubMed] [Google Scholar]
- Eisenhardt A, Scherag A, Jockel KH, Reis H, Rubben H, et al. Lack of association of the genotype in the GNAS Fok I polymorphism and prostate cancer. Urol Int. 2011;87:80–6. doi: 10.1159/000325398. [DOI] [PubMed] [Google Scholar]
- Adany R, Heimer R, Caterson B, Sorrell JM, Iozzo RV. Altered expression of chondroitin sulfate proteoglycan in the stroma of human colon carcinoma. Hypomethylation of PG-40 gene correlates with increased PG-40 content and mRNA levels. J Biol Chem. 1990;265:11389–96. [PubMed] [Google Scholar]
- Gu Y, Zhang S, Wu Q, Xu S, Cui Y, et al. Differential expression of decorin, EGFR and cyclin D1 during mammary gland carcinogenesis in TA2 mice with spontaneous breast cancer. J Exp Clin Cancer Res. 2010;29:6. doi: 10.1186/1756-9966-29-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skandalis SS, Labropoulou VT, Ravazoula P, Likaki-Karatza E, Dobra K, et al. Versican but not decorin accumulation is related to malignancy in mammographically detected high density and malignant-appearing microcalcifications in non-palpable breast carcinomas. BMC Cancer. 2011;11:314. doi: 10.1186/1471-2407-11-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L. Small leucine-rich proteoglycans in kidney disease. J Am Soc Nephrol. 2011;22:1200–7. doi: 10.1681/ASN.2010050570. [DOI] [PubMed] [Google Scholar]
- Hu Y, Sun H, Owens RT, Wu J, Chen YQ, et al. Decorin suppresses prostate tumor growth through inhibition of epidermal growth factor and androgen receptor pathways. Neoplasia. 2009;11:1042–53. doi: 10.1593/neo.09760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AG, Liu J, Yuan Y, Gopalakrishnan VK, Johansson SL, et al. Expression of biomarkers modulating prostate cancer angiogenesis: differential expression of annexin II in prostate carcinomas from India and USA. Mol Cancer. 2003;2:34. doi: 10.1186/1476-4598-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonn L, Ananthanarayanan V, Gann PH. Evidence for field cancerization of the prostate. Prostate. 2009;69:1470–9. doi: 10.1002/pros.20983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel B, Metsuyanim S, Schmidt-Ott KM, Fridman E, Jacob-Hirsch J, et al. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res. 2006;66:6040–9. doi: 10.1158/0008-5472.CAN-05-4528. [DOI] [PubMed] [Google Scholar]
- Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, et al. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem. 2003;278:8826–36. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- Ayesh B, Matouk I, Ohana P, Sughayer MA, Birman T, et al. Inhibition of tumor growth by DT-A expressed under the control of IGF2 P3 and P4 promoter sequences. Mol Ther. 2003;7:535–41. doi: 10.1016/s1525-0016(03)00056-x. [DOI] [PubMed] [Google Scholar]
- Warzee B, Mesnil C, Hober D, Caloone D, Pirottin D, et al. Viral induction of Zac1b through TLR3- and IRF3-dependent pathways. Mol Immunol. 2010;48:119–27. doi: 10.1016/j.molimm.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Shibui T, Higo Y, Tsutsui TW, Uchida M, Oshimura M, et al. Changes in expression of imprinted genes following treatment of human cancer cell lines with non-mutagenic or mutagenic carcinogens. Int J Oncol. 2008;33:351–60. [PubMed] [Google Scholar]
- Wang ZA, Shen MM. Revisiting the concept of cancer stem cells in prostate cancer. Oncogene. 2011;30:1261–71. doi: 10.1038/onc.2010.530. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction. Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Murrell A. Genomic imprinting and cancer: from primordial germ cells to somatic cells. Sci World J. 2006;6:1888–910. doi: 10.1100/tsw.2006.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda A, Wang CJ, Cheong R, Timp W, Onyango P, et al. Enhanced sensitivity to IGF-II signaling links loss of imprinting of IGF2 to increased cell proliferation and tumor risk. Proc Natl Acad Sci USA. 2007;104:20926–31. doi: 10.1073/pnas.0710359105. [DOI] [PMC free article] [PubMed] [Google Scholar]