Abstract
Prostate cancer (PCa) results from a multistep process. This process includes initiation, which occurs through various aging events and multiple insults (such as chronic infection, inflammation and genetic instability through reactive oxygen species causing DNA double-strand breaks), followed by a multistep process of progression. These steps include several genetic and epigenetic alterations, as well as alterations to the chromatin structure, which occur in response to the carcinogenic stress-related events that sustain proliferative signaling. Events such as evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, and activating invasion and metastasis are readily observed. In addition, in conjunction with these critical drivers of carcinogenesis, other factors related to the etiopathogenesis of PCa, involving energy metabolism and evasion of the immune surveillance system, appear to be involved. In addition, when cancer spread and metastasis occur, the ‘tumor microenvironment' in the bone of PCa patients may provide a way to sustain dormancy or senescence and eventually establish a ‘seed and soil' site where PCa proliferation and growth may occur over time. When PCa is initiated and progression ensues, significant alterations in nuclear size, shape and heterochromatin (DNA transcription) organization are found, and key nuclear transcriptional and structural proteins, as well as multiple nuclear bodies can lead to precancerous and malignant changes. These series of cellular and tissue-related malignancy-associated events can be quantified to assess disease progression and management.
Keywords: active surveillance, cancer metastasis, cancer progression, digital image analysis, molecular biomarkers, morphological biomarkers, nuclear morphometry, nuclear proteins and nuclear structure, prostate cancer
Nuclear morphometry, nucleomics and prostate cancer progression
Prostate cancer (PCa) is the second most common cause of cancer-specific mortality among men in the United States, with an anticipated 217 730 newly diagnosed cases and 32 050 deaths in 2010.1 It has been estimated that up to 50% of screening-detected PCa cases would never have been diagnosed in the absence of screening. However, the benefits of PCa screening remain unclear; the burdens of overdiagnosis and overtreatment associated with early detection are well known.2, 3, 4, 5 Even with major improvements in diagnosis and treatment modalities for PCa, we still fail to identify and successfully treat the aggressive malignancies that lead to death. Hence, the remaining clinical dilemmas for the management of PCa patients are the early diagnostic features of those patients requiring definitive treatment and the identification of aggressive PCa that requires additional therapy after definitive treatment to prevent recurrence and progression to death.6, 7, 8
The nucleus is uniquely organized, with highly ordered compartmentalization (including chromatin (euchromatin and heterochromatin), several nuclear bodies, and nucleosomes). This ordered nuclear structure is instrumental to the maintenance of normal cellular functions such as cell division, proliferation, transcription and translation as well as to the cellular response to stress and disease.9, 10, 11, 12, 13, 14 These complex and ordered cellular processes involve the regulation of RNA, DNA, histone protein modification, nuclear membrane proteins and nuclear matrix proteins which are important to the maintenance of chromatin structure, nuclear body dynamics, and transcription/translation cellular functions. However, when cancer is initiated, alterations in nuclear size, shape, heterochromatin (DNA transcription) organization, key nuclear proteins and nuclear bodies occur owing to changes in key molecular and regulatory pathways, and these alterations lead to precancerous and malignant changes. For example, once cancer is initiated through various insults (such as chronic infection, inflammation, aging and genetic instability owing to DNA double-strand breaks (DSBs)), there is a multistep process of progression, which may include several genetic, epigenetic and nuclear structural alterations that occur in response to the ensuing carcinogenic stress-related events.6, 7, 8, 9, 10, 11, 12, 13, 14 One critical stressful event in carcinogenesis is the occurrence of DNA DSBs and the resultant repair process. DSBs arise through replication errors and exposure to exogenous and endogenous genotoxic agents such as ionizing radiation and reactive oxygen species, respectively.15 Genetic instability in cancer occurs in the form of somatic mutations, deletions, translocations, amplifications, copy number variations or single nucleotide polymorphisms, which are quantifiable as balanced alterations (approximately the same amount of chromosomal material observed after the event) or unbalanced alterations (there is a major loss and/or gain of chromosomal material). In the latter case, such changes can result in major alterations in nuclear bodies (such as number, location or structure), chromosome structure and organization, and changes to both of their normal locations.16, 17, 18
Similar to other cancers, the six hallmarks of cancer occur in PCa development. These hallmarks include sustained proliferative signaling, evasion of growth suppressors, resistance to cell death, replicative immortality, induction of angiogenesis, and activation of invasion and metastasis.14 The initiation events for PCa include aging, diet, inflammation and genomic instability resulting in neoplasia.19, 20, 21, 22, 23 In conjunction with these critical drivers of carcinogenesis, other factors related to the etiopathogenesis of cancer involve energy metabolism24, 25, 26 and evasion of the immune surveillance system.27, 28, 29 When cancer spread and metastasis occur, the ‘tumor microenvironment' in the bone of PCa patients may allow cells to remain dormant or senescent and eventually allow the establishment of a ‘seed and soil' site where PCa proliferation and growth may occur over time.29, 30 These molecular alterations can be evaluated with a variety of protocols that detect the numerous changes in chromosome structure and organization in cancer and other diseases. These protocols include nucleic acid sequencing, real-time PCR, methylation-specific qRT-PCR, comparative genomic hybridization, chromosome staining, fluorescence in situ hybridization, genotyping and cloning. Many of these molecular tools or modifications are currently being applied as diagnostic and prognostic tools for oncology research.
The organization and function of the nucleus depends on the efficient packaging of chromatin, the spatial geometry of the chromosomes and the organization of the nuclear matrix and its interactions with actin, lamins, nucleolus and other nuclear bodies (such as PMLs, Polycomb, Cajal and Speckles).17, 18, 31, 32 These structural elements are normally proficiently organized to provide the required physiological nuclear functions related to RNA, DNA and protein biosynthesis and regulation. In 1974, Berezney and Coffey identified the importance of a new class of nuclear proteins, the nuclear matrix proteins, which play a key role in the maintenance of nuclear structure (shape, size and chromatin organization) and function in normal cells and cancer cells.33 Numerous advances have been made in the understanding of how the nuclear matrix structure is organized with respect to interactions with the chromatin and inner nuclear membrane structures such as lamins, emerin, matrix attachment region and other nuclear membrane proteins that impact nuclear structure in cancer.13, 34, 35, 36 Another critical class of nuclear proteins is the high mobility group N (HMGN) protein family. HMGN1 has a particularly important role in inducing chromatin alterations in response to DSBs originating from reactive oxygen species, ultraviolet radiation and ionizing irradiation. When such DSBs occur in the DNA, the nucleosomes and several transcription factors (p300, γH2AX, CSA, XAB2 and modified histones) as well as the DSB-sensing factors, ataxia telangiectasia mutated, CAPF and HMGN1 are recruited to correct the DNA damage.10, 15, 37 This repair process is not always perfect because occasionally there are alterations in DNA that initiate carcinogenesis, and if the causes of initiation persist, cancer progression ensues.
There is an excellent review by Bianco-Miotto et al.,38 which covers a portion of the genetic and epigenetic alterations that occur in PCa development and progression. In addition, other molecular events have been demonstrated in PCa, and initiators such as chronic infections, inflammation, aging and genetic instability are clearly involved and result in amplifications, mutations, deletions, copy number variation (CNV) and single nucleotide polymorphisms.38, 39, 40 This review is focused on the assessment of nuclear structure and alterations in nuclear morphometry, which can be accurately quantified by digital image analysis via computer-assisted imaging systems (CAIS).35, 36, 38, 39 In addition, there are new tools for studying the tissue and nuclear architecture by quantitative histomorphometry, which calculates spatial topological texture features by using graphics tools and newly developed multimodal data algorithms. Hence, the overall biological and clinical translational contributions of the use of nuclear morphometry using CAIS in PCa will be addressed in men with indolent PCa or aggressive PCa.
Evolution of nuclear morphometry technology
Manual planimetry
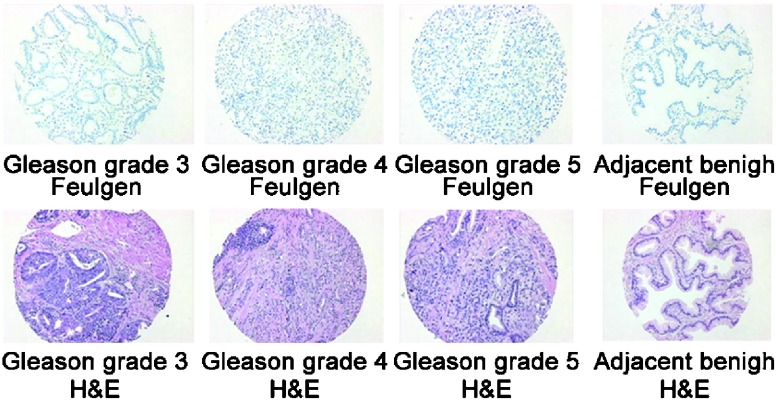
The history of pathologic cellular diagnosis dates back to the mid-1800s with the efforts of Virchow40 and Beale,41 where the importance of a microscopic interpretation of the cancer cell and the nucleus was first characterized as being diagnostic. The importance of nuclear structure in cancer diagnosis and prognosis made its first major advance with the use of histochemical stains such as hematoxylin and eosin (H&E) and the Papanicolaou (Pap stain) for staining fixed human tissue for cytology and cell morphology, respectively. The microscope and several improvements in lens and lighting over the years allowed for the detailed observation of nuclear size, shape and chromatin texture in cells, which clearly indicated abnormalities in cancer cells. Hematoxylin was demonstrated to form a dye–metal complex with arginine-rich ‘basic (cationic) nucleoproteins' such as histones of the nucleus. Eosin dye is acidic in nature and tends to bind to more eosinophilic cellular structures (cytoplasm, collagen and muscle fibers) producing various shades of pink. Combining H&E permitted the opportunity to study nuclear structure and its internal organization. The Feulgen staining reagent specifically and quantitatively binds to DNA in cellular material. The reagent binds to DNA by uncovering the free aldehyde groups in DNA during the acid hydrolysis process, which then reacts with the Feulgen reagent via a Schiff-Base interaction to form a stable, colored compound (blue) that absorbs light at 560 nm.42, 43 Figure 1 illustrates the H&E as well as Feulgen DNA-stained cancer and benign-appearing (cancer-adjacent) tissue microarray (TMA) spots.
Figure 1.
Illustration of the hematoxylin and eosin (H&E) and the Feulgen DNA-stained cancer and benign-appearing histology preparation (cancer-adjacent area) of prostate tissue microarray (TMA) spots.
This review will focus on the nucleus and the quantitative methods used to assess its structure in PCa. The methodology will briefly cover flow cytometry (FCM), digitizing nuclear size and shape by tracing and focus on the application of quantitative image analysis utilizing a microscope and computer-assisted software to study nuclear size, shape and chromatin texture. These protocols may be manual, semi-automated or completely automated. Notably, it is also feasible to conduct a quantitative biomarker image analysis by applying a fluorescent or chromogen-based antibody-directed biomarker analysis in pathologic tissues or cells.
Manual digital image (planimetry) analysis technology
In 1982, Diamond et al.44 were the first to utilize Graphpad software and a Zeiss Planapochromatic ×100 oil immersion objective with a ×12.5 ocular (magnification, ×1250) to trace 300 malignant and benign nuclei from each PCa case. With this approach, they easily separated the benign (normal) nuclei from the malignant nuclei. Next, they compared nuclear size and shape in all stage B PCa cases that had long-term follow-up and determined that they could distinguish those with a good prognosis from those with a poor prognosis (metastasis) with high accuracy (P<0.005). Defining a circle as 1.0, they calculated the nuclear roundness factor as follows: our roundness factor=(C/2π)/(A/π)1/2 (C=circumference and A=area), where as the circularity form factor =4πA/c2.
The nuclear roundness factor technology successfully identified men with a 14- to 15-year survival-free metastasis from those that did not. Subsequently, Dr Donald Coffey's laboratory and Dr Mitchell Benson compared the use of FCM (where the nuclei were labeled with acridine orange) to measure light scatter (forward and perpendicular) with the nuclear roundness factor performed on the same nuclei to assess tumor aggressiveness and heterogeneity of several well- to poorly-differentiated rat Dunning prostate tumor cell lines. The correlation between FCM and nuclear roundness variance (NRV) was exceptional.45, 46 These two key papers from 1984 demonstrated that measuring shape and size by FCM correlated well with NRV (the formula is NRV=radius (circumference)/radius (area) (R/r)=(P/2π)/(A/π)1/2) (P=perimeter). Furthermore, these data indicated that perpendicular light scatter by FCM correlated with the two critical biological properties of PCa pathogenesis: tumor aggressiveness and tumor heterogeneity. Also in 1984, Epstein et al.47 confirmed the clinical value of the irregularity of nuclear shape or NRV in 19 Stage A2 human PCa tissues (eight cases with a good prognosis and 11 with a poor prognosis) and could separate these two groups better than the Gleason grade. Later, others using a commercially available hardware and software program validated the clinical value of NRV measurements with a Zeiss inverted IM microscope (Carl Zeiss, Inc., Thornwood, NY, USA) equipped with a Zeiss planapachromatic ×100 oil immersion objective at a total magnification of ×2440. The images were analyzed with the DynaCELL Motility Morphometry Measurement workstation (JAW Associates, Inc., Annapolis, MD, USA). With this method, measurements differed by less than 5% among examiners, and the authors confirmed that this variable readily predicts progressive disease and mortality of PCa.48, 49, 50 Finally, Veltri et al.51 showed that the accuracy of NRV is significantly higher than the Gleason score to predict metastasis and PCa-specific death in men with long-term follow-up (median follow-up: 19 years). Therefore, NRV is better able to identify the aggressive phenotypes of PCa than the Gleason score and is associated with an increase in the irregularity of the nuclear border. Partin et al.52 were able to combine nuclear morphometry (NRV) with the Gleason score, clinical stage and age to create the ‘Prognostic Factor Score' to predict three risk groups based on Kaplan–Meier plots. Therefore, NRV alone and in combination with other clinical and pathologic features not only acts as a quantitative measure of pleomorphisms, which reflects PCa heterogeneity, but also predicts PCa progression when combined with other parameters.
Quantification of nuclear size, shape and chromatin organization using computer-assisted digital image analysis
The use of quantitative nuclear structure to assess prostate histology and predict PCa outcomes became a focus in the late 1980s. Digital image analysis and computers capable of accurately measuring nuclear structure and calculating algorithms based on combining significant features were able to provide integrated solutions. A number of approaches have been evaluated in an attempt to replace the current subjective visual approach of grading PCa. In fact, experienced pathologists have used the Gleason system to diagnose and prognosticate PCa since the 1970s with a great deal of success. This system has been markedly improved and enhanced to improve accuracy, reduce interpretation errors and ensure the reproducibility of prognostic information based on the efforts of many experts in the field of urology.53, 54, 55 However, Kuroiwa et al.56 reported a 54.8% concordance rate of the Gleason scores between a local and central review. In addition, there was an undergrading rate of 25.9% and an overgrading rate of 19.2% at a local review based upon data collected from 50 institutions on 2015 patients with T1c-3 PCa who underwent radical prostatectomy (RP) between 1997 and 2005.
The Gleason System for PCa histopathological grading is based on how benign the cancer tissue looks when viewed under a low power microscope (×10–×20) by an expert pathologist. Less dangerous prostate tumors have an appearance of normal glandular tissue; aggressive tumors that are more likely to invade and metastasize differ from normal tissue owing to a loss of normal glandular architecture (size, shape and organization), as well as other histological features, including changes in the cell nuclear structure. To assign a Gleason score, the pathologist first looks for a dominant (primary) pattern of cell growth or grade (the area where the cancer is most prominent) and then looks for a less widespread pattern or grade (secondary), and gives each one a grade number. The Gleason score is the sum of the dominant, or primary, tissue pattern grade (representing the majority of tumor) and the less dominant, or secondary, tissue pattern grade (assigned to the minority of the tumor). Today, pathologists tend to describe a Gleason score of 5 or 6 as a low-grade cancer, 7 (3+4 or 4+3) as medium-grade, and 8, 9 or 10 as high-grade cancer. Occasionally, a pathologist may note a small area of a higher grade pattern in a biopsy or RP specimens known as a ‘tertiary pattern' and may record this result.55 A lower-grade cancer tends to grow more slowly and is less likely to spread than a cancer with a higher grade. Some limitations for the Gleason score system involve interpretations when comparing a biopsy to RP specimens, reproducibility and difficulty in diagnosing small acinar atypical lesions.
Gleason grade/score and image analysis
Initially, attempts were made to improve the prognostic capabilities of PCa grading by visually fusing features using a multivariate analysis method; three variables were selected to create a new prognostic grading system: tumor tissue architecture, nuclear anaplasia and presence or absence of mitoses.57 The authors claim that their new system demonstrated improvements over the routine Gleason scoring system in evaluating prognosis as assessed by Kaplan–Meier survival plots. However, this approach was not routinely adapted for the pathological analysis of PCa clinical specimens. Several other laboratories have used nuclear morphometry to study the Gleason grade at the nuclear and tissue architectural level. The Gleason grading system (tissue and cellular changes indicative of cancer) and tumor stage (pathologic extent of disease inside the gland and if it has spread outside the gland) have served as independent and clinically significant ‘prognostic factors' which can predict biochemical recurrence, metastasis and overall patient survival. PCa glands and cells that appear to be healthy cells (benign) are given a low Gleason grade, and cancer cells that look less like healthy cells are given a higher Gleason grade. To assign a Gleason score, the pathologist first looks for a dominant (primary) pattern of cell growth or grade (area where the cancer is most prominent) and then looks for a less widespread pattern or grade (secondary) of growth, and gives each one a grade number. The Gleason score is the sum of the dominant or primary cancer tissue pattern grade (representing the majority of tumor; see the embedded figure) and the less dominant or secondary cancer tissue pattern grade (assigned to the minority of the tumor), resulting in a Gleason score or sum ranging from 2 to 10. Today, urologists describe a Gleason score of 6 as a low-grade cancer, 7 (3+4 or 4+3) as medium-grade, and 8, 9 or 10 as high-grade cancer. A lower-grade cancer grows more slowly and is less likely to spread than a cancer with a higher grade. Hence, this important subjective parameter of pathological diagnosis has resulted in the application of diverse approaches using various CAIS as well as methods for high-dimensional data analysis to assess tissue-based morphological and molecular parameters to develop prognostic decision support tools such as algorithms, nomograms and Correlation & Differential Dependency Networks.58, 59, 60, 61
One commercial approach used the Bacus CAS digital CAIS.62 The Bacus imaging system evolved from CAS-100 to CAS-200 and eventually to the BLISS CAIS, which could scan, store and perform quantitative image analysis on tissue slides.63 The BLISS system was capable of scanning TMAs and surgical slides, conducting quantitative image analyses for immunohistochemistry, DNA content and nuclear morphometry and then transporting the images and data via the internet. The CAS software was also capable of conducting DNA ploidy and nuclear morphometry analyses.64 Numerous researchers utilized the CAS system to quantify DNA ploidy, and in some institutions, nuclear morphometry was performed on Feulgen-stained tissue preparations. The CAS-200 imaging system has a morphometry software package that includes 40 nuclear features measurements that are expressed as equations based on the pixel maps captured from the nuclei analyzed.63, 64, 65 Several image analysis companies offered nuclear morphometry software programs; the features are similar whether it is light microscope-based (CAS200, AutoCyte and BLISS) or fluorescence-based (Chromavision).
Bacus used the BLISS imaging system and the CAS-200 quantitative DNA software program (40 features) to develop a nuclear grading morphometric calculation (continuous variable) to monitor a chemoprevention treatment regimen.62, 63 He used the mean and standard deviation (s.d.) of the 40 CAS-200 nuclear-feature measurements and transformed measurements of normal, preneoplastic and neoplastic nuclei into Z-scores scaled in s.d. units (Z-units). By this method, the nuclear grades of normal epithelia are always distributed about a mean value of zero. Hence, the individual Z-scores can be weighted by a coefficient and summed to obtain the final nuclear grade. These weighting coefficients are obtained by a Fisher linear discriminate analysis of the n-dimensional mean differences of pre-invasive (intraepithelial neoplasia) or invasive neoplastic nuclei compared with the reference population of normal nuclei.63 This Z-scoring grading system was able to differentiate breast DCIS grades 1, 2 and 3. In addition, this approach could discriminate CIN-3 lesions from normal and detected changes pre- and post-DMFO chemopreventive treatment.
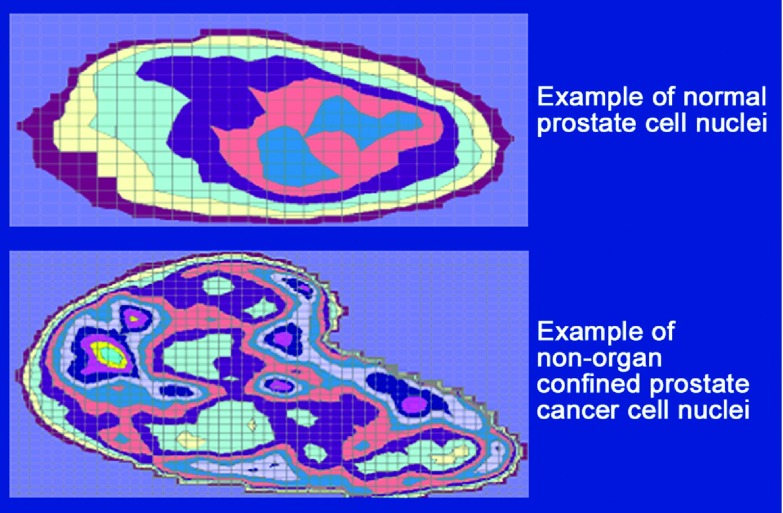
Veltri et al.65 used the AutoCyte Pathology Workstation (TriPath Inc., Burlington, NC, USA) digital imaging system and Feulgen-stained prostatic nuclei (Figure 2) to study the Gleason grades in RP specimens. A Gleason grade TMA prepared by Dr Jonathan I Epstein, a pathologist at Johns Hopkins Hospital, contained sets of four cores (0.6 mm) per case of Gleason grade 3, Gleason grade 4 and Gleason grade 5 PCa. The concept was to use the AutoCyte nuclear features shown in Table 1 on pools of 1100 nuclei from these three Gleason grades to determine the optimal variables to discriminate the three Gleason grades (3, 4 and 5) based on the Multivariate Logistic Regression analysis. Nine CAS-200 features (perimeter, area, Feret X, Feret Y, maximum Feret, intensity, median optical density, s.d. of gray values and DNA ploidy) were selected for Gleason grade comparisons. The results yielded areas of receiver operating characteristic (ROC) curves that distinguished differences among benign cancer-adjacent nuclei and Gleason grade 3 (ROC–AUC=0.78); Gleason grade 4 (ROC–AUC=0.86) and Gleason grade 5 (ROC–AUC=0.88) with accuracies of 73%, 78% and 80%, respectively.65 By calculating the predictive probability plots, we were able to note significant variations (marked heterogeneity) among the three Gleason grade patterns. Additionally, using the CAS-200 system, Venkataraman et al.66 could separate Gleason grade 3 and Gleason grade 4 by evaluating prostate biopsies with an 85% classification accuracy using a three-nuclear CAS-200 feature set (minimum diameter, angularity and sum optical density) as determined by a decision-tree analysis. The latter feature set provides one approach to determine Gleason grade based on quantitative nuclear morphometry and provides data to improve our understanding of morphological variability in nuclear structure and its role in PCa heterogeneity among the Gleason grades.
Figure 2.
Digital image analysis of Feulgen-stained prostate nuclei. These artificially colored images are based on the pixel maps obtained from the AutoCyte Pathology Workstation of the two representative cells that are illustrated above.
Table 1. Nuclear morphometric descriptors of important parameters in biochemical recurrence in the AutoCyte Pathology Workstation and quantitative nuclear grade (QNG) signature. Reproduced from Veltri et al.101.
| QUIC DNA morphometric measurement | Measurement type | β coefficient | P>z |
|---|---|---|---|
| Perimeter | Size/shape | ||
| Area | Size/shape | −0.0490319 | 0.004 |
| Circular form factor | Size/shape | ||
| Feret X | Size/shape | ||
| Feret Y | Size/shape | ||
| Minimum Feret | Size/shape | ||
| Maximum Feret | Size/shape | ||
| Excess of gray values | DNA content | ||
| Skewness of gray values | DNA content | ||
| Standard deviation (s.d.) of gray values | DNA content | −0.0762125 | 0.010 |
| Maximum gray value | DNA content | ||
| Minimum gray value | DNA content | ||
| Intensity | DNA content | 6.07×10−9 | 0.010 |
| Minimum optical density (o.d.) | DNA content | ||
| Maximum o.d. | DNA content | ||
| Median o.d. | DNA content | ||
| s.d. o.d. | DNA content | ||
| Skewness of o.d. | DNA content | 5.849436 | 0.048 |
| Excess of o.d. | DNA content | −0.0249485 | 0.034 |
| DNA ploidy | DNA content | 17.29464 | 0.001 |
| Transmission | Texture | ||
| Variance | Texture | ||
| Sum average | Texture | 2603.313 | 0.021 |
| Sum entropy | Texture | −11225.53 | 0.024 |
| Sum variance | Texture | ||
| Cluster shade | Texture | 0.0270699 | 0.012 |
| Diagonal moment | Texture | ||
| Sum of homogeneity | Texture | −29869.61 | 0.017 |
| Correlation | Markovian | −643.6912 | 0.037 |
| Difference moment | Markovian | ||
| Inverse difference moment | Markovian | 41413.18 | 0.018 |
| Sum entropy | Markovian | ||
| Entropy | Markovian | ||
| Information measure A | Markovian | ||
| Information measure B | Markovian | ||
| Maximal correlation coefficient | Markovian | ||
| Coefficient of variation | Markovian | ||
| Peak transition probability | Markovian | 16359.42 | 0.048 |
| Diagonal moment | Markovian | ||
| Second diagonal moment | Markovian | −838.5605 | 0.013 |
Abbreviation: QUIC, quantitative immunohistochemistry.
Using the AutoCyte imaging technology and the QUIC-DNA software package, Isharwal et al.67 demonstrated that the %DNA index has an independent prognostic value to predict progression, metastasis and PCa-specific survival in men with long-term follow-up (median follow-up: 17 years). Next, Isharwal et al.68 showed that DNA content measured in the benign cancer-adjacent and the cancer area of the diagnostic biopsy can predict unfavorable biopsy conversion (i.e., a recommendation for curative intervention) among men enrolled in an ‘active surveillance' program. Finally, in further support of the use of DNA content as a morphometric biomarker, Isharwal et al.69 demonstrated that DNA could serve as a ‘surrogate' to the Gleason score to predict PCa pathological stage. Although several nuclear features including nuclear size and shape parameters were significantly able to predict PCa pathologic stage, DNA content was the best predictor. Further, replacing the Gleason score with DNA content in each perioperative model showed equivalent or improvement in the accuracy of predicting the PCa pathological stage. The DNA ploidy in PCa and in other tumor types has demonstrated clinical potential, provided that a commercially proven system is employed to generate reproducible results.70, 71, 72, 73, 74
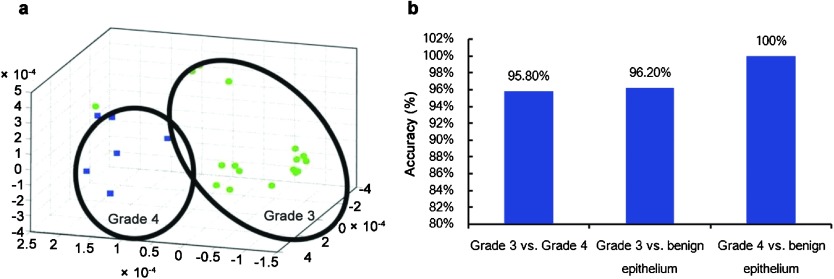
Another modern computational approach to quantify the Gleason score utilizes the capturing of digitized tissue histology images and developing multimodal data algorithms, which fuses several variable clinical and pathological types (categorical (nominal, ordinal and dichotomous); discrete, continuous and quantitative). These variables originate from routine diagnostic parameters and images of benign, pre-invasive and/or malignant tissue architecture bioinformatics and molecular biomarker parameters, which are used to predict PCa patient outcomes such as postoperative stage, clinical recurrence and patient survival. The computational methodology currently involves complex high dimensional histomorphological imaging data and develops mathematical solutions based upon a variety of machine learning devices (i.e., Support Vector Machines, Artificial Neural Networks, Genetic Algorithms, etc.).75, 76, 77, 78, 79, 80, 81 Note that these innovative approaches do not focus solely on the nucleus; rather, they assess the key molecular alterations and histological glandular, cellular and nuclear changes in prostate tissue architecture during malignant progression to create an objective measure instead of a subjective visual interpretation of changes (i.e., the Gleason score) in PCa. Several methods are used to calculate the alterations in tissue glandular architecture and the host immune responses following the initiation and progression of PCa (Table 2). These methods utilize spatial topological texture features (i.e., fractals, wavelet and multiwavelet transforms, Voronoi Diagram, Delaunay Triangulation and Minimum Spanning Tree).80 One group applying a Support Vector Machine has been able to separate Gleason grade 3 vs. Gleason grade 4 with 95.8% accuracy, Gleason grade 3 vs. benign epithelium with 96.2% accuracy and Gleason grade 4 vs. benign epithelium with 100% accuracy (Figure 3). Recently, Ali et al.82 used a novel ‘adaptive active contour scheme' that combines boundary and region-based energy terms with a shape prior to a multilevel set formulation to separate Gleason grades 3–5 in a TMA containing 40 PCa cases (two images per case). Using a total of seven nuclear features (area overlap ratio, average radial ratio, compactness, convexity, mean nuclear area, mean nuclear perimeter, mean nuclear diameter and principal component analysis) from each of the segmented nuclei, they could differentiate Gleason grade patterns 3 vs. 4 in the reduced embedding space with a classification accuracy of 84%. Applying a different approach (a leave-one-out approach), Lee and Huang83 used fractal dimensions and two classifiers to process a total of 182 pathological images with a resolution of 512×384 pixels (34 images were Gleason grade 3 and 46 images Gleason grade 4); one classifier used a Bayesian approach and the other a k-nearest neighbor approach. The Gleason grades were estimated with a 92.86% accuracy using the Bayes classifier and an 89.01% accuracy using the k-nearest neighbor classifier. In summary, using a variety of nuclear morphometry approaches, the nuclear size, shape and chromatin organization (including DNA) are often valuable features for constructing solutions relating to the current visual interpretation of the Gleason grade/score. These innovative image and data analysis approaches are methods that may be automatable and will create new objective methods to interpret the Gleason grade and/or score and, when combined with clinical and pathological variables, will be able to augment PCa outcome predictions employing decision-support tools.80, 81, 82, 83 A limitation at this time is that these new models for computing novel solutions to serve as surrogates for the Gleason grade/score have been used only rarely for predicting PCa outcomes.
Table 2. Summary of image-derived features used to characterize architectural arrangement and morphological appearance of prostate cancer nuclei. The relationship of graph and morphological features to visual attributes used for diagnostic decision-making by pathologists is also shown. Modified from Madabhushi et al.102.
| Feature class | Derived attributes | Relevance to histology |
|---|---|---|
| Voronoi tessellation | Number of nodes, number of edges, cyclomatic number, number of triangles, number of k-walks, spectral radius, eigenexponent, Randic index, area, roundness factor, area disorder, roundness factor homogeneity | Tissue architecture and arrangement of nuclei |
| Delaunay triangulation | Number of nodes, edge length, degree, number of edges, cyclomatic number, number of triangles, number of k-walks, spectral radius, eigenexponent, Wiener index, eccentricity, Randic index, fractal dimension | |
| Minimum spanning tree | Number of nodes, edge length, degree, number of neighbors, Wiener index, eccentricity, Randic index, Balaban index, fractal dimension | |
| Nuclear morphology | Margin spicularity, fractal dimension, height to width ratio, roundness factor, area overlap ratio, area disorder, perimeter, diameter | Nuclear, size boundary, appearance |
Figure 3.
Dimensionality reduction and manifold learning. Reproduced from Madabhushi et al.102 Non-linear dimensionality reduction used to classify prostate adenocarcinoma into Gleason grade 3 and grade 4 patterns. (a) Low dimensional embedding of the high dimensional attribute space via local linear embedding of 20 images representing prostate cancer grades 3 (circles) and 4 (squares). Each image is displayed as a point in 3D eigenspace. The clustering clearly shows very good discrimination between these two classes, which clinically is the most challenging problem in terms of Gleason grading. (b) Bar plots reflecting the classification accuracy obtained via a supervised classifier in distinguishing between pairs of tissue classes—grade 3/4, grade 3 vs. benign epithelium, and grade 4 vs. benign epithelium via a Support Vector Machine classifier. Note that in every case, the classification accuracy is over 90%.
Predicting outcomes—applying nuclear morphometry
Postoperative stage
The prediction of pathological stage based upon pre-treatment biopsy results using tissue morphometry has used routine clinical and biomarker bioinformatics. By combining biopsy serum prostate-specific antigen (PSA), the Gleason score and clinical stage or digital rectal exam, Partin and his colleagues84, 85, 86, 87, 88 revised a series of tables to predict the likelihood of four pathological outcomes. Others have used these variables as well as molecular biomarkers to predict outcomes for PCa patients.70, 71, 72 Badalament et al.72 combined a nuclear morphometric signature using the CAS-200 instrument with serum PSA to predict stage with an AUC–ROC =86% (sensitivity=85.7% specificity=71.3%). This was at a time in the diagnosis and prognosis of PCa when such predictions were less accurate. A limitation of this early algorithm was the number of nuclear features available and the stringency for the Multivariate Logistic Regression modeling. However, when the model was applied to incoming biopsy specimens, the algorithm performed within 5% of specifications.
Veltri et al.89 studied the biopsies of 557 consecutive men that underwent RP at Johns Hopkins Hospital from October 1998 to January 2000. Combining quantitative nuclear grade (QNG), the Gleason score and complexed PSA density (complete model) yielded an area under the ROC curve of AUC–ROC=82.4% (sensitivity=73.5% specificity=83%) to predict non-organ-confined PCa. Hence, this model confirmed that when QNG is combined with the Gleason score and PSA, an improved prediction of pathological stage is possible.
Biochemical recurrence
In 1996, Veltri et al.64 used the CAS-200 imaging system with only 38 nuclear morphometric descriptors to predict PCa biochemical progression. The patient cohort included 115 patients with clinically localized PCa, and the mean follow-up period in 70/115 patients without disease progression was 10.4±1.7 years. Using backward stepwise multiple linear regression at a variable feature selection stringency of P<0.05, the variances of 11/38 of the nuclear morphometric descriptors were found to be significant for biochemical progression (P<0.00001; AUC–ROC=86% sensitivity=78% specificity=83%). Furthermore, the QNG and the postoperative Gleason score, when combined, created a Multivariate Logistic Regression model for the prediction of biochemical progression, yielding an AUC–ROC =92% and having a sensitivity of 89% and specificity of 84%. These two parameters (QNG and Gleason score) separated the 115 patients into three statistically significant ‘risk groups' (from low to high) based upon Kaplan–Meier analysis. Next, our laboratory90 proposed a possible mechanism involving histone acetyltransferase p300 (p300 HAT) in PCa, which was previously described by Debes et al.91 as a basis for alterations in cancer cell nuclear structure and the biomarker (p300) does predict PCa biochemical progression. Employing an NCI Cooperative Prostate Cancer Tissue Resource TMA of 92 cases with ong-term follow-up (56 non-recurrences and 36 recurrences), we demonstrated that nuclear features, i.e., circular form factor (rho=−0.26; P=0.012) and minimum Feret (rho=−0.21; P=0.048) exhibited significant correlations with p300 protein expression. Finally, p300 expression was able to predict biochemical recurrence as a continuous or a dichotomous variable and can be combined with the Gleason score for biochemical recurrence risk stratification. In addition, studies in our laboratory revealed that the Gleason score and pathological stage correlated to several DNA content measurements supporting prior and current research.67, 68, 92, 93, 94, 95 More image-based morphometry translational research needs to be performed to understand the molecular mechanisms that alter nuclear structure during PCa progression.
Metastasis and PCa-specific survival
Predicting aggressive PCa is based on having a sufficient sample size and long-term follow-up data for the successful application of nuclear morphometry as a variable in addition to routine pathological and clinical variables. Khan et al.92 in our laboratory predicted progression to metastasis and/or PCa mortality in 227 RP specimens ‘marked' by pathologist Dr Epstein by employing the AutoCyte Pathology Workstation (TriPath Inc., Burlington, NC, USA.) and QUIC-DNA morphology software. They compared the Gleason score and lymph node status to QNG for partitioning the patient cohort into three risk groups to predict local recurrence and/or mortality; the QNG demonstrated improved accuracy for both outcomes over the Gleason score and lymph node status. The QNG variable proved to be a strong predictor of PCa progression to metastasis and/or mortality in this study. Next, Veltri et al.93 employed the same imaging technology and used a TMA with cancer and adjacent-benign areas to evaluate the use of QNG alone and with pathological and clinical variables to predict metastasis and PCa death. Both the cancer epithelial and the benign-adjacent nuclear area QNG solutions could predict metastasis and/or PCa death, and the Kaplan–Meier plots partitioned the patients into three groups with very significant log ranks. Furthermore, it was possible to combine QNG with the Gleason score and lymph node status to generate improvements in the prediction of biochemical recurrence as well as distant metastasis and/or PCa death. The only other single variable capable of predicting PCa progression outcomes was NRV using the JAW nuclear tracing hardware and software.48, 49 Partin et al.50 studied 100 PCa cases with long-term follow-up and were able to combine NRV (captured 150 nuclei/case), clinical stage, Gleason score and age to create the ‘Prognostic Factor Score' to predict disease progression and/or PCa death. Unfortunately, the JAW apparatus is no longer commercially available, and it took approximately 2 h to capture nuclei for each case. However, the potential value of measuring NRV accurately with a rapid, automated imaging system would now be commercially useful. In addition, other research using QNG and quantitative nuclear morphometry as well as other imaging technology consistently have demonstrated that shape, size and DNA content are predictive of PCa progression to metastasis and death67, 70 and therefore require validation for possible implementation in the future.
Active surveillance (AS) for prostate cancer and CAIS
A current dilemma in PCa management is overdetection and overtreatment because of PSA screening, which detects cases that would have remained undetected and caused no morbidity in the absence of screening.3 AS with delayed curative intervention has been proposed as an alternative to immediate surgery for low-grade, low-stage (indolent) tumors in an effort to reduce unnecessary treatment for prostate cancer.5, 96 Epstein et al.97 proposed a PSA density of <0.15 ng ml−1 cm−3 and favorable diagnostic needle-biopsy characteristics (i.e., a Gleason score <7, two or fewer cores involved with cancer, ≤50% of any core involved with cancer) as criteria to identify low-grade, low-stage tumors that can be followed with serial measurements of PSA and repeated biopsies without immediate intervention until reclassification requires definitive treatment. Makarov et al.98 identified 75 cases that qualified for AS (30 men required reclassification upon annual follow-up) to evaluate nuclear morphometry using the AutoCyte system and created a QNG signature of 12 nuclear morphometric descriptors. The QNG signature had an ROC–AUC of 87% with a sensitivity of 82%, specificity of 70% and accuracy of 75% to predict reclassification using the diagnostic AS biopsy. Therefore, QNG can predict AS failure using the diagnostic biopsy and might be improved in the future by adding specific molecular biomarkers. Next, Isharwal et al.68 demonstrated that DNA content could predict AS failure when either the benign cancer-adjacent or cancer areas were measured. Recently, our laboratory99 combined DNA content with the measurement of serum [−2]ProPSA to predict the likelihood for biopsy reclassification at the annual surveillance biopsy exam among men enrolled in an AS program. Therefore, nuclear morphometry and specific molecular biomarkers can be combined to improve the accuracy of predicting the failure of AS candidates at the time of diagnosis and safely manage men who choose this option.
What does the future hold for nuclear morphometry?
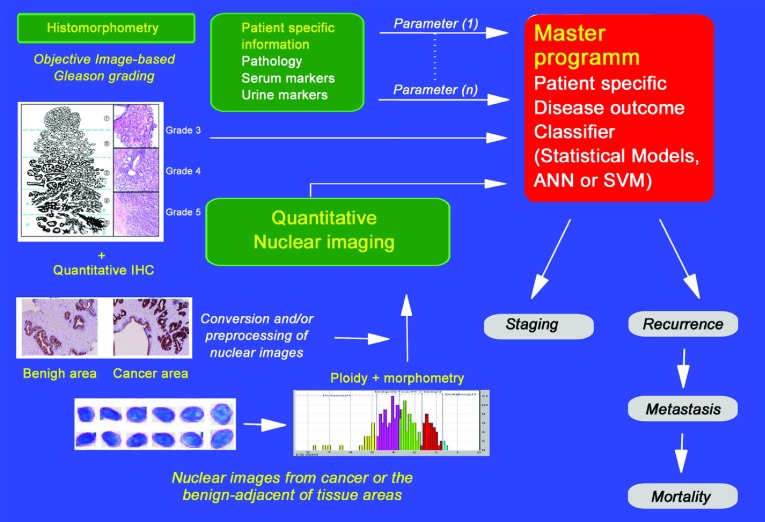
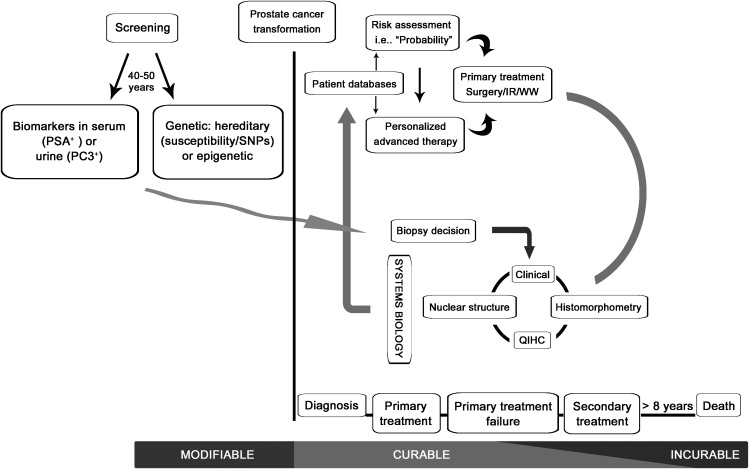
The application of bioengineering computational sciences to quantify nuclear structure and tissue architecture, as well as the development of automated processes for this quantification, will set the stage for improved imaging systems and software to calculate solutions for the diagnosis and prognosis of PCa. Significant challenges need to be met: (i) there is as yet no strong commercial partner to engineer the hardware with the correct specifications for the microscope, camera and software; (ii) direct collaboration with urologists and pathologists is required to ensure that the appropriate clinical dilemmas are addressed; (iii) validation studies are needed to verify the individual clinical applications; and (iv) FDA-approved trials must be carried out to establish specific claims. Currently, our research partners compute alterations in tissue glandular architecture and the host immune responses following the initiation and progression of PCa, and they utilize spatial topological texture features (such as fractals, wavelet and multiwavelet transforms, Voronoi Diagram, Delaunay Triangulation and Minimum Spanning Tree) to assess the tissue and nuclear architecture.80, 82 Finally, we now have the advanced imaging systems and the bio-informatic tools necessary to permit high-dimensional data of all types to be combined, allowing the generation of computational solutions that can be readily validated in appropriate clinical settings (Figures 4 and 5). However, to benefit patient management, these new tools will require automation and technical and clinical validation in multisite studies for various specific outcomes of PCa, such as active surveillance, biochemical recurrence, metastasis, and survival.100
Figure 4.
Automated imaging technology for the diagnosis and prognostic evaluation of prostate cancer. ANN, Artificial Neural Networks; QIHC, quantitative immunohistochemistry; SVM, Support Vector Machines.
Figure 5.
Personalized concept to decision making for prostate cancer. IR, irradiation; QIHC, quantitative immunohistochemistry; WW, watchful waiting or active surveillance for very low or low risk prostate cancers observed on a diagnostic biopsy.
The authors declare no competing financial interests.
References
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Etzioni R, Penson DF, Legler JM, di Tommaso D, Boer R, et al. Overdiagnosis due to prostate-specific antigen screening: lessons from U.S. prostate cancer incidence trends. J Natl Cancer Inst. 2002;94:981–90. doi: 10.1093/jnci/94.13.981. [DOI] [PubMed] [Google Scholar]
- Draisma G, Boer R, Otto SJ, van der Cruijsen IW, Damhuis RA, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95:868–78. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- Albertsen PC. Treatment of localized prostate cancer: when is active surveillance appropriate. Nat Rev Clin Oncol. 2010;7:394–400. doi: 10.1038/nrclinonc.2010.63. [DOI] [PubMed] [Google Scholar]
- Bastian PJ, Carter BH, Bjartell A, Seitz M, Stanislaus P, et al. Insignificant prostate cancer and active surveillance: from definition to clinical implications. Eur Urol. 2009;55:1321–30. doi: 10.1016/j.eururo.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA. 2005;293:2095–101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, et al. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. TJ Urol. 2003;169:517–23. doi: 10.1097/01.ju.0000045749.90353.c7. [DOI] [PubMed] [Google Scholar]
- Freedland SJ, Humphreys EB, Mangold LA, Eisenberger M, Dorey FJ, et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA. 2005;294:433–9. doi: 10.1001/jama.294.4.433. [DOI] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev. 2004;4:677–87. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- Bartova E, Krejci J, Harnicarova A, Galiova G, Kozubek S. Histone modifications and nuclear architecture: a review. J Histochem Cytochem. 2008;56:711–21. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonisch C, Nieratschker SM, Orfanos NK, Hake SB. Chromatin proteomics and epigenetic regulatory circuits. Expert Rev Proteomics. 2008;5:105–19. doi: 10.1586/14789450.5.1.105. [DOI] [PubMed] [Google Scholar]
- Goncalves Dos Santos Silva A, Sarkar R, Harizanova J, Guffei A, Mowat M, et al. Centromeres in cell division, evolution, nuclear organization and disease. J Cell Biochem. 2008;104:2040–58. doi: 10.1002/jcb.21766. [DOI] [PubMed] [Google Scholar]
- Rynearson AL, Sussman CR. Nuclear structure, organization, and oncogenesis. J Gastrointest Cancer. 2011;42:112–7. doi: 10.1007/s12029-011-9253-5. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–7. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–73. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Nelson WG, de Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349:366–81. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–64. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- Nelson WG, DeWeese TL, DeMarzo AM. The diet, prostate inflammation, and the development of prostate cancer. Cancer Metastasis Rev. 2002;21:3–16. doi: 10.1023/a:1020110718701. [DOI] [PubMed] [Google Scholar]
- Platz EA, de Marzo AM. Epidemiology of inflammation and prostate cancer. J Urol. 2004;171:S36–40. doi: 10.1097/01.ju.0000108131.43160.77. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–62. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–40. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Parsa AT. Cancer and the complement cascade. Mol Cancer Res. 2010;8:1453–65. doi: 10.1158/1541-7786.MCR-10-0225. [DOI] [PubMed] [Google Scholar]
- Rajarubendra N, Lawrentschuk N, Bolton DM, Klotz L, Davis ID. Prostate cancer immunology—an update for Urologists. BJU Int. 2011;107:1046–51. doi: 10.1111/j.1464-410X.2010.09820.x. [DOI] [PubMed] [Google Scholar]
- Sprenger CC, Plymate SR, Reed MJ. Aging-related alterations in the extracellular matrix modulate the microenvironment and influence tumor progression. Int J Cancer. 2010;127:2739–48. doi: 10.1002/ijc.25615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley RR, Fidler IJ. The seed and soil hypothesis revisited—the role of tumor–stroma interactions in metastasis to different organs. Int J Cancer. 2011;128:2527–35. doi: 10.1002/ijc.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsov S, Schafer G, Stasyk T, Fuchsberger C, Bonn GK, et al. Proteomics profiling of microdissected low- and high-grade prostate tumors identifies Lamin A as a discriminatory biomarker. J Proteome Res. 2011;10:259–68. doi: 10.1021/pr100921j. [DOI] [PubMed] [Google Scholar]
- Getzenberg RH, Pienta KJ, Huang EY, Coffey DS. Identification of nuclear matrix proteins in the cancer and normal rat prostate. Cancer Res. 1991;51:6514–20. [PubMed] [Google Scholar]
- Elcock LS, Bridger JM. Exploring the effects of a dysfunctional nuclear matrix. Biochem Soc Trans. 2008;36:1378–83. doi: 10.1042/BST0361378. [DOI] [PubMed] [Google Scholar]
- He S, Dunn KL, Espino PS, Drobic B, Li L, et al. Chromatin organization and nuclear microenvironments in cancer cells. J Cell Biochem. 2008;104:2004–15. doi: 10.1002/jcb.21485. [DOI] [PubMed] [Google Scholar]
- Pezo RC, Singer RH. Nuclear microenvironment in cancer diagnosis and treatment. J Cell Biochem. 2008;104:1953–63. doi: 10.1002/jcb.21353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G. HMGNs, DNA repair and cancer. Biochi Biophys Acta. 2010;1799:80–5. doi: 10.1016/j.bbagrm.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco-Miotto T, Chiam K, Buchanan G, Jindal S, Day TK, et al. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev. 2010;19:2611–22. doi: 10.1158/1055-9965.EPI-10-0555. [DOI] [PubMed] [Google Scholar]
- Xu J, Zheng SL, Isaacs SD, Wiley KE, Wiklund F, et al. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc Natl Acad Sci USA. 2010;107:2136–40. doi: 10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virchow R. Philadelphia, PA; J B Lippincott; 1863. Cellular Pathology as Based upon Physiological and Pathological Histology. [DOI] [PubMed] [Google Scholar]
- Beale L. Examination of sputum from a case of cancer of the pharynx and the adjacent parts. Arch Med Lond. 1860;2:44. [Google Scholar]
- Gill JE, Jotz MM. Further observations on the chemistry of pararosaniline-Feulgen staining. Histochemistry. 1976;46:147–60. doi: 10.1007/BF02462739. [DOI] [PubMed] [Google Scholar]
- Schulte E, Wittekind D. Standardization of the Feulgen-Schiff technique. Staining characteristics of pure fuchsin dyes; a cytophotometric investigation. Histochemistry. 1989;91:321–31. doi: 10.1007/BF00493008. [DOI] [PubMed] [Google Scholar]
- Diamond DA, Berry SJ, Umbricht C, Jewett HJ, Coffey DS. Computerized image analysis of nuclear shape as a prognostic factor for prostatic cancer. Prostate. 1982;3:321–32. doi: 10.1002/pros.2990030402. [DOI] [PubMed] [Google Scholar]
- Benson MC, McDougal DC, Coffey DS. The application of perpendicular and forward light scatter to assess nuclear and cellular morphology. Cytometry. 1984;5:515–22. doi: 10.1002/cyto.990050513. [DOI] [PubMed] [Google Scholar]
- Benson MC, McDougal DC, Coffey DS. The use of multiparameter flow cytometry to assess tumor cell heterogeneity and grade prostate cancer. Prostate. 1984;5:27–45. doi: 10.1002/pros.2990050104. [DOI] [PubMed] [Google Scholar]
- Epstein JI, Berry SJ, Eggleston JC. Nuclear roundness factor. A predictor of progression in untreated Stage A2 prostate cancer. Cancer. 1984;54:1666–71. doi: 10.1002/1097-0142(19841015)54:8<1666::aid-cncr2820540830>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Mohler JL, Partin AW, Epstein JI, Lohr WD, Coffey DS. Nuclear roundness factor measurement for assessment of prognosis of patients with prostatic carcinoma. II. Standardization of methodology for histologic sections. J Urol. 1988;139:1085–90. doi: 10.1016/s0022-5347(17)42792-3. [DOI] [PubMed] [Google Scholar]
- Mohler JL, Partin AW, Lohr WD, Coffey DS. Nuclear roundness factor measurement for assessment of prognosis of patients with prostatic carcinoma. I. Testing of a digitization system. J Urol. 1988;139:1080–4. doi: 10.1016/s0022-5347(17)42791-1. [DOI] [PubMed] [Google Scholar]
- Partin AW, Walsh AC, Pitcock RV, Mohler JL, Epstein JI, et al. A comparison of nuclear morphometry and Gleason grade as a predictor of prognosis in stage A2 prostate cancer: a critical analysis. J Urol. 1989;142:1254–8. doi: 10.1016/s0022-5347(17)39049-3. [DOI] [PubMed] [Google Scholar]
- Veltri RW, Isharwal S, Miller C, Epstein JE, Partin AW. Nuclear roundness variance predicts prostate cancer progression, metastasis and death: a prospective evaluation with up to 25 years of follow-up after radical prostatectomy. Prostate. 2010;70:1333–9. doi: 10.1002/pros.21168. [DOI] [PubMed] [Google Scholar]
- Partin AW, Steinberg GD, Pitcock RV, Wu L, Piantadosi S, et al. Use of nuclear morphometry, gleason histologic scoring, clinical stage, and age to predict disease-free survival among patients with prostate cancer. Cancer. 1992;70:161–8. doi: 10.1002/1097-0142(19920701)70:1<161::aid-cncr2820700126>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Gleason DF. Classification of prostatic carcinomas. Cancer Chemother Rep. 1966;50:125–8. [PubMed] [Google Scholar]
- Gleason DF. Histologic grading of prostate cancer: a perspective. Hum Pathol. 1992;23:273–9. doi: 10.1016/0046-8177(92)90108-f. [DOI] [PubMed] [Google Scholar]
- Epstein JI. An update of the Gleason grading system. J Urol. 2010;183:433–40. doi: 10.1016/j.juro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- Kuroiwa K, Shiraishi T, Ogawa O, Usami M, Hirao Y, et al. Discrepancy between local and central pathological review of radical prostatectomy specimens. J Urol. 2010;183:952–7. doi: 10.1016/j.juro.2009.11.024. [DOI] [PubMed] [Google Scholar]
- Schroeder FH, Hop WC, Blom JH, Mostofi FK. Grading of prostatic cancer: III. Multivariate analysis of prognostic parameters. Prostate. 1985;7:13–20. doi: 10.1002/pros.2990070103. [DOI] [PubMed] [Google Scholar]
- Diamond J, Anderson NH, Bartels PH, Montironi R, Hamilton PW. The use of morphological characteristics and texture analysis in the identification of tissue composition in prostatic neoplasia. Hum Pathol. 2004;35:1121–31. doi: 10.1016/j.humpath.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Montironi R, Cheng L, Lopez-Beltran A, Mazzucchelli R, Scarpelli M, et al. Decision support systems for morphology-based diagnosis and prognosis of prostate neoplasms: a methodological approach. Cancer. 2009;115:3068–77. doi: 10.1002/cncr.24345. [DOI] [PubMed] [Google Scholar]
- Madabhushi A, Agner S, Basavanhally A, Doyle S, Lee G. Computer-aided prognosis: Predicting patient and disease outcome via quantitative fusion of multi-scale, multi-modal data. Comput Med Imaging Graph. 2011;35:506–14. doi: 10.1016/j.compmedimag.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Zhang B, Li H, Riggins RB, Zhan M, Xuan J, et al. Differential dependency network analysis to identify condition-specific topological changes in biological networks. Bioinformatics. 2009;25:526–32. doi: 10.1093/bioinformatics/btn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacus JW, Grace LJ. Optical microscope system for standardized cell measurements and analyses. Appl Opt. 1987;26:3280–93. doi: 10.1364/AO.26.003280. [DOI] [PubMed] [Google Scholar]
- Bacus JW, Boone CW, Bacus JV, Follen M, Kelloff GJ, et al. Image morphometric nuclear grading of intraepithelial neoplastic lesions with applications to cancer chemoprevention trials. Cancer Epidemiol Biomarkers Prev. 1999;8:1087–94. [PubMed] [Google Scholar]
- Veltri RW, Partin AW, Epstein JE, Marley GM, Miller CM, et al. Quantitative nuclear morphometry, Markovian texture descriptors, and DNA content captured on a CAS-200 Image analysis system, combined with PCNA and HER-2/neu immunohistochemistry for prediction of prostate cancer progression. J Cell Biochem. 1994;19:249–58. [PubMed] [Google Scholar]
- Veltri RW, Marlow C, Khan MA, Miller MC, Epstein JI, et al. Significant variations in nuclear structure occur between and within Gleason grading patterns 3, 4, and 5 determined by digital image analysis. Prostate. 2007;67:1202–10. doi: 10.1002/pros.20614. [DOI] [PubMed] [Google Scholar]
- Venkataraman G, Rycyna K, Rabanser A, Heinze G, Baesens BM, et al. Morphometric signature differences in nuclei of Gleason pattern 4 areas in Gleason 7 prostate cancer with differing primary grades on needle biopsy. J Urol. 2009;181:88–93. doi: 10.1016/j.juro.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Isharwal S, Miller MC, Epstein JI, Mangold LA, Humphreys E, et al. Prognostic value of Her-2/neu and DNA index for progression, metastasis and prostate cancer-specific death in men with long-term follow-up after radical prostatectomy. Int J Cancer. 2008;123:2636–43. doi: 10.1002/ijc.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isharwal S, Makarov DV, Carter HB, Epstein JI, Partin AW, et al. DNA content in the diagnostic biopsy for benign-adjacent and cancer-tissue areas predicts the need for treatment in men with T1c prostate cancer undergoing surveillance in an expectant management programme. BJU Int. 2010;105:329–33. doi: 10.1111/j.1464-410X.2009.08791.x. [DOI] [PubMed] [Google Scholar]
- Isharwal S, Miller MC, Epstein JI, Mangold LA, Humphreys E, et al. DNA Ploidy as surrogate for biopsy Gleason score for preoperative organ versus nonorgan-confined prostate cancer prediction. Urology. 2009;73:1092–7. doi: 10.1016/j.urology.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri RW, Partin AW, Miller CM.Quantitative Nuclear Grade (QNG): the clinical applications of the quantitative measurement of nuclear structure using image analysisIn: Kelloff GJ, Hawk ET, Sigman CC, editors. Cancer chemoprevention Totowa, NJ; Humana Press; 2005pp 97–108. [Google Scholar]
- Cordon-Cardo C, Kotsianti A, Verbel DA, Teverovskiy M, Capodieci P, et al. Improved prediction of prostate cancer recurrence through systems pathology. J Clin Invest. 2007;117:1876–83. doi: 10.1172/JCI31399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badalament RA, Miller MC, Peller PA, Young DC, Bahn DK, et al. An algorithm for predicting nonorgan confined prostate cancer using the results obtained from sextant core biopsies with prostate specific antigen level. J Urol. 1996;156:1375–80. [PubMed] [Google Scholar]
- Venkataraman G, Heinze G, Holmes EW, Ananthanarayanan V, Bostwick DG, et al. Identification of patients with low-risk for aneuploidy: comparative discriminatory models using linear and machine-learning classifiers in prostate cancer. Prostate. 2007;67:1524–36. doi: 10.1002/pros.20629. [DOI] [PubMed] [Google Scholar]
- Carmichael MJ, Veltri RW, Partin AW, Miller MC, Walsh PC, et al. Deoxyribonucleic acid ploidy analysis as a predictor of recurrence following radical prostatectomy for stage T2 disease. J Urol. 1995;153:1015–9. [PubMed] [Google Scholar]
- Potter SR, Miller MC, Mangold LA, Jones KA, Epstein JI, et al. Genetically engineered neural networks for predicting prostate cancer progression after radical prostatectomy. Urology. 1999;54:791–5. doi: 10.1016/s0090-4295(99)00328-3. [DOI] [PubMed] [Google Scholar]
- Reckwitz T, Potter SR, Snow PB, Zhang Z, Veltri RW, et al. Artificial neural networks in urology: Update 2000. Prostate Cancer Prostatic Dis. 1999;2:222–6. doi: 10.1038/sj.pcan.4500374. [DOI] [PubMed] [Google Scholar]
- Chun FK, Karakiewicz PI, Briganti A, Walz J, Kattan MW, et al. A critical appraisal of logistic regression-based nomograms, artificial neural networks, classification and regression-tree models, look-up tables and risk-group stratification models for prostate cancer. BJU Int. 2007;99:794–800. doi: 10.1111/j.1464-410X.2006.06694.x. [DOI] [PubMed] [Google Scholar]
- Shariat SF, Karakiewicz PI, Margulis V, Kattan MW. Inventory of prostate cancer predictive tools. Curr Opin Urol. 2008;18:279–96. doi: 10.1097/MOU.0b013e3282f9b3e5. [DOI] [PubMed] [Google Scholar]
- Karakiewicz PI, Hutterer GC. Predictive models and prostate cancer. Nat Clin Pract. 2008;5:82–92. doi: 10.1038/ncpuro0972. [DOI] [PubMed] [Google Scholar]
- Gurcan MN, Boucheron L, Can A, Madabhushi A, Rajpoot N, et al. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–71. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser K, Kayser G, Metze K. The concept of structural entropy in tissue-based diagnosis. Anal Quant Cytol Histol. 2007;29:296–308. [PubMed] [Google Scholar]
- Ali S, Madabhushi A.Active Contour for Overlap Resolution using Watershed BASED initialization (ACOReW): applications to histopathologyIn: Proceedings of the 8th IEEE International Symposium on Biomedical Imaging: From Nano to Macro; 30 March–2 April 2011; Chicago, IL, USA. IEEE: Piscataway, NJ, USA, 2011pp614–7.
- Lee CH, Huang PW.Classification for pathological prostate images based on fractal analysisIn: Proceedings of the 2008 Congress on Image and Signal Processing; 27–30 May; Sanya, China. IEEE: Piscataway, NJ, USA, Vol. 3, pp113–37.
- Partin AW, Yoo J, Carter HB, Pearson JD, Chan DW, et al. The use of prostate specific antigen, clinical stage and Gleason score to predict pathological stage in men with localized prostate cancer. J Urol. 1993;150:110–4. doi: 10.1016/s0022-5347(17)35410-1. [DOI] [PubMed] [Google Scholar]
- Partin AW, Kattan MW, Subong EN, Walsh PC, Wojno KJ, et al. Combination of prostate-specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer. A multi-institutional update. JAMA. 1997;277:1445–51. [PubMed] [Google Scholar]
- Partin AW, Mangold LA, Lamm DM, Walsh PC, Epstein JI, et al. Contemporary update of prostate cancer staging nomograms (Partin Tables) for the new millennium. Urology. 2001;58:843–8. doi: 10.1016/s0090-4295(01)01441-8. [DOI] [PubMed] [Google Scholar]
- Makarov DV, Trock BJ, Humphreys EB, Mangold LA, Walsh PC, et al. Updated nomogram to predict pathologic stage of prostate cancer given prostate-specific antigen level, clinical stage, and biopsy Gleason score (Partin tables) based on cases from 2000 to 2005. Urology. 2007;69:1095–101. doi: 10.1016/j.urology.2007.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Isharwal S, Haese A, Chun FK, Makarov DV, et al. Prediction of patient-specific risk and percentile cohort risk of pathological stage outcome using continuous prostate-specific antigen measurement, clinical stage and biopsy Gleason score. BJU Int. 2011;107:1562–69. doi: 10.1111/j.1464-410X.2010.09692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri RW, Miller MC, Mangold LA, O'Dowd GJ, Epstein JI, et al. Prediction of pathological stage in patients with clinical stage T1c prostate cancer: the new challenge. J Urol. 2002;168:100–4. [PubMed] [Google Scholar]
- Isharwal S, Miller C, Marlow C, Makarov DV, Partin AW, et al. p300 (Histone Acetyltransferase) biomarker predicts prostate cancer biochemical recurrence and correlates with changes in epithelia nuclear size and shape. Prostate. 2008;68:1097–104. doi: 10.1002/pros.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debes JD, Sebo TJ, Heemers HV, Kipp BR, Haugen DL, et al. p300 modulates nuclear morphology in prostate cancer. Cancer Res. 2005;65:708–12. [PubMed] [Google Scholar]
- Khan MA, Walsh PC, Miller MC, Bales WD, Epstein JI, et al. Quantitative alterations in nuclear structure predict prostate carcinoma distant metastasis and death in men with biochemical recurrence after radical prostatectomy. Cancer. 2003;98:2583–91. doi: 10.1002/cncr.11852. [DOI] [PubMed] [Google Scholar]
- Veltri RW, Khan MA, Miller MC, Epstein JI, Mangold LA, et al. Ability to predict metastasis based on pathology findings and alterations in nuclear structure of normal-appearing and cancer peripheral zone epithelium in the prostate. Clin Cancer Res. 2004;10:3465–73. doi: 10.1158/1078-0432.CCR-03-0635. [DOI] [PubMed] [Google Scholar]
- Veltri RW, Miller MC, Partin AW, Coffey DS, Epstein JI. Ability to predict biochemical progression using Gleason score and a computer-generated quantitative nuclear grade derived from cancer cell nuclei. Urology. 1996;48:685–91. doi: 10.1016/S0090-4295(96)00370-6. [DOI] [PubMed] [Google Scholar]
- Isharwal S, Miller MC, Epstein JI, Mangold LA, Humphreys E, et al. DNA ploidy as surrogate for biopsy Gleason score for preoperative organ versus nonorgan-confined prostate cancer prediction. Urology. 2009;73:1092–7. doi: 10.1016/j.urology.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, et al. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–83. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JI, Walsh PC, Carmichael M, Brendler CB. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA. 1994;271:368–74. [PubMed] [Google Scholar]
- Makarov DV, Marlow C, Epstein JI, Miller MC, Landis P, et al. Using nuclear morphometry to predict the need for treatment among men with low grade, low stage prostate cancer enrolled in a program of expectant management with curative intent. Prostate. 2008;68:183–9. doi: 10.1002/pros.20679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isharwal S, Makarov DV, Sokoll LJ, Landis P, Marlow C, et al. ProPSA and diagnostic biopsy tissue DNA content combination improves accuracy to predict need for prostate cancer treatment among men enrolled in an active surveillance program. Urology. 2011;77:763.e1–6. doi: 10.1016/j.urology.2010.07.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–8. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri RW, Miller MC, Isharwal S, Marlow C, Makarov DV, et al. Prediction of prostate-specific antigen recurrence in men with long-term follow-up post-prostatectomy using quantitative nuclear morphometry. Cancer Epidemiol Biomarkers Prev. 2008;17:102–10. doi: 10.1158/1055-9965.EPI-07-0175. [DOI] [PubMed] [Google Scholar]
- Madabhushi A, Doyle S, Lee G, Basavanhally A, Monaco J, Masters S, et al. Integrated diagnostics: a conceptual framework with examples. Clin Chem Lab Med. 2010;48:989–98. doi: 10.1515/CCLM.2010.193. [DOI] [PubMed] [Google Scholar]