Abstract
The introduction of serum prostate-specific antigen (PSA) in the 1980s has dramatically altered and benefited the initial diagnosis of prostate cancer. However, the widespread use of PSA testing has resulted in overdetection and overtreatment of potentially indolent disease. Thus, a clinical dilemma today in the management of prostate cancer is to discern men with aggressive disease who need definitive treatment from men whose disease are not lethal. Although several serum and tissue biomarkers have been evaluated during the past decade, improved markers are still needed to enhance the accuracy, with which patients at risk can be discerned and treated more aggressively. The cancer/testis antigens (CTAs) are a group of proteins that are restricted to the testis in the normal adult, but are aberrantly expressed in several types of cancers. Because of their restricted expression pattern, the CTAs represent attractive biomarker candidates for cancer diagnosis/prognosis. Furthermore, several studies to date have reported the differential expression of CTAs in prostate cancer. Here, we review recent developments that demonstrate the potential of the CTAs as biomarkers to discern the aggressive phenotype of prostate cancer.
Keywords: cancer/testis antigens, DNA microarrays, prostate cancer, prostate carcinoma tumor antigen
Introduction
Prostate cancer is one of the most common cancers and among the leading causes of cancer-related death in the industrialized nations of the world, and its incidence and mortality have been increasing in developing countries as well.1 In 2011, approximately 240 890 new cases of prostate cancer will be diagnosed in the United States of America and 33 720 men will die from the disease.2 The introduction of serum prostate-specific antigen (PSA) in the 1980s has dramatically altered and benefited the initial diagnosis of men with prostate cancer. As a result of PSA screening, the lifetime risk of prostate cancer diagnosis has increased to 16% whereas the lifetime risk of dying from the disease is only 3.4%,3 indicating that intensive screening is likely to increase the detection of disease that may not be lethal. It is now obvious that overdetection of potentially indolent disease by the widespread use of PSA testing has resulted in overtreatment of the disease.4 Thus, a clinical dilemma today in the management of prostate cancer is to distinguish men with aggressive disease who need definitive treatment from men whose disease does not require such intervention.
Prognostic indicators of localized prostate cancer
Currently, at the time of diagnosis, most prostate cancer cases present as localized disease and are preferentially treated by radical prostatectomy or radiation therapy with curative intent. During the last decade, ‘stage migration' has been observed in the Western world, which is characterized by a significant shift towards localized, well-differentiated tumors at radical prostatectomy.5, 6 This phenomenon might be related to the widespread use of PSA for prostate cancer screening or perhaps a recent change in prostate cancer biology.7 Thus, prostate cancer detected by PSA alone is often characterized by small-size and low-grade tumors in relatively younger male populations. In fact, it is reported that around 30% of cancers treated with radical prostatectomy in the United States of America are ‘insignificant' tumors.8 On the other hand, nearly 30% are reported to experience an isolated increase in serum PSA with long-term follow-up.9, 10, 11, 12, 13, 14, 15 Han et al.16 reported that the incidence of biochemical recurrence was 17% in 2091 consecutive men who underwent radical prostatectomy and pelvic lymphadenectomy for clinically localized prostate cancer, and that the distant metastasis in these patients was 34%. Therefore, it is important for physicians and patients to know the likelihood of disease progression following radical prostatectomy.
Many clinical variables like Gleason score, tumor stage, margin status and PSA concentration, in various combinations, have been used for prediction of disease outcome.9, 10, 11, 12, 13, 14, 15, 17, 18, 19, 20 Although these clinicopathological variables are useful in predicting biochemical recurrence, these prognostic indicators do not accurately predict clinical outcome for individual patients.
Aside from these clinicopathological factors, several biomarkers have been evaluated during the past decade, including serum markers (hK2, uPA/uPAR, TGF-β1, IL-6), a urine marker (PCA3), and tissue markers such as proliferation index (Ki-67), cell cycle-related markers (p27, p21 and p16), apoptosis-related markers (p53, EZH2 and BCl-2), angiogenesis marker (microvessel density), genetic and epigenetic markers (TMPRSS2-ERG fusion, GSTP1 and APC) that have met with various degrees of success (reviewed by Agrawal et al.,21 Makarov et al.,22 Netto23 and Lopergolo et al.24). Although such molecular prognostic factors that complement classic clinicopathological factors may be useful, improved markers are still needed to enhance the accuracy with which patients at risk can be discerned and treated more aggressively.
Cancer/testis antigens (CTAs)
Beginning in the early 1990s, several tumor-associated antigen genes including the CTAs were identified that exhibited tumor-specific expression. The CTAs are a group of proteins that are typically restricted to the testis in the normal adult but are aberrantly expressed in several types of cancers.25 To date, more than 200 genes encoding CTAs have been identified that can be broadly divided into two groups based on their chromosomal location: the cancer/testis (CT)-X antigens located on the X chromosome and non-X CT antigens located on various autosomes. In the normal testis, the CT-X genes are generally expressed in the spermatogonia, the proliferating germ cells. In contrast, expression of non-X CT genes appears more dominant in the later stages of germ cell differentiation, such as in spermatocytes.26
According to the number of CT genes expressed and their expression frequency, tumor types can be grouped into three groups, namely: (i) high CT expressers including bladder cancer, melanoma, and non-small cell lung cancer; (ii) moderate CT expressers including breast cancer and prostate cancer; and (iii) low CT expressers including renal cell carcinoma and colon cancer.25 As a result of their restricted expression in an immune-privileged organ, the CTAs are widely recognized as attractive immunotherapeutic targets and several clinical trials with promising results have been reported.27 On the other hand, the CTAs also appear to serve as unique biomarkers for cancer diagnosis/prognosis due to their restricted expression patterns. Furthermore, members of the CT-X antigens in particular are typically associated with advanced cancer with poorer outcomes.28, 29, 30, 31, 32 Two dedicated databases published recently, CTpedia (http://www.cta.lncc.br/index.php) that catalogues the known CTAs33 and ACTAbase (http://actabase.jhu.edu) that also houses microarray gene expression data in addition, serve as excellent resources for detailed information regarding the CTAs (Parekh et al., in preparation).
CTAs as potential biomarkers in prostate cancer
Despite the attention the CTAs have received as unique biomarkers in several types of cancers, until recently, not much was known about their usefulness in prostate cancer. Earlier studies employing conventional and real-time PCR assays had showed that NY-ESO-1 mRNA was detected in 38% of tumor specimens, and 27% of stage C, 33% of stage D1 and 43% of stage D2 prostate cancers were positive, respectively.34 Further, NY-ESO-1 expression was also found in biopsies from 3% of localized prostate cancer and 15% of hormone refractory prostate cancer patients by immunohistochemistry.35 Lethe et al.36 reported that expression of MAGE-A1 was observed in 10.8% of carcinoma samples, whereas multi-MAGE-A and NY-ESO-1/LAGE-1 stained 85.9% and 84.8% of samples using immunohistochemistry, suggesting that a panel of CTAs rather than individual ones maybe more valuable as biomarkers. Recently, Smith et al.37 showed that SSX protein expression was restricted mainly to metastatic prostate cancer and not expressed in any primary prostate cancer using immunohistochemistry.
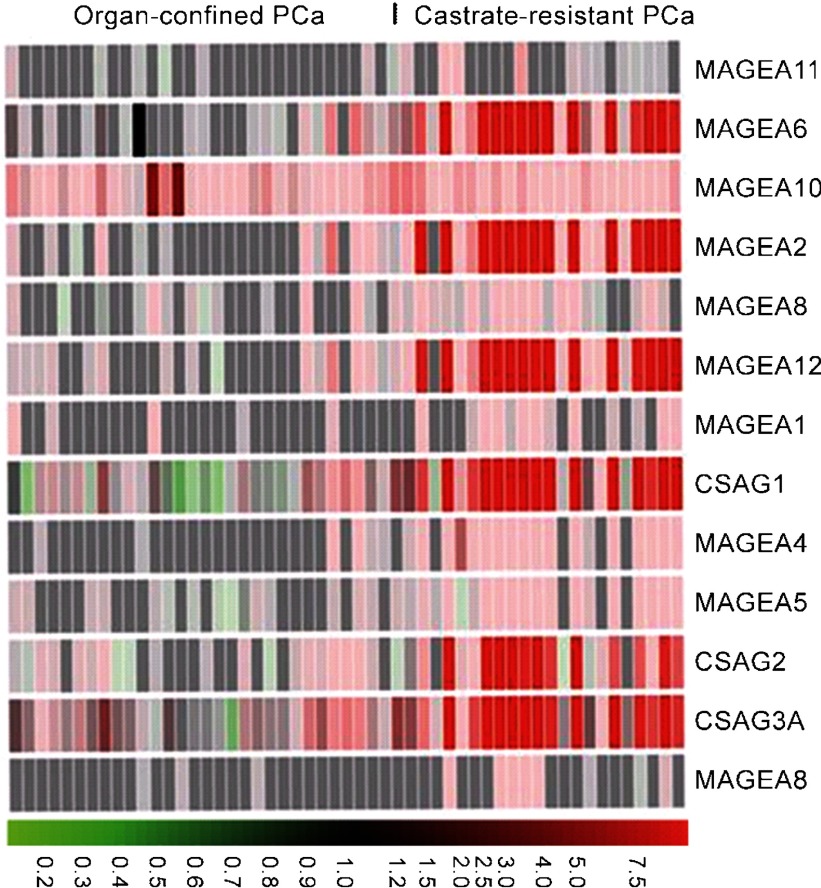
While these observations demonstrating the differential expression of specific CTAs during prostate cancer progression showed promise, they fell short of inspiring confidence in their potential use as biomarkers. Recently, Suyama et al.38 used a custom DNA microarray that was tiled with DNA probes representing a significant portion of all known CTAs to discern CTA expression patterns in prostate cancer. These studies revealed that: (i) numerous CTAs are upregulated in prostate cancer; (ii) a majority of them are CT-X antigens; and (iii) while several CT-X antigens from MAGEA/chondrosarcoma-associated gene (CSAG) subfamilies are coordinately upregulated in castrate-resistant prostate cancer but not in primary prostate cancer (Figure 1), PAGE4 is highly upregulated in primary prostate cancer, but is silent in castrate-resistant prostate cancer. These observations raised the possibility that the CTAs could potentially be used as biomarkers to distinguish men with aggressive disease who need treatment from men with indolent disease not requiring immediate intervention in prostate cancer.
Figure 1.
The MAGEA/CSAG cancer/testis antigen subfamilies are coordinately upregulated in castrate-resistant prostate cancer. Primary prostate cancer samples were obtained from radical prostatectomy while castrate-resistant prostate cancer were obtained from ‘rapid' autopsies and propagated as xenografts. RNA samples were run on a whole genome microarray. Heat map shows the predominant upregulation of MAGEA genes as well as the CSAG genes. The color display is on a 10-fold scale and red indicates overexpression relative to BPH. Green indicates downregulation. Adapted from Ref. 38. PCa, prostate cancer.
CTAs as potential biomarkers to discern aggressive phenotype of prostate cancer
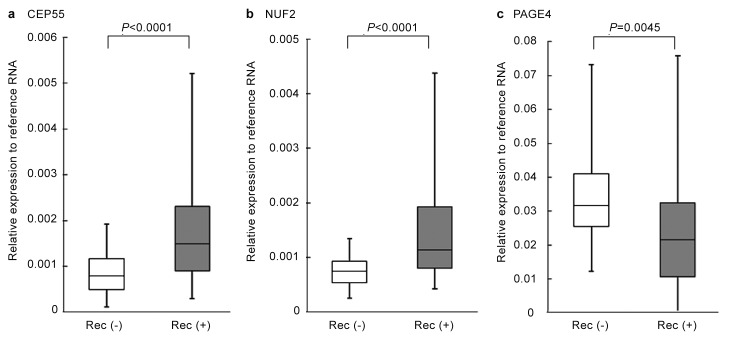
Inspired by this intriguing possibility, Shiraishi et al.39 devised a strategy to further explore the CTAs as potential biomarkers that can predict prostate cancer progression. We first mined publicly available microarray data on CTA expression in prostate cancer samples from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) in conjunction with our own data that we had previously obtained using a custom CT microarray38 and selected a panel of candidate CTAs for investigation. The representative microarray data (GEO, accession no. 1439) of the selected CTAs are listed in Table 1. A multiplex real-time PCR assay was developed and it was demonstrated that from this initial panel, four non-X CT antigens (CEP55, NUF2, PBK and TTK) were upregulated and as expected, the CT-X antigen, PAGE4, was downregulated in patients with recurrent prostate cancer after radical prostatectomy (Figure 2). Furthermore, Kaplan–Meier curves revealed that higher levels of expression of CEP55 and NUF2 were significantly correlated with shorter biochemical recurrence-free time. In contrast, higher expression of PAGE4 was significantly correlated with longer biochemical recurrence-free time (Figure 3). Further, with the exception of TTK, the other CTAs were significantly correlated with prostatectomy Gleason score, but none were correlated with age, preoperative PSA and tumor stage. Although Gleason score is believed to be one of the strongest predictors of recurrence, it tends to be subjective. Therefore, this quantitative multiplex real-time PCR assay developed by Shiraishi et al.39 could provide an objective assessment that is quantitative and consistent across institutions. Thus, this CTA-based ‘gene signature' appeared to have the potential to be a ‘molecular' Gleason score.
Table 1. Differential expression of the cancer/testis antigens in benign prostate, primary prostate cancer and metastatic prostate cancer.
| GEOa accession no. 1439 | Benign prostate(n=6) | Primary prostate cancer(n=7) | Metastatic prostate cancer(n=6) |
|---|---|---|---|
| SSX2 | 0.0 | 0.0 | 436.9 |
| CSAG2 | 0.0 | 0.0 | 1018.2 |
| MAGEA2 | 0.0 | 0.0 | 229.0 |
| MAGEA12 | 0.0 | 0.0 | 1139.7 |
| CEP55 | 0.0 | 56.0 | 640.2 |
| NUF2 | 14.1 | 32.3 | 1788.6 |
| PBK | 30.1 | 120.3 | 1411.5 |
| TTK | 83.8 | 216.0 | 1187.1 |
| PAGE4 | 6732.5 | 5702.8 | 0.0 |
| TBP | 608.7 | 577.6 | 882.2 |
| ACTB | 42572.0 | 41320.7 | 38735.7 |
GEO; The Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
Figure 2.
CTA expression in recurrent and non-recurrent prostate cancer. CTA expression of (a) CEP55, (b) NUF2 and (c) PAGE4 in clinically localized prostate cancer with recurrence (Rec (+)) (n=43) and without recurrence (Rec (−)) (n=29), were examined using multiplexed quantitative real time PCR. Adapted from Ref. 39. For details of the method, see Ref. 39. CTA, cancer/testis antigen.
Figure 3.
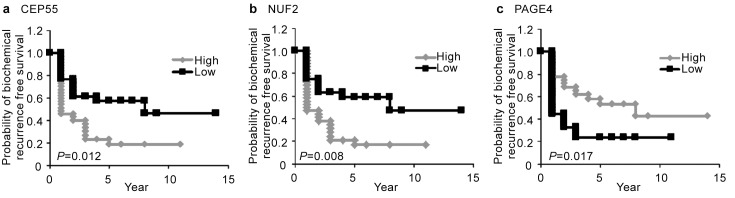
Kaplan–Meier analyses. Kaplan–Meier curves showing biochemical recurrence-free survival against time after radical prostatectomy stratified by the mRNA expression of (a) CEP55, (b) NUF2 and (c) PAGE4 (high versus low groups dichotomized by median value). Adapted from Ref. 39. For details of the method, see Ref. 39.
One of the advantages of using CTAs as biomarkers for prostate cancer is their potential as immunotherapeutic targets. Cancer immunotherapy is emerging as a promising modality for castration- and chemotherapy-resistant prostate cancer. Indeed, many CTAs are immunogenic and their use as therapeutic cancer vaccines is being evaluated. Thus, a CTA-based biomarker can provide information not only for predicting disease recurrence, but also suggesting treatment options, that can be personalized to the patient.
Despite the promise, several limitations to this study were outlined. The sample number was limited (n=72) and these patients were not consecutively and prospectively collected for this study. Furthermore, a high-risk cohort was used as a result of selection of specimens with large-volume tumors appropriate for frozen tissue collection, not reflecting contemporary, newly screened radical prostatectomy population.40, 41, 42 Furthermore, NY-ESO-1 which has been reported to be commonly expressed in later stages of the disease was not included in this study, because we did not observe its upregulation using our limited sample set. Although, in this study there was no significant difference in the CT-X antigens (SSX2, CSAG2, MAGEA2 and MAGEA12) between patients with or without recurrence, von Boehmer et al.43 recently demonstrated that the CT-X antigen MAGE-C2/CT10 may be a predictor of biochemical recurrence after radical prostatectomy, even though its expression was detected only in 3.3% of primary prostate cancer samples. Therefore, larger and contemporary cohorts presenting newly diagnosed radical prostatectomy population across multiple institutions are required for further validation of these results using a larger set of CTAs including CT-X antigens such as NY-ESO-1, SSX family and MAGEA family. This approach can be performed employing more robust mRNA-based assays such as the nCounter Gene Expression Assay (NanoString Technologies, Seattle, WA, USA), which is designed to provide a highly multiplexed method (up to 800 transcripts in one tube) and is reported to achieve superior gene expression quantification results when compared to real-time PCR. Taken together, accumulating evidences suggest that the CTAs have good potential to predict the risk of disease progression after radical prostatectomy.
Conclusion and future directions
Heterogeneity in prostate cancer is one of the major problems in developing reliable biomarkers. Therefore, a combination of markers is more likely to provide improved prognostication and treatment stratification rather than a single marker. In fact, there has already been some success in the identification of subtypes of prostate cancer based on derived sets of signature gene clusters.44, 45 Numerous studies reviewed here strongly suggest that CTAs are good candidates to establish a gene signature that can predict prostate cancer progression as a result of their advanced stage specific and coordinate expression pattern. In the future, CTA gene signatures might complement clinicopathological factors in the molecular differential diagnosis of aggressive and indolent prostate cancer.
Acknowledgments
This work was supported by NCI SPORE Grant 2P50CA058236–16 and the Bernard L. Schwartz Scholar Award by the Patrick C Walsh Cancer Research Fund (PK).
A provisional patent application covering this invention has been filed on behalf of Prakash Kulkarni, Takahito Suyama, Takumi Shiraishi and Robert H Getzenberg by Johns Hopkins University.
References
- Haas GP, Delongchamps N, Brawley OW, Wang CY, de la Roza G. The worldwide epidemiology of prostate cancer: perspectives from autopsy studies. Can J Urol. 2008;15:3866–71. [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- Ries LA, Wingo PA, Miller DS, Howe HL, Weir HK, et al. The annual report to the nation on the status of cancer, 1973–1997, with a special section on colorectal cancer. Cancer. 2000;88:2398–424. doi: 10.1002/(sici)1097-0142(20000515)88:10<2398::aid-cncr26>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, et al. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–83. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson RA, Stanford JL. Population-based prostate cancer trends in the United States: patterns of change in the era of prostate-specific antigen. World J Urol. 1997;15:331–5. doi: 10.1007/BF01300179. [DOI] [PubMed] [Google Scholar]
- Galper SL, Chen MH, Catalona WJ, Roehl KA, Richie JP, et al. Evidence to support a continued stage migration and decrease in prostate cancer specific mortality. J Urol. 2006;175:907–12. doi: 10.1016/S0022-5347(05)00419-2. [DOI] [PubMed] [Google Scholar]
- Polascik TJ, Oesterling JE, Partin AW. Prostate specific antigen: a decade of discovery—what we have learned and where we are going. J Urol. 1999;162:293–306. doi: 10.1016/s0022-5347(05)68543-6. [DOI] [PubMed] [Google Scholar]
- Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, et al. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–7. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- Pound CR, Partin AW, Epstein JI, Walsh PC. Prostate-specific antigen after anatomic radical retropubic prostatectomy. Patterns of recurrence and cancer control. Urol Clin North Am. 1997;24:395–406. doi: 10.1016/s0094-0143(05)70386-4. [DOI] [PubMed] [Google Scholar]
- Catalona WJ, Smith DS. 5-year tumor recurrence rates after anatomical radical retropubic prostatectomy for prostate cancer. J Urol. 1994;152:1837–42. doi: 10.1016/s0022-5347(17)32397-2. [DOI] [PubMed] [Google Scholar]
- Catalona WJ, Smith DS. Cancer recurrence and survival rates after anatomic radical retropubic prostatectomy for prostate cancer: intermediate-term results. J Urol. 1998;160:2428–34. doi: 10.1097/00005392-199812020-00012. [DOI] [PubMed] [Google Scholar]
- Ohori M, Goad JR, Wheeler TM, Eastham JA, Thompson TC, et al. Can radical prostatectomy alter the progression of poorly differentiated prostate cancer. J Urol. 1994;152:1843–9. doi: 10.1016/s0022-5347(17)32398-4. [DOI] [PubMed] [Google Scholar]
- Trapasso JG, deKernion JB, Smith RB, Dorey F. The incidence and significance of detectable levels of serum prostate specific antigen after radical prostatectomy. J Urol. 1994;152:1821–5. doi: 10.1016/s0022-5347(17)32394-7. [DOI] [PubMed] [Google Scholar]
- Zincke H, Oesterling JE, Blute ML, Bergstralh EJ, Myers RP, et al. Long-term (15 years) results after radical prostatectomy for clinically localized (stage T2c or lower) prostate cancer. J Urol. 1994;152:1850–7. doi: 10.1016/s0022-5347(17)32399-6. [DOI] [PubMed] [Google Scholar]
- Han M, Partin AW, Pound CR, Epstein JI, Walsh PC. Long-term biochemical disease-free and cancer-specific survival following anatomic radical retropubic prostatectomy. The 15-year Johns Hopkins experience. Urol Clin North Am. 2001;28:555–65. doi: 10.1016/s0094-0143(05)70163-4. [DOI] [PubMed] [Google Scholar]
- Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, et al. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169:517–23. doi: 10.1097/01.ju.0000045749.90353.c7. [DOI] [PubMed] [Google Scholar]
- Kattan MW, Eastham J. Algorithms for prostate-specific antigen recurrence after treatment of localized prostate cancer. Clin Prostate Cancer. 2003;1:221–6. doi: 10.3816/cgc.2003.n.003. [DOI] [PubMed] [Google Scholar]
- Stephenson AJ, Scardino PT, Eastham JA, Bianco FJ, Jr, Dotan ZA, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol. 2005;23:7005–12. doi: 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattan MW, Vickers AJ, Yu C, Bianco FJ, Cronin AM, et al. Preoperative and postoperative nomograms incorporating surgeon experience for clinically localized prostate cancer. Cancer. 2009;115:1005–10. doi: 10.1002/cncr.24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat SF, Karakiewicz PI, Roehrborn CG, Kattan MW. An updated catalog of prostate cancer predictive tools. Cancer. 2008;113:3075–99. doi: 10.1002/cncr.23908. [DOI] [PubMed] [Google Scholar]
- Agrawal S, Dunsmuir WD. Molecular markers in prostate cancer. Part I: predicting lethality. Asian J Androl. 2009;11:14–21. doi: 10.1038/aja.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov DV, Loeb S, Getzenberg RH, Partin AW. Biomarkers for prostate cancer. Annu Rev Med. 2009;60:139–51. doi: 10.1146/annurev.med.60.042307.110714. [DOI] [PubMed] [Google Scholar]
- Netto GJ. Molecular diagnostics in urologic malignancies: a work in progress. Arch Pathol Lab Med. 2011;135:610–21. doi: 10.5858/2010-0727-RAIR.1. [DOI] [PubMed] [Google Scholar]
- Lopergolo A, Zaffaroni N. Biomolecular markers of outcome prediction in prostate cancer. Cancer. 2009;115:3058–67. doi: 10.1002/cncr.24346. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Brichard VG, Lejeune D. GSK's antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine. 2007;25 Suppl 2:B61–71. doi: 10.1016/j.vaccine.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res. 2005;11:8055–62. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- Velazquez EF, Jungbluth AA, Yancovitz M, Gnjatic S, Adams S, et al. Expression of the cancer/testis antigen NY-ESO-1 in primary and metastatic malignant melanoma (MM)-correlation with prognostic factors. Cancer Immun. 2007;7:11. [PMC free article] [PubMed] [Google Scholar]
- Andrade VC, Vettore AL, Felix RS, Almeida MS, Carvalho F, et al. Prognostic impact of cancer/testis antigen expression in advanced stage multiple myeloma patients. Cancer Immun. 2008;8:2. [PMC free article] [PubMed] [Google Scholar]
- Napoletano C, Bellati F, Tarquini E, Tomao F, Taurino F, et al. MAGE-A and NY-ESO-1 expression in cervical cancer: prognostic factors and effects of chemotherapy. Am J Obstet Gynecol. 2008;198:99.e1–7. doi: 10.1016/j.ajog.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Grigoriadis A, Caballero OL, Hoek KS, da Silva L, Chen YT, et al. CT-X antigen expression in human breast cancer. Proc Natl Acad Sci USA. 2009;106:13493–8. doi: 10.1073/pnas.0906840106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816–9. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada T, Noguchi Y, Satoh S, Ono T, Saika T, et al. NY-ESO-1 mRNA expression and immunogenicity in advanced prostate cancer. Cancer Immun. 2003;3:10. [PubMed] [Google Scholar]
- Fossa A, Berner A, Fossa SD, Hernes E, Gaudernack G, et al. NY-ESO-1 protein expression and humoral immune responses in prostate cancer. Prostate. 2004;59:440–7. doi: 10.1002/pros.20025. [DOI] [PubMed] [Google Scholar]
- Hudolin T, Juretic A, Spagnoli GC, Pasini J, Bandic D, et al. Immunohistochemical expression of tumor antigens MAGE-A1, MAGE-A3/4, and NY-ESO-1 in cancerous and benign prostatic tissue. Prostate. 2006;66:13–8. doi: 10.1002/pros.20312. [DOI] [PubMed] [Google Scholar]
- Smith HA, Cronk RJ, Lang JM, McNeel DG. Expression and immunotherapeutic targeting of the SSX family of cancer-testis antigens in prostate cancer. Cancer Res. 2011;71:6785–95. doi: 10.1158/0008-5472.CAN-11-2127. [DOI] [PubMed] [Google Scholar]
- Suyama T, Shiraishi T, Zeng Y, Yu W, Parekh N, et al. Expression of cancer/testis antigens in prostate cancer is associated with disease progression. Prostate. 2010;70:1778–87. doi: 10.1002/pros.21214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi T, Terada N, Zeng Y, Suyama T, Luo J, et al. Cancer/testis antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy. J Transl Med. 2011;9:153. doi: 10.1186/1479-5876-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Partin AW, Mangold LA, Epstein JI, Walsh PC. Probability of biochemical recurrence by analysis of pathologic stage, Gleason score, and margin status for localized prostate cancer. Urology. 2003;62:866–71. doi: 10.1016/s0090-4295(03)00674-5. [DOI] [PubMed] [Google Scholar]
- Hull GW, Rabbani F, Abbas F, Wheeler TM, Kattan MW, et al. Cancer control with radical prostatectomy alone in 1,000 consecutive patients. J Urol. 2002;167:528–34. doi: 10.1016/S0022-5347(01)69079-7. [DOI] [PubMed] [Google Scholar]
- Roehl KA, Han M, Ramos CG, Antenor JA, Catalona WJ. Cancer progression and survival rates following anatomical radical retropubic prostatectomy in 3,478 consecutive patients: long-term results. J Urol. 2004;172:910–4. doi: 10.1097/01.ju.0000134888.22332.bb. [DOI] [PubMed] [Google Scholar]
- von Boehmer L, Keller L, Mortezavi A, Provenzano M, Sais G, et al. MAGE-C2/CT10 protein expression is an independent predictor of recurrence in prostate cancer. PLoS ONE. 2011;6:e21366. doi: 10.1371/journal.pone.0021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney KL, Sinnott JA, Fall K, Pawitan Y, Hoshida Y, et al. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J Clin Oncol. 2011;29:2391–6. doi: 10.1200/JCO.2010.32.6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–55. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]