Abstract
We investigated the importance of HMGN5, a nuclear protein that binds to nucleosomes, unfolds chromatin, and affects transcription, in the LNCaP prostate cancer cell line. We also examined the molecular mechanisms that promote apoptosis of LNCaP cells after infection with small interfering RNA (siRNA) targeting HMGN5 (siRNA-HMGN5). The androgen-dependent LNCaP human prostate cancer cells were infected with siRNA-HMGN5. Apoptosis was detected using the Annexin V-PE/7-AAD double staining and the terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) assays. Mitochondrial membrane potential was measured by JC-1 staining. HMGN5 and GAPDH mRNA expression were determined using real-time PCR. Bcl-2 and other apoptosis-related protein levels were determined by Western blot analysis. Caspase activity was measured by cleavage of the caspase substrate. Infection with siRNA targeting HMGN5 efficiently and specifically reduced the HMGN5 expression in LNCaP cells. The downregulation of HMGN5 induced remarkable apoptosis of LNCaP cells and resulted in the reduction of mitochondrial membrane potential. The induction of cell apoptosis was accompanied by the upregulation of Bax, the Bax/Bcl-2 ratio and the activation of caspase3. The HMGN5-targeted siRNA was effective in downregulating the expression of HMGN5 in androgen-dependent prostate cancer cells and inducing cell apoptosis via the regulation of a caspase-related mitochondrial pathway and Bcl-2 family proteins. This study suggests that HMGN5 may be a potential molecular target with therapeutic relevance for the treatment of prostate cancer.
Keywords: apoptosis, HMGN5, mitochondrial pathway, prostate cancer, siRNA
Introduction
Prostate cancer is one of the most frequently diagnosed cancers and a major cause of cancer related deaths in males worldwide.1 Most patients are now diagnosed with low-risk, clinically localized cancer that can be treated effectively with surgery and radiation.2 Nevertheless, a number of patients' conditions could progress despite surgery, radiation and androgen deprivation therapy. Furthermore, advanced prostate cancer still remains a significant treatment challenge.3
It is gratifying to observe that research dedicated to understanding the biology of prostate cancer has increased exponentially. Apoptosis is now recognized as a critical process for the normal development of multicellular organisms and plays a major role in homeostasis. The disruption of this homeostasis can lead to the development of cancer.4 The pathways of cellular proliferation and apoptosis appear to be linked to minimize the occurrence of neoplasia. Deregulation of apoptotic signaling pathways may suggest a mechanism to promote tumorigenesis by preventing the elimination of abnormal cells through checkpoint control.5
HMGN5, previously known as NBP-45, GARP45 and NSBP1, is a new member of the HMGN protein family. The gene coding for HMGN5 is located at the Xq13.3 region of the human genome (XD in the mouse genome). HMGN5 is localized to the nucleus and contains a functional nucleosome-binding domain and negatively charged C-terminus.6, 7 The C-terminus of HMGN5 is unusually long and affects the cellular localization and architectural properties of the protein. HMGN5 proteins move notably rapidly throughout the nucleus and transcription changes induced by HMGN5 are linked to its ability to interact with nucleosomes.8
Aberrant expression of HMGN proteins is associated with developmental defects, hypersensitivity to stress and increased tumorigenic potential.9 Upregulation of HMGN5 proteins was observed in human prostate cancer,10 glioma cells,11 squamous-cell carcinoma,12 as well as adenocarcinoma in aging mice,13 suggesting a role of HMGN5 in tumorigenesis. We have shown that the expression of HMGN5 was significantly upregulated in prostatic carcinoma, suggesting the possible role of HMGN5 in the development and metastasis of prostate cancer.14 Furthermore, it has been demonstrated that downregulation of HMGN5 decreases the proliferation of prostate cancer cells both in vitro and in vivo.10, 15, 16
Due to the diversity of advanced prostate cancers and its capacity to adapt to changing conditions, combining targeted agents in an effort to inhibit multiple pathways would be the best approach.17 Downregulation of anti-apoptotic proteins by RNA interference is a promising tool for the prevention of tumor growth.18 We constructed a lentiviral vector system containing small interfering RNA (siRNA) sequences that target the human HMGN5 transcript. Infection of DU145, an androgen-independent metastatic prostate cancer cell line, showed that HMGN5 played a role in the cell cycle, cell proliferation and apoptosis in androgen-independent prostate cancer cells.15 However, the gene-silencing effects of HMGN5 RNA interference (RNAi) on androgen-dependent cells and the possible mechanisms of inducing apoptosis are not well known yet.
Therefore, this study was designed to investigate the anti-cancer potential of siRNA targeting HMGN5 in androgen-dependent LNCaP cells and the possible molecular mechanisms of HMGN5.
Materials and methods
Cell culture
All cell culture was performed according to the previously described protocols.15, 19 The RWPE-1, DU145, PC-3 and LNCaP cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). The RWPE-1 cells were grown in Keratinocyte serum-free medium supplemented with 0.05 mg ml−1 bovine pituitary extract and 5 ng ml−1 epidermal growth factor (Invitrogen, Carlsbad, CA, USA) in 5% CO2 atmosphere at 37 °C. The LNCaP, DU145 and PC3 cells were cultured in RPMI 1640, and supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum (Invitrogen).
siRNA lentiviral vector infection
HMGN5 siRNA sequences and constructions of lentivirus were identical to those used in previous studies.15 The most effective double-stranded HMGN5-targeted siRNAs, PscSI616 (5′-CACAGCCTTTCTTTAGCAT-3′), were synthesized by GeneChem Corporation (Shanghai, China). A double-stranded siRNA with no sequence homology to any known human genes, Psc-NC (5′-TTCTCCGAACGTGTCACGT-3′), was also generated as a negative control. For gene silencing, purified lentivirus (PscSI616) was added to cells at a multiplicity of infection of 50 for 8 h and was washed twice with medium. Infection with a multiplicity of infection of 50 resulted in >90% infection of LNCaP cells after 72 h, as monitored by GFP expression. Therefore, we used a multiplicity of infection of 50 for the lentivirus in all of the experiments, as it yielded optimal knockdown of the gene in the required time. Control cells were infected with a negative control siRNA (Psc-NC), as a vector control according to the same protocol. A third set of infections was performed with no siRNA (mock).
Real-time PCR
RNA from cultured cells was isolated using a TRIzol reagent (Invitrogen) by following the manufacturer's protocol. The integrity of the RNA was checked using the Nano Drop (Agilent Technologies, Santa Clara, CA, USA). First-strand cDNA was generated using 2 µg of total RNA via MMLV-reverse transcriptase. Real-time PCR was performed using an Applied Biosystems 7300 Fast Real-time PCR System (SYBR Green PCR Master Mix; Applied Biosystems, Foster City, CA, USA). The primer sequences for real-time PCR of HMGN5 were as follows: 5′-GCAGTCAGGCAGTGACTGCCTTCG-3′ (forward) and 5′-CCCTTTTCTGTGGCATCTTC-3′ (reverse). The primers for human GAPDH were as follows: 5′-CAGTCAGCCGCATCTTCTTTT-3′ (forward) and 5′-GTGACCAGGCGCCCAATAC-3′ (reverse). All reactions were performed in triplicate, and a negative control lacking cDNA was included. Human GAPDH was used to normalize the data for quantification of HMGN5 mRNA using the delta-delta Ct method.
Cell viability assay
Cell viability was determined using the WST-8 assay (Keygen, Shanghai, China). The WST-8 assay was used according to the manufacturer's instructions. LNCaP cells were seeded into a 96-well plate at a concentration of 1×106 cells ml−1 and cultured for 24 h. After 24 h of infection, 10 µl of WST-8 solution was added to each well, and the plates were incubated for 4 h at 37 °C. The absorbance value was measured at 450 nm using a 96-well spectrophotometer (BioRad, Inc., Hercules, CA, USA).
Quantitation of apoptosis
Flow cytometry.
Cells were cultured in six-well plates at a concentration of 2×105 cells per well and infected with lentivirus after culturing for 24 h. After 72 h of infection, cells were stained by Annexin V-PE and 7-AAD in binding buffer using the Annexin V-PE/7-AAD Kit (Keygen), as previously described.20 After incubation at room temperature for 15 min, cells were analyzed using a BD FACStar flow cytometer (Becton Dickinson, San Jose, CA, USA).
Mitochondrial damage measurement.
JC-1 dye was used to evaluate mitochondrial damage. LNCaP cells infected with HMGN5-targeted siRNA were incubated with 5 g ml−1 JC-1 (Cayman, Ann Arbor, MI, USA) at 37 °C for a further 30 min. After washing the cells twice with phosphate-buffered saline (PBS), the cells were imaged using laser confocal fluorescence microscopy. The incorporated JC-1 was excited at 488 nm and detected at both 590 nm (orange) and 527 nm (green) fluorescence using a confocal laser-scanning microscope (Fluoview FV500; Olympus, Tokyo, Japan). In fluorescence microscopy images, orange dot-like images reflecting JC-1 aggregation within the mitochondria were observed in cells with high mitochondrial membrane potential. In the cells with low mitochondrial membrane potential, the orange color changed into diffuse green fluorescence, reflecting the monomeric state of JC-1.21
Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling (TUNEL) assay.
The TUNEL technique was performed using the In Situ Cell Death Detection Kit Fluorescein (Roche Diagnostic, Mannheim, Germany) according to the manufacturer's instructions. Cells were seeded on cover slips in six-well plates (5×104 per cell). Twenty-four h later, LNCaP cells were infected with lentiviral vectors expressing HMGN5-targeted siRNA. In brief, after an indicated duration of infection, cells were fixed using 10% buffered formalin, washed with PBS, permeabilized in 0.2% Triton X-100, washed again in PBS, and equilibrated with the equilibration buffer. The labeling reactions were carried out by addition of the rTdT enzyme for 1 h. The signals were subsequently detected by incubation of the cells with streptavidin HRP and DAB components until a light brown background was visualized. The apoptotic index was determined at ×200 magnification as the proportion of TUNEL-positive cells relative to the total number of cells per section.
Caspase activity
To quantify caspase3 activity, 20 000 cells ml−1 were plated onto a 96-well plate, and after 24-h adherence and medium replacement, LNCaP cells were infected with lentivirus for 72 h. Caspase3 activity was assessed using the caspase3/caspase7 Luminescent Assay Kit (Promega, Madison, WI, USA). Photon flux values were measured using a luminescent meter.
Western blot analysis
Whole cell lysates from cultured cells were prepared by homogenization in a lysis buffer containing phosphatase and protease inhibitors. Western blot assays were performed according to the previously described protocols.15 A specific antibody against HMGN5 (NSBP1) was obtained from the Prostate Disease Prevention and Cure Center of Jilin University (Jilin, China). Anti-Bcl-2, Bcl-xl, Bax, Bid, PARP, AIF, ENDO G and caspase3 antibodies (Cell Signal Tech, Beverly, MA, USA) and an anti-β-actin polyclonal antibody (sc-1616-R; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used as primary antibodies. After incubation with an HRP-conjugated secondary antibody, immunoreactive proteins were visualized using the ECL detection system. Semi-quantitative analysis of the results was enumerated by densitometry of the bands (Quantity One; BioRad, Inc.).
Statistical analysis
Data are expressed as mean±s.e.m. All data groups were analyzed by one-way ANOVA using the software SPSS v13.0 (SPSS Inc., Chicago, IL, USA). A P value <0.05 was used to determine the statistical significance when interpreting the results.
Results
High expression of HMGN5 in prostate cancer cell lines
The expression of HMGN5 in prostate cell lines was determined using real-time PCR and western blot analysis. As shown in Figure 1a and b, HMGN5 is highly expressed in LNCaP, PC-3 and DU145 cell lines compared to normal prostate derived epithelial cell line (RWPE-1) (P<0.05). Among these cell lines, we chose LNCaP (androgen-dependent) as our study object.
Figure 1.
The expression profile of HMGN5 in different prostate cell lines and siRNA-HMGN5 expression in LNCaP cells. (a) The expression level of HMGN5 in RWPE-1 cells was treated as the baseline, and GAPDH was used as the internal control (*P<0.05 compared to RWPE-1 cell line). (b) Western blots for HMGN5 in RWPE-1, PC-3, LNCaP and DU145 cell lines. β-actin was used as an internal control, semiquantitative analysis of the results was enumerated by densitometry of the bands. (c) The expression level of HMGN5 mRNA in LNCaP cells. GAPDH was used as an internal control to normalize the data for quantification of HMGN5 mRNA (*P<0.05 compared to the mock or the control group). (d) Western blot of HMGN5 in LNCaP cells. β-actin was used as an internal control. Corresponding densitometry of each band is presented in a bar graph. Each assay was performed in duplicate (*P<0.05 compared to the mock or the control group). mRNA, messenger RNA.
HMGN5-targeted siRNA reduced HMGN5 expression in LNCaP cells
The gene-silencing effect mediated by siRNA specifically targeting HMGN5 in LNCaP cells was confirmed by real-time PCR and Western blot analysis. Approximately 72 h after infection, HMGN5 transcript (Figure 1c) and HMGN5 protein levels (Figure 1d) were reduced remarkably in infected cells. This HMGN5 gene silencing was reproducible and specific, because HMGN5-targeted siRNA failed to knockdown the expression of an unrelated protein, β-actin. A semiquantitative analysis of the Western blot assay results showed that HMGN5-targeted siRNA (siRNA-HMGN5) decreased the expression levels of HMGN5 in LNCaP cells by approximately six fold (Figure 1d).
HMGN5-targeted siRNA decreased cell proliferation
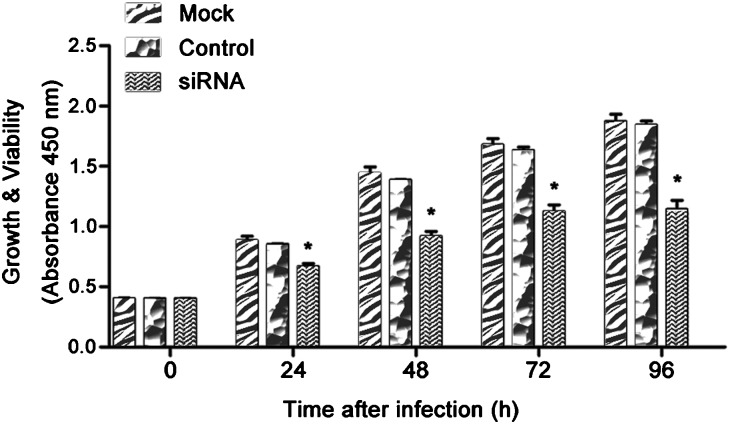
Following knockdown of the HMGN5 gene with siRNA, we measured the viability of infected LNCaP cells. Compared to control cultures, cultures infected with siRNA-HMGN5 showed significantly decreased cell viability. The cell viability was shown in Figure 2 (72 h, 70.7% of the control group, P<0.05; 96 h, 68.2% of the control group, P<0.05). These studies showed that knockdown of HMGN5 resulted in decreased proliferation and survival of LNCaP cells.
Figure 2.
The siRNA-HMGN5 treatment decreased cell proliferation and increased apoptosis. LNCaP cells were infected with siRNA, and the cell viability was assessed using the CCK-8 assay at five time points. Each sample was tested in triplicate (*P<0.05 compared to the mock or the control group). siRNA, small interfering RNA.
HMGN5-targeted siRNA induced cell apoptosis
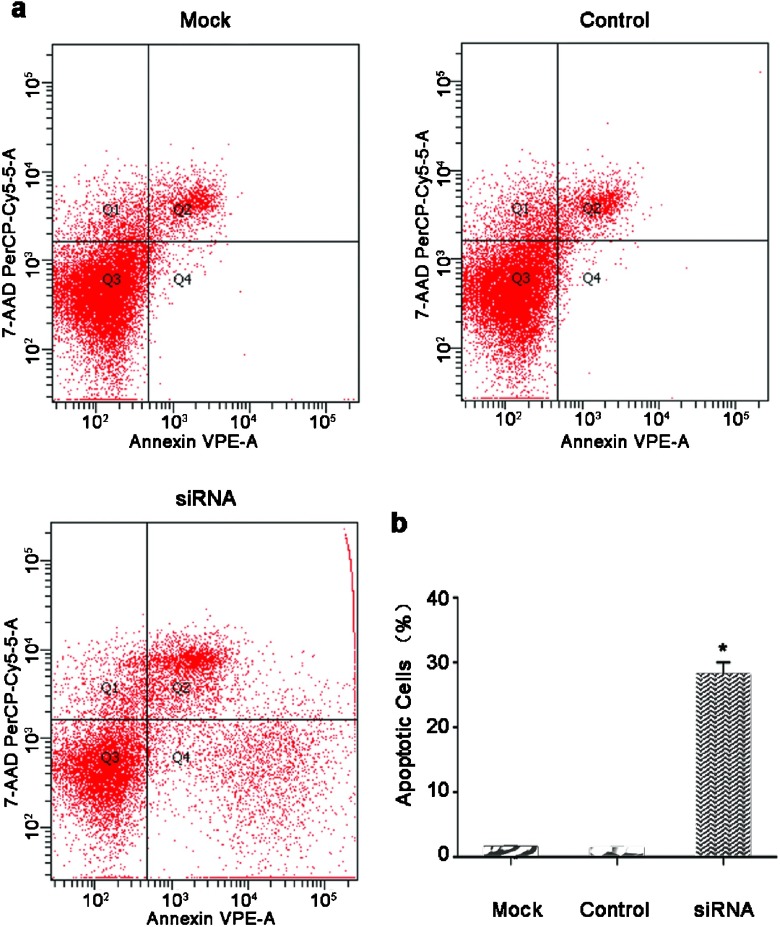
The siRNA-HMGN5-infected and mock-infected LNCaP cells were stained with Annexin V-PE and 7-AAD to distinguish between apoptotic cells and cells undergoing necrotic death. Stained cells were analyzed by flow cytometry (Figure 3a). The data showed that siRNA-HMGN5 infection of LNCaP cells resulted in increased apoptosis (1.4% for the mock and 1.3% for the control group vs. 28.1% for the siRNA-HMGN5 group) (Figure 3b). Significantly lower levels of apoptosis were observed in control-infected and mock-infected cells compared with siRNA-HMGN5-infected cells (P<0.05).
Figure 3.
siRNA-HMGN5 induced apoptosis in LNCaP cells. (a) Cells were trypsinized and stained with Annexin V-PE/7-AAD followed by analysis using flow cytometry. Early apoptotic cells (Annexin V-PE positive and 7-AAD negative) are in the right lower quadrant. The percentage of Annexin V-PE/7-AAD stained cells was determined by flow cytometry. (b) Early apoptosis is expressed as a percentage of the total number of cells counted. The results are expressed as mean±s.e.m. from three separate experiments (*P<0.05 compared to the mock or the control group). siRNA, small interfering RNA.
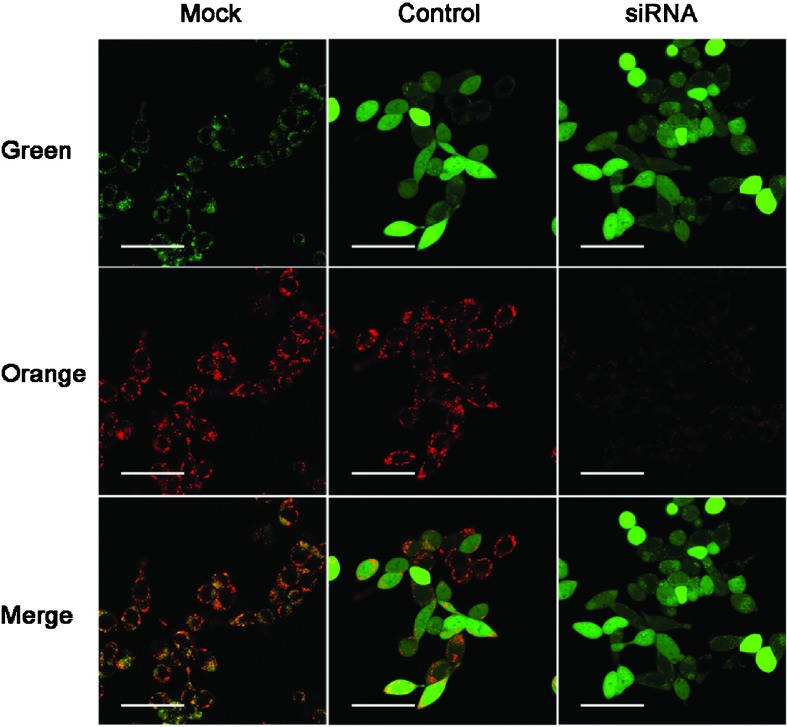
To assess mitochondrial damage, JC-1 staining was performed and showed that infection with siRNA-HMGN5 decreased mitochondrial membrane potential. The obvious decrease of orange fluorescence indicated mitochondrial dysfunction (Figure 4).
Figure 4.
Fluorescence microscopy of JC-1-stained LNCaP cells. Change in mitochondrial membrane potential was determined by flow cytometry analysis using the JC-1 dye (scale bars=10 µm). The lentiviral vector stably expressed the siRNA and a marker (a GFP–RFP fusion protein) that resulted in the green channel, the control group and the siRNA group being much brighter than the mock group. Only the orange channel was considered. siRNA, small interfering RNA.
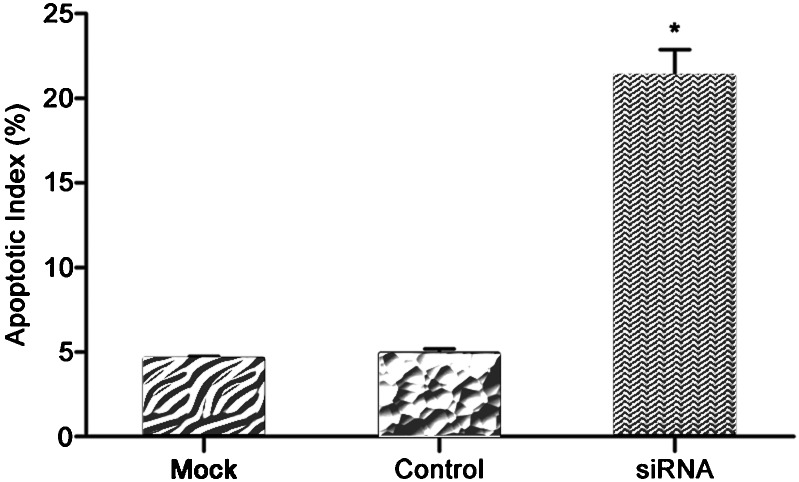
Furthermore, compared to the control treatment, siRNA-HMGN5 infection resulted in an increased number of apoptotic LNCaP cells (TUNEL-positive); the apoptosis index score was significantly higher in infected LNCaP cells (4.5% for the mock and 4.8% for the control group vs. 21.25% for the siRNA-HMGN5 group, respectively (P<0.05, Figure 5).
Figure 5.
In TUNEL assay of siRNA-HMGN5 infection in LNCaP cells, apoptotic cells were counted in randomly selected fields. Each bar represents the apoptotic index of the corresponding group from three independent experiments (*P<0.05 compared to the mock or the control group). siRNA, small interfering RNA.
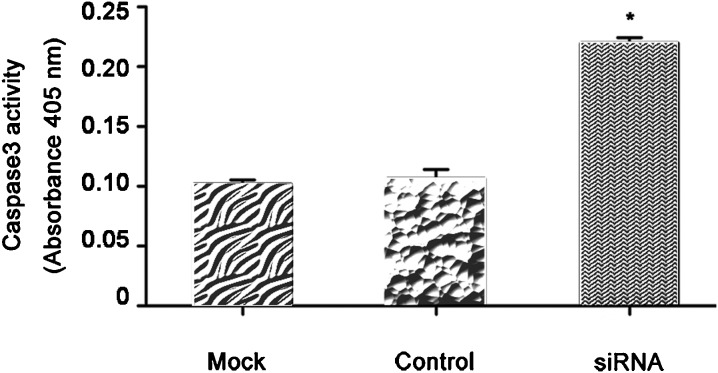
HMGN5-targeted siRNA activated caspase activity
To determine whether caspase3 is activated in siRNA-HMGN5 infected LNCaP cells, we used a chromogenic caspase3 assay. The results show that siRNA-HMGN5 infection of LNCaP cells induced activation of caspase3 compared to mock-infected or control cells (P<0.05, Figure 6).
Figure 6.
siRNA-HMGN5 activated caspase activity in LNCaP cells. The LNCaP cell protein extracts were analyzed for caspase3 levels, which were represented as absorbance values. Each value is mean ± SEM of six independent observations (*P<0.05 compared to the mock or the control group).
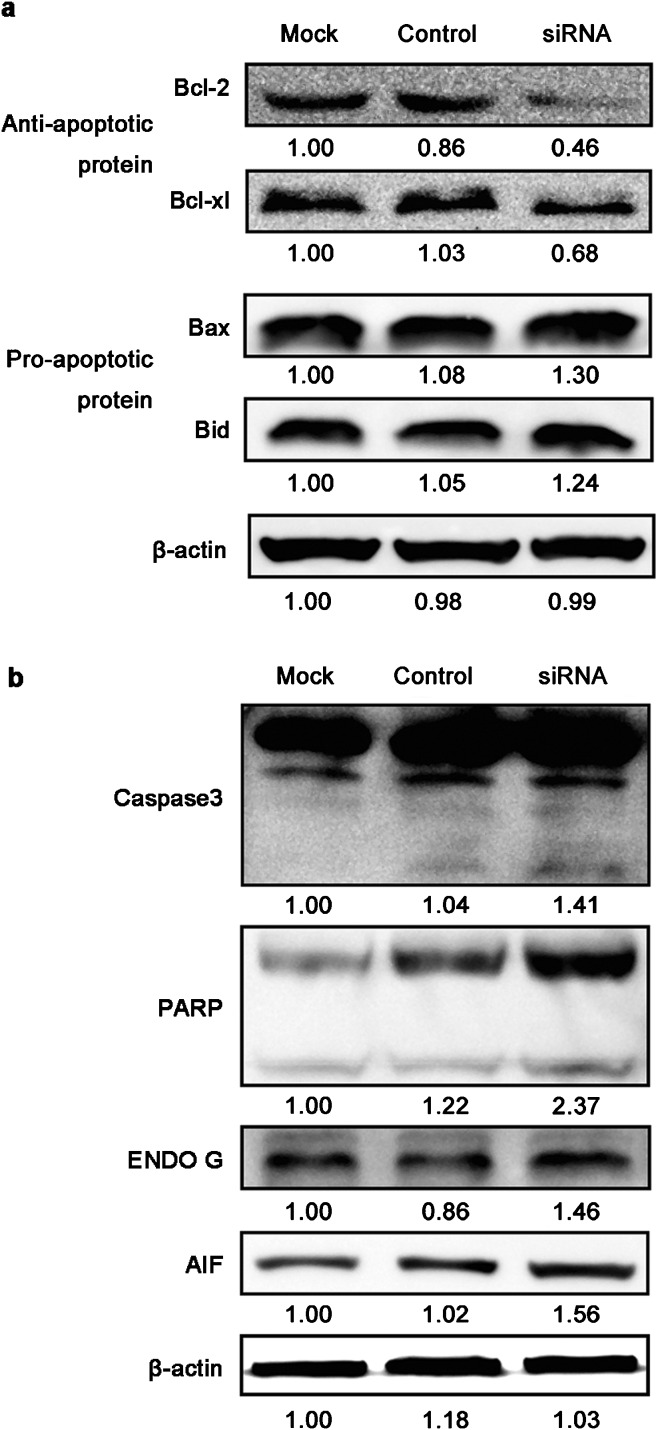
The effect of HMGN5-targeted siRNA on the expression of apoptosis-associated proteins in LNCaP cells
In order to verify whether the siRNA-HMGN5-induced apoptotic cell death observed was a result of expression changes of apoptosis associated proteins, a series of Western blot assays were conducted. The siRNA-HMGN5 infection increased the levels of Bax, Bid, caspase3, PARP, AIF and ENDO G, but decreased the levels of Bcl-2 and Bcl-xl, which mediate the apoptosis of LNCaP cells (Figure 7).
Figure 7.
The effect of siRNA-HMGN5 on the expression of apoptosis associated proteins in LNCaP cells. LNCaP cells were infected with siRNA-HMGN5, and representative blots are shown of (a) Bcl-2, Bcl-xl, Bax, Bid and (b) Caspase3, PARP, ENDO G and AIF. β-actin served as an internal control. The values were enumerated by densitometry of the bands. siRNA, small interfering RNA.
Discussion
In this study, we described the use of a lentiviral siRNA vector targeting HMGN5 expression in the LNCaP cell line, an in vitro prostate cancer model system. Results from our study showed that HMGN5 is highly expressed in prostate cancer cells compared to normal prostate-derived epithelial cells (RWPE-1). This finding is in agreement with reports of HMGN5 expression in other cancers. Several therapeutic strategies have been used to downregulate gene expression in cancer cells. Among these approaches, the RNAi-based techniques for silencing specific genes are used more widely.22 The combination of RNAi and lentiviral transgenesis has been demonstrated to result in efficient and stable knockdown of target genes both in vitro and in vivo.23
Disruption of HMGN5 results in potent anti-tumor effects in an in vivo mouse model using DU145 cells (androgen-independent prostate cancer cells),15 suggesting that targeting of HMGN5 inhibits DU145 cell proliferation in vivo. Because the expression of HMGN5 in LNCaP cells (androgen-dependent prostate cancer cells) was much higher than in DU145 cells, we hypothesized that downregulating the expression of HMGN5 by an RNAi approach would affect the apoptosis of LNCaP cells. As expected, results from our studies showed that this approach decreased the levels of transcribed mRNA and HMGN5 protein levels in infected LNCaP cells, thereby leading to increased cellular apoptosis, as indicated by Annexin V staining and TUNEL assays.
New insights into the mechanisms of action and cellular functions of HMG proteins are emerging from imaging studies of their interactions with chromatin in living cells and from analyses of organisms in which their expression levels have been altered. HMGN5 is widely expressed in mouse tissues, affects the binding of linker histones to chromatin and destabilizes the compact structure of the chromatin fiber. Most likely, these changes in chromatin architecture lead to alterations in the cellular transcription profile.8 In this study, we focus on the roles of HMGN5 in modulating cellular proliferation and apoptosis.
Apoptosis is a controlled physiological process that is inhibited in cancer cells. Many molecules have been reported that are linked with the regulation of apoptosis, of which bcl-2 and bax genes are thought to play major roles in determining cell survival or death after apoptotic stimuli.24 Several high-throughput studies have confirmed that Bcl-2 is overexpressed in clinically localized prostate cancer compared with benign prostatic tissue.25 Apoptosis is regulated through a diverse range of cellular signals. Overall, these signals can be divided into two pathways, namely, the extrinsic and the intrinsic apoptotic pathways. The extrinsic apoptotic pathway is mediated by death receptors. The activation of death receptors can set in motion the caspase cascade, such as caspase8, and finally lead to the activation of caspase3. The intrinsic apoptotic pathway is initiated by the loss of mitochondrial membrane potential and the subsequent release of cytochrome c from the mitochondria into the cytosol. This release of cytochrome c results in the activation of caspase9, which leads to the activation of caspase3.26
Caspase3 activation is a critical event in cancer cell death. In our studies, we found that the knockdown of HMGN5 elevated caspase3 activity in LNCaP cells. Furthermore, siRNA-HMGN5 reduced mitochondrial membrane potential and led to the release of AIF and ENDO G through the mitochondria-mediated intrinsic pathway.
In this study, we demonstrated that siRNA-HMGN5 promoted the levels of Bax and Bid but suppressed the levels of Bcl-2 and Bcl-xl (Figure 7a). Our Western blot assays (Figure 7a) also indicated that siRNA-HMGN5 increases apoptosis in LNCaP cells through the mitochondria-mediated intrinsic pathway release of AIF and ENDO G. These molecules may have a relationship with the alterations in the cellular transcription profile caused by the downregulation of HMGN5. Further research is required to examine this potential fully.
Our results also show that the apoptosis caused by siRNA-HMGN5 is more severe in androgen-dependent cell lines than in previous studies. This phenomenon suggests that there may be connections between HMGN5 and the androgen receptor. Because HMGN5 continuously modulates the local and global architecture of the chromatin fiber and affects the binding of regulatory factors to their nucleosomal targets, based on previous studies, we believe that HMGN5 may either increase the transcriptional activity of the receptor-hormone complex or just affect the basal promoter activity of the androgen receptor gene.27 The relationship of the androgen receptor with HMGN5 should be investigated in our future studies.
Although the function of HMGN5 in prostate cancer cells is not well known, it has been previously established that HMGN5 plays an important role in maintaining the integrity of the chromatin fiber in cells, and its aberrant expression is deleterious for cell fate. HMGN5 also has a potential association with cancer progression. Currently, there are no drugs available that suppress HMGN5. If direct antagonists prove elusive, an alternative therapeutic strategy might be developed to target the HMGN5 suppressor and/or its unique co-regulators. Therefore, targeting HMGN5 could be a beneficial therapeutic approach for prostate cancer treatment.
In summary, according to the current results, we concluded that HMGN5 functions as an oncogene by suppressing apoptosis of prostate cancer cells. HMGN5 knockdown in LNCaP cells resulted in decreased cell viability and remarkably increased cell apoptosis. Cells that were manipulated in this way underwent cell death via apoptosis. Analysis of the apoptotic mediators suggests the involvement of caspase3 activation, the mitochondrial pathway and the Bcl-2 family in HMGN5 gene silencing. Therefore, silencing the HMGN5 gene by siRNA could be a therapeutic approach for prostate cancer treatment.
Author contributions
XYZ carried out the molecular genetic studies, participated in the sequence alignment and drafted the manuscript. ZQG and MZ cultured cells. SQJ and NJ participated in the sequence alignment. XSL participated in the design of the study and performed the statistical analysis. LQZ conceived of the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This study is supported by grants from the National Natural Science Foundation of China (No. 30672099) and the Beijing Natural Science Foundation of China (No. 7122183).
The authors have no competing financial interests regarding this article.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Cooperberg MR, Moul JW, Carroll PR. The changing face of prostate cancer. J Clin Oncol. 2005;23:8146–51. doi: 10.1200/JCO.2005.02.9751. [DOI] [PubMed] [Google Scholar]
- Bracarda S, de Cobelli O, Greco C, Prayer-Galetti T, Valdagni R, et al. Cancer of the prostate. Crit Rev Oncol Hematol. 2005;56:379–96. doi: 10.1016/j.critrevonc.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat Rev Cancer. 2009;9:501–7. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- Shirakawa H, Landsman D, Postnikov YV, Bustin M. NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J Biol Chem. 2000;275:6368–74. doi: 10.1074/jbc.275.9.6368. [DOI] [PubMed] [Google Scholar]
- King LM, Francomano CA. Characterization of a human gene encoding nucleosomal binding protein NSBP1. Genomics. 2001;71:163–73. doi: 10.1006/geno.2000.6443. [DOI] [PubMed] [Google Scholar]
- Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, et al. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell. 2009;35:642–56. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman M, Malicet C, Bustin M. HMGN5/NSBP1: a new member of the HMGN protein family that affects chromatin structure and function. Biochim Biophys Acta. 2010;1799:86–92. doi: 10.1016/j.bbagrm.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhou LQ, Song G. Effect of nucleosomal binding protein 1 in androgen-independent prostatic carcinoma. Zhonghua Yi Xue Za Zhi. 2008;88:657–60. Chinese. [PubMed] [Google Scholar]
- Qu J, Yan R, Chen J, Xu T, Zhou J, et al. HMGN5: a potential oncogene in gliomas. J Neurooncol. 2011;104:729–36. doi: 10.1007/s11060-011-0558-9. [DOI] [PubMed] [Google Scholar]
- Green J, Ikram M, Vyas J, Patel N, Proby CM, et al. Overexpression of the Axl tyrosine kinase receptor in cutaneous SCC-derived cell lines and tumours. Br J Cancer. 2006;94:1446–51. doi: 10.1038/sj.bjc.6603135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WY, Newbold R, Mardilovich K, Jefferson W, Cheng RY, et al. Persistent hypomethylation in the promoter of nucleosomal binding protein 1 (Nsbp1) correlates with overexpression of Nsbp1 in mouse uteri neonatally exposed to diethylstilbestrol or genistein. Endocrinology. 2008;149:5922–31. doi: 10.1210/en.2008-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Zhou LQ, Weng M, He Q, He ZS, et al. Expression of nucleosomal binding protein 1 in normal prostate benign prostate hyperplasia, and prostate cancer and significance thereof. Zhonghua Yi Xue Za Zhi. 2006;86:1962–5. Chinese. [PubMed] [Google Scholar]
- Jiang N, Zhou LQ, Zhang XY. Downregulation of the nucleosome-binding protein 1 (NSBP1) gene can inhibit the in vitro and in vivo proliferation of prostate cancer cells. Asian J Androl. 2010;12:709–17. doi: 10.1038/aja.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LQ, Song G, He ZS, Hao JR, Na YQ. Effects of inhibiting nucleosomal binding protein 1 on proliferation of human prostate cancer cells. Zhonghua Yi Xue Za Zhi. 2007;87:404–8. Chinese. [PubMed] [Google Scholar]
- Fizazi K, Sternberg CN, Fitzpatrick JM, Watson RW, Tabesh M. Role of targeted therapy in the treatment of advanced prostate cancer. BJU Int. 2010;105:748–67. doi: 10.1111/j.1464-410X.2010.09236.x. [DOI] [PubMed] [Google Scholar]
- Putral LN, Gu W, McMillan NA. RNA interference for the treatment of cancer. Drug News Perspect. 2006;19:317–24. doi: 10.1358/dnp.2006.19.6.985937. [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez AB, Bajo AM, Isabel Arenas M, Sanchez-Chapado M, Prieto JC, et al. Vasoactive intestinal peptide (VIP) induces malignant transformation of the human prostate epithelial cell line RWPE-1. Cancer Lett. 2010;299:11–21. doi: 10.1016/j.canlet.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Zimmermann M, Meyer N. Annexin V/7-AAD staining in keratinocytes. Methods Mol Biol. 2011;740:57–63. doi: 10.1007/978-1-61779-108-6_8. [DOI] [PubMed] [Google Scholar]
- Sung DK, Chang YS, Kang S, Song HY, Park WS, et al. Comparative evaluation of hypoxic-ischemic brain injury by flow cytometric analysis of mitochondrial membrane potential with JC-1 in neonatal rats. J Neurosci Methods. 2010;193:232–8. doi: 10.1016/j.jneumeth.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Bantounas I, Phylactou LA, Uney JB. RNA interference and the use of small interfering RNA to study gene function in mammalian systems. J Mol Endocrinol. 2004;33:545–57. doi: 10.1677/jme.1.01582. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Kang MH, Reynolds CP. Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res. 2009;15:1126–32. doi: 10.1158/1078-0432.CCR-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nariculam J, Freeman A, Bott S, Munson P, Cable N, et al. Utility of tissue microarrays for profiling prognostic biomarkers in clinically localized prostate cancer: the expression of BCL-2, E-cadherin, Ki-67 and p53 as predictors of biochemical failure after radical prostatectomy with nested control for clinical and pathological risk factors. Asian J Androl. 2009;11:109–18. doi: 10.1038/aja.2008.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–6. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, et al. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–87. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]