Abstract
Among bird species, the most studied major histocompatibility complex (MHC) is the chicken MHC. Although the number of studies on MHC in free-ranging species is increasing, the knowledge on MHC variation in species closely related to chicken is required to understand the peculiarities of bird MHC evolution. Here we describe the variation of MHC class IIB (MHCIIB) exon 2 in a population of the Grey partridge (Perdix perdix), a species of high conservation concern throughout Europe and an emerging galliform model in studies of sexual selection. We found 12 alleles in 108 individuals, but in comparison to other birds surprisingly many sites show signatures of historical positive selection. Individuals displayed between two to four alleles both on genomic and complementary DNA, suggesting the presence of two functional MHCIIB loci. Recombination and gene conversion appear to be involved in generating MHCIIB diversity in the Grey partridge; two recombination breakpoints and several gene conversion events were detected. In phylogenetic analysis of galliform MHCIIB, the Grey partridge alleles do not cluster together, but are scattered through the tree instead. Thus, our results indicate that the Grey partridge MHCIIB is comparable to most other galliforms in terms of copy number and population polymorphism.
Introduction
Genes of the major histocompatibility complex (MHC) encode for molecules which bind small fragments of peptides and present these on cell surfaces for recognition by immune cells. Recognition of the presented fragment as foreign (i.e. of pathogenic origin), triggers a whole cascade of immune reactions [1], [2]. The evolutionary pressure of parasite burden has been suggested to contribute to the extraordinary polymorphism of MHC genes, which belong among the most variable genes of vertebrate genomes known to date [1], [3], [4]. Apart from pathogen pressure, sexual selection has been shown to influence MHC class I and class II polymorphism by a number of studies in mammals (e.g. [5]–[7]), fish (e.g. [8], [9]), amphibians [10], reptiles [11], [12], and in birds [13]–[20]. The link between MHC polymorphism and mate choice arises because (i) “good” MHC genes confer better viability to offspring [21], [22], or (ii) MHC polymorphism might help avoiding kin mating (reviewed in [23]).
Among birds, the most studied and best described MHC is that of the chicken. It consists of two unlinked clusters of genes: the B-locus and the Y-locus (also called Rfp-Y). Both contain class I and II genes [24]–[26]. However, the B-locus contains highly polymorphic and expressed genes showing strong signals of historical balancing selection, while the Y-locus appears to be less expressed, holds mostly pseudogenes and genes with to date unclear function (e.g. [27]–[32]). The chicken MHC differs from mammalian MHC by the presence of only one dominantly expressed molecule of class I and one of class II, a feature which coined the term ‘minimal essential MHC’ [33], [34]. Other characteristic features of the chicken MHC include its small physical size, dense organisation with short introns, lack of redundancy, and low number of other class I and class II genes with very few pseudogenes [34]–[36]. Some other galliform birds have been shown to share the simple genetic organisation of chicken MHC, such as the pheasant (Phasianus colchicus) [37] or black grouse (Tetrao tetrix) [32], each with two copies of MHC class IIB (MHCIIB). Other galliforms possess a slightly higher number of loci. For example, there are three MHCIIB loci in turkey (Meleagris gallopavo) [38] and several class I and class II genes in the Japanese quail (Coturnix japonica), including pseudogenes [39]–[41]. However, similarly to chicken in the Japanese quail only a fraction of the present class I and class II genes are dominantly expressed [39], [40]. In avian species outside the galliforms, the genetic complexity of MHC class I and class II might vary from low copy numbers, e.g. in owls [42], [43] or great snipe (Gallinago media) [44], to passerines displaying the most extreme patterns of polymorphism, typically with high levels of gene duplication (e.g. [45]–[50]) and extensive pseudogenes [51]. Although many of the duplicated genes have been shown to transcribe to cDNA (e.g. [49]), little is known about the levels of expression and hence true functional diversity of class I and class II genes in passerines.
It remains unclear whether the compact organization of MHC genes with rather few genes in some birds is ancestral, and how the variability in complexity evolves. The comparison of MHC structure and variation across the galliform birds is therefore necessary in order to analyse the patterns and processes involved in MHC evolution of these birds. Here we studied the structure, variation and evolutionary mechanisms acting on MHCIIB in the Grey partridge (Perdix perdix), a galliform bird that is diverged from the chicken by 33.7 million years (based on TimeTree, [52]).
The Grey partridge is a typical farmland inhabitant and game bird. The species used to be common in Central and Eastern Europe, but since the change in agricultural strategy in the 1950 s its populations have been rapidly declining [53], [54]. Our study population originates from the Czech Republic, where the species is bred in captivity for later release of offspring into the wild. Thus, as part of the conservation programme, it is crucial to study adaptive genetic variation in this species and MHC genes have been used as very suitable markers for assessing the status of endangered populations (e.g. [55], [56]). Additionally, the Grey partridge is a socially monogamous species, with females being particularly choosy [57], and, therefore, the MHC markers can be used to test the presence and mechanism of MHC-based mate choice.
Materials and Methods
Ethics Statement
The individuals used in this study were sampled in 2010 at a private breeding farm in Letonice, Czech Republic under the permission of the owner. The Grey partridge is not specifically protected in the Czech Republic and all manipulations with animals and methods of sacrifice were approved by the ethic committee of the Academy of Sciences of the Czech Republic (permit number 147/2007 to JB, who is holder of the certificate of competency on Protection Animals against Cruelty, reg. no. V/1/2005/05).
Study Species, Sampling, DNA and RNA Extraction
The Grey partridge (Perdix perdix) is a sedentary, socially monogamous and territorial galliform species bound to open countryside. In total 108 adult individuals (54 females and 54 males, i.e. all breeding pairs in the farm) were sampled prior to nesting, and ca 1 ml of blood was taken by wing venipuncture and stored in 96% ethanol at −20°C until DNA extraction.
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit, Qiagen (Hilden, Germany). Spleen from two sacrificed individuals and blood from additional four individuals were sampled in order to analyse MHC expression. These samples were stored in RNAlater (Qiagen) at −80°C, and RNA was extracted using the RNeasy Plus Mini kit (Qiagen). RNA was transcribed to cDNA using the Transcriptor First Strand cDNA Synthesis kit (Roche) according to the manufacturer’s instructions.
Design of Species-Specific Primers
In MHCIIB molecules, the amino acids being directly in touch with bound peptides (i.e. the β1 domain) are encoded by exon 2. To obtain initial sequences of Grey partridge MHC we used primers that were published for other galliform species (an overview of tested primers and their positions is given in File S1). We aligned our initial Grey partridge sequence with chicken, ring-necked pheasant, Japanese quail, helmeted guineafowl, black grouse and Indian peafowl and we designed two new primer pairs in conserved regions. For analysis of cDNA, we used primers BLB-fw7126 (5′-GTG CTG GTG GCA CTG CTG G-3′) and BLB-rev7776 (5′-CGT TCT GCA TCA CGT CCG TGG-3′) situated in exon 1 and exon 3, respectively. For high-throughput genotyping on genomic DNA, we designed primers BLBintr1-Fw (5′-TGC CCG CAG CGT TCT TCC TC-3′) and BLBintr2-Rev (5′-TCA CCT TGG GCT CCA CTG CG-3′) situated at the border of intron 1 with exon 2, and the border of intron 2 with exon 3, respectively (File S1).
MHC Genotyping - PCR, CE-SSCP, Cloning and Sequencing
Grey partridge MHCIIB region displays high GC content, and particularly introns 1 and 3 contain long repetitive GC stretches (data not shown). Thus, we applied PCR chemistry designed for GC-rich target sequences. MHCIIB exon 2 was amplified by PCR in a final volume of 10 µl with the following conditions: 1x Kapa2G GC buffer (including MgCl2 in final concentration of 1.5 mM), 0.2 µM of each forward (BLBintr1-Fw) and reverse (BLBintr2-Rev) primer, 0.2 mM mixed dNTPs, 0.5 units of Kapa2G Robust HotStart polymerase (Kapa Biosystems), and a final adjustment with ddH2O to 9 µl. 1 µl of DNA (ca 50 ng/µl) was then added to the master-mix. Thermal cycling was performed on the Master-cycler ep (Eppendorf) with an initial denaturation at 94°C for 10 min, followed by 30 cycles of each 94°C (30 sec), 62°C (30 sec) and 72°C (40 sec). A final step of 72°C for 8 min was introduced to finish elongation. PCR products were visualized on agarose gel stained by GoldView™ (Ecoli).
All individuals were genotyped by capillary electrophoresis single strand conformation polymorphism (CE-SSCP) analysis. For this purpose, exon 2 was amplified by PCR as described above, but using fluorescently labelled primers (BLBintr1-Fw: 6-FAM; BLBintr2-Rev: NED). CE-SSCP was performed on a 3130 Genetic Analyzer (Applied Biosystems) as described in [48]. After testing several electrophoresis run temperatures, the best resolution was achieved using a run temperature of 22°C. Electropherograms were analysed in GeneMapper v3.7 (Applied Biosystems).
CE-SSCP displays different alleles by visualizing each sequence variant by a unique shape and position on the electropherogram. In order to obtain nucleotide sequences for all alleles present in the studied population (i.e. all alleles visualized on CE-SSCP), we cloned the amplicons of 24 individuals based on their CE-SSCP genotypes. Amplicons for bacterial cloning were purified using the MinElute PCR purification kit (Qiagen), ligated to the pJET1.2/blunt vector and transformed using the CloneJet™ PCR cloning kit (Fermentas) according to the manufacturer’s instructions. Colonies were screened for inserts of the expected length (cca 400 bp) by PCR (using the vector primers pJET1.2-Fw and pJET1.2-Rev) and gel electrophoresis. Positive clones were then screened using CE-SSCP as described above. In total we screened 220 clones. The CE-SSCP profiles of the clones were compared to the CE-SSCP profiles of individuals’ uncloned amplicons, in order to (i) avoid sequencing of obvious PCR artefacts, (ii) make sure we capture all the alleles of an individual and (iii) avoid multiple repeated sequencing of the same alleles. On average we screened eight clones per individual. For some individuals as much as 23 clones had to be screened before all their alleles displayed by CE-SSCP were captured. Selected clones were then sequenced using the BigDye Terminator Sequencing kit v3.1 (Applied Biosystems) and the primer pJET1.2 Reverse Sequencing Primer (Fermentas).
Expression Analysis
To verify whether sequences obtained from gDNA using intronic primers are transcribed and hence potentially functional, we analysed cDNA of 6 individuals. The whole MHCIIB exon 2 was amplified using primers BLB-fw7126 (in exon 1) and BLB-rev7776 (in exon 3) with the same PCR conditions as described above. The purified PCR products were then cloned as described above and 3 to 27 clones per individual were Sanger sequenced. Two of these six individuals were screened also for MHCIIB variation at gDNA as described above.
Analysis of Recombination, Historical Selection, and Gene Conversion
Sequences were edited in BioEdit Sequence Alignment Editor v7.0.5.3 [58] and aligned using the ClustalW algorithm [59]. Primers and intronic sequences were trimmed from clone sequences. As the first nucleotide of the starting amino acid of exon 2 is situated at the end of exon 1, the sequences were cut to 267 bp (two bases in the 5′-end and one base in the 3′-end) to fit the reading frame for further analysis. To avoid analysis of PCR artefacts, we only considered exon 2 variants as ‘true’ alleles if they occurred in at least two individuals or two independent PCRs of the same individual.
Recombination breakpoints were detected using the GARD algorithm in the HyPhy package (available at http://www.datamonkey.org) [60]. Historical selection was analysed by likelihood ratio modelling in the program CodeML, which is a component of the PAML 4 program suite [61]. In this study we compared models allowing for positive selection (M2a and M8) with those assuming no selection (M1a and M7) [62], [63]. The tested models differ in the number and type of included parameters that are based on ω ratio, i.e. the ratio of non-synonymous mutations (dN) versus synonymous mutations (dS). Comparison of nested models (M1a with M2a, and M7 with M8, respectively) was obtained using likelihood ratio test statistics; i.e. 2(Lb-La) was compared with χ2 distribution with Pb-Pa degrees of freedom (La, Lb – log-likelihood values for each compared model; Pa, Pb – number of parameters for each compared model). The Bayes empirical Bayes (BEB) method was used to calculate posterior probabilities for site classes in models M2a and M8. If the posterior probabilities for some sites are significant (ω>1), those sites are inferred to be under positive selection. We also calculated the number of synonymous and non-synonymous substitutions per site, as an overall mean value for all sequences with a standard error estimated by bootstrapping (1000 replicates). The analysis was conducted separately for antigen-binding sites (ABS) and non-ABS derived from the ABS of human HLA-DRB1 [64] in MEGA v5.03 [65].
Occurrence of gene conversion events in MHC IIB exon 2 was tested using GENECONV version 1.81a [66], with 10,000 permutations. The analysis is based on a pairwise comparison of aligned alleles. G-scale value was set to 0, not allowing for any mismatches within the gene conversion tracts. Gene conversion events were considered significant if the global P value was lower than 0.05.
Phylogenetic Analysis
To analyze the relationships among exon 2 sequences of the Grey partridge and other galliform species, we downloaded available MHCIIB and MHCIIY sequences of 14 and five galliform species, respectively, from GenBank (File S2). We used the MHCIIB (DAB1) sequence of the barn owl (GenBank acc. no. EF641260) [42] as outgroup. Phylogenetic analysis of the alignment was performed by maximum likelihood (ML) approach using the PhyML 3.0 online web server [67]. The BIONJ distance-based tree was used as the starting tree and GTR+G as the substitution model. Clade support was tested by 1,000 bootstraps.
Results
MHCIIB Intra-Population Variation and Expression
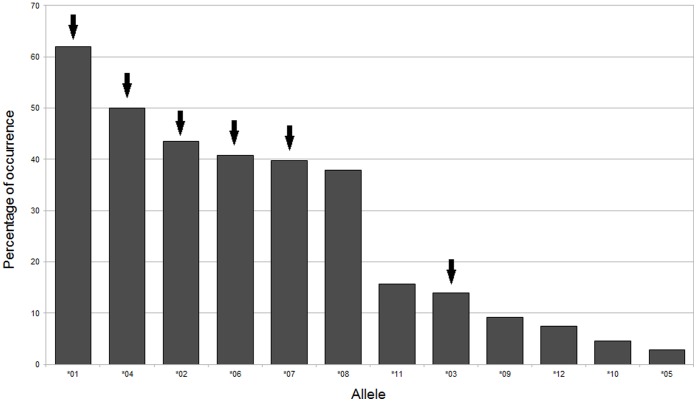
In total, 12 different exon 2 variants were identified using CE-SSCP, cloning, and sequencing (sequences are deposited in GenBank, under accession numbers: KF007890-KF007901). Individuals displayed between two to four variants on gDNA. Of the 108 Grey partridge individuals, 17 displayed two alleles, 44 displayed three and 47 displayed four alleles, suggesting the presence of at least two MHCIIB loci in the Grey partridge. The assignment of sequences to specific loci based on phylogenetic analysis was not possible and, for simplicity, we thus call MHCIIB variants as ’alleles‘. In chicken, MHCIIB is called BLB. However, we cannot identify whether the genes we amplify in the Grey partridge are orthologous to BLB. Therefore, following the nomenclature suggestions of Klein et al. [68] we call our alleles MhcPepe-DAB*01–12, but for simplicity throughout this paper the alleles are referred to as Pepe*01-Pepe*12. The most abundant allele, Pepe*01 was present in 67 individuals out of 108 (62%), and five other alleles were relatively common, occurring in more than one third of the birds (Fig. 1). The rarest allele, Pepe*05, was only detected in 3 individuals.
Figure 1. Frequency of occurrence (in %) of MHCIIB exon 2 alleles in 108 individuals, sorted from highest to lowest values.
Arrows indicate the alleles found in gDNA as well as cDNA.
All sequences translate into amino-acid sequences without stop codons or indels shifting the reading frame (Fig. 2). Six of the alleles (Pepe*01, *02, *03, *04, *06 and *07) were sequenced also from cDNA of six individuals and cDNA genotypes were identical with the genotypes obtained from genomic DNA of the same individuals, suggesting that both MHCIIB loci are expressed.
Figure 2. Putative amino acid sequence alignment.
The alignment starts at the 2nd amino acid position encoded by the exon 2 in MHCIIB. Sequences of chicken (Gaga_BLB1-haplB19 and Gaga_BLB2-haplB19, GenBank accession no. AB426151), barn owl (Tyal_DAB1-allele02, EU442602) and little spotted kiwi (Apow_DAB*allele08, HQ639687) [90] are shown for comparison. Dots represent identity to the sequence on top. Asterisks (*) indicate the amino acid positions under positive selection (based on CodeML analysis, model M8, posterior probability >0.95). Plus signs (+) mark the antigen-binding sites (ABS) in humans [64]. Tracts 1–7 depict codons contained in conversion tracts as given in Table 3. Black triangles represent the placement of two recombination breakpoints as detected by GARD.
Historical Positive Selection, Recombination and Gene Conversion
Out of the 270 nucleotide positions in exon 2, 63 positions were variable among the 12 alleles. Non-synonymous substitutions per site (dN) outnumbered the synonymous ones (dS) when considering all codons of the sequence (Table 1). Based on the Z-test of positive selection, this difference was significant only for presumable ABS sites (Z-test, ZdN-dS = 2.156, P = 0.017), but not for non-ABS sites (Z-test, ZdN-dS = 0.401, P = 0.344) (Table 1).
Table 1. The number of synonymous (dS) and non-synonymous (dN) substitutions per site for ABS, non-ABS and all sites in exon 2 of MHC class IIB (mean±SE); the value of Z-test of positive selection and probability (P) that positive selection acts on these sites.
| dN±SE | dS±SE | dN/dS | Z-test | P | |
| ABS | 0.303±0.075 | 0.134±0.058 | 2.261 | 2.156 | 0.017 |
| non-ABS | 0.066±0.017 | 0.056±0.025 | 1.179 | 0.401 | 0.344 |
| all sites | 0.120±0.020 | 0.078±0.023 | 1.538 | 1.610 | 0.055 |
Maximum likelihood models also show that positive selection is acting on the MHCIIB in the Grey partridge (Table 2). Both alternative models which take into account the positive selection fitted our data significantly better than the basic models of neutral evolution (M2a versus M1a: df = 2, Test statistic = 49.552, p<0.001; M8 versus M7: df = 2, Test statistic = 50.382, p<0.001). Model M8 identified 21 positively selected sites (17 with >99% probability and additional 4 sites with >95% probability; Table 2); 13 of the positively selected sites in M8 correspond to the ABS residues known from humans (Table 2, File S3).
Table 2. Results of maximum likelihood models for testing historical selection on exon 2 of MHC class IIB in the Grey partridge.
| model | P | log-likelihood (L) | parameter estimates | positively selected sites |
| M1a | 2 | –969.408 | p 0 = 0.575, p 1 = 0.425 | not allowed |
| M2a | 4 | –944.632 | p 0 = 0.573, p 1 = 0.101, p 2 = 0.326, ω 2 = 8.281 | 5, 7, 9, 22, 23, 26, 33, 43, 50, 52, 56, 61, 63, 66, 67, 68, 69, 70, 81, 82 |
| M7 | 2 | –969.871 | p = 0.016, q = 0.023 | not allowed |
| M8 | 4 | –944.680 | p 0 = 0.674, p 1 = 0.326, p = 0.187, q = 0.617, ω = 8.372 | 5, 7, 9, 22, 23, 26, 33, 43, 50, 52, 56, 61, 63, 66, 67, 68, 69, 70, 71, 81, 82 |
The maximum likelihood models were performed in CodeML integrated in PAML package v4 [61]. P indicates a number of parameters considered by the model, ω is a parameter based on dN/dS ratio (selection parameter), p n is a proportion of sites in a specific ω site class, p and q are the indicators of shape of β distribution; models M2a and M8 estimated the positively selected sites of exon 2 by Bayes Empirical Bayes procedure [63]. Sites positively selected with probability >99% are in bold, sites with p>95% are in italics, underlined sites correspond to the ABS residues in humans [64].
Test of recombination revealed two breakpoints (at 79 bp and 178 bp; Fig. 2). The evidence was given by AICc (i.e. the small-sample-size corrected version of Akaike Information Criterion), which achieved an improvement of ∼ 129 points over the model without recombination. Moreover, both sites were tested for topological incongruence and one site (178 bp) proved to be significant (KH-test, P = 0.01).
Gene conversion analysis in GENECONV showed several significant gene conversion events among the Grey partridge MHCIIB exon 2 sequences (summarised in Table 3 and Fig. 2). Conversion tracts vary in size between 37 to 79 bp. Only one of the 12 exon 2 sequences described in this study shares no fragment with other sequences (Pepe*03).
Table 3. Gene conversion events among 12 nucleotide sequences of MHCIIB exon 2 in the Grey partridge.
| Tract no. | Seq 1 | Seq 2 | Simulated P value | Start | End | Length | No. of Polymorphisms | Total Differences |
| 1 | Pepe*09 | Pepe*12 | 0.0033 | 96 | 174 | 79 | 22 | 23 |
| Pepe*02 | Pepe*09 | 0.024 | 96 | 150 | 55 | 12 | 33 | |
| 2 | Pepe*07 | Pepe*11 | 0.013 | 105 | 155 | 51 | 13 | 32 |
| Pepe*05 | Pepe*11 | 0.027 | 105 | 155 | 51 | 13 | 31 | |
| Pepe*06 | Pepe*11 | 0.027 | 105 | 155 | 51 | 13 | 31 | |
| 3 | Pepe*06 | Pepe*12 | 0.0169 | 126 | 162 | 37 | 14 | 30 |
| Pepe*07 | Pepe*12 | 0.0169 | 126 | 162 | 37 | 14 | 30 | |
| Pepe*05 | Pepe*12 | 0.0292 | 126 | 162 | 37 | 14 | 29 | |
| Pepe*04 | Pepe*08 | 0.0354 | 126 | 174 | 49 | 18 | 23 | |
| 4 | Pepe*01 | Pepe*12 | 0.0064 | 133 | 177 | 45 | 17 | 27 |
| 5 | Pepe*08 | Pepe*12 | 0.0006 | 165 | 238 | 74 | 25 | 23 |
| 6 | Pepe*02 | Pepe*08 | 0.0277 | 165 | 196 | 32 | 11 | 35 |
| 7 | Pepe*04 | Pepe*09 | 0.0017 | 195 | 241 | 47 | 18 | 28 |
| Pepe*09 | Pepe*10 | 0.0038 | 195 | 241 | 47 | 18 | 27 |
Gene conversion tracts as identified by GENECONV [66]. Tract no. as depicted in Fig. 2. Simulated P value is based on 10,000 permutations; Start and End indicate the first and last nucleotide position of the conversion tract, respectively; Fragment Length is the length of the conversion tract; No. of Polymorphisms is the number of sites within conversion tract which are polymorphic in the alignment of all alleles; Total Differences is the number of mismatches between the two compared sequences. Sequence pairs are sorted based on the starting position of the conversion tract.
Phylogenetic Analysis of MHCIIB Sequences in Galliform Birds
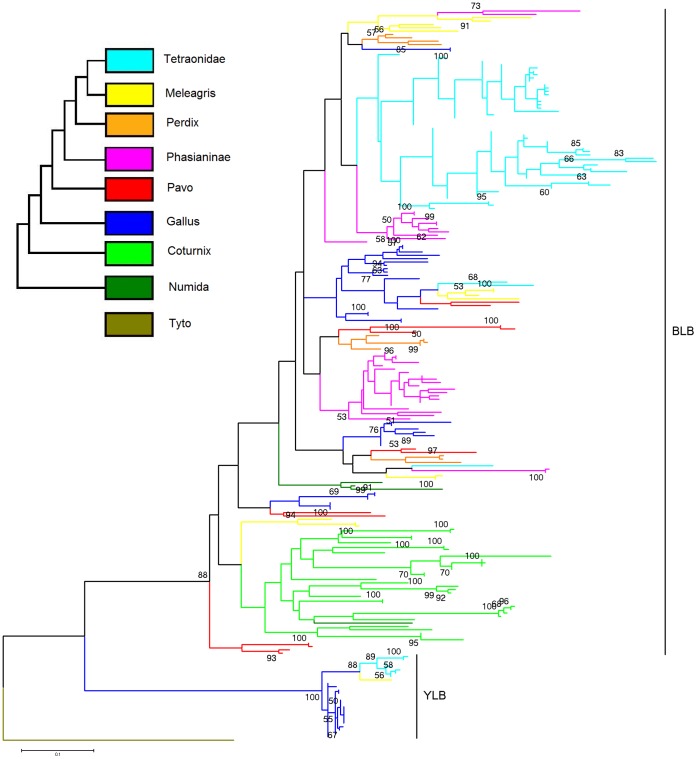
In the ML tree of MHC class II sequences of galliform species, the MHCIIB sequences are clearly separated from MHCIIY (Fig. 3). Grey partridge alleles do not cluster together, but are scattered through the tree instead (Fig. 3). Most of the clades, however, are not or only weakly supported, which is probably caused by short length of the sequence and presence of balancing selection as well as concerted evolution typical for avian MHC. It is not possible to assign sequences of exon 2 to specific DAB loci based on the phylogenetic tree.
Figure 3. Maximum likelihood tree of exon 2 MHCIIB and MHCIIY sequences across galliform species.
Different colours indicate taxa, whose neutral phylogeny is shown in small inserted figure (based on [91] and [92]). The sequence of DAB1 of the barn owl (Acc. no. EF641260) was used as an outgroup. Only bootstrap values >50 are shown. For the complete list of sequences used, with GenBank accession numbers and references see File S2.
Discussion
In this study, we characterized MHCIIB exon 2 variation in a population of the Grey partridge, using an approach combining PCR, cloning, sequencing and CE-SSCP genotyping. We found 12 alleles in total, which is comparable to the MHCIIB diversity in red jungle fowl (Gallus gallus), where ten alleles were identified in a captive population of 80 birds [69]. Thus, our results show further evidence that variation in terms of number of alleles per species is lower in galliforms than in other groups of birds. For comparison, 50 alleles were found in 175 great snipes (Gallinago media) sampled over several years from a relatively large geographical region [70], 103 alleles at a single locus in 121 lesser kestrels (Falco naumanni) from a wide geographical range [71] and 194 exon 2 sequences were found in 237 individuals of collared flycatcher (Ficedula albicollis; [51]).
In our study, individuals displayed between two to four alleles. The maximum of four alleles per individual, isolated from both genomic and complementary DNA, and the absence of signatures of pseudogenes (i.e. stop codons or frameshift mutations) suggests the presence of two presumably functional MHCIIB loci in the Grey partridge. This number is equal to the number of known functional MHCIIB genes observed in chicken [34], black grouse [32] and pheasant [37]. Other closely related galliforms display slightly higher number of loci, with turkey (M. gallopavo) displaying three [72] and greater prairie-chicken (Tympanachus cupido) displaying four MHCIIB loci [73] (Table 4).
Table 4. Review of MHC structure in galliform species.
| Species | Code | N loci | gDNA | cDNA | Y locus | Gene Names | References |
| Coturnix japonica | Coja | 7 | + | + | ? | DAB1, DBB1, DCB1, DDB1,DEB1, DFB1, DGB1 | [39]–[41] |
| Gallus gallus | Gaga | 2 | + | + | + | BLB1, BLB2 | [24], [26], [34], [40], [69] |
| Gallus lafayetii | Gala | ? | + | − | ? | − | [94] |
| Meleagris gallopavo | Mega | 3 | + | + | + | − | [72], [82] |
| Numida meleagris | Nume | ? | + | + | ? | − | [95] |
| Pavo cristatus | Pacr | 3 | + | − | ? | − | [96] |
| Perdix perdix | Pepe | 2 | + | + | ? | DAB | this study |
| Phasianus colchicus | Phco | 2 | + | + | + | DAB1, DAB2 | [13], [37], [40], [81] |
| Tetrao tetrix | Tete | 2 | + | + | + | − | [32] |
| Tympanuchus cupido | Tycu | 4 | + | − | + | − | [73] |
Most galliform species display low number of MHCIIB loci, except the migratory Japanese Quail (Coturnix japonica). The second genetically unlinked MHC region called Y locus has been found in half of them. Columns gDNA and cDNA show what kind of template was used in the studies. Species are ordered alphabetically. Used symbols:+yes, - no, ? yet unknown.
CE-SSCP genotyping method has by some studies been shown to underestimate MHC variation [74], [75]. This is particularly true in case of highly duplicated genes where locus-specific amplification is not possible, such as passerine MHC class I [76]. However, the method has been successfully applied in a wide range of species where MHC class I and class II display moderate levels of polymorphism, such as mammals (e.g. [56], [77], [78]) and galliform birds [79]. While studies using next-generation sequencing of MHC in passerines have reported highly duplicated loci with almost 200 alleles of both MHCIIB [80] and MHCI [76] from single populations, such extensive variation is not expected in galliforms. Hence, we believe that the applied method is suitable for MHCIIB genotyping in the Grey partridge, particularly when combined with cloning and Sanger sequencing.
A feature typical for the structure of chicken MHC and some other closely related species is the presence of a second, unlinked cluster of MHC genes, the Y locus [27], [32], [81], [82] (Table 4). This cluster consists of genes with yet unclear functional significance [32], [81], but some of the MHC genes situated in the Y region have been shown to be expressed [31], [82], [83]. We tested many combinations of primers, including those that specifically amplify Y loci in other species [32], but were unable to isolate sequences of Y loci in the present study (Fig. 3). It is possible that if the Grey partridge, like several closely related species, has a Y locus, it is distinct enough in sequence such that it is not amplified with any of the primers used here. More powerful approaches, such as whole-genome sequencing, or screening of BAC libraries might be necessary to rule out the presence of Y loci in the Grey partridge (see for example [84], where only whole genome sequencing confirmed the complete absence of MHC class II in the cod).
Positive selection and gene conversion appear to have prominent roles in the evolution of MHCIIB diversity in the Grey partridge. We identified widespread traces of positive selection in exon 2, with 21 out of 89 tested sites showing ω significantly higher than 1. This level of historical positive selection is extraordinarily high compared to other studies of avian species analysed by the same methods. For example in the barn owl (T. alba) seven sites showed signs of positive selection [42], 12 sites in the blue petrel (Halobaena caerulea) [85], and 10 sites in the house sparrow (Passer domesticus) [86]. Moreover, recombination and gene conversion were shown to significantly affect sequence diversity in the Grey partridge. Fifteen out of 21 sites inferred to be under positive selection lay within conversion tracts (Fig. 2). Gene conversion processes within the MHC create polymorphism much faster than point mutations by reshuffling polymorphic fragments among alleles, thus creating new haplotypes [87], [88]. Variation at avian MHC has been shown to originate at least partially from gene conversion (e.g. [37]). Gene conversion can be an important driver of allelic diversity especially in populations that have undergone bottleneck and suffered from depleted MHC variation. In one such population of the Berthelot’s pipit, gene conversion created diversity by one order of magnitude faster than generated by point mutations [89]. It is highly probable that the processes of gene conversion, recombination and historical selection all act in concert, and their respective contribution to the generation and maintenance of sequence diversity thus might often be difficult to tell apart.
Our study showed that even the captive Grey partridge population display MHCIIB variation that is comparable to other galliform species. The European population of this species decreased by more than 80% since 1980 [54] and conservation programs are often using captive animals to support natural populations. Here we provide first data showing that the studied captive population could have a good chance to withstand pathogens after release. Results of this study will also be useful in testing the alternative hypotheses of MHC-based mate choice in the Grey partridge, which is an emerging model in the research of sexual selection [57].
Supporting Information
List of all primers tested for PCR amplification of MHCIIB in the Grey partridge and their schematic overview.
(DOC)
Overview of all MHCIIB sequences of Galliformes used in the phylogenetic analysis, with references and GenBank accession numbers.
(DOC)
Posterior means of ω for each codon of MHCIIB exon 2 in the Grey partridge, calculated as the average of ω over the 11 site classes, weighted by the posterior probabilities under the random-sites model M8 (β and ω). The posterior probabilities were computed by the Bayes empirical Bayes procedure in the program CodeML implemented in the PAML3.14 package.
(DOCX)
Acknowledgments
We are grateful to D. Rymešová, K. Kotasová, O. Tomášek, B. Gabrielová and M. Šálek for help with sample collection and P. Hájková for help in the lab. We thank R. Burri for help with the gene conversion analysis and extensive discussions and two anonymous reviewers for their comments on the previous version of the paper.
Funding Statement
The study was supported by the Czech Science Foundation, project no. 206/08/1281 and with institutional support RVO:68081766. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Klein J (1986) Natural history of the major histocompatibility complex. New York: John Wiley and Sons. 775 p.
- 2.Janeway CA, Travers P, Walport M, Schlomchik MJ (2004) Immunobiology. New York: Garland Science. 800 p.
- 3. Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16: 363–377. [DOI] [PubMed] [Google Scholar]
- 4. Piertney SB, Oliver MK (2006) The evolutionary ecology of the major histocompatibility complex. Heredity 96: 7–21 doi:10.1038/sj.hdy.6800724 [DOI] [PubMed] [Google Scholar]
- 5. Schwensow N, Fietz J, Dausmann K, Sommer S (2008) MHC-associated mating strategies and the importance of overall genetic diversity in an obligate pair-living primate. Evol Ecol 22: 617–636 doi:10.1007/s10682-007-9186-4 [Google Scholar]
- 6. Schwensow N, Eberle M, Sommer S (2008) Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proc R Soc Lond B Biol Sci 275: 555–564 doi:10.1098/rspb.2007.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Setchell JM, Vaglio S, Abbott KM, Moggi-Cecchi J, Boscaro F, et al. (2010) Odour signals major histocompatibility complex genotype in an Old World monkey. Proc R Soc Lond B Biol Sci 278: 274–280 doi:10.1098/rspb.2010.0571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eizaguirre C, Yeates SE, Lenz TL, Kalbe M, Milinski M (2009) MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol Ecol 18: 3316–3329 doi:×10.1111/j.1365-294×.2009.04243.x [DOI] [PubMed] [Google Scholar]
- 9. Agbali M, Reichard M, Bryjova A, Bryja J, Smith C (2010) Mate choice for nonadditive genetic benefits correlate with MHC dissimilarity in the rose bitterling (Rhodeus ocellatus). Evolution 64: 1683–1696 doi:10.1111/j.1558-5646.2010.00961.x [DOI] [PubMed] [Google Scholar]
- 10. Bos DH, Williams RN, Gopurenko D, Bulut Z, Dewoody JA (2009) Condition dependent mate choice and a reproductive disadvantage for MHC-divergent male tiger salamanders. Mol Ecol 18: 3307–3315 doi:–10.1111/j.1365–294X.2009.04242.x [DOI] [PubMed] [Google Scholar]
- 11. Olsson M, Madsen T, Nordby J, Wapstra E, Ujvari B, et al. (2003) Major histocompatibility complex and mate choice in sand lizards. Proc R Soc Lond B Biol Sci 270: S254–S256 doi:10.1098/rsbl.2003.0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miller HC, Moore JA, Nelson NJ, Daugherty CH (2009) Influence of major histocompatibility complex genotype on mating success in a free-ranging reptile population. Proc R Soc Lond B Biol Sci 276: 1695–1704 doi:10.1098/rspb.2008.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. von Schantz T, Wittzell H, Göransson G, Grahn M (1997) Mate choice, male condition dependent ornamentation and MHC in the pheasant. Hereditas 127: 133–140. [Google Scholar]
- 14. Freeman-Gallant CR, Meguerdichian M, Wheelwright NT, Sollecito SV (2003) Social pairing and female mating fidelity predicted by restriction fragment length polymorphism similarity at the major histocompatibility complex in a songbird. Mol Ecol 12: 3077–3083 doi:10.1046/j.1365-294X.2003.01968.x [DOI] [PubMed] [Google Scholar]
- 15. Ekblom R, Sæther A, Grahn M, Fiske P, Kålås JA, et al. (2004) Major histocompatibility complex variation and mate choice in a lekking bird, the great snipe (Gallinago media). Mol Ecol 13: 3821–3828 doi:10.1111/j.1365-294X.2004.02361.x [DOI] [PubMed] [Google Scholar]
- 16. Richardson DS, Komdeur J, Burke T, von Schantz T (2005) MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc R Soc Lond B Biol Sci 272: 759–767 doi:10.1098/rspb.2004.3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bonneaud C, Chastel O, Federici P, Westerdahl H, Sorci G (2006) Complex Mhc-based mate choice in a wild passerine. Proc R Soc Lond B Biol Sci 273: 1111–1116 doi:10.1098/rspb.2005.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brouwer L, Barr I, van de Pol M, Burke T, Komdeur J, et al. (2010) MHC-dependent survival in a wild population: evidence for hidden genetic benefits gained through extra-pair fertilizations. Mol Ecol 19: 3444–3455 doi:10.1111/j.1365-294X.2010.04750.x [DOI] [PubMed] [Google Scholar]
- 19. Griggio M, Biard C, Penn DJ, Hoi H (2011) Female house sparrows “count on” male genes: experimental evidence for MHC-dependent mate preference in birds. BMC Evol Biol 11: 44 doi:10.1186/1471-2148-11-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Promerová M, Vinkler M, Bryja J, Poláková R, Schnitzer J, et al. (2011) Occurrence of extra-pair paternity is connected to social male’s MHC-variability in the scarlet rosefinch Carpodacus erythrinus . J Avian Biol 42: 5–10 doi:10.1111/j.1600-048X.2010.05221.x [Google Scholar]
- 21. von Schantz T, Wittzell H, Goransson G, Grahn M, Persson K (1996) MHC genotype and male ornamentation: genetic evidence for the Hamilton-Zuk model. Proc Biol Sci 263: 265–271. [DOI] [PubMed] [Google Scholar]
- 22. Cutrera AP, Fanjul MS, Zenuto RR (2012) Females prefer good genes: MHC-associated mate choice in wild and captive tuco-tucos. Anim Behav 83: 847–856 doi:10.1016/j.anbehav.2012.01.006 [Google Scholar]
- 23. Penn DJ, Potts WK (1999) The evolution of mating preferences and major histocompatibility complex genes. Am Nat 153: 145–164. [DOI] [PubMed] [Google Scholar]
- 24. Briles WE, Goto RM, Auffray C, Miller MM (1993) A polymorphic system related to but genetically independent of the chicken major histocompatibility complex. Immunogenetics 37: 408–414 doi:10.1007/BF00222464 [DOI] [PubMed] [Google Scholar]
- 25. Miller MM, Goto RM, Taylor RL Jr, Zoorob R, Auffray C, et al. (1996) Assignment of Rfp-Y to the chicken major histocompatibility complex/NOR microchromosome and evidence for high-frequency recombination associated with the nucleolar organizer region. Proc Natl Acad Sci USA 93: 3958–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller MM, Bacon LD, Hala K, Hunt HD, Ewald SJ, et al. (2004) 2004 Nomenclature for the chicken major histocompatibility (B and Y) complex. Immunogenetics 56: 261–279 doi:10.1007/s00251-004-0682-1 [DOI] [PubMed] [Google Scholar]
- 27. Zoorob R, Behar G, Kroemer G, Auffray C (1990) Organization of a functional chicken class-II B-gene. Immunogenetics 31: 179–187. [DOI] [PubMed] [Google Scholar]
- 28. Zoorob R, Bernot A, Renoir DM, Choukri F, Auffray C (1993) Chicken major histocompatibility complex class-II B-genes – analysis of interallelic and interlocus sequence variance. Eur J Immunol 23: 1139–1145 doi:10.1002/eji.1830230524 [DOI] [PubMed] [Google Scholar]
- 29. Hunt HD, Fulton JE (1998) Analysis of polymorphisms in the major expressed class I locus (B-FIV) of the chicken. Immunogenetics 47: 456–467. [DOI] [PubMed] [Google Scholar]
- 30. Rogers S, Shaw I, Ross N, Nair V, Rothwell L, et al. (2003) Analysis of part of the chicken Rfp-Y region reveals two novel lectin genes, the first complete genomic sequence of a class I α-chain gene, a truncated class II β-chain gene, and a large CR1 repeat. Immunogenetics 55: 100–108 doi:10.1007/s00251-003-0553-1 [DOI] [PubMed] [Google Scholar]
- 31. Hunt HD, Goto RM, Foster DN, Bacon LD, Miller MM (2006) At least one YMHCI molecule in the chicken is alloimmunogenic and dynamically expressed on spleen cells during development. Immunogenetics 58: 297–307 doi:10.1007/s00251-005-0074-1 [DOI] [PubMed] [Google Scholar]
- 32. Strand T, Westerdahl H, Höglund J, Alatalo RV, Siitari H (2007) The Mhc class II of the Black grouse (Tetrao tetrix) consists of low numbers of B and Y genes with variable diversity and expression. Immunogenetics 59: 725–734 doi:10.1007/s00251-007-0234-6 [DOI] [PubMed] [Google Scholar]
- 33. Kaufman J, Völk H, Wallny HJ (1995) A ‘minimal essential Mhc’ and an ‘unrecognized Mhc’: two extremes in selection for polymorphism. Immunol Rev 143: 63–88. [DOI] [PubMed] [Google Scholar]
- 34. Kaufman J, Milne S, Göbel TWF, Walker BA, Jacob JP, et al. (1999) The chicken B locus is a minimal essential major histocompatibility complex. Nature 401: 923–925 doi:10.1038/44856 [DOI] [PubMed] [Google Scholar]
- 35. Shaw I, Powell TJ, Marston DA, Baker K, van Hateren A, et al. (2007) Different evolutionary histories of the two classical class I genes BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J Immunol 178: 5744–5752. [DOI] [PubMed] [Google Scholar]
- 36. Hess CM, Edwards SV (2002) The evolution of major histocompatibility genes in birds. Bioscience 52: 423–431 doi:[];10.1641/0006-3568(2002)052[0423:TEOTMH]2.0.CO;2 [Google Scholar]
- 37. Wittzell H, Bernot A, Auffray C, Zoorob R (1999) Concerted evolution of two Mhc class II B loci in pheasants and domestic chicken. Mol Biol Evol 16: 479–490. [DOI] [PubMed] [Google Scholar]
- 38. Chaves LD, Faile GM, Krueth SB, Hendrickson JA, Reed KM (2010) Haplotype variation, recombination, and gene conversion within the turkey MHC-B locus. Immunogenetics 62: 465–477 doi:10.1007/s00251-010-0451-2 [DOI] [PubMed] [Google Scholar]
- 39. Shiina T, Shimizu S, Hosomichi K, Kohara S, Watanabe S, et al. (2004) Comparative genomic analysis of two avian (Quail and Chicken) MHC regions. J Immunol 172: 6751–6763. [DOI] [PubMed] [Google Scholar]
- 40. Shiina T, Hosomichi K, Hanzawa K (2006) Comparative genomics of the poultry major histocompatibility complex. Anim Sci J 77: 151–162 doi:10.1111/j.1740-0929.2006.00333.x [Google Scholar]
- 41. Hosomichi K, Shiina T, Suzuki S, Tanaka M, Shimizu S, et al. (2006) The major histocompatibility complex (Mhc) class IIB region has greater genomic structural flexibility and diversity in the quail than the chicken. BMC Genomics 7: 322–335 doi:10.1186/1471-2164-7-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burri R, Niculita-Hirzel H, Roulin A, Fumagalli L (2008) Isolation and characterization of major histocompatibility complex (MHC) class II B genes in the Barn owl (Aves: Tyto alba). Immunogenetics 60: 543–550 doi:10.1007/s00251-008-0308-0 [DOI] [PubMed] [Google Scholar]
- 43. Burri R, Salamin N, Studer RA, Roulin A, Fumagalli L (2010) Adaptive divergence of ancient gene duplicates in the avian MHC class II β. Mol Biol Evol 27: 2360–2374 doi:10.1093/molbev/msq120 [DOI] [PubMed] [Google Scholar]
- 44. Ekblom R, Grahn M, Höglund J (2003) Patterns of polymorphism in the MHC class II of a non-passerine bird, the great snipe (Gallinago media). Immunogenetics 54: 734–741 doi:10.1007/s00251-002-0503-3 [DOI] [PubMed] [Google Scholar]
- 45. Westerdahl H, Wittzell H, von Schantz T (1999) Polymorphism and transcription of MHC class I genes in a passerine bird, the great reed warbler. Immunogenetics 49: 158–170 doi:10.1007/s002510050477 [DOI] [PubMed] [Google Scholar]
- 46. Westerdahl H, Wittzell H, von Schantz T (2000) MHC diversity in two passerine birds: no evidence for a minimal essential MHC. Immunogenetics 52: 92–100 doi:10.1007/s002510000256 [DOI] [PubMed] [Google Scholar]
- 47. Hess CM, Gasper J, Hoekstra H, Hill C, Edwards SV (2000) MHC class II pseudogene and genomic signature of a 32-kb cosmid in the House Finch (Carpodacus mexicanus). Genome Res 10: 13–23 doi:10.1101/gr.10.5.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Promerová M, Albrecht T, Bryja J (2009) Extremely high MHC class I variation in a population of a long-distance migrant, the Scarlet Rosefinch (Carpodacus erythrinus). Immunogenetics 61: 451–461 doi:10.1007/s00251-009-0375-x [DOI] [PubMed] [Google Scholar]
- 49. Anmarkrud JA, Johnsen A, Bachmann L, Lifjeld JT (2010) Ancestral polymorphism in exon 2 of bluethroat (Luscinia svecica) MHC class II B genes. J Evol Biol 23: 1206–1217 doi:10.1111/j.1420-9101.2010.01999.x [DOI] [PubMed] [Google Scholar]
- 50. Sepil I, Moghadam HK, Huchard E, Sheldon BC (2012) Characterization and 454 pyrosequencing of Major Histocompatibility Complex class I genes in the great tit reveal complexity in a passerine system. BMC Evol Biol 12: 68 doi:10.1186/1471-2148-12-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zagalska-Neubauer M, Babik W, Stuglik M, Gustafsson L, Cichon M, et al. (2010) 454 sequencing reveals extreme complexity of the class II major histocompatibility complex in the collared flycatcher. BMC Evol Biol 10: 395 doi:10.1186/1471-2148-10-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hedges SB, Dudley J, Kumar S (2006) TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22: 2971–2972 doi:10.1093/bioinformatics/btl505 [DOI] [PubMed] [Google Scholar]
- 53. Leo GA, Focardi S, Gatto M, Cattadori IM (2004) The decline of the Grey Partridge in Europe: comparing demographies in traditional and modern agricultural landscapes. Ecol Model 177: 313–335 doi:10.1016/j.ecolmodel.2003.11.017 [Google Scholar]
- 54.EBCC (2012) Pan-European Common Bird Monitoring Scheme. Available: http://www.ebcc.info/index.php?ID=457. Accessed 2012 Feb 28.
- 55. Radwan J, Biedrzycka A, Babik W (2010) Does reduced MHC diversity decrease viability of vertebrate populations? Biol Conserv 143: 537–544 doi:10.1016/j.biocon.2009.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Říčanová Š, Bryja J, Cosson J-F, Gedeon C, Choleva L, et al. (2011) Depleted genetic variation of the European ground squirrel in Central Europe in both microsatellites and the major histocompatibility complex gene: implications for conservation. Conserv Genet 12: 1115–1129 doi:10.1007/s10592-011-0213-1 [Google Scholar]
- 57. Beani L, Dessì-Fulgheri F (1995) Mate choice in the grey partridge, Perdix perdix: role of physical and behavioural male traits. Anim Behav 49: 347–356 doi:10.1006/anbe.1995.0047 [Google Scholar]
- 58. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 59. Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acid Res 22: 4673–4680 doi:10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW (2006) Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol 23: 1891–1901 doi:10.1093/molbev/msl051 [DOI] [PubMed] [Google Scholar]
- 61. Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 62. Yang Z, Swanson WJ, Vacquier VD (2000) Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol 17: 1446–1455. [DOI] [PubMed] [Google Scholar]
- 63. Yang Z, Wong WSW, Nielsen R (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22: 1107–1118 doi:10.1093/molbev/msi097 [DOI] [PubMed] [Google Scholar]
- 64. Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, et al. (1993) Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364: 33–39 doi:10.1038/364033a0 [DOI] [PubMed] [Google Scholar]
- 65. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28: 2731–2739 doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sawyer SA (1999) GENECONV: a computer package for the statistical detection of gene conversion. Distributed by the author, Department of Mathematics, Washington University in St. Louis. Available: http://www.math.wustl.edu/~sawyer/geneconv/
- 67. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 68. Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, et al. (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31: 217–219. [DOI] [PubMed] [Google Scholar]
- 69. Worley K, Gillingham M, Jensen P, Kennedy LJ, Pizzari T, et al. (2008) Single locus typing of MHC class I and class II B loci in a population of red jungle fowl. Immunogenetics 60: 233–247 doi:10.1007/s00251-008-0288-0 [DOI] [PubMed] [Google Scholar]
- 70. Ekblom R, Sæther SA, Jacobsson P, Fiske P, Sahlman T, et al. (2007) Spatial pattern of MHC class II variation in the great snipe (Gallinago media). Mol Ecol 16: 1439–1451. [DOI] [PubMed] [Google Scholar]
- 71. Alcaide M, Edwards SV, Negro JJ, Serrano D, Tella JL (2008) Extensive polymorphism and geographical variation at a positively selected MHC class II B gene of the lesser kestrel (Falco naumanni). Mol Ecol 17: 2652–2665 doi:10.1111/j.1365-294X.2008.03791.x [DOI] [PubMed] [Google Scholar]
- 72. Chaves LD, Krueth SB, Reed KM (2009) Defining the turkey MHC: sequence and genes of the B locus. J Immunol 183: 6530–6537 doi:10.4049/jimmunol.0901310 [DOI] [PubMed] [Google Scholar]
- 73. Eimes JA, Bollmer JL, Dunn PO, Whittingham LA, Wimpee C (2010) Mhc class II diversity and balancing selection in greater prairie-chickens. Genetica 138: 265–271 doi:10.1007/s10709-009-9417-4 [DOI] [PubMed] [Google Scholar]
- 74. Bryja J, Galan M, Charbonnel N, Cosson J-F (2005) Analysis of major histocompatibility complex class II gene in water voles using capillary electrophoresis-single stranded conformation polymorphism. Mol Ecol Notes 5: 173–176 doi:10.1111/j.1471-8286.2004.00855.x [Google Scholar]
- 75. Babik W (2010) Methods for MHC genotyping in non-model vertebrates. Mol Ecol Resour 10: 237–251 doi:10.1111/j.1755-0998.2009.02788.x [DOI] [PubMed] [Google Scholar]
- 76. Promerová M, Babik W, Bryja J, Albrecht T, Stuglik M, et al. (2012) Evaluation of two approaches to genotyping major histocompatibility complex class I in a passerine – CE-SSCP and 454 pyrosequencing. Mol Ecol Resour 12: 285–292 doi:10.1111/j.1755-0998.2011.03082.x [DOI] [PubMed] [Google Scholar]
- 77. Goüy de Bellocq J, Suchentrunk F, Baird SJE, Schaschl H (2009) Evolutionary history of an MHC gene in two leporid species: characterisation of MHC-DQA in the European brown hare and comparison with the European rabbit. Immunogenetics 61: 131–144 doi:10.1007/s00251-008-0349-4 [DOI] [PubMed] [Google Scholar]
- 78. Čížková D, Goüy de Bellocq J, Baird SJE, Piálek J, Bryja J (2011) Genetic structure and contrasting selection pattern at two major histocompatibility complex genes in wild house mouse populations. Heredity 106: 727–740 doi:10.1038/hdy.2010.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Baratti M, Ammannati M, Magnelli C, Massolo A, Dessì-Fulgheri F (2010) Are large wattles related to particular MHC genotypes in the male pheasant? Genetica 138: 657–665 doi:10.1007/s10709-010-9440-5 [DOI] [PubMed] [Google Scholar]
- 80. Radwan J, Zagalska-Neubauer M, Cichon M, Sendecka J, Kulma K, et al. (2012) MHC diversity, malaria and lifetime reproductive success in collared flycatchers. Mol Ecol 21: 2469–2479 doi:10.1111/j.1365-294X.2012.05547.x [DOI] [PubMed] [Google Scholar]
- 81. Wittzell H, von Schantz T, Zoorob R, Auffray C (1995) Rfp-Y-like sequences assort independently of pheasant Mhc genes. Immunogenetics 42: 68–71. [DOI] [PubMed] [Google Scholar]
- 82. Reed KM, Bauer MM, Monson MS, Benoit B, Chaves LD, et al. (2011) Defining the turkey MHC: identification of expressed class I- and IIB-like genes independent of the MHC-B. Immunogenetics 63: 753–771 doi:10.1007/s00251-011-0549-1 [DOI] [PubMed] [Google Scholar]
- 83. Afanassieff M, Goto RM, Ha J, Sherman MA, Zhong L, et al. (2001) At least one class I gene in restriction fragment pattern-Y (Rfp-Y), the second MHC gene cluster in the chicken, is transcribed, polymorphic, and shows divergent specialization in antigen binding region. J Immunol 166: 3324–3333. [DOI] [PubMed] [Google Scholar]
- 84. Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrøm M, et al. (2011) The genome sequence of Atlantic cod reveals a unique immune system. Nature 477: 207–210 doi:10.1038/nature10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Strandh M, Lannefors M, Bonadonna F, Westerdahl H (2011) Characterization of MHC class I and II genes in a subantarctic seabird, the blue petrel, Halobaena caerulea (Procellariiformes). Immunogenetics 63: 653–666 doi:10.1007/s00251-011-0534-8 [DOI] [PubMed] [Google Scholar]
- 86. Borg AA, Pedersen SA, Jensen H, Westerdahl H (2011) Variation in MHC genotypes in two populations of house sparrow (Passer domesticus) with different population histories. Ecol Evol 1: 145–159 doi:10.1002/ece3.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ohta T (1991) Role of diversifying selection and gene conversion in evolution of major histocompatibility complex loci. Proc Natl Acad Sci USA 88: 6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ohta T (1995) Gene conversion vs point mutation in generating variability at the antigen recognition site of major histocompatibility complex loci. J Mol Evol 41: 115–119. [DOI] [PubMed] [Google Scholar]
- 89. Spurgin LG, van Oosterhout C, Illera JC, Bridgett S, Gharbi K, et al. (2011) Gene conversion rapidly generates major histocompatibility complex diversity in recently founded bird populations. Mol Ecol 20: 5213–5225 doi:–10.1111/j.1365–294X.2011.05367.x [DOI] [PubMed] [Google Scholar]
- 90. Miller HC, Bowker-Wright G, Kharkrang M, Ramstad K (2011) Characterisation of class II B MHC genes from a ratite bird, the little spotted kiwi (Apteryx owenii). Immunogenetics 63: 223–233 doi:10.1007/s00251-010-0503-7 [DOI] [PubMed] [Google Scholar]
- 91. Eo SH, Bininda-Emonds ORP, Carroll JP (2009) A phylogenetic supertree of the fowls (Galloanserae, Aves). Zool Scr 38: 465–481 doi:10.1111/j.1463-6409.2008.00382.x [Google Scholar]
- 92. Shen Y, Liang L, Sun Y, Yue B, Yang X, et al. (2010) A mitogenomic perspective on the ancient, rapid radiation in the Galliformes with an emphasis on the Phasianidae. BMC Evol Biol 10: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426. [DOI] [PubMed] [Google Scholar]
- 94. Lambourne MD, Si W, Niemiec PK, Read LR, Kariyawasam S, et al. (2005) Identification of novel polymorphisms in the B-LB locus of Gallus lafayettei. Anim Genet 36: 435–462 doi: 10.1111/j.1365-2052.2005.01332.x [DOI] [PubMed] [Google Scholar]
- 95. Singh SK, Mehra S, Kumar V, Shukla SK, Tiwari A, et al. (2010) Sequence variability in the BLB2 region among guinea fowl and other poultry species. Int J Genet Mol Biol 2: 048–051. [Google Scholar]
- 96. Hale ML, Verduijn MH, Moller AP, Wolff K, Petrie M (2009) Is the peacock’s train an honest signal of genetic quality at the major histocompatibility complex? J Evol Biol 22: 1284–1294 doi:10.1111/j.1420-9101.2009.01746.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all primers tested for PCR amplification of MHCIIB in the Grey partridge and their schematic overview.
(DOC)
Overview of all MHCIIB sequences of Galliformes used in the phylogenetic analysis, with references and GenBank accession numbers.
(DOC)
Posterior means of ω for each codon of MHCIIB exon 2 in the Grey partridge, calculated as the average of ω over the 11 site classes, weighted by the posterior probabilities under the random-sites model M8 (β and ω). The posterior probabilities were computed by the Bayes empirical Bayes procedure in the program CodeML implemented in the PAML3.14 package.
(DOCX)