Abstract
Background
Artesunate/sulphadoxine-pyrimethamine (AS/SP) has been the first-line treatment for falciparum malaria in Sudan since 2004. The impact of this combination on anti-malarial resistance-associated molecular markers has not been investigated. In this study, an evaluation of the efficacy and prevalence of drug resistance alleles (pfcrt, pfmdr1, pfdhfr and pfdhps) eight years after the adoption of AS/SP in eastern Sudan is reported.
Methods
A 28-day follow-up efficacy trial of AS/SP was conducted in eastern Sudan during the 2012 transmission season. Blood smears were collected from patients on days 0, 1, 2, 3, 7, 14, 21 and 28. Blood spots on filter paper were obtained pre-treatment and on the day the patient was parasite positive by microscopy. Genotyping of alleles was performed by qPCR (pfcrt 72–76 and pfmdr1 copy number) and direct sequencing of pfmdr1, pfdhfr and pfdhps.
Results
Sixty-three patients out of 68 (93%) completed the 28-day follow-up, adequate clinical, and parasitological response occurred in 90.5% and 85.3% of the patients in the per-protocol and intent-to-treat analyses, respectively. PCR corrected per-protocol efficacy was 93.7%. The enrolment prevalence of pfcrt-CVMNK was 30.2% and pfmdr1-N86 was 40.3%. The pfmdr1 haplotype NFD occurred in 32.8% of pre-treatment samples and was significantly higher than previous reports (Fisher’s exact p = 0.0001). The pfdhfr-51I/108N combination occurred in all sequenced isolates and 59R was observed in a single individual. pfdhps substitutions 436A, 437G, 540E, 581G and 613S were observed at 7.8, 77.3, 76.9%, 33.8% and 0.0%, respectively. Treatment failures were associated with the pfdhps haplotype SGEGA at these five codons (OR 7.3; 95% CI 0.65 - 368; p = 0.048).
Conclusion
The decrease of CQR associated genotypes reflects the formal policy of complete removal of CQ in Sudan. However, the frequency of markers associated with SP failure is increasing in this study area and may be contributing to the treatment efficacy falling below 90%. Further monitoring of AS/SP efficacy and of post-treatment selection of pfdhfr and pfdhps alleles in vivo is required to inform future treatment guidelines.
Keywords: Plasmodium falciparum, pfdhfr, Pfdhps, Pfcrt, Pfmdr1, Artesunate, Sulphadoxine-pyrimethamine, Sudan
Background
Malaria continues to affect over two million people worldwide with more than 600,000 reported deaths in 2010, most of these in sub-Saharan Africa [1]. There are increasing efforts to control, eliminate or eradicate malaria in many countries [2], however these are hindered by ineffective treatment and vector control strategies that cannot be sustained in economically disadvantageous countries.
Malaria in Sudan causes morbidities and mortality among all age groups and was responsible for more than 95,000 hospital admissions in 2011 [3]. Plasmodium falciparum causes over 95% of malaria infections and drug resistance to CQ and SP is well established [4-7]. The wide-scale deployment of artemisinin combination therapy (ACT) across malaria-endemic zones is intended to counteract the emergence and spread of resistance [8]. Artesunate-sulphadoxine/pyrimethamine (AS/SP) and artemether-lumefantrine (AL) became the first- and second-line of treatment, respectively, for uncomplicated P. falciparum malaria in 2004 [9]. Recently, the efficacy of AS/SP has been questioned among practicing physicians with some patients returning with recurring infections. The in vivo efficacy of SP in eastern Sudan was reported at 82% [10], similar to Tanzania (86%) [11] and high levels of resistance to SP in East Africa [12,13] may render the short half-life artesunate partner unprotected. Furthermore the emergence of slow clearing parasites following artesunate treatment in South East Asia [14,15] has prompted closer monitoring of the efficacy of ACT and necessitated extensive research on molecular markers of resistance to artemisinins and partner drugs.
Specific haplotypes of the pfcrt gene at positions 72 – 76 (CVMNK; wild haplotype and CVIET; mutant Asian/African haplotype; mutant SVMNT South American haplotype) are well-established markers for resistance to CQ [16,17]. In eastern Sudan the prevalence of the pfcrt-76T increased from 54% in the early 1990’s [18,19] to more than 90% in the 2000’s [7,20,21]. However, the pfcrt-76K allele has returned to high prevalence in parts of Africa where CQ was discontinued, such as Malawi [22,23], Kenya [24,25], Senegal, [26] Tanzania [27,28] and Mozambique [29].
Pfmdr1 haplotypes N86, 184F and 1246D have been selected by artemether-lumefantrine in eastern Sudan [30] as well as several parts of Africa [31-34]. These haplotypes were also found to increase the risk of treatment failure with AL [35]. Amplification of pfmdr1 which is associated with decreased sensitivity to artemisinins in South East Asia [36,37] has recently been observed in eastern Sudan [30] and in neighbouring east African countries [38,39]. Therefore, pfmdr1 may continue to be under the influence of ACT currently employed in Sudan.
Resistance to pyrimethamine is due to a set of sequential mutations in the dihydrofolate reductase gene (dhfr) [40,41]. The serine to asparagine at position 108 is essential with additional mutations at positions N51I, C59R and I164L leading to higher resistance levels [42,43]. Mutations in the pfdhps gene (S436A, G437A, K540E, A581G and A613S) encoding the target enzyme for sulphadoxine are also predictors of treatment failure in vivo[44]. It has been recommended that SP should be discontinued in areas where the prevalence of the pfdhps-540E allele is more than 50% [12]. In eastern Sudan, the prevalence of the pfdhfr CICNI haplotype at codons 50, 51, 59, 108 and 164 has been reported at 90% [20]. The pfdhps haplotype SGEAA (436/437/540/581/613) was higher than 80% in the current study area prior to the introduction of AS/SP [20]. These two alleles may be maintained in the population by the strong selection pressure imposed by SP as an ACT component.
In this study, the prevalence of molecular markers in four genes (pfcrt, pfmdr1, pfdhfr and pfdhps) associated with drug resistance following the wide scale deployment of AS/SP in eastern Sudan is investigated and an updated clinical efficacy of AS/SP is reported.
Methods
Patients enrolment
Patients were recruited between January and March 2012 at Fatima Eldiaaig Health Centre in Kassala City in eastern Sudan, which is characterized by moderate perennial malaria transmission with a peak in January [45]. Febrile patients (auxiliary temperature of ≥37.5°C) from all age groups with microscopically confirmed uncomplicated P. falciparum mono-infection and a parasite count of a minimum of 1,000 a sexual parasites/μl were asked to participate and those who consented were enrolled. Patients with concomitant illnesses and pregnant women were excluded from the study. A structured questionnaire for socio- demographic information and medical history was completed for each patient by a physician. The study was performed as per the WHO guidelines for anti-malarial drug efficacy surveillance methods [46].
Sample collection and laboratory diagnosis
Finger prick samples were taken from all participants, thick smears were prepared and stained with 10X Giemsa stain and slides were read under a 100X oil immersion field. Parasite density was obtained by counting asexual parasites against 200 leucocytes and parasite density was calculated assuming an average of 6,000 leucocytes/μl. Slides were read by two experienced microscopists: a slide was considered negative if no parasites were detected in 100 high powerfields. Blood spots were collected on Whatman® No. 3 filter paper, air dried and stored in self-sealing bags for DNA analysis pre-treatment and on the day of treatment failure.
Treatment and follow up
All participants were given, under observation, artesunate/sulphadoxine-pyrimethamine(Artecospe, Guilin pharmaceutical, China) comprising 4 mg/kg artesunate on days 0–2 as a single daily dose and a single dose of SP (25 mg/kg sulphadoxine, 1.25 mg/kg pyrimethamine) on day 0. For children the dose was adjusted by weight and the tablets were dissolved in water for oral administration. A full dose of the drug was repeated in case of vomiting after 60 minutes and half the dose was repeated if the patient vomited between 30 to 60 minutes after drug administration.
Patients were asked to return to the health centre on days 1, 2, 3, 7, 14, 21 and 28 or if they remained unwell, on each visit axillary temperature was measured and a blood smear was taken. Patients were asked about any adverse effects of the drugs such as nausea, vomiting, abdominal pain, itching and rashes: these symptoms were considered drug induced if they were not reported on enrolment.
Patients who remained microscopy negative for asexual parasitaemia of P. falciparum throughout the follow up period were considered to have achieved adequate clinical and parasitological response (ACPR). Treatment failure was classified as early treatment failure (ETF), late parasitological failure (LPF) or late clinical failure (LCF) according to the WHO guidelines [46]. Those who failed treatment were given AL (Coartem®) as per the Sudanese National Malaria Control Programme guidelines.
DNA extraction and loci detection
Parasite DNA was extracted from dried filter paper in 96-deep-well plates by the Chelex® extraction method [47] with modifications for 96-well plates [34]. The pfcrt loci at codons 72 – 76 were detected by a Taqman assay as described previously [7]. Briefly, 5 μl of DNA were added to the master mix containing 0.3 μM of primers CRTD1 and CRTD2 [48] and 0.1 μM of each probe (crt76-CVMNK, crt76-CVIET and crt76-SVMNT).
Regions of pfmdr1 encompassing polymorphisms at positions N86Y, Y184F, C1034S, D1042N and D1246Y were amplified with previously reported primers [34]. Polymorphic sites in pfdhfr N51I, C59R, S108N, I164L, pfdhps A436, A437G, K540E, A581G and A613S were amplified as previously described [49] and directly sequenced.
Amplification of the pfmdr1 gene was investigated by a qPCR assay as previously described [50] employing the Taqman® Universal Mastermix. DNA from clones 3D7 and Dd2 were employed as controls for single and multiple copy number, respectively. Each sample and control was run in triplicate.
Msp1 and msp2 polymorphic markers were amplified as described previously [51] to confirm the origin of recurring parasites in those patients that were microscopically positive during the follow up.
Ethical considerations
The study received ethical clearance from the Health Research Board at Ministry of Health, Kassala State. Written informed consent was obtained from each patient or child guardian.
Statistical analysis
The intention-to-treat analysis included all enrolled patients who met the inclusion criteria and took at least one full dose of AS/SP. Patients lost to follow-up or withdrawn from the study were considered to be treatment failures. The per protocol (PP) analysis included data for patients who had completed the follow-up. Those patients lost to the follow-up or who were withdrawn because of protocol violations were excluded from the PP analysis. Clinical data were entered in MS-Excel and analysed in SPSS software (SPSS Inc., Chicago, IL, USA). The means (age, weight, temperature and parasite count) were calculated for all patients. The difference in frequency of haplotypes observed in 2003 [20,52], 2006 [30], and in this study were analysed by Fisher’s exact test (STATA Statistical Software: Release 8.1, 2003, Stata Corp., College Station, TX). Odds ratios were calculated where appropriate. The difference was assumed significant if the p-value ≤ 0.05.
Results
A total of 453 patients who came to the clinic with fever and reporting any malaria-like symptoms were screened for P. falciparum infection. Sixty-eight patients fulfilled the enrolment criteria and consented to participate in the study, sixty-three (92.6%) of whom completed 28-days of follow up. Patient age, weight, temperature, parasite density and gender are summarized in Table 1. Mean time to resolving clinical symptoms such as fever (axillary temperature above 37°C), malaise and nausea was 2.7 days (range 2 – 7 days) and was not a risk for failing treatment.
Table 1.
Characteristics of study participants
| Variable | Mean [range] |
|---|---|
| Age (years) |
25.8 [2 – 70] |
| Weight (Kg) |
44.9 [6 – 104] |
| Temperature °C |
38.3 [37.5 – 40.7] |
| Geometric mean parasite density parasites/μl |
4167.1 [1,080 – 200,000] |
| Sex: female (%) male (%) | 44.4% 55. 6% |
On day 1; 12.7% (8/63) patients were febrile and 20.6% (13/63) patients were parasitaemic. However, all patients cleared their parasitaemia by day 3 with one patient remaining febrile (temperature 38°C). Gametocytaemia was observed in 3.2% (2/63) of patients at recruitment. Adverse effects of the drugs such as nausea, headache, cough, dizziness, abdominal pain and diarrhoea were reported in 34.9% (22/63) of patients between day 19 and 28. In the intent-to-treat analysis where all patients who were enrolled were included in the analysis, the efficacy of AS/SP was 85.3%. While in the per-protocol analysis the efficacy was 90.5%. Two patients failed treatment at day 21, one failed treatment at day 26 and two failed treatment at day 28. The PCR corrected efficacy was 93.6% where 4/5 of treatment failures were classified as recrudescing parasites. Factors such as pre-treatment parasite density, axillary temperature and age did not differ significantly between those who failed treatment and those who were successfully treated.
Expansion of wild type pfcrt and pfmdr1 haplotypes following withdrawal of CQ
The wild type pfcrt haplotype CVMNK has significantly recovered from 10% in 2003 [52] to 30.2% (Fishers exact p < 0.001) in this study. . The N86 allele has also increased significantly from 14% [52] to 40.3% (Fisher’s exact p = 0.0059). The prevalence of the wild type Y184 was 15.6% but this was not significantly different from 2006 (Fisher’s exact p = 0.16), while there was a significant decrease in mutant 1246Y (Fisher’s exact p = 0.018) (Table 2) [30]. Pfmdr1 haplotypes at positions 86/184/1246 were less diverse than 2006. The major pfmdr1 haplotype in this study was YFD observed in 51.6% of pre-treatment samples. NFD has significantly increased in prevalence from 5.6% in 2006 to 32.8% in 2012 (Fishers exact p = 0.0001) (Figure 1). The NFD haplotype was detected in 3/5 post-treatment samples and 2 samples were YFD. All the isolates analysed in this study carried the wild type alleles at positions S1034 and N1042 and a single copy of the pfmdr1 gene (mean 1.2; range 0.6 to 1.5).
Table 2.
Changes in pre-treatment frequencies of pfcrt and pfmdr1 haplotypes between 2003 and 2012 in eastern Sudan
| pfcrt/pfmdr1 allele | 2003 % | 2006 % | 2012 % | p-value 2003/2006 |
|---|---|---|---|---|
|
CVIET |
93[20] 90[52] |
|
74.6 |
0.000/0.080 |
|
CVMNK |
10[52] |
|
30.2 |
0.010 |
|
CVMNK/CVIET |
24[52] |
|
4.8 |
0.012 |
|
N86 |
14[52] |
12.1[30] |
40.3 |
0.006/0.0001 |
|
86Y |
84[52] |
85[30] |
55.2 |
0.006/0.000 |
|
86 N/Y |
2[52] |
3[30] |
4.5 |
0.629/0.691 |
|
184F |
|
74[30] |
80.6 |
0.165 |
|
184 Y/F |
|
0[30] |
4.5 |
0.074 |
|
1246Y |
|
6[30] |
0.0 |
0.755 |
| 1246 D/Y | 3[30] | 0.0 | 0.270 |
Figure 1.
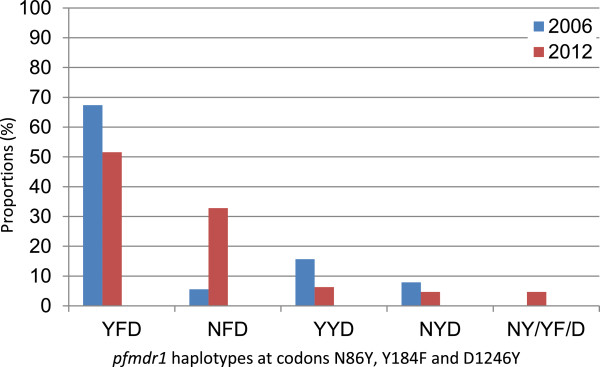
Difference in proportions of pfmdr1 haplotypes in eastern Sudan between 2006 and 2012.
Changes in antifolate resistance markers
For the pfdhfr gene a total of 59 samples were successfully sequenced for positions 50 to 164. All of which carried the 51I and 108N mutant alleles. This is a significant increase from previous years (Fisher’s exact p = 0.041). One sample (1.7%) harboured the mutant 59R and all of the samples were wild-type at position 164. Thus the predominant haplotype for pfdhfr in eastern Sudan is 51I/C59/108N.
A total of 65 pre-treatment samples were successfully genotyped at the pfdhps positions 430 to 613. A novel polymorphism at position L516F was detected in one sample. The prevalence of the 581G allele significantly increased since the 2003 study from 14% to 33.8% (Fisher’s exact p = 0.0005), while the 437G decreased slightly from 89% to 77% (Fisher’s exact p = 0.0134) [20]. In pre-treatment samples, the 581G allele was strongly linked to the 540E (p = 0.0009) and the 437G (p = 0.0018). The prevalence of S436A, A437G, L516F, K540E and A613S are listed in Table 3.
Table 3.
Prevalence of pfdhfr and pfdhps alleles in Sudan before and after adoption of ACT
| pfdhfr/pfdhps allele | 2003 % | 2012 % | p-value [[20],[52]] |
|---|---|---|---|
|
51I/108N |
90[20] 84[52] |
100 |
0.004/0.001 |
|
59R |
1[20] 0[52] |
1.7 |
0.477/1.000 |
|
436A |
0[52] |
7.8 |
0.069 |
|
437G |
89[20] 0[52] |
77.3 |
0.013/0.000 |
|
516F |
|
1.7* |
|
|
K540 |
20[52] |
21.5 |
0.000 |
|
540E |
79[20] |
76.9 |
0.860 |
|
540K/E |
80[52] |
1.5 |
0.000 |
|
581G |
14[20] 10[52] |
33.8 |
0.001/0.007 |
|
581A/G |
10[52] |
1.5 |
0.202 |
| A613 | 100[20,52] | 100 | 1.000 |
Pfdhps sequencing was successful for all five treatment failures, collected at day 21 (2), day 26 (1) and day 28 (2). The haplotype across the five polymorphic codons 436, 437, 540, 581 and 613 are shown for these two groups in Figure 2. Whereas all five samples collected at the time of treatment failure carried the 540E substitution as expected, all of them also carried the 437G and 581G. Thus, the pfdhps haplotype SGEGA, at these five polymorphic positions, was significantly more common in parasites detected after treatment (OR 7.3; 95% CI 0.65 - 368; p = 0.048) whether these parasites were recrudescing or re-infecting parasites. Further, the presence of this pfdhps genotype in the pre-treatment infection was a significant risk factor for later treatment failure (relative risk 7.3, 95% CI 0.87 - 61.6; p = 0.030) the wide confidence intervals indicate that these observations need detailed exploration in larger studies.
Figure 2.
Proportions of pfdhps haplotypes before and after treatment with AS/SP in Kassala State 2012.
Discussion
Plasmodium falciparum carrying mutations associated with drug resistance may be less fit to survive in the absence of drug pressure. The continuous use of CQ monotherapy for decades has lead to almost saturation of the P. falciparum population in eastern Sudan carrying the mutant pfcrt allele CVIET [7]. Lower drug pressure is reflected by more wild type pfcrt where transmission is higher and perennial in central Sudan [53]. Moreover SP has been used intensively in patients where CQ failed. This has resulted in the high prevalence of the pfdhfr/pfdhps mutants (51I/108N/437G/540E) in the late 1990’s and early 2000’s [18,20,54]. The adoption of AS/SP in eastern Sudan in 2004 has lead to considerable decrease in malaria cases in the country [9,55-57]. The clinical efficacy of SP monotherapy in Sudan was approximately 68% prior to introduction of AS/SP [58]. Therefore, the wisdom of deploying this SP containing combination has been debated among the malaria research community in Sudan e.g. [59]. The base-line efficacy of AS/SP in eastern Sudan has been reported between 98 - 100% [56,60-62], while we have reported a base-line PCR corrected efficacy of 93% [57]. In the present study the in vivo per-protocol PCR corrected efficacy of AS/SP over 28 days is 90.5%. All patients cleared their microscopic parasitaemia by day 3. Approximately 10% of patients exhibited recrudescing parasites as confirmed by msp1 and msp2 genotyping during the study. It is important to emphasize the relatively lower transmission season in eastern Sudan compared to other parts of East Africa. Therefore, to definitely establish whether these post-treatment infections were true recrudescence or re-infections requires genotyping at more polymorphic loci.
A limitation to our data is the lack of parallel PCR data for days 3 and 14. Sub-microscopic parasites at these time points have been detected in previous studies from eastern Sudan [7,30]. Detection of parasites on day 3 is an early warning of slow clearance by artemisinin derivatives observed in South East Asia [14,15]. A decline in the efficacy of ACT e.g. AL and DHA/PIP has been reported from Kenya [63]. Sub-microscopic parasitaemia has previously been observed following AL treatment in eastern Sudan [30]. While an artemisinin resistant phenotype has not been reported in Africa [64], the history of anti-malarial resistance suggests that artemisinin resistance is likely to spread to East Africa despite the global efforts in its containment.
In the present study, the prevalence of molecular markers eight years after the adoption of ACT is compared to previous reports from the same study area of eastern Sudan. In this area IPTp is recommended for all pregnant women, as in other parts of Sudan.
The prevalence of a double mutant pfdhfr allele (51I/108N) has reached 100%, which clearly demonstrates the effect of drug pressure on this locus. The observation that contrasts with other African countries is the slow evolution of pfdhfr in this region where the double mutant 51–108 allele has widely spread while the wild type C59 has been maintained. However it is in agreement with a recent report from Yemen [65]. The 59R is associated with increased resistance to pyrimethamine, therefore absence of this mutation may be a good indicator for the in vivo efficacy of SP in this region. The pfdhfr 59R has been previously reported at low frequency in Khartoum, central Sudan in 1996/1997 [66] and Nuba mountains in western Sudan in 2003 [67]. However the 51I/108N haplotype has been observed at low frequencies in Kenya and Cameroon in the early 2000s where the pfdhfr-51I/59R/108N haplotype currently predominates [68,69]. These studies employed microsatellite markers which demonstrated the divergence of origin of the double (51I/108N) and triple mutants within the same geographic location. An earlier study in eastern Sudan has shown that microsatellites around pfdhfr were stable over four consecutive seasons [18]. Thus, the pfdhfr allele 51I/108N may have been imported into this area earlier and has been maintained within a limited parasite population in a low transmission area of eastern Sudan. However, it would be of interest to examine pfdhfr sequences and microsatellite markers from other regions of Sudan with different transmission intensities to test this hypothesis.
Interestingly in this study, a small decrease in pfdhps-437G (Fisher’s exact p = 0.0134) is observed compared to previous reports [20]. The 581G allele has significantly increased to 33% and has previously been associated with SP failure in eastern Sudan [20] emphasizing the importance of close monitoring of the parasite population in this region. For the first time, a particular pfdhps allele, SGEGA, is significantly associated with treatment failure in patients receiving AS/SP for clinical malaria. However of 23 pretreatment infections harbouring SGEGA, only four recrudesced. The increase of SGEGA has been previously reported from Tanzania in cross sectional surveys following several years of SP use [27]. The potential of this haplotype as a single marker of drug efficacy for this anti-malarial combination should be further investigated.
As previously reported from other African countries [22,24,28], there is a significant decrease in CQR associated genotypes pfcrt-76T/pfmdr1-86Y. Wild type genotypes at these two loci have significantly recovered, which provides supporting evidence that CQ use has been successfully reduced [70]. Interestingly the multiple copy pfmdr1 genotype previously reported from eastern Sudan [30] was not observed here. This genotype is associated with resistance to mefloquine (MQ) in South East Asia [50] and may not spread in the absence of MQ pressure, which is the case in Sudan. In addition pfmdr1amplification has failed to be established and spread in Africa, possibly due to lack of the use of MQ across the continent.
In Sudan SP is recommended for iPTp and the availability of SP as an over-the-counter drug maintains the pressure on the parasite population. The efficacy of AS/SP is reaching the current threshold for anti-malarial policy change of 90%. Continuous use of SP as a partner to artesunate may be the cause of declined AS/P efficacy. In addition discontinuation of SP in areas where the population prevalence of pfdhps-540E is above 50% is also recommended [12,71]. Monitoring of SP resistance associated loci, and in particular the SGEGA pfdhps haplotype identified here, is required to provide informed treatment guidelines in the near future. In addition the emergence of slow parasite clearance in South East Asia warrants close monitoring of the efficacy of ACT and molecular markers associated with resistance to partner drugs.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NBG carried out the molecular genetic studies, the sequence alignment, analysed the results and drafted the manuscript. TMA carried out the in vivo study, analysed the clinical data and drafted the manuscript. SA participated in the molecular genetic studies and analyses. CJS designed the molecular study, performed the statistical association analysis, contributed to the manuscript and critically revised it, IA conceived of the study, participated in its design and coordination and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Nahla B Gadalla, Email: nahla.gadalla@nih.gov.
Tajeldin M Abdallah, Email: tajeldinmm@yahoo.com.
Sharanjeet Atwal, Email: sharan939@hotmail.com.
Colin J Sutherland, Email: colin.sutherland@lshtm.ac.uk.
Ishag Adam, Email: ishagadam@hotmail.com.
Acknowledgements
We are indebted to the study participants for their cooperation through the study. We would like to thank Rebekah Burrow and Don van Schalkwyk for genotyping msp1/msp2. The in vivo study was supported by Kassala University. Genotyping of the samples was supported by the EU- FP7 MALACTRES consortium. CJS is supported by Public Health England.
References
- WHO. World Malaria Report 2011. Geneva: World Health Organization; 2011. [Google Scholar]
- Feachem RG, Phillips AA, Hwang J, Cotter C, Wielgosz B, Greenwood BM, Sabot O, Rodriguez MH, Abeyasinghe RR, Ghebreyesus TA, Snow RW. Shrinking the malaria map: progress and prospects. Lancet. 2010;376:1566–1578. doi: 10.1016/S0140-6736(10)61270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World Malaria Report 2012. Geneva: World Health Organization; 2012. [Google Scholar]
- Omer AH. Response of Plasmodium falciparum in Sudan to oral chloroquine. Am J Trop Med Hyg. 1978;27:853–857. doi: 10.4269/ajtmh.1978.27.853. [DOI] [PubMed] [Google Scholar]
- Bayoumi RA, Babiker HA, Ibrahim SM, Ghalib HW, Saeed BO, Khider S, Elwasila M, Karim EA. Chloroquine-resistant Plasmodium falciparum in eastern Sudan. Acta Trop. 1989;46:157–165. doi: 10.1016/0001-706x(89)90032-6. [DOI] [PubMed] [Google Scholar]
- Ibrahim ME, Awad-el-Kariem FM, el Hassan IM, el Mubarak ER. A case of Plasmodium falciparum malaria sensitive to chloroquine but resistant to pyrimethamine/sulfadoxine in Sennar, Sudan. Trans R Soc Trop Med Hyg. 1991;85:446. doi: 10.1016/0035-9203(91)90210-p. [DOI] [PubMed] [Google Scholar]
- Gadalla NB, Elzaki SE, Mukhtar E, Warhurst DC, El-Sayed B, Sutherland CJ. Dynamics of pfcrt alleles CVMNK and CVIET in chloroquine-treated Sudanese patients infected with Plasmodium falciparum. Malar J. 2010;9:74. doi: 10.1186/1475-2875-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ. Antimalarial drug resistance. J Clin Invest. 2004;113:1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik EM, Mohamed TA, Elmardi KA, Mowien RM, Elhassan AH, Elamin SB, Mannan AA, Ahmed ES. From chloroquine to artemisinin-based combination therapy: the Sudanese experience. Malar J. 2006;5:65. doi: 10.1186/1475-2875-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam I, Ibrahim MH, Ae IA, Elbashir MI. Efficacy of sulfadoxin pyrimethamine for uncomplicated Plasmodium falciparum malaria in a small sample of Sudanese children. East Mediterr Health J. 2004;10:309––314. [PubMed] [Google Scholar]
- Mugittu K, Abdulla S, Falk N, Masanja H, Felger I, Mshinda H, Beck HP, Genton B. Efficacy of sulfadoxine-pyrimethamine in Tanzania after two years as first-line drug for uncomplicated malaria: assessment protocol and implication for treatment policy strategies. Malar J. 2005;4:55. doi: 10.1186/1475-2875-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo I, Roper C. Following the path of most resistance: dhps K540E dispersal in African Plasmodium falciparum. Trends Parasitol. 2010;26:447–456. doi: 10.1016/j.pt.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Sridaran S, McClintock SK, Syphard LM, Herman KM, Barnwell JW, Udhayakumar V. Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malar J. 2010;9:247. doi: 10.1186/1475-2875-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Steketee RW, Plowe CV. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet. 2001;358:890–891. doi: 10.1016/S0140-6736(01)06040-8. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Muhsin AM, Mackinnon MJ, Ali E, el Nassir BA, Suleiman S, Ahmed S, Walliker D, Babiker HA. Evolution of drug-resistance genes in Plasmodium falciparum in an area of seasonal malaria transmission in Eastern Sudan. J Infect Dis. 2004;189:1239–1244. doi: 10.1086/382509. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J Infect Dis. 2001;183:1535–1538. doi: 10.1086/320195. [DOI] [PubMed] [Google Scholar]
- A-Elbasit IE, Khalil IF, Elbashir MI, Masuadi EM, Bygbjerg IC, Alifrangis M, Giha HA. High frequency of Plasmodium falciparum CICNI/SGEAA and CVIET haplotypes without association with resistance to sulfadoxine/pyrimethamine and chloroquine combination in the Daraweesh area, in Sudan. Eur J Clin Microbiol Infect Dis. 2008;27:725–732. doi: 10.1007/s10096-008-0499-1. [DOI] [PubMed] [Google Scholar]
- Tagelsir N, Ibrahim Z, Medani A, Salih O, Hamad A, Giha H, El-Agib A, Khan B, Saeed N, Ibrahim M. High frequency of Plasmodium falciparum PfCRT K76T and PfpghN86Y in patients clearing infection after chloroquine treatment in the Sudan. Acta Trop. 2006;97:19–25. doi: 10.1016/j.actatropica.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimde AA, Kouriba B, Taylor TE, Plowe CV. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- Mita T, Kaneko A, Lum JK, Bwijo B, Takechi M, Zungu IL, Tsukahara T, Tanabe K, Kobayakawa T, Bjorkman A. Recovery of chloroquine sensitivity and low prevalence of the Plasmodium falciparum chloroquine resistance transporter gene mutation K76T following the discontinuance of chloroquine use in Malawi. Am J Trop Med Hyg. 2003;68:413–415. [PubMed] [Google Scholar]
- Mang'era CM, Mbai FN, Omedo IA, Mireji PO, Omar SA. Changes in genotypes of Plasmodium falciparum human malaria parasite following withdrawal of chloroquine in Tiwi, Kenya. Acta Trop. 2012;123:202–207. doi: 10.1016/j.actatropica.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, Sasi P, Marsh K, Borrmann S, Mackinnon M, Nzila A. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye M, Faye B, Tine R, Ndiaye JL, Lo A, Abiola A, Dieng Y, Ndiaye D, Hallett R, Alifrangis M, Gaye O. Assessment of the molecular marker of Plasmodium falciparum chloroquine resistance (Pfcrt) in Senegal after several years of chloroquine withdrawal. Am J Trop Med Hyg. 2012;87:640–645. doi: 10.4269/ajtmh.2012.11-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifrangis M, Lusingu JP, Mmbando B, Dalgaard MB, Vestergaard LS, Ishengoma D, Khalil IF, Theander TG, Lemnge MM, Bygbjerg IC. Five-year surveillance of molecular markers of Plasmodium falciparum antimalarial drug resistance in Korogwe District, Tanzania: accumulation of the 581G mutation in the P. falciparum dihydropteroate synthase gene. Am J Trop Med Hyg. 2009;80:523–527. [PubMed] [Google Scholar]
- Kamugisha E, Bujila I, Lahdo M, Pello-Esso S, Minde M, Kongola G, Naiwumbwe H, Kiwuwa S, Kaddumukasa M, Kironde F, Swedberg G. Large differences in prevalence of Pfcrt and Pfmdr1 mutations between Mwanza, Tanzania and Iganga, Uganda-a reflection of differences in policies regarding withdrawal of chloroquine? Acta Trop. 2011;121:148–151. doi: 10.1016/j.actatropica.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Thomsen TT, Madsen LB, Hansson HH, Tomas EV, Charlwood D, Bygbjerg IC, Alifrangis M. Rapid selection of Plasmodium falciparum chloroquine resistance transporter gene and multidrug resistance gene-1 haplotypes associated with past chloroquine and present artemether-lumefantrine use in Inhambane District, southern Mozambique. Am J Trop Med Hyg. 2013;88:536–541. doi: 10.4269/ajtmh.12-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla NB, Adam I, Elzaki SE, Bashir S, Mukhtar I, Oguike M, Gadalla A, Mansour F, Warhurst D, El-Sayed BB, Sutherland CJ. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrob Agents Chemother. 2011;55:5408–5411. doi: 10.1128/AAC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- Thomsen TT, Ishengoma DS, Mmbando BP, Lusingu JP, Vestergaard LS, Theander TG, Lemnge MM, Bygbjerg IC, Alifrangis M. Prevalence of single nucleotide polymorphisms in the Plasmodium falciparum multidrug resistance gene (Pfmdr-1) in Korogwe District in Tanzania before and after introduction of artemisinin-based combination therapy. Am J Trop Med Hyg. 2011;85:979–983. doi: 10.4269/ajtmh.2011.11-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJ, Mutabingwa TK, Sutherland CJ, Hallett RL. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg M, Ferreira PE, Tarning J, Ursing J, Ngasala B, Bjorkman A, Martensson A, Gil JP. Plasmodium falciparum drug resistance phenotype as assessed by patient antimalarial drug levels and its association with pfmdr1 polymorphisms. J Infect Dis. 2013;207:842–847. doi: 10.1093/infdis/jis747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Alam MT, Sem R, Shah NK, Susanti AI, Lim P, Muth S, Maguire JD, Rogers WO, Fandeur T, Barnwell JW, Escalante AA, Wongsrichanalai C, Ariey F, Meshnick SR, Udhayakumar V. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J Infect Dis. 2010;201:1551–1560. doi: 10.1086/651949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, van Vugt M, Brockman A, Hutagalung R, Nair S, Nash D, Singhasivanon P, Anderson TJ, Krishna S, White NJ, Nosten F. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis. 2006;42:1570–1577. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren G, Bjorkman A, Gil JP. Amodiaquine resistance is not related to rare findings of pfmdr1 gene amplifications in Kenya. Trop Med Int Health. 2006;11:1808–1812. doi: 10.1111/j.1365-3156.2006.01742.x. [DOI] [PubMed] [Google Scholar]
- Ngasala BE, Malmberg M, Carlsson AM, Ferreira PE, Petzold MG, Blessborn D, Bergqvist Y, Gil JP, Premji Z, Bjorkman A, Martensson A. Efficacy and effectiveness of artemether-lumefantrine after initial and repeated treatment in children <5 years of age with acute uncomplicated Plasmodium falciparum malaria in rural Tanzania: a randomized trial. Clin Infect Dis. 2011;52:873–882. doi: 10.1093/cid/cir066. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snewin VA, England SM, Sims PF, Hyde JE. Characterisation of the dihydrofolate reductase-thymidylate synthetase gene from human malaria parasites highly resistant to pyrimethamine. Gene. 1989;76:41–52. doi: 10.1016/0378-1119(89)90006-1. [DOI] [PubMed] [Google Scholar]
- Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirawaraporn W, Yongkiettrakul S, Sirawaraporn R, Yuthavong Y, Santi DV. Plasmodium falciparum: asparagine mutant at residue 108 of dihydrofolate reductase is an optimal antifolate-resistant single mutant. Exp Parasitol. 1997;87:245–252. doi: 10.1006/expr.1997.4221. [DOI] [PubMed] [Google Scholar]
- Triglia T, Menting JG, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci USA. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeidan YE, Elbashir MI, El Rayah el A, Adam I. Epidemiology of malaria in New Halfa, an irrigated area in eastern Sudan. East Mediterr Health J. 2005;11:499–504. [PubMed] [Google Scholar]
- WHO. Methods for surveillance of antimalarial drug efficacy. Geneva: World Health Organization; 2009. [Google Scholar]
- Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- Pearce RJ, Drakeley C, Chandramohan D, Mosha F, Roper C. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrob Agents Chemother. 2003;47:1347–1354. doi: 10.1128/AAC.47.4.1347-1354.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- Osman ME, Mockenhaupt FP, Bienzle U, Elbashir MI, Giha HA. Field-based evidence for linkage of mutations associated with chloroquine (pfcrt/pfmdr1) and sulfadoxine-pyrimethamine (pfdhfr/pfdhps) resistance and for the fitness cost of multiple mutations in P. falciparum. Infect Genet Evol. 2007;7:52–59. doi: 10.1016/j.meegid.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Menegon M, Talha AA, Severini C, Elbushra SM, Mohamedani AA, Malik EM, Mohamed TA, Wernsdorfer WH, Majori G, Nour BY. Frequency distribution of antimalarial drug resistance alleles among Plasmodium falciparum isolates from Gezira State, central Sudan, and Gedarif State, eastern Sudan. Am J Trop Med Hyg. 2010;83:250–257. doi: 10.4269/ajtmh.2010.09-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil I, Alifrangis M, Ronn AM, Gabar HA, Jelinek T, Satti GM, Bygbjerg IC. Pyrimethamine/sulfadoxine combination in the treatment of uncomplicated falciparum malaria: relation between dihydropteroate synthase/dihydrofolate reductase genotypes, sulfadoxine plasma levels, and treatment outcome. Am J Trop Med Hyg. 2002;67:225–229. doi: 10.4269/ajtmh.2002.67.225. [DOI] [PubMed] [Google Scholar]
- Adam I, IE AE, Idris SM, Malik EM, Elbashir MI. A comparison of the efficacy of artesunate plus sulfadoxine-pyrimethamine with that of sulfadoxine-pyrimethamine alone, in the treatment of uncomplicated, Plasmodium falciparum malaria in eastern Sudan. Ann Trop Med Parasitol. 2005;99:449–455. doi: 10.1179/136485905X36299. [DOI] [PubMed] [Google Scholar]
- Mohamed AO, Eltaib EH, Ahmed OA, Elamin SB, Malik EM. The efficacies of artesunate-sulfadoxine-pyrimethamine and artemether-lumefantrine in the treatment of uncomplicated, Plasmodium falciparum malaria, in an area of low transmission in central Sudan. Ann Trop Med Parasitol. 2006;100:5–10. doi: 10.1179/136485906X86239. [DOI] [PubMed] [Google Scholar]
- Mukhtar EA, Gadalla NB, El-Zaki SE, Mukhtar I, Mansour FA, Babiker A, El-Sayed BB. A comparative study on the efficacy of artesunate plus sulphadoxine/pyrimethamine versus artemether-lumefantrine in eastern Sudan. Malar J. 2007;6:92. doi: 10.1186/1475-2875-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A-Elbasit I, Elbashir MI, Khalil IF, Alifrangis M, Giha HA. The efficacy of sulfadoxine-pyrimethamine alone and in combination with chloroquine for malaria treatment in rural Eastern Sudan: the interrelation between resistance, age and gametocytogenesis. Trop Med Int Health. 2006;11:604–612. doi: 10.1111/j.1365-3156.2006.01616.x. [DOI] [PubMed] [Google Scholar]
- Giha HA. Artemisinin derivatives for treatment of uncomplicated Plasmodium falciparum malaria in Sudan: too early for too much hope. Parasitol Res. 2009;106:549–552. doi: 10.1007/s00436-009-1700-x. [DOI] [PubMed] [Google Scholar]
- Elamin SB, Malik EM, Abdelgadir T, Khamiss AH, Mohammed MM, Ahmed ES, Adam I. Artesunate plus sulfadoxine-pyrimethamine for treatment of uncomplicated Plasmodium falciparum malaria in Sudan. Malar J. 2005;4:41. doi: 10.1186/1475-2875-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah MT, Mohammed MM, Himeidan YE, Malik EM, Elbashir MI, Adam I. A randomized comparison of sulphadoxine-pyrimethamine and combination of sulphadoxine pyrimethamine with chloroquine in the treatment of uncomplicated falciparum malaria in Eastern Sudan. Saudi Med J. 2005;26:147–148. [PubMed] [Google Scholar]
- Van den Broek I, Amsalu R, Balasegaram M, Hepple P, Alemu E, Hussein el B, Al-Faith M, Montgomery J, Checchi F. Efficacy of two artemisinin combination therapies for uncomplicated falciparum malaria in children under 5 years, Malakal, Upper Nile, Sudan. Malar J. 2005;4:14. doi: 10.1186/1475-2875-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann S, Sasi P, Mwai L, Bashraheil M, Abdallah A, Muriithi S, Fruhauf H, Schaub B, Pfeil J, Peshu J, Hanpithakpong W, Rippert A, Juma E, Tsofa B, Mosobo M, Lowe B, Osier F, Fegan G, Lindegårdh N, Nzila A, Peshu N, Mackinnon M, Marsh K. Declining responsiveness of Plasmodium falciparum infections to artemisinin-based combination treatments on the Kenyan coast. PLoS One. 2011;6:e26005. doi: 10.1371/journal.pone.0026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiga AW, Fofana B, Sagara I, Dembele D, Dara A, Traore OB, Toure S, Sanogo K, Dama S, Sidibe B, Kone A, Thera MA, Plowe CV, Doumbo OK, Djimde AA. No evidence of delayed parasite clearance after oral artesunate treatment of uncomplicated falciparum malaria in Mali. Am J Trop Med Hyg. 2012;87:23–28. doi: 10.4269/ajtmh.2012.12-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hamidhi S, Mahdy M, Al-Hashami Z, Al-Farsi H, Al-mekhlafi A, Idris M, Beja-Pereira A, Babiker H. Genetic diversity of Plasmodium falciparum and distribution of drug resistance haplotypes in Yemen. Malar J. 2013;12:244. doi: 10.1186/1475-2875-12-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil IF, Ronn AM, Alifrangis M, Gabar HA, Jelinek T, Satti GM, Bygbjerg IC. Response of Plasmodium falciparum to cotrimoxazole therapy: relationship with plasma drug concentrations and dihydrofolate reductase and dihydropteroate synthase genotypes. Am J Trop Med Hyg. 2005;73:174–177. [PubMed] [Google Scholar]
- Hamour S, Melaku Y, Keus K, Wambugu J, Atkin S, Montgomery J, Ford N, Hook C, Checchi F. Malaria in the Nuba Mountains of Sudan: baseline genotypic resistance and efficacy of the artesunate plus sulfadoxine-pyrimethamine and artesunate plus amodiaquine combinations. Trans R Soc Trop Med Hyg. 2005;99:548–554. doi: 10.1016/j.trstmh.2004.10.003. [DOI] [PubMed] [Google Scholar]
- McCollum AM, Basco LK, Tahar R, Udhayakumar V, Escalante AA. Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob Agents Chemother. 2008;52:4089–4097. doi: 10.1128/AAC.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum AM, Schneider KA, Griffing SM, Zhou Z, Kariuki S, Ter-Kuile F, Shi YP, Slutsker L, Lal AA, Udhayakumar V, Escalante AA. Differences in selective pressure on dhps and dhfr drug resistant mutations in western Kenya. Malar J. 2012;11:77. doi: 10.1186/1475-2875-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah TM, Ali AA, Bakri M, Gasim GI, Musa IR, Adam I. Efficacy of artemether-lumefantrine as a treatment for uncomplicated Plasmodium vivax malaria in eastern Sudan. Malar J. 2012;11:404. doi: 10.1186/1475-2875-11-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RJ, Pota H, Evehe MS, Bâ e-H, Mombo-Ngoma G, Malisa AL, Ord R, Inojosa W, Matondo A, Diallo DA, Mbacham W, van den Broek IV, Swarthout TD, Getachew A, Dejene S, Grobusch MP, Njie F, Dunyo S, Kweku M, Kweku M, Owusu-Agyei S, Chandramohan D, Bonnet M, Guthmann JP, Clarke S, Barnes KI, Streat E, Katokele ST, Uusiku P, Agboghoroma CO, Elegba OY, Cissé B, A-Elbasit IE, Giha HA, Kachur SP, Lynch C, Rwakimari JB, Chanda P, Hawela M, Sharp B, Naidoo I, Roper C. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]



