Abstract
Inflammatory bowel diseases (Crohn’s disease (CD); ulcerative colitis (UC)) are chronic immunologically mediated diseases of the gut. Advances in genetics have revolutionized our understanding of the pathogenesis of these conditions with 163 risk loci identified, encompassing a variety of immunologic functions. There is substantial heterogeneity in the natural history of these diseases with respect to disease onset, course, and progression to complications. There are also significant variations in response to therapies, and susceptibility to therapy-related and disease-related complications. An important need in the field is to identify predictors of disease course, complications, and likelihood of response and adverse events to allow for targeted therapeutic decision making. The genotype of an individual in constant and non-modifiable, and thus could potentially fulfill the role of important predictors of these outcomes. In this review, we discuss the existing literature on the prediction of various disease phenotypes in CD and UC utilizing underlying genotype. We also identify gaps in the literature and suggest future directions for research. There is need for large, multi-institution, and international collaborative consortia with efficient and detailed cohort accrual, phenotypic definition, genotyping, and dynamic assessments of external (for example, diet) and internal (microbiome) environment to allow us to progress towards personalized and precision medicine in the management of these complex diseases.
Keywords: Crohn’s disease, ulcerative colitis, genetics, complications, surgery
Inflammatory bowel diseases (IBD; Crohn’s disease (CD), ulcerative colitis (UC)) affect an estimated 1.4 million Americans1, 2. Recent advances have significantly improved our understanding of the pathogenesis of these chronic inflammatory disorders. Through the use of genome wide association scans (GWAS), a total of 163 distinct genetic risk loci have been identified that contribute to risk of either disease with 110 loci shared between both3, 4. The identified loci encompass a spectrum of functions including innate immunity, maintenance of epithelial barrier function, regulation of adaptive immunity, autophagy, endoplasmic reticulum stress, epithelial restitution, pathogen sensing, and modulation of activation thresholds for adaptive immunity3, 4. In particular, recent analyses by the Immunochip consortium have revealed significant overlap between IBD risk loci and susceptibility to mycobacterial infection highlighting the key role of host-microbial interaction in influencing disease pathogenesis3. However, despite significant advances in our understanding of the genetic contribution to the pathogenesis of these diseases, much less is known about the influence of genetics on the natural history of disease. The past decade has also witnessed tremendous advances in the therapy of IBD. The availability of monoclonal antibodies to tumor necrosis factor α (TNF-α) has significantly increased our ability to achieve disease remission, reduce the need for hospitalizations or surgery, and improve patients’ quality of life5. In addition, recent trials have demonstrated that it is possible to achieve complete mucosal healing, raising the possibility of potentially moderating the natural history of IBD5, 6. Furthermore, drugs under development attack novel targets including integrin, janus kinases, and IL-12/IL-23 allowing for the potential to select therapeutic agents based on dominant mechanism of underlying inflammation in different patient subsets. Variations in cytokine profiles among individuals with IBD, and their potential implication for the underlying pathogenesis including microbial reactivity could also potentially allow for such therapeutic targeting. In this review, we discuss the existing literature on the prediction of various disease phenotypes in CD and UC by underlying genotype. We also identify gaps in the literature and propose future directions for research.
The need for “personalizing” IBD care
Both CD and UC are characterized by remarkable heterogeneity in their natural history, and response to therapy. A majority of patients with CD present with inflammatory disease, but over time will develop stricturing or penetrating complications, particularly those with small bowel involvement2, 7. While a minority of patients have perianal disease, this phenotype is associated with considerable morbidity and is a marker for more aggressive disease behavior8. In UC, 10% of those with left-sided colitis progress to pancolitis over 20 years. The extent of colitis is associated with risk of surgery and approximately one-third of all patients may eventually need colectomy for management of refractory disease or disease-related complications such as dysplasia or cancer9.
Despite the significant advances rendered by existing (and novel) therapies, there is a similar substantial heterogeneity in likelihood of response. Over a quarter of the patients may not achieve even an initial response to anti-TNF therapies and an additional 10–15% lose response every year10. In addition, there remain the significant risks associated with immunosuppression including the risk of infections and treatment-related malignancies. Such risks are not uniformly distributed across all users. Our ability to accurately predict adverse events associated with treatment remains inadequate. Equally important is the ability to identify non-responders to specific treatments, allowing for early use of therapies with alternate modalities of action, thus potentially reducing cumulative bowel damage. Recent strategy trials have highlighted that use of biologic anti-TNF therapies earlier in the disease course6 or in combination with azathioprine in newly diagnosed disease11 may allow for superior rates of clinical and endoscopic remission. However, safety concerns suggest the need for caution prior to the wide adoption of such strategies to all patients. In addition, the high costs associated with newer therapies makes it imprudent from a societal view point to apply the same treatment paradigm to all patients regardless of their risk of disease complications, likelihood of response or adverse events related to treatment. Thus, a key need in the field is the ability to identify patients who are most likely to have aggressive disease (or conversely, those least likely to develop complications). In addition, the ability to predict both response and adverse outcomes related to therapy offers the promise of being able to tailor therapy to maximize likelihood of response and minimize adverse events.
Genotype-phenotype correlations in IBD
A majority of the existing studies examining the effect of genotype on IBD phenotype have been correlative studies where statistical methods are applied to explore the association between the occurrence of one (or more) genotype(s) more frequently in individuals with a phenotype of interest compared to controls without that phenotype. While this method is susceptible to biases and confounding, nevertheless these studies have yielded some consistent associations with disease phenotype reviewed below and have suggested other hypotheses that haven’t been replicated.
Crohn’s disease phenotypes and genetic associations
Disease location
Several genetic variants have been associated with disease location in CD. In a UK cohort of 244 Caucasian patients with CD, carriage of the NOD2 variant allele, in particular the Leu1007fsX1008 mutation, was associated with a four-fold increase in likelihood of ileal CD12. Similar but weaker trends were observed for the two other common NOD2 polymorphisms. All patients who were compound heterozygotes or homozygotes had ileal disease (OR 30.3, p<0.0001)12. However, a subsequent study by Abreu et al. suggested that the association between NOD2 and ileal location was specific to the fibrostenosing phenotype13. The rate of carriage of NOD2 was similar in those with small bowel disease without fibrostenosing complications as the overall CD cohort. Mutations in the autophagy pathway have also been associated with ileal CD but less consistently so. Prescott et al. identified an association between the T300A mutation at the ATG16L1 locus and ileal location of CD, with carriage of the rare allele conferring a two-fold elevation in risk14. Similarities in the effect of risk alleles on disease phenotypes may also offer clues regarding overlapping functional consequences of the genetic variants. For example, both NOD2 and autophagy variants have been associated with specific cellular phenotypes, in particular, distinct paneth cell granule distribution, defective paneth cell function and impaired microbial clearance15, 16. The overlapping function may be inferred from synergistic effects on disease risk and behavior. Variants in the Wnt pathway transcription factor (TCF-4)17 and hetero- or homozygosity for the +1059G/C polymorphism in the C-reactive protein gene18 have also been associated with terminal ileal involvement in CD.
Fewer associations have been identified with non-ileal CD. In a German study, the rs7574865 SNP in the STAT4 gene was associated with colonic CD19. The HLA-DRB1 (DR3) allele has been associated with colonic CD (11.5% vs. 0.4%)20 while the TNF receptor superfamily polymorphism TNFRSF1B was inversely associated with colonic involvement independent of NOD221. Homozygosity for the TCF4 transcription factor in the Wnt pathway17 and NOD2 (Leu1007fsX1008) homozygosity have been associated upper gastrointestinal involvement in CD while a promoter region polymorphism in the macrophage migration inhibitor factor (MIF) appears to confer protection against upper gut involvement22. The above studies also highlight that different allelic variants within a specific gene, for example NOD2, may have varying consequences, both at macroscopic and cellular functional levels, and it is important for future studies to have an adequate sample size to be able to tease out the effect of these individual variants.
Perianal disease
Unlike its association with internal fistulizing complications, NOD2 variants do not predict perianal disease or associated complications13 or may, in fact, be inversely associated23, 24. In the study by Annese et al., NOD2 homozygosity was associated with perianal disease. Among the individual SNPs, R702W mutation was positively associated (OR 1.48) while the G908R mutation was inversely associated (OR 0.56) with perianal disease25. In a Belgian study, the NOD2 variant was associated with a reduction in risk for perianal disease (OR 0.56, 95%CI 0.38 – 0.83)23. In the meta-analysis by Adler et al., the cumulative weight of evidence suggested a 60% reduction in likelihood of perianal complications in those with carriage of the G908R mutation (RR 0.40, 95% CI 0.19 – 0.83)24. Other mutations that have been associated with perianal fistulae include the presence of a C-allele at CDKAL1 rs690842523 and IRGM 26 though replication has been less robust. The overall burden of genetic polymorphisms has not been shown to correlate with the occurrence of perianal CD27.
Disease complications - Penetrating / Stricturing disease
The greatest interest in the clinical application of genetics in CD has been for the prediction of complicated disease. Studies have differed in their definition of complicated disease with some including only either the penetrating or stricturing phenotypes while others have used a composite endpoint including both. In one of the earliest studies, Abreu et al. identified that just under half the patients with fibrostenosing CD carried at least one NOD2 variant allele (46%), significantly exceeding the rate of carriage in those without this complication (24%)13. Similarly, Annese et al. found the effect of NOD2 variant allele carriage to be associated with an increased risk of fibrostenosing disease independent of its effect on disease location25. However, in other studies, it has been more challenging to tease out the independent effect of NOD2 carriage on disease behavior from disease location. In the study by Henckaerts et al., while the presence of at least one NOD2 variant was associated with increased risk for stricturing disease, this did not retain statistical significance after correcting for ileal location, male gender and duration of follow-up23. In an elegant meta-analysis, Adler et al. reviewed the literature examining this association and demonstrated a dose-dependent effect. The presence of one variant NOD2 allele was associated only weakly with complicated disease (RR 1.08, 95% CI 0.96 – 1.21)24. Carriage of two NOD2 variant alleles increased the RR to 1.41 (95%CI 1.26 – 1.57). However, NOD2 mutations had poor sensitivity and a low positive likelihood ratio conferring only weak utility in clinical practice. Among the three common NOD2 mutations, the G908R variant was the only allele independently associated with complicated disease in a dominant model (RR 1.33, 95%CI 1.11 – 1.60). However, in a homozygous model, the Leu1007fsX1008 achieved statistical significance (RR 1.30, 95%CI 1.10 – 1.54). Examining stricturing and penetrating phenotypes separately, the G908R allele was the only mutation that was statistically significant for both associations24. The reason for the differing effects of the various NOD2 polymorphisms is unclear. NOD2 consists of a NOD region linked to a leucine-rich repeat (LRR) region in its C-terminal side and two caspase recruitment domains (CARD) at its N-terminal28. While 30 different polymorphisms have been described within the gene, three (R702W, G908R, and Leu1007fsX1008) account for the vast majority of the mutations. There is limited data describing the functional consequence of each mutation, but there is evidence that there may be variations in effect. For example, the frameshift mutation is specifically associated with a reduction in ileal defensin production28. Such functional variations could lead to different effects on disease phenotype.
Other genetic variants have also been associated with the complicated phenotype, though the replication has been less robust than for NOD2. Homozygosity at the rs1363670 G allele located in the vicinity of the IL12B gene conferred a five-fold increase in risk for stricturing disease23, and a shorter time to strictures. In the same study, the IRGM rs4958847 G allele conferred a strong risk (OR 9.22) for non-perianal penetrating disease23. In addition to prediction of complicated disease, there is also clinical utility to variants that may predict a milder course of CD, a group where the traditional step-up therapy may be appropriate. However, few such genetic variants have been identified. In a Canadian cohort, the TNFRSF1A +36 variant allele was negatively associated with the stricturing phenotype independent of the NOD2 status21.
While most studies have examined the contribution of individual genetic alleles to complicated disease, it is plausible that there exists an additive or multiplicative interaction between the various loci such that while the individual contribution of each to complicated disease may be weak, the cumulative burden of risk loci may explain considerably more of the heterogeneity in disease phenotype. To answer this question, Weersma et al. genotyped 1684 CD patients at 5 loci (NOD2, ATG16L1, IL23R, DLG5, IBD5). In addition to the individual SNPs, the overall lumber of mutations also directly correlated with likelihood of complicated disease (Ptrend = 0.0008)27. Despite the number of distinct risk loci for CD and UC now at 163, many of the genetic variants can be mapped to a much smaller number of specific pathways4. Though the effect of individual variants may be weak, there may be a stronger effect size for concurrent mutations affect different steps within the same pathway. As our understanding of the functional consequences of the various risk alleles increases, further analyses should attempt to group genes within the same pathway, and in addition to examining the effect of individual SNPs, examine the association between functional pathways and disease phenotype and complications.
Need for surgery
Consistent with its association with complicated CD, NOD2 variants have been alleles most frequently associated with need for surgery in adult and pediatric CD. Alvarez-Lobos identified a three-fold increase in risk independent of its association with stricturing disease29. Annese et al. also found a 50% increase in risk of bowel resection in those with carriage of at least one mutant NOD2 allele25. In the meta-analysis incorporating 17 relevant studies, the presence of one or more mutant NOD2 alleles was associated with a 50% increase in risk for surgery (RR 1.58, 95% CI 1.38–1.80)24. In contrast to its effect on complicated disease, the excess risk of surgery was similar in magnitude among those who possessed one (RR 1.48) or two (RR 1.49) mutant alleles24. The pooled sensitivity of one NOD2 mutant allele was 0.31 while that of two mutant alleles was lower at 0.27. It also remains to be clearly defined if the effect of NOD2 on surgery is independent of its association with ileal disease or the fibrostenosing phenotype. In a Belgian cohort, carriage of at least one NOD2 variant was associated with higher risk for IBD-related surgery (OR 1.4)23. However, adjusting for ileal involvement and development of strictures neutralized this. NOD2 has also been associated with increased risk of surgery earlier in the disease course in pediatric CD30, 31.
Other genetic variants have also been associated with the need for intestinal surgery in CD. Sehgal et al. genotyped a cohort of 66 patients with ileocolonic Crohn’s disease who underwent ileocolectomy for 83 IBD-associated SNPs; the average number of resections per patient was 1.7 with a mean duration of disease of 14.7 years. Only one SNP is the IRGM gene (rs4958847) was significant after correcting for multiple observation testing, both in the additive and dominant models32. Patients with the mutant allele had a mean interval between resections of 6.9 years compared to 11.4 years among those with the wild-type (p = 0.007). NOD2 was not associated with surgery in this study possibly due to the fact that the cohort was restricted to those with ileocolonic location, a phenotype that correlates significantly with NOD2. In the study by Weersma et al., increasing number of mutations at the five risk alleles genotyped demonstrated a direct correlation with the likelihood of intestinal resection (Ptrend = 0.02)27.
There is less data examining whether genotype predicts repeat resections in those with prior surgery, a predictive power that would have important clinical utility by allowing for selection of patients for tailored post-operative prophylaxis. The NOD2 variants have been variably associated with risk of repeat surgery. Alvarez-Lobos et al. identified a three-fold risk for repeat resection and earlier time to repeat resection for NOD2 carriers29, but other studies failed to find a similar association 33.
Response to therapy - Immunomodulators
The widest use of genetics to predict treatment response or treatment-related AE in CD is the use of the thiopurines methyl transferase (TPMT) genotype and enzyme activity to guide response to treatment with thiopurines (azathioprine, 6-mercaptopurine). The most common variant alleles associated with reduced TPMT activity include TPMT*2, TPMT*3A, TPMT*3B, and TPMT*3C. Approximately 4–11% of individuals are heterozygous while 0.3% of the population is homozygous for these variant alleles. A recent systematic review identified the sensitivity of the genotype in determining the enzymatic activity to be 80%34. Individuals who are heterozygotes for the reduced enzyme activity alleles had a four-fold increase in risk for leukopenia34. Homozygotes had a substantially greater incidence of myelotoxicity but the effect size estimates were imprecise due to small numbers. Testing for TPMT activity prior to initiating thiopurines has been shown to be cost-effective35. In contrast to its utility in predicting myelosuppression, TPMT has limited role in predicting response to thiopurines. However, a polymorphism in the inosine triphosphate pyrophosphatase (ITPA), the c.94 C > A variant has been associated with non-response to thiopurine therapy36.
Response to therapy - Biologic anti-TNF agents
Given the central role of TNF-α in systemic and intestinal inflammation, it has been hypothesized that several of the pathways involved in CD pathogenesis or related to the TNF- α may also play an important role in predicting response to therapy directed against it using monoclonal antibodies (infliximab, adalimumab, certolizumab pegol). However, results have been divergent and inconsistent with weak effects that haven’t been replicated in independent cohorts. Initial studies suggested that TNF-α and TNF-receptor (TNFR) polymorphisms may predict response to infliximab. In the study by Taylor and colleagues, individuals homozygous for the TNF-α/lymphotoxin A (LTA) polymorphism were non-responders to infliximab37. A Belgian cohort subsequently demonstrated an association between TNFR1A36G polymorphism and response to infliximab with a lower rate of response in those carrying the G-allele. However, neither of these associations was replicated in subsequent cohorts38–40. Similarly, Louis et al. initially identified an association between a polymorphism in the Fc receptor III a (FcYRIIIa), a mediator of antibody-mediated cell cytotoxicity and non-response to infliximab41, but a subsequent analysis of the ACCENT I trial failed to replicated this association42. Several authors have investigated the potential role of NOD2 polymorphisms and response to infliximab and have failed to identify a correlation39, 43, 44. In contrast, the homozygous variant of the IBD5 locus (5q31) was associated with a three-fold increase in likelihood of non-response to infliximab45. Induction of apoptosis is a key mechanism by which infliximab and adalimumab exert their anti-inflammatory effect; thus, apoptosis related genes may also influence response to therapy. To examine this hypothesis, Hlavaty and colleagues genotyped 287 consecutive patients on infliximab with refractory luminal or fistulizing CD for 21 apoptosis related genes46. Fas-ligand TT genotype and caspase-9 CC/CT genotypes were associated with lower rates of response to infliximab46. In a subsequent study, the authors developed an apoptotic pharmacogenetic index (API) that ranged from 0 (low apoptotic response) to 3 (high apoptotic response). Response in both luminal and fistulizing CD increased with increasing apoptosis scores. Individuals with an API ≤ 1 had remission rates of 39.5% and 28.6% for luminal and fistulizing CD respectively compared to 56.1% and 44.9% in those with an API of 2. However, the effect of low apoptotic index could be neutralized by the addition of azathioprine. The remission rate in those with API < 1on infliximab monotherapy was 15.8%, increasing to 63.2% in those who were also on azathioprine, a rate comparable to those with higher apoptotic index47. Thus genetic predictors may be incorporated into individualized therapeutic decision making.
Ulcerative colitis phenotypes and genetic associations
Disease extent
In contrast to the wealth of data examining phenotypic correlates of genetic variants in CD, there is much less data associating genotype with extensive, severe or refractory disease in UC. In one such study, Annese et al. genotyped 114 UC patients and 102 CD patients for various HLA-DRB1 alleles20. While the HLA-DRB1 genotype itself was not associated with risk of CD or UC after applying Bonferroni correction, the HLA-DRB1 (DR13) allele was strongly associated with the presence of pancolitis20. However, even in patients with pancolitis, the occurrence of the allele was infrequent (5%). The HLA DRB1 (DR3) allele was found more commonly in those with distal UC though its occurrence within this cohort also remained infrequent (12.5%). In the Scottish population, a polymorphism in the multi-drug resistance gene (MDR1) locus was associated with extensive colitis48.
Need for Colectomy
While most studies have adopted a candidate-gene approach to identify risk factors for severe disease, Haritunians et al. performed an unbiased GWAS analysis comparing 324 patients with medically refractory UC to 537 controls with treatment responsive disease49. A risk score that incorporated 46 SNPs explained 48% of the variance for colectomy risk with a sensitivity and specificity of 79% and 86% respectively. Patients in the lowest quartile of the risk score were unlikely to require surgery while those in the highest quartile nearly universally required colectomy. Relevant SNPs that were significant in this analysis included the TNFSF15 (TL1A), a known UC susceptibility locus, other putative loci including IL-10 (1q32.1), IL-12B (5q33.3), 12q15 (INFG/IL-26), and KIF1A. In addition, the previously described associations with various major histocompatibility complex (MHC) loci were replicated. Several of the other loci implicated had functions related to bacterial-host interactions (BICD1), epithelial barrier integrity (MAGI1), and Th17 regulation (retinoid-related orphan receptor)49.
Response to therapy
Studies on prediction of response to therapy in UC have focused on corticosteroids and infliximab. Glucocorticoids (GC) exert their action by passive diffusion across the plasma membrane, binding to the cytosolic GC receptor, subsequent nuclear translocation, and activation of the target genes50. Approximately half of the UC patients initiated steroids achieve immediate and long-term response at one year, but approximately one-quarter each become steroid dependent or require surgery51. The most widely studied polymorphism in the context of resistance to steroids has been at the MDR1 locus. Over-expression of MDR1 results in increased p-glycoprotein mediated efflux of corticosteroids and consequent resistance to steroids. MDR1 polymorphisms themselves have been variably associated with UC. From a French cohort, Daniel et al. identified an association between the TT genotype of exon 21 MDR1 polymorphism and lack of response to cyclosporine in patients with steroid refractory UC52. Similar refractoriness to therapy was demonstrated in an independent cohort by Potocnik and colleagues53. In rectal biopsies, MDR1 expression appears reduced in inflamed tissue compared to non-inflamed tissue or from controls. MDR1 expression level also correlates inversely with response to treatment with higher levels of MDR1 gene expression in those with therapy responsive disease or in long-term remission54. Dubinsky et al. conducted an unbiased GWAS analysis of therapeutic response to IFX in pediatric CD and UC pooled together55. Among the susceptibility loci, only the BRWD1 locus was associated with non-response to IFX; three other pharmacogenetic loci – TACR1, FAM19A4 and PHACTR3 were also associated with non-response to IFX56. Duraes et al. identified an association between mutations in the autophagy loci - ATG16L1, ITLN1, IRGM - with response to corticosteroids, immunosuppressants, and biologic therapy57.
Recent studies have used unbiased RNA expression studies using microarray technology applied to colon biopsies obtained before and after administration of infliximab to identify potential predictors of treatment response. Data from two independent cohorts of UC patients receiving infliximab, a Belgian cohort (n=24) and participants in the ACT trials (n=22) was used to develop a probe set that would allow for prediction of response to 1–3 doses of infliximab58. A total of 74 probe sets comprising 53 known genes were differentially expressed between responders and non-responders. The top five differentially expressed genes were all involved in the adaptive immune response and included osteoprotegerin, stanniocalcin-1, prostaglandin-endoperoxide synthase 2, IL-13Rα2 and IL-1158. In contrast, homozygous carriers of the UC risk-increasing IL23-R variants were associated with increased rates of response to IFX than those homozygous for the risk-decreasing variants59.
Phenotypes common to CD and UC
Age of onset
Epidemiologic evidence suggests that patients with early onset disease may have a stronger genetic contribution to their disease pathogenesis. Individuals with younger age at diagnosis of CD or UC are more likely to have an affected family member than those diagnosed at a later age60, 61. However, literature examining the contribution of genotype in determining age of onset of CD or UC has yielded inconsistent results. In the cohort by Abreu et al., carriage of NOD2 variants was not associated with earlier age of onset of CD13. Ahmad et al. similarly did not find a significant association between NOD2 carriage and age of diagnosis once correction for multiple comparison testing had been applied12. In contrast, in the study by Annese et al., carriers of at least one NOD2 mutation had a younger age of diagnosis than those with wild-type alleles25. The same study also identified a dose-dependent effect; NOD2 homozygotes had a younger age of diagnosis (24 years) than heterozygotes (29 years) or those with no risk alleles (33 years, p=0.001)25. Another group demonstrated an association between the 3020insC polymorphism at the NOD2 locus and early onset disease62, while the IBD5 locus63 and the STAT4 loci19 have been associated with earlier onset in some studies. In addition to individual SNPs, it has been hypothesized that an overall increase in the number of CD-risk alleles may correlate with age of onset. In the study by Weersma et al., individuals with onset of disease at an age younger than 40 years had a greater number of variant alleles compared to those with later onset disease (Ptrend=0.028)27. However, most studies have adopted a candidate gene approach to answer this question with few studies large enough to perform an unbiased GWAS for early onset disease. Two such studies, together comprising over 2000 CD and 1000 UC patients found a substantial overlap in the distribution of risk alleles between pediatric- and adult-onset IBD with the majority of risk alleles for adult CD being replicated in the pediatric group55. However, two SNPs, rs3024505 (in the region of IL10) and rs917997 (IL18R1) were significantly associated with CD in the pediatric but not adult population.
Extra-intestinal symptoms
Extra-intestinal manifestations (EIM) are common in patients with IBD, occurring in 25–40% of patients. The most common EIM is arthropathy involving either the axial (1–26%) or the appendicular skeleton (5–20%). In addition, ankylosing spondylitis (AS) can occur in 3–10% of patients with IBD and is associated with HLA-B27 positivity though the strength of association is weaker than in non-IBD AS64, 65. The known IBD risk alleles have not demonstrated a strong association with the occurrence of EIM. NOD2 carriage or the total number of variant risk alleles did not correlate with the presence of EIM20, 27. In contrast, HLA-linked variants have demonstrated stronger correlation with the occurrence of EIM. The HLA DRB1 (DR13) allele was associated with EIM in UC while HLA DRB1 (DR3) allele was associated with EIM in CD. In a British study of 976 UC and 483 CD patients, HLA-B*58, HLA-B*27 and HLA-DRB1*0103 were associated with uveitis, an EIM associated with CD or UC and occurring occasionally in patients with IBD where it correlates with active bowel disease. Prior studies had identified the latter two alleles to be associated with peripheral arthritis as well. Mendoza et al. demonstrated an association between the Fc receptor-like 3 gene (FcRL3) promoter variant and peripheral arthritis in CD66. Dermatologic manifestations are the next most common EIM, occurring in 3–24% of patients and predominantly taking the form of either erythema nodosum (EN) or pyoderma gangrenosum (PG). Orchard et al. demonstrated an association between a polymorphisms in the TNF-α promoter (-1031C) and EN; this mutation was twice as common in cases (67%) as controls (37%)64. However replication studies are lacking, largely due to lack of cohorts of sufficient size.
Primary sclerosing cholangitis (PSC) occurs in 2–10% of patients with IBD and also shares some common environmental risk factors with current smoking conferring protection against both PSC and UC. Recent advances in genetics have improved our understanding of this disease through GWAS from multicenter genetics consortia. Karlsen et al. identified two SNPs near HLA-B locus at chromosome 6p21 to be associated with risk of PSC67. In other studies, other UC-associated loci demonstrated independent association with PSC suggesting a shared pathogenic mechanism68, 69.
Genotype as a window to susceptibility to environmental influences
Environmental factors that may play an important role in IBD pathogenesis include smoking, infections, diet, use of non-steroidal anti-inflammatory agents, hormone agents, antibiotics, stress, and vitamin D status. However, there is limited understanding of whether susceptibility to the effect of such environmental factors relates to underlying genetic proneness to develop IBD or polymorphisms in other pathways involved in metabolism. Studies have demonstrated an interaction between polymorphisms in the cytochrome P-450 or glutathione transferase and cigarette smoking in mediating risk of lung cancer and rheumatoid arthritis, respectively70, 71. In animal models, a GRK4 polymorphism mediates salt sensitivity and blood pressure response to salt intake72. Hutter et al. demonstrated that the effect of the EIF3H/UTP23 polypmorphism (8q23) on colorectal cancer risk was influenced by vegetable consumption, with a stronger effect at higher levels of vegetable consumption73. Mutations in GPR120 influence sensing of dietary fat and susceptibility to obesity and regulation of energy balance74. Whether similar genetic polymorphism influence susceptibility to the deleterious (in CD) or protective effect (in UC) of cigarette smoke and other environmental factors has not been studied but remains an important area of research. Examination of gene-environment interactions often require detailed environmental exposure assessments which impact ability to assemble large cohorts or willingness for participation. However, novel methods in environmental exposure assessment including the concept of ‘top-down’ assessment of an individuals lifetime history of both external and internal environmental exposures (the “exposome”) using biomarker assessments from single biospecimens offers considerable promise in extending our ability to quantify and study environmental influences in the context of microbiota and genetics75.
Epigenetics
Recent studies have been published exploring the emerging role of epigenetics in disease risk and phenotype. In a study by Lin and colleagues, seven CpG sites were differentially methylated between IBD and controls76. In addition, CpG methylation was also different at specific sites between UC and CD. Examination of effect of epigenetics on IBD phenotype has been mainly limited to colorectal cancer where CpG island methylation was found less commonly than in sporadic colorectal cancer77.
Personalizing IBD therapy – Pitfalls and Challenges
The realm where personalized medicine has achieved widest success has been in oncology. Molecular characterization led to the classification of glioblastoma multiforme into distinct disease subtypes based on specific mutations in EGFR, NF1, and PDFGR/IDH1 respectively. These subtypes correlate with age of diagnosis, prognosis, and response to treatment78. A similar personalized medicine approach has demonstrated utility for the selection of chemotherapeutic regimens based on underlying genetic data. Between 20–30% of breast cancer tumor tissue over-express human epithelial growth factor receptor (HER2); such tumors have superior outcomes on treatment with traztusumab (herceptin), a monoclonal antibody against HER2 whereas the drug is ineffective in those without overexpression of the receptor79. Similar, while the IRESSA trial of gefitinib (EGFR inhibitor) in non-small cell lung cancer showed no benefit, the subgroup of patients with patients with a somatic EGFR mutation benefitted from drug administration80. Recent studies have highlighted the role of interleukin-28B polymorphisms in predicting viral kinetics and response to interferon treatment in hepatitis C81 providing further support for the principle of incorporating genomic medicine into clinical practice. However, several challenges to development and adoption of a similar approach in IBD care remain.
First, single center studies often lack sufficient numbers of patients with uncommon variants or phenotypes. Second, there are several existing large genetic IBD consortia that have contributed significantly towards the identification of the 163 genetic risk loci for CD or UC. But such consortia with detailed genetic information often lack detailed information on disease sub-phenotypes. More importantly, there is also missing, inaccurate or only approximate information on potentially important environmental factors. In contrast, cohorts that are rich in phenotypic data lack associated biosamples for genetic analysis. Structured clinical trials that allow for the best opportunity to accurately assess therapeutic response without the biases of observational assessment also often lack the ability to examine genetic information. Third, current studies have mostly employed an ‘association’ approach where statistical methods are employed to demonstrate an association between candidate or unbiased SNPs and disease phenotypes at varying levels of statistical significance. However, candidate gene approaches run the risk of a high number of false positive findings, and often ignore inter-relationship between various SNPs affecting the same pathway. As our understanding of the function associated with the various genetic polymorphisms grows incrementally, there is need for associating clinical cohorts with fresh biosamples for functional analysis to develop a more informed approach to selection of relevant genes or pathways.
Implications for future study design
There are a few key principles in the design of future studies examining genotype-phenotype studies in IBD. First, there is the need for large enough samples sizes that would allow for examination of the effect of uncommon variants, as well as accurately define the effect sizes for known associations while controlling for potential confounders. Several recently assembled cohorts offer insights into how this may be done efficiently and cost-effectively. The Type 1 diabetes exchange (http://t1dxregistry.jaeb.org/) is a registry of 67 participating clinics that collect structured clinical and laboratory information with the data warehoused in a common data repository. Through such multicenter collaboration, over 25,000 patients with type 1 diabetes have been assembled within 20 months. The Crohn’s and Colitis Foundation of America (CCFA) Partners registry (https://cgibd.med.unc.edu/ccfapartners/index.php), a similar registry, has recruited nearly 12,000 CD or UC patients who have self-reported detailed clinical information. In addition, patient self-report in such registries also allows for ascertainment of detailed and repeated environmental exposure data as well as key patient reported outcomes. Finally, multi-institution networks such as the eMERGE consortium (https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page) are leveraging the power of bioinformatics and natural language processing to mine vast numbers of electronic medical records assembled as part of routine care to accurately determine disease phenotype. This allows for efficient assembly of extremely large cohorts, and when linked to biospecimen repositories or discarded blood samples, allows for large scale genotype-phenotype analysis.
Second, most of the effect sizes identified have been relatively modest. While 163 alleles appear to modify risk of CD or UC, most of the relative risks are between 1–1.8, suggesting small effect sizes. Cumulative genetic burden of disease appears to demonstrate a stronger association. The odds ratios for genetic polymorphisms in predicting disease course, and in particular, therapy response are likely to be larger given that such processes have not been subject to the principle of natural selection. As more information on this become available, we will know if individual SNPs (like IL-28 polymorphisms in hepatitis C) will be key predictors, or if a more inclusive ‘genetic panel’ approach to stratify disease and therapy will be required. A pre-requisite for this is that in addition to examining individual SNPs, analyses also be undertaken to group polymorphisms into pathways based on their functional consequences. This would allow for examination of whether pathway-level effect sizes are stronger than that of individual SNPs.
Third, evolution of disease behavior is a dynamic process involving varying environmental influences. For example, stress, smoking, diet, and antibiotic exposure have all been proposed to influence disease course either through their direct effects on immune cell function or through their effect on intestinal microbiota. There may be genetic variants that do not directly influence disease behavior, but may exert their effect through modulation of susceptibility to specific environmental influences. Thus, dynamic measures of putative environmental influences, should be incorporated where possible, to allow for examination of gene-environment interactions.
Finally, a key requirement for the widespread adoption of genomic medicine is the cost-effectiveness of such an approach, particularly when effect sizes are modest. Genomic sequencing has become markedly less expensive over the past several years, and is likely to continue to get cheaper as more efficient and large-scale operations become available. This will allow for ready incorporation of such platforms, first within the setting of clinical trials, to provide the background that therapeutic decisions can be made based on underlying genotype, and subsequently into routine clinical practice.
Conclusions
In conclusion, while genetics has tremendously improved our understanding of the pathogenesis of CD and UC, it has only modestly allowed for the ability to predict complicated disease. The most widely replicated association is between fibrostenosing ileal CD and NOD2 variants alleles. Other associations have been demonstrated less consistently and merit replication in larger cohorts. One limitation has been the relative paucity of large cohorts with detailed phenotypic, genetic, and environmental (e.g. smoking) information suggesting the need for multicenter collaborative cohorts. In addition, a majority of the phenotype-genotype data stems from Caucasian cohorts in the Western hemisphere. As several of the genetic risk alleles have failed to demonstrate a similar association in Asian cohorts, there is also the need to replicate the genotype-phenotype correlative studies within diverse populations. The utility of genotype in predicting response to therapy has been less widely studied with some of the earlier reported associations failing to be replicated in subsequent studies. Pharmacogenetics SNPs may be useful in predicting likelihood of response to therapy. There is also need to determine if genetic predictors can be utilized to predict likelihood of adverse events related to therapy. This would allow for a more personalized approach to management of IBD incorporating predictors of disease progression as well as a tailored approached to therapeutic decision making.
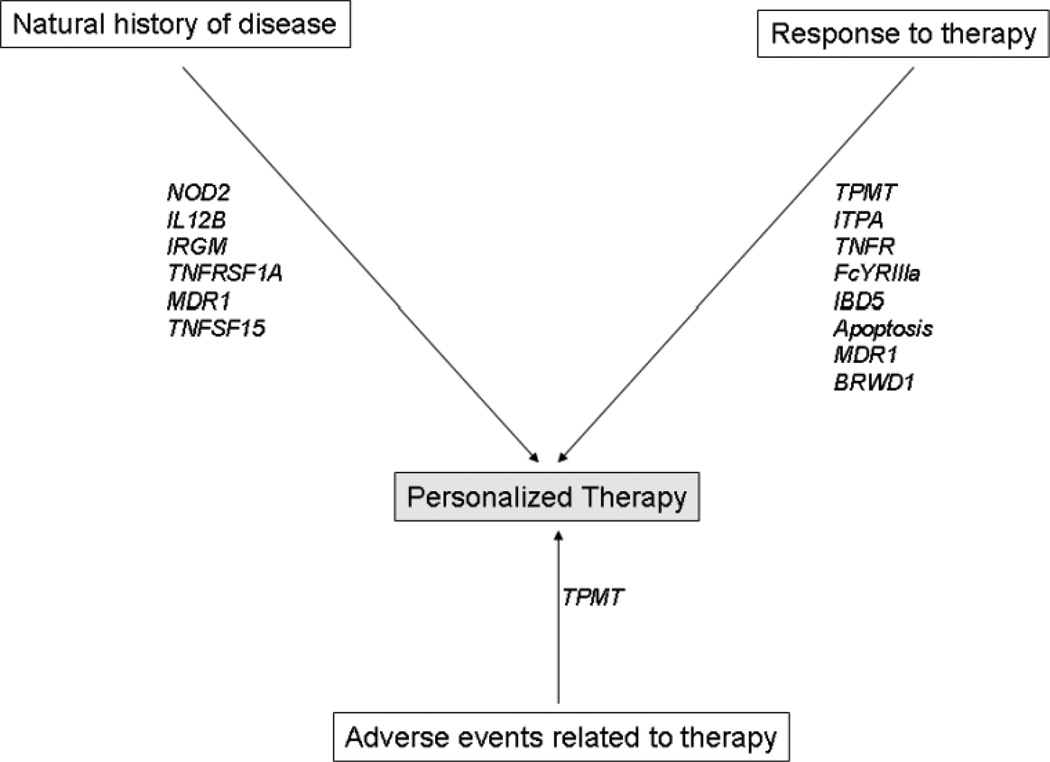
Figure 1.
Conceptual framework for use of genetics to personalize management of inflammatory bowel diseases
* SNPs described in the figure are those where there exists ≥ 1 study in the literature demonstrating an association with the outcome of interest.
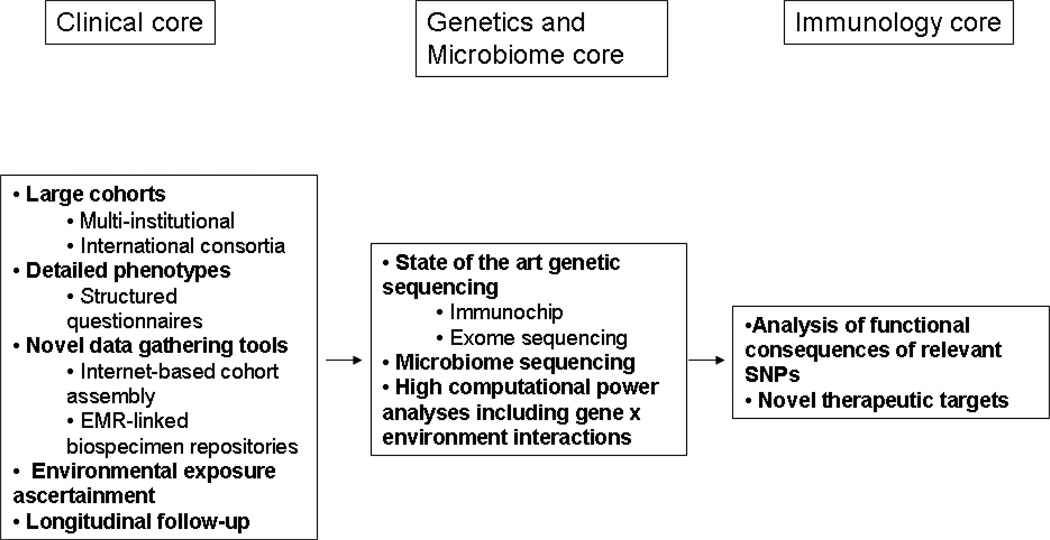
Figure 2.
Proposed structural framework for study design examining genotype-phenotype relationships in IBD
Acknowledgments
Source of funding: This work is supported by the National Institutes of Health (NIH) (P30 DK043351) to the Center for Study of Inflammatory Bowel Diseases. Ananthakrishnan is supported in part by a grant from the National Institutes of Health (K23 DK097142). Xavier is supported by grants U01 DK062432 & R01 DK064869 from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: None
REFERENCES
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 3.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern D, Hun KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amineinejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Cohain A, Cichon S, D'Amato M, De Jong DJ, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Gieger C, Karlsen J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro MD, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor K, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, S Z, Zhang B, Zhang CK, Zhao H, Consortium TIIG, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly M, Franke A, Parkes M, Vermeire S, Barret JC, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012 doi: 10.1038/nature11582. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Haens GR, Panaccione R, Higgins PD, Vermeire S, Gassull M, Chowers Y, Hanauer SB, Herfarth H, Hommes DW, Kamm M, Lofberg R, Quary A, Sands B, Sood A, Watermayer G, Lashner B, Lemann M, Plevy S, Reinisch W, Schreiber S, Siegel C, Targan S, Watanabe M, Feagan B, Sandborn WJ, Colombel JF, Travis S. The London Position Statement of the World Congress of Gastroenterology on Biological Therapy for IBD With the European Crohn's and Colitis Organization: When to Start, When to Stop, Which Drug to Choose, and How to Predict Response? Am J Gastroenterol. 2011 doi: 10.1038/ajg.2010.392. [DOI] [PubMed] [Google Scholar]
- 6.D'Haens G, Baert F, van Assche G, Caenepeel P, Vergauwe P, Tuynman H, De Vos M, van Deventer S, Stitt L, Donner A, Vermeire S, Van de Mierop FJ, Coche JC, van der Woude J, Ochsenkuhn T, van Bodegraven AA, Van Hootegem PP, Lambrecht GL, Mana F, Rutgeerts P, Feagan BG, Hommes D. Early combined immunosuppression or conventional management in patients with newly diagnosed Crohn's disease: an open randomised trial. Lancet. 2008;371:660–667. doi: 10.1016/S0140-6736(08)60304-9. [DOI] [PubMed] [Google Scholar]
- 7.Cosnes J, Cattan S, Blain A, Beaugerie L, Carbonnel F, Parc R, Gendre JP. Long-term evolution of disease behavior of Crohn's disease. Inflamm Bowel Dis. 2002;8:244–250. doi: 10.1097/00054725-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Beaugerie L, Seksik P, Nion-Larmurier I, Gendre JP, Cosnes J. Predictors of Crohn's disease. Gastroenterology. 2006;130:650–656. doi: 10.1053/j.gastro.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 9.Langholz E, Munkholm P, Davidsen M, Nielsen OH, Binder V. Changes in extent of ulcerative colitis: a study on the course and prognostic factors. Scand J Gastroenterol. 1996;31:260–266. doi: 10.3109/00365529609004876. [DOI] [PubMed] [Google Scholar]
- 10.Gisbert JP, Panes J. Loss of response and requirement of infliximab dose intensification in Crohn's disease: a review. Am J Gastroenterol. 2009;104:760–767. doi: 10.1038/ajg.2008.88. [DOI] [PubMed] [Google Scholar]
- 11.Colombel JF, Sandborn WJ, Reinisch W, Mantzaris GJ, Kornbluth A, Rachmilewitz D, Lichtiger S, D'Haens G, Diamond RH, Broussard DL, Tang KL, van der Woude CJ, Rutgeerts P. Infliximab, azathioprine, or combination therapy for Crohn's disease. N Engl J Med. 362:1383–1395. doi: 10.1056/NEJMoa0904492. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad T, Armuzzi A, Bunce M, Mulcahy-Hawes K, Marshall SE, Orchard TR, Crawshaw J, Large O, de Silva A, Cook JT, Barnardo M, Cullen S, Welsh KI, Jewell DP. The molecular classification of the clinical manifestations of Crohn's disease. Gastroenterology. 2002;122:854–866. doi: 10.1053/gast.2002.32413. [DOI] [PubMed] [Google Scholar]
- 13.Abreu MT, Taylor KD, Lin YC, Hang T, Gaiennie J, Landers CJ, Vasiliauskas EA, Kam LY, Rojany M, Papadakis KA, Rotter JI, Targan SR, Yang H. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology. 2002;123:679–688. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 14.Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, Bagnall R, Mirza MM, Sanderson J, Forbes A, Mansfield JC, Lewis CM, Schreiber S, Mathew CG. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 15.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HWt. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. 1641 e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koslowski MJ, Kubler I, Chamaillard M, Schaeffeler E, Reinisch W, Wang G, Beisner J, Teml A, Peyrin-Biroulet L, Winter S, Herrlinger KR, Rutgeerts P, Vermeire S, Cooney R, Fellermann K, Jewell D, Bevins CL, Schwab M, Stange EF, Wehkamp J. Genetic variants of Wnt transcription factor TCF-4 (TCF7L2) putative promoter region are associated with small intestinal Crohn's disease. PLoS One. 2009;4:e4496. doi: 10.1371/journal.pone.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thalmaier D, Dambacher J, Seiderer J, Konrad A, Schachinger V, Pfennig S, Otte JM, Crispin A, Goke B, Ochsenkuhn T, Lohse P, Brand S. The +1059G/C polymorphism in the C-reactive protein (CRP) gene is associated with involvement of the terminal ileum and decreased serum CRP levels in patients with Crohn's disease. Aliment Pharmacol Ther. 2006;24:1105–1115. doi: 10.1111/j.1365-2036.2006.03093.x. [DOI] [PubMed] [Google Scholar]
- 19.Glas J, Seiderer J, Nagy M, Fries C, Beigel F, Weidinger M, Pfennig S, Klein W, Epplen JT, Lohse P, Folwaczny M, Goke B, Ochsenkuhn T, Diegelmann J, Muller-Myhsok B, Roeske D, Brand S. Evidence for STAT4 as a common autoimmune gene: rs7574865 is associated with colonic Crohn's disease and early disease onset. PLoS One. 2010;5:e10373. doi: 10.1371/journal.pone.0010373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Annese V, Piepoli A, Latiano A, Lombardi G, Napolitano G, Caruso N, Cocchiara E, Accadia L, Perri F, Andriulli A. HLA-DRB1 alleles may influence disease phenotype in patients with inflammatory bowel disease: a critical reappraisal with review of the literature. Dis Colon Rectum. 2005;48:57–64. doi: 10.1007/s10350-004-0747-0. discussion 64-5. [DOI] [PubMed] [Google Scholar]
- 21.Waschke KA, Villani AC, Vermeire S, Dufresne L, Chen TC, Bitton A, Cohen A, Thomson AB, Wild GE. Tumor necrosis factor receptor gene polymorphisms in Crohn's disease: association with clinical phenotypes. Am J Gastroenterol. 2005;100:1126–1133. doi: 10.1111/j.1572-0241.2005.40534.x. [DOI] [PubMed] [Google Scholar]
- 22.Dambacher J, Staudinger T, Seiderer J, Sisic Z, Schnitzler F, Pfennig S, Hofbauer K, Konrad A, Tillack C, Otte JM, Diebold J, Goke B, Ochsenkuhn T, Lohse P, Brand S. Macrophage migration inhibitory factor (MIF)-173G/C promoter polymorphism influences upper gastrointestinal tract involvement and disease activity in patients with Crohn's disease. Inflamm Bowel Dis. 2007;13:71–82. doi: 10.1002/ibd.20008. [DOI] [PubMed] [Google Scholar]
- 23.Henckaerts L, Van Steen K, Verstreken I, Cleynen I, Franke A, Schreiber S, Rutgeerts P, Vermeire S. Genetic risk profiling and prediction of disease course in Crohn's disease patients. Clin Gastroenterol Hepatol. 2009;7:972–980. doi: 10.1016/j.cgh.2009.05.001. e2. [DOI] [PubMed] [Google Scholar]
- 24.Adler J, Rangwalla SC, Dwamena BA, Higgins PD. The prognostic power of the NOD2 genotype for complicated Crohn's disease: a meta-analysis. Am J Gastroenterol. 2011;106:699–712. doi: 10.1038/ajg.2011.19. [DOI] [PubMed] [Google Scholar]
- 25.Annese V, Lombardi G, Perri F, D'Inca R, Ardizzone S, Riegler G, Giaccari S, Vecchi M, Castiglione F, Gionchetti P, Cocchiara E, Vigneri S, Latiano A, Palmieri O, Andriulli A. Variants of CARD15 are associated with an aggressive clinical course of Crohn's disease--an IG-IBD study. Am J Gastroenterol. 2005;100:84–92. doi: 10.1111/j.1572-0241.2005.40705.x. [DOI] [PubMed] [Google Scholar]
- 26.Latiano A, Palmieri O, Cucchiara S, Castro M, D'Inca R, Guariso G, Dallapiccola B, Valvano MR, Latiano T, Andriulli A, Annese V. Polymorphism of the IRGM gene might predispose to fistulizing behavior in Crohn's disease. Am J Gastroenterol. 2009;104:110–116. doi: 10.1038/ajg.2008.3. [DOI] [PubMed] [Google Scholar]
- 27.Weersma RK, Stokkers PC, van Bodegraven AA, van Hogezand RA, Verspaget HW, de Jong DJ, van der Woude CJ, Oldenburg B, Linskens RK, Festen EA, van der Steege G, Hommes DW, Crusius JB, Wijmenga C, Nolte IM, Dijkstra G. Molecular prediction of disease risk and severity in a large Dutch Crohn's disease cohort. Gut. 2009;58:388–395. doi: 10.1136/gut.2007.144865. [DOI] [PubMed] [Google Scholar]
- 28.Strober W, Watanabe T. NOD2, an intracellular innate immune sensor involved in host defense and Crohn's disease. Mucosal Immunol. 2011;4:484–495. doi: 10.1038/mi.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Lobos M, Arostegui JI, Sans M, Tassies D, Plaza S, Delgado S, Lacy AM, Pique JM, Yague J, Panes J. Crohn's disease patients carrying Nod2/CARD15 gene variants have an increased and early need for first surgery due to stricturing disease and higher rate of surgical recurrence. Ann Surg. 2005;242:693–700. doi: 10.1097/01.sla.0000186173.14696.ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kugathasan S, Collins N, Maresso K, Hoffmann RG, Stephens M, Werlin SL, Rudolph C, Broeckel U. CARD15 gene mutations and risk for early surgery in pediatric-onset Crohn's disease. Clin Gastroenterol Hepatol. 2004;2:1003–1009. doi: 10.1016/s1542-3565(04)00452-5. [DOI] [PubMed] [Google Scholar]
- 31.Lacher M, Helmbrecht J, Schroepf S, Koletzko S, Ballauff A, Classen M, Uhlig H, Hubertus J, Hartl D, Lohse P, von Schweinitz D, Kappler R. NOD2 mutations predict the risk for surgery in pediatric-onset Crohn's disease. J Pediatr Surg. 2010;45:1591–1597. doi: 10.1016/j.jpedsurg.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 32.Sehgal R, Berg A, Polinski JI, Hegarty JP, Lin Z, McKenna KJ, Stewart DB, Poritz LS, Koltun WA. Mutations in IRGM are associated with more frequent need for surgery in patients with ileocolonic Crohn's disease. Dis Colon Rectum. 55:115–121. doi: 10.1097/DCR.0b013e31823ccea8. [DOI] [PubMed] [Google Scholar]
- 33.Maconi G, Colombo E, Sampietro GM, Lamboglia F, D'Inca R, Daperno M, Cassinotti A, Sturniolo GC, Ardizzone S, Duca P, Porro GB, Annese V. CARD15 gene variants and risk of reoperation in Crohn's disease patients. Am J Gastroenterol. 2009;104:2483–2491. doi: 10.1038/ajg.2009.413. [DOI] [PubMed] [Google Scholar]
- 34.Booth RA, Ansari MT, Loit E, Tricco AC, Weeks L, Doucette S, Skidmore B, Sears M, Sy R, Karsh J. Assessment of thiopurine S-methyltransferase activity in patients prescribed thiopurines: a systematic review. Ann Intern Med. 2011;154:814–823. doi: 10.7326/0003-4819-154-12-201106210-00009. W-295-8. [DOI] [PubMed] [Google Scholar]
- 35.Dubinsky MC, Reyes E, Ofman J, Chiou CF, Wade S, Sandborn WJ. A cost-effectiveness analysis of alternative disease management strategies in patients with Crohn's disease treated with azathioprine or 6-mercaptopurine. Am J Gastroenterol. 2005;100:2239–2247. doi: 10.1111/j.1572-0241.2005.41900.x. [DOI] [PubMed] [Google Scholar]
- 36.Zabala-Fernandez W, Barreiro-de Acosta M, Echarri A, Carpio D, Lorenzo A, Castro J, Martinez-Ares D, Pereira S, Martin-Granizo I, Corton M, Carracedo A, Barros F. A pharmacogenetics study of TPMT and ITPA genes detects a relationship with side effects and clinical response in patients with inflammatory bowel disease receiving Azathioprine. J Gastrointestin Liver Dis. 2011;20:247–253. [PubMed] [Google Scholar]
- 37.Taylor KD, Plevy SE, Yang H, Landers CJ, Barry MJ, Rotter JI, Targan SR. ANCA pattern and LTA haplotype relationship to clinical responses to anti-TNF antibody treatment in Crohn's disease. Gastroenterology. 2001;120:1347–1355. doi: 10.1053/gast.2001.23966. [DOI] [PubMed] [Google Scholar]
- 38.Dideberg V, Louis E, Farnir F, Bertoli S, Vermeire S, Rutgeerts P, De Vos M, Van Gossum A, Belaiche J, Bours V. Lymphotoxin alpha gene in Crohn's disease patients: absence of implication in the response to infliximab in a large cohort study. Pharmacogenet Genomics. 2006;16:369–373. doi: 10.1097/01.fpc.0000204993.91806.b1. [DOI] [PubMed] [Google Scholar]
- 39.Louis E, Vermeire S, Rutgeerts P, De Vos M, Van Gossum A, Pescatore P, Fiasse R, Pelckmans P, Reynaert H, D'Haens G, Malaise M, Belaiche J. A positive response to infliximab in Crohn disease: association with a higher systemic inflammation before treatment but not with-308 TNF gene polymorphism. Scand J Gastroenterol. 2002;37:818–824. [PubMed] [Google Scholar]
- 40.Mascheretti S, Hampe J, Kuhbacher T, Herfarth H, Krawczak M, Folsch UR, Schreiber S. Pharmacogenetic investigation of the TNF/TNF-receptor system in patients with chronic active Crohn's disease treated with infliximab. Pharmacogenomics J. 2002;2:127–136. doi: 10.1038/sj.tpj.6500091. [DOI] [PubMed] [Google Scholar]
- 41.Louis E, El Ghoul Z, Vermeire S, Dall'Ozzo S, Rutgeerts P, Paintaud G, Belaiche J, De Vos M, Van Gossum A, Colombel JF, Watier H. Association between polymorphism in IgG Fc receptor IIIa coding gene and biological response to infliximab in Crohn's disease. Aliment Pharmacol Ther. 2004;19:511–519. doi: 10.1111/j.1365-2036.2004.01871.x. [DOI] [PubMed] [Google Scholar]
- 42.Louis EJ, Watier HE, Schreiber S, Hampe J, Taillard F, Olson A, Thorne N, Zhang H, Colombel JF. Polymorphism in IgG Fc receptor gene FCGR3A and response to infliximab in Crohn's disease: a subanalysis of the ACCENT I study. Pharmacogenet Genomics. 2006;16:911–914. doi: 10.1097/01.fpc.0000230421.12844.fd. [DOI] [PubMed] [Google Scholar]
- 43.Mascheretti S, Hampe J, Croucher PJ, Nikolaus S, Andus T, Schubert S, Olson A, Bao W, Folsch UR, Schreiber S. Response to infliximab treatment in Crohn's disease is not associated with mutations in the CARD15 (NOD2) gene: an analysis in 534 patients from two multicenter, prospective GCP-level trials. Pharmacogenetics. 2002;12:509–515. doi: 10.1097/00008571-200210000-00002. [DOI] [PubMed] [Google Scholar]
- 44.Vermeire S, Louis E, Rutgeerts P, De Vos M, Van Gossum A, Belaiche J, Pescatore P, Fiasse R, Pelckmans P, Vlietinck R, Merlin F, Zouali H, Thomas G, Colombel JF, Hugot JP. NOD2/CARD15 does not influence response to infliximab in Crohn's disease. Gastroenterology. 2002;123:106–111. doi: 10.1053/gast.2002.34172. [DOI] [PubMed] [Google Scholar]
- 45.Urcelay E, Mendoza JL, Martinez A, Fernandez L, Taxonera C, Diaz-Rubio M, de la Concha EG. IBD5 polymorphisms in inflammatory bowel disease: association with response to infliximab. World J Gastroenterol. 2005;11:1187–1192. doi: 10.3748/wjg.v11.i8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hlavaty T, Pierik M, Henckaerts L, Ferrante M, Joossens S, van Schuerbeek N, Noman M, Rutgeerts P, Vermeire S. Polymorphisms in apoptosis genes predict response to infliximab therapy in luminal and fistulizing Crohn's disease. Aliment Pharmacol Ther. 2005;22:613–626. doi: 10.1111/j.1365-2036.2005.02635.x. [DOI] [PubMed] [Google Scholar]
- 47.Hlavaty T, Ferrante M, Henckaerts L, Pierik M, Rutgeerts P, Vermeire S. Predictive model for the outcome of infliximab therapy in Crohn's disease based on apoptotic pharmacogenetic index and clinical predictors. Inflamm Bowel Dis. 2007;13:372–379. doi: 10.1002/ibd.20024. [DOI] [PubMed] [Google Scholar]
- 48.Ho GT, Nimmo ER, Tenesa A, Fennell J, Drummond H, Mowat C, Arnott ID, Satsangi J. Allelic variations of the multidrug resistance gene determine susceptibility and disease behavior in ulcerative colitis. Gastroenterology. 2005;128:288–296. doi: 10.1053/j.gastro.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Haritunians T, Taylor KD, Targan SR, Dubinsky M, Ippoliti A, Kwon S, Guo X, Melmed GY, Berel D, Mengesha E, Psaty BM, Glazer NL, Vasiliauskas EA, Rotter JI, Fleshner PR, McGovern DP. Genetic predictors of medically refractory ulcerative colitis. Inflamm Bowel Dis. 2010;16:1830–1840. doi: 10.1002/ibd.21293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Iudicibus S, Franca R, Martelossi S, Ventura A, Decorti G. Molecular mechanism of glucocorticoid resistance in inflammatory bowel disease. World J Gastroenterol. 2011;17:1095–1108. doi: 10.3748/wjg.v17.i9.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faubion WA, Jr, Loftus EV, Jr, Harmsen WS, Zinsmeister AR, Sandborn WJ. The natural history of corticosteroid therapy for inflammatory bowel disease: a population-based study. Gastroenterology. 2001;121:255–260. doi: 10.1053/gast.2001.26279. [DOI] [PubMed] [Google Scholar]
- 52.Daniel F, Loriot MA, Seksik P, Cosnes J, Gornet JM, Lemann M, Fein F, Vernier-Massouille G, De Vos M, Boureille A, Treton X, Flourie B, Roblin X, Louis E, Zerbib F, Beaune P, Marteau P. Multidrug resistance gene-1 polymorphisms and resistance to cyclosporine A in patients with steroid resistant ulcerative colitis. Inflamm Bowel Dis. 2007;13:19–23. doi: 10.1002/ibd.20046. [DOI] [PubMed] [Google Scholar]
- 53.Potocnik U, Ferkolj I, Glavac D, Dean M. Polymorphisms in multidrug resistance 1 (MDR1) gene are associated with refractory Crohn disease and ulcerative colitis. Genes Immun. 2004;5:530–539. doi: 10.1038/sj.gene.6364123. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto-Furusho JK, Villeda-Ramirez MA, Fonseca-Camarillo G, Sanchez-Munoz F, Dominguez-Lopez A, Barreto-Zuniga R, Uribe M. High gene expression of MDR1 (ABCB1) is associated with medical treatment response and long-term remission in patients with ulcerative colitis. Inflamm Bowel Dis. 2010;16:541–542. doi: 10.1002/ibd.21016. [DOI] [PubMed] [Google Scholar]
- 55.Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P, Kim CE, Muise A, Wang K, Glessner JT, Saeed S, Zhang H, Frackelton EC, Hou C, Flory JH, Otieno G, Chiavacci RM, Grundmeier R, Castro M, Latiano A, Dallapiccola B, Stempak J, Abrams DJ, Taylor K, McGovern D, Silber G, Wrobel I, Quiros A, Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmuda MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwillam R, Tremelling M, Delukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ, Heyman MB, Ferry GD, Kirschner B, Lee J, Essers J, Grand R, Stephens M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dubinsky MC, Mei L, Friedman M, Dhere T, Haritunians T, Hakonarson H, Kim C, Glessner J, Targan SR, McGovern DP, Taylor KD, Rotter JI. Genome wide association (GWA) predictors of anti-TNFalpha therapeutic responsiveness in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1357–1366. doi: 10.1002/ibd.21174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duraes C, Machado JC, Portela F, Rodrigues S, Lago P, Cravo M, Ministro P, Marques M, Cremers I, Freitas J, Cotter J, Tavares L, Matos L, Medeiros I, Sousa R, Ramos J, Deus J, Caldeira P, Chagas C, Duarte MA, Goncalves R, Loureiro R, Barros L, Bastos I, Cancela E, Moraes MC, Moreira MJ, Vieira AI, Magro F. Phenotype-genotype profiles in Crohn's disease predicted by genetic markers in autophagy-related genes (GOIA study II) Inflamm Bowel Dis. 2012 doi: 10.1002/ibd.23007. [DOI] [PubMed] [Google Scholar]
- 58.Arijs I, Li K, Toedter G, Quintens R, Van Lommel L, Van Steen K, Leemans P, De Hertogh G, Lemaire K, Ferrante M, Schnitzler F, Thorrez L, Ma K, Song XY, Marano C, Van Assche G, Vermeire S, Geboes K, Schuit F, Baribaud F, Rutgeerts P. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58:1612–1619. doi: 10.1136/gut.2009.178665. [DOI] [PubMed] [Google Scholar]
- 59.Jurgens M, Laubender RP, Hartl F, Weidinger M, Seiderer J, Wagner J, Wetzke M, Beigel F, Pfennig S, Stallhofer J, Schnitzler F, Tillack C, Lohse P, Goke B, Glas J, Ochsenkuhn T, Brand S. Disease activity, ANCA, and IL23R genotype status determine early response to infliximab in patients with ulcerative colitis. Am J Gastroenterol. 2010;105:1811–1819. doi: 10.1038/ajg.2010.95. [DOI] [PubMed] [Google Scholar]
- 60.Paul T, Birnbaum A, Pal DK, Pittman N, Ceballos C, LeLeiko NS, Benkov K. Distinct phenotype of early childhood inflammatory bowel disease. J Clin Gastroenterol. 2006;40:583–586. doi: 10.1097/00004836-200608000-00004. [DOI] [PubMed] [Google Scholar]
- 61.Polito JM, 2nd, Childs B, Mellits ED, Tokayer AZ, Harris ML, Bayless TM. Crohn's disease: influence of age at diagnosis on site and clinical type of disease. Gastroenterology. 1996;111:580–586. doi: 10.1053/gast.1996.v111.pm8780560. [DOI] [PubMed] [Google Scholar]
- 62.de Ridder L, Weersma RK, Dijkstra G, van der Steege G, Benninga MA, Nolte IM, Taminiau JA, Hommes DW, Stokkers PC. Genetic susceptibility has a more important role in pediatric-onset Crohn's disease than in adult-onset Crohn's disease. Inflamm Bowel Dis. 2007;13:1083–1092. doi: 10.1002/ibd.20171. [DOI] [PubMed] [Google Scholar]
- 63.Russell RK, Drummond HE, Nimmo ER, Anderson NH, Noble CL, Wilson DC, Gillett PM, McGrogan P, Hassan K, Weaver LT, Bisset WM, Mahdi G, Satsangi J. Analysis of the influence of OCTN1/2 variants within the IBD5 locus on disease susceptibility and growth indices in early onset inflammatory bowel disease. Gut. 2006;55:1114–1123. doi: 10.1136/gut.2005.082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orchard TR, Chua CN, Ahmad T, Cheng H, Welsh KI, Jewell DP. Uveitis and erythema nodosum in inflammatory bowel disease: clinical features and the role of HLA genes. Gastroenterology. 2002;123:714–718. doi: 10.1053/gast.2002.35396. [DOI] [PubMed] [Google Scholar]
- 65.Orchard TR, Thiyagaraja S, Welsh KI, Wordsworth BP, Hill Gaston JS, Jewell DP. Clinical phenotype is related to HLA genotype in the peripheral arthropathies of inflammatory bowel disease. Gastroenterology. 2000;118:274–278. doi: 10.1016/s0016-5085(00)70209-5. [DOI] [PubMed] [Google Scholar]
- 66.Mendoza JL, Lana R, Martin MC, de la Concha EG, Urcelay E, Diaz-Rubio M, Abreu MT, Mitchell AA. FcRL3 gene promoter variant is associated with peripheral arthritis in Crohn's disease. Inflamm Bowel Dis. 2009;15:1351–1357. doi: 10.1002/ibd.20895. [DOI] [PubMed] [Google Scholar]
- 67.Karlsen TH, Franke A, Melum E, Kaser A, Hov JR, Balschun T, Lie BA, Bergquist A, Schramm C, Weismuller TJ, Gotthardt D, Rust C, Philipp EE, Fritz T, Henckaerts L, Weersma RK, Stokkers P, Ponsioen CY, Wijmenga C, Sterneck M, Nothnagel M, Hampe J, Teufel A, Runz H, Rosenstiel P, Stiehl A, Vermeire S, Beuers U, Manns MP, Schrumpf E, Boberg KM, Schreiber S. Genome-wide association analysis in primary sclerosing cholangitis. Gastroenterology. 138:1102–1111. doi: 10.1053/j.gastro.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 68.Janse M, Lamberts LE, Franke L, Raychaudhuri S, Ellinghaus E, Muri Boberg K, Melum E, Folseraas T, Schrumpf E, Bergquist A, Bjornsson E, Fu J, Jan Westra H, Groen HJ, Fehrmann RS, Smolonska J, van den Berg LH, Ophoff RA, Porte RJ, Weismuller TJ, Wedemeyer J, Schramm C, Sterneck M, Gunther R, Braun F, Vermeire S, Henckaerts L, Wijmenga C, Ponsioen CY, Schreiber S, Karlsen TH, Franke A, Weersma RK. Three ulcerative colitis susceptibility loci are associated with primary sclerosing cholangitis and indicate a role for IL2, REL and CARD9. Hepatology. 2011;53:1977–1985. doi: 10.1002/hep.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Melum E, Franke A, Schramm C, Weismuller TJ, Gotthardt DN, Offner FA, Juran BD, Laerdahl JK, Labi V, Bjornsson E, Weersma RK, Henckaerts L, Teufel A, Rust C, Ellinghaus E, Balschun T, Boberg KM, Ellinghaus D, Bergquist A, Sauer P, Ryu E, Hov JR, Wedemeyer J, Lindkvist B, Wittig M, Porte RJ, Holm K, Gieger C, Wichmann HE, Stokkers P, Ponsioen CY, Runz H, Stiehl A, Wijmenga C, Sterneck M, Vermeire S, Beuers U, Villunger A, Schrumpf E, Lazaridis KN, Manns MP, Schreiber S, Karlsen TH. Genome-wide association analysis in primary sclerosing cholangitis identifies two non-HLA susceptibility loci. Nat Genet. 43:17–19. doi: 10.1038/ng.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keenan BT, Chibnik LB, Cui J, Ding B, Padyukov L, Kallberg H, Bengtsson C, Klareskog L, Alfredsson L, Karlson EW. Effect of interactions of glutathione S-transferase T1, M1, and P1 and HMOX1 gene promoter polymorphisms with heavy smoking on the risk of rheumatoid arthritis. Arthritis Rheum. 2010;62:3196–3210. doi: 10.1002/art.27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103:1342–1346. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanada H, Jones JE, Jose PA. Genetics of salt-sensitive hypertension. Curr Hypertens Rep. 2011;13:55–66. doi: 10.1007/s11906-010-0167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hutter CM, Chang-Claude J, Slattery ML, Pflugeisen BM, Lin Y, Duggan D, Nan H, Lemire M, Rangrej J, Figueiredo JC, Jiao S, Harrison TA, Liu Y, Chen LS, Stelling DL, Warnick GS, Hoffmeister M, Kury S, Fuchs CS, Giovannucci E, Hazra A, Kraft P, Hunter DJ, Gallinger S, Zanke BW, Brenner H, Frank B, Ma J, Ulrich CM, White E, Newcomb PA, Kooperberg C, LaCroix AZ, Prentice RL, Jackson RD, Schoen RE, Chanock SJ, Berndt SI, Hayes RB, Caan BJ, Potter JD, Hsu L, Bezieau S, Chan AT, Hudson TJ, Peters U. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012;72:2036–2044. doi: 10.1158/0008-5472.CAN-11-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ichimura A, Hirasawa A, Poulain-Godefroy O, Bonnefond A, Hara T, Yengo L, Kimura I, Leloire A, Liu N, Iida K, Choquet H, Besnard P, Lecoeur C, Vivequin S, Ayukawa K, Takeuchi M, Ozawa K, Tauber M, Maffeis C, Morandi A, Buzzetti R, Elliott P, Pouta A, Jarvelin MR, Korner A, Kiess W, Pigeyre M, Caiazzo R, Van Hul W, Van Gaal L, Horber F, Balkau B, Levy-Marchal C, Rouskas K, Kouvatsi A, Hebebrand J, Hinney A, Scherag A, Pattou F, Meyre D, Koshimizu TA, Wolowczuk I, Tsujimoto G, Froguel P. Dysfunction of lipid sensor GPR120 leads to obesity in both mouse and human. Nature. 2012;483:350–354. doi: 10.1038/nature10798. [DOI] [PubMed] [Google Scholar]
- 75.Lioy PJ, Rappaport SM. Exposure science and the exposome: an opportunity for coherence in the environmental health sciences. Environ Health Perspect. 2011;119:A466–A467. doi: 10.1289/ehp.1104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin Z, Hegarty JP, Cappel JA, Yu W, Chen X, Faber P, Wang Y, Kelly AA, Poritz LS, Peterson BZ, Schreiber S, Fan JB, Koltun WA. Identification of disease-associated DNA methylation in intestinal tissues from patients with inflammatory bowel disease. Clin Genet. 2011;80:59–67. doi: 10.1111/j.1399-0004.2010.01546.x. [DOI] [PubMed] [Google Scholar]
- 77.Olaru AV, Cheng Y, Agarwal R, Yang J, David S, Abraham JM, Yu W, Kwon JH, Lazarev M, Brant SR, Marohn MR, Hutcheon DF, Harpaz N, Meltzer SJ, Mori Y. Unique patterns of CpG island methylation in inflammatory bowel disease-associated colorectal cancers. Inflamm Bowel Dis. 2012;18:641–648. doi: 10.1002/ibd.21826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 80.Thatcher N, Chang A, Parikh P, Rodrigues Pereira J, Ciuleanu T, von Pawel J, Thongprasert S, Tan EH, Pemberton K, Archer V, Carroll K. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 81.Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, Afdhal NH, Jacobson IM, Esteban R, Poordad F, Lawitz EJ, McCone J, Shiffman ML, Galler GW, Lee WM, Reindollar R, King JW, Kwo PY, Ghalib RH, Freilich B, Nyberg LM, Zeuzem S, Poynard T, Vock DM, Pieper KS, Patel K, Tillmann HL, Noviello S, Koury K, Pedicone LD, Brass CA, Albrecht JK, Goldstein DB, McHutchison JG. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129. doi: 10.1053/j.gastro.2010.04.013. e18. [DOI] [PubMed] [Google Scholar]