Summary
Transcription of the mammalian genome is pervasive but productive transcription outside protein-coding genes is limited by unknown mechanisms1. In particular, although RNA polymerase II (RNAPII) initiates divergently from most active gene promoters, productive elongation occurs primarily in the sense coding direction2–4. Here we show that asymmetric sequence determinants flanking gene transcription start sites (TSS) control promoter directionality by regulating promoter-proximal cleavage and polyadenylation. We find that upstream antisense RNAs (uaRNAs) are cleaved and polyadenylated at poly (A) sites (PAS) shortly after their initiation. De novo motif analysis reveals PAS signals and U1 snRNP (U1) recognition sites as the most depleted and enriched sequences, respectively, in the sense direction relative to the upstream antisense direction. These U1 and PAS sites are progressively gained and lost, respectively, at the 5′ end of coding genes during vertebrate evolution. Functional disruption of U1 snRNP activity results in a significant increase in promoter-proximal cleavage events in the sense direction with slight increases in the antisense direction. These data suggests that a U1-PAS axis characterized by low U1 recognition and high density of PAS in the upstream antisense region reinforces promoter directionality by promoting early termination in upstream antisense regions whereas proximal sense PAS signals are suppressed by U1 snRNP. We propose that the U1-PAS axis limits pervasive transcription throughout the genome.
Two potential mechanisms for suppressing transcription elongation in the upstream antisense region of gene TSS include inefficient release of paused RNAPII and/or early termination of transcription. RNAPII pauses shortly after initiation downstream of the gene TSS and the paused state is released by the recruitment and activity of p-TEFb5. A detailed characterization of several uaRNAs in mouse embryonic stem cells (mESCs) suggested that p-TEFb is recruited similarly in both sense and antisense directions6, and in human cells, elongating RNAPII (phosphorylated at serine 2 in the C-terminal domain) occupies the proximal upstream transcribed region7. These data argue that the upstream antisense RNAPII complex undergoes the initial phase of elongation but likely terminates early due to an unknown mechanism.
To globally test whether upstream antisense transcripts undergo early termination (compared to coding mRNA) by a canonical PAS-dependent cleavage mechanism, we mapped by deep sequencing the 3′-ends of polyadenylated RNAs in mESCs8. For most protein-coding genes, transcription termination is triggered by cleavage of the nascent RNA upon recognition of a PAS whose most essential feature is an AAUAAA sequence or a close variant located about 10–30 nucleotides upstream of the cleavage site9. We sequenced two cDNA libraries and obtained over 230 million reads, of which 114 million mapped uniquely to the genome with at most two mismatches. We developed a computational pipeline to identify 835,942 unique 3′-ends (cleavage sites) whose poly (A) tails are likely to be added post-transcriptionally and are also associated with the canonical PAS hexamer or its common variants (Supplementary Fig. 1, see Methods).
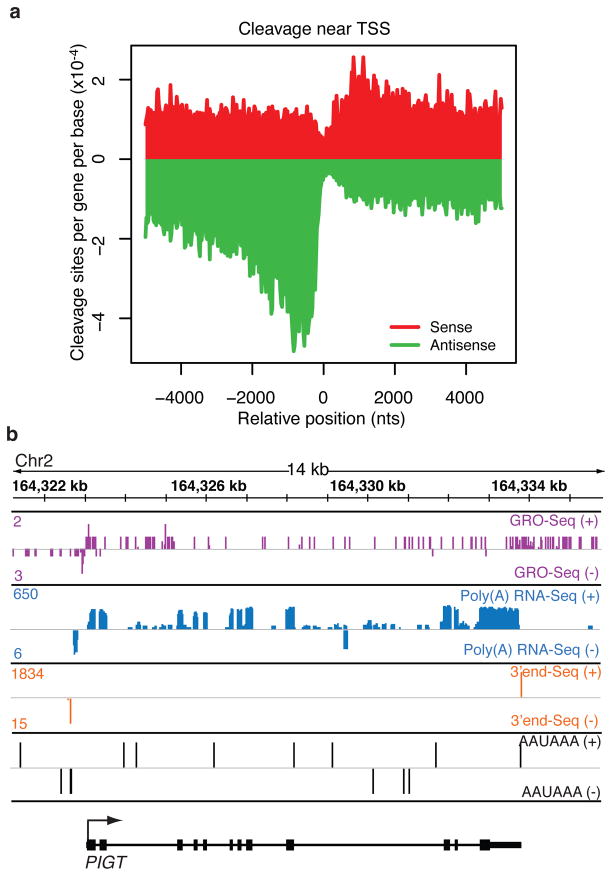
To investigate whether uaRNAs are terminated by PAS-dependent mechanisms, we focused our analysis on cleavage sites proximal to gene TSS and at least 5 kilobases (kb) away from known gene transcription end sites (TES). Interestingly, in the upstream antisense region we observed a 2-fold higher number of cleavage sites compared to the downstream sense sites flanking protein-coding gene TSS (Fig. 1a). The peak of the upstream antisense cleavage sites is about 700 bases from the coding gene TSS. This observation suggests that upstream antisense transcripts are frequently terminated by PAS-directed cleavage shortly after initiation, a trend we also observe in various tissues of mouse and human10 (Supplementary Fig. 2). Inspection of gene tracks at the PIGT locus reveals upstream antisense cleavage shortly after a PAS (AATAAA) less than 400 bases from the PIGT TSS, whereas in the sense direction cleavage is confined to the TES (Fig. 1b). Similar patterns were observed for subsets of promoters (promoters without nearby genes, Global Run-On Sequencing (GRO-seq) defined divergent promoters, and Chromatin immunoprecipitation sequencing (ChIP-seq) defined RNAPII-occupied promoters), or for high confidence cleavage sites, cleavage reads, and cleavage clusters (Supplementary Fig. 3). Of all divergent promoters, nearly half (48%) produce PAS-dependent upstream antisense cleavage events within 5 kb of coding gene TSS, compared to 33% downstream of the TSS. We validated several of these promoter proximal sense and antisense cleavage sites using Rapid Amplification of 3′ cDNA ends (3′-RACE) (Supplementary Fig. 4).
Figure 1.
Promoter-proximal PAS-dependent termination of uaRNA. a, Metagene plot of sense (red) or antisense (green) unique cleavage sites flanking coding gene TSS. The number of unique cleavage sites per gene per base in each 25 bp bin across 5 kb upstream and downstream of the TSS is plotted. Mean cleavage density of first 2 kb: sense/antisense = 1.45/3.10. b, Genome browser view from the PIGT locus (shown in black on the + strand) displaying the following tracks with + strand (top) and − strand (bottom) represented: GRO-Seq (purple)11, Poly (A)+ RNA-Seq (blue)12, 3′end RNA-Seq (orange), and PAS (AAUAAA, black). For each gene track, the x-axis represents the linear sequence of genomic DNA. The numbers on the top left corner represent the maximum read density on each track.
Similar to annotated cleavage sites at TES of genes, these upstream antisense cleavage sites are associated with the PAS located at the expected position, about 22 nucleotides upstream the cleavage site (Supplementary Fig. 5a–b)13,14. Moreover, the nucleotide sequence composition flanking the cleavage sites resembles that of TES of genes (Supplementary Fig. 5c–e) including a downstream U-rich region15,16. To determine whether members of the canonical cleavage and polyadenylation machinery bind specifically to uaRNA cleavage sites, we analyzed available cross-linking immunoprecipitation (CLIP) sequencing datasets for 10 canonical 3′ end processing factors, including CPSF-160, CPSF-100, CPSF-73, CPSF-30, Fip1, CstF-64, CstF-64τ, CF Im25, CF Im59, and CF Im68 along with poly (A) 3′-end sequencing data generated in HEK293 cells17. We detect specific binding of all 10 factors at uaRNA cleavage sites with positional profiles identical or very similar to that of mRNA cleavage sites (Supplementary Fig. 6). These results indicate the poly (A) tails that we analyzed are products of PAS-dependent cleavage and polyadenylation, rather than either a priming artifact or PAS-independent polyadenylation representing a transient signal for RNA degradation18–20.
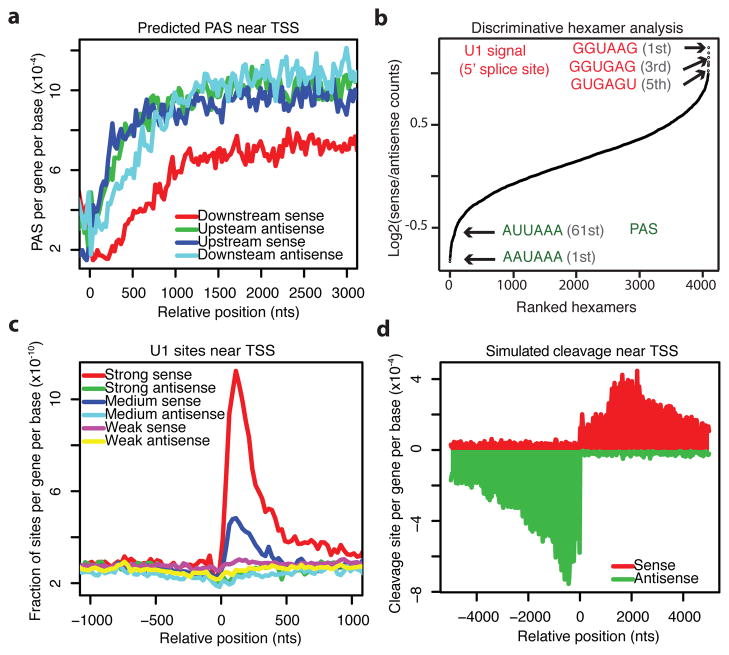
As a first step to understand the molecular mechanism underlying the cleavage bias, we examined the frequency of PAS in a 6 kb region on the four strands flanking the coding gene TSS. We observed an approximately 33% depletion of the canonical AATAAA PAS hexamer specifically downstream of the TSS on the coding strand of genes as compared to the other regions (Fig. 2a). Since this 33% depletion is unlikely to explain the 2-fold cleavage bias observed (see simulation results in Supplementary Fig. 8a), we searched for additional discriminative 6-mer sequence signals in an unbiased manner. All 4096 hexamers were ranked by enrichment in the first 1 kb of the sense strand of genes relative to the corresponding upstream antisense region (Fig. 2b). Interestingly, we identified the PAS as the most depleted sequence in sense genes relative to the upstream antisense region of gene TSS. In addition, we identified 5′ splice site related sequences (or sequences recognized by U1 referred to as U1 sites) as the most enriched hexamers in sense genes (Fig. 2b) relative to antisense regions. This includes the consensus GGUAAG (first) that is perfectly complementary to the 5′ end of the U1 snRNA, as well as GGUGAG (third) and GUGAGU (fifth), which represent common 5′ splice site sequences (with the first GU in each motif located at the intron start). Consistent with the hexamer enrichment analysis, a metagene plot displaying an unbiased prediction of strong, medium, and weak U1 sites (see Methods) revealed strong enrichment of U1 signals in the first 500 bps downstream of the TSS, with essentially only background levels observed in all other regions and a small depletion in the upstream antisense direction (Fig. 2c).
Figure 2.
Asymmetric distribution of PAS and U1 signals flanking coding gene TSS. a. Number of AATAAA sites per gene per base in each 25 bp bin within a 3 kb region flanking gene TSS on the downstream sense (red), downstream antisense (light blue), upstream antisense (green), and upstream sense (dark blue) strands. b, Rank of all 4096 hexamers by enrichment (log2 ratio) in the first 1 kb of all coding genes in the sense direction relative to 1 kb in the upstream antisense direction of the TSS. c, Density of predicted 5′ splice sites within a 1 kb region flanking gene TSS. Strong, medium, and weak 5′ splice sites are defined in Methods. d, Metagene plot of simulated cleavage sites around gene TSS. The first unprotected PAS (AAUAAA) that is not within 1 kb downstream of a strong U1 site for all coding genes is plotted. Mean cleavage density of first 2 kb: sense/antisense = 2.08/4.99.
The asymmetric distribution of U1 sites and PAS sites flanking the TSS could potentially explain the biased cleavage pattern shown in Fig. 1a if the U1 complex suppresses cleavage and polyadenylation near a U1 site, as has been observed in various species including human and mouse21–23. Consistent with this model, we observed a depletion of cleavage sites, especially frequent cleavage sites, downstream of strong U1 sites (Supplementary Fig. 7a). Focusing on the upstream antisense direction, the presence of proximal PAS sites (within 1 kb of coding gene TSS) is significantly associated with shorter uaRNAs (p < 1e-15), whereas the presence of proximal U1 sites is significantly associated with longer uaRNAs but only in the presence of proximal PAS sites (p < 0.0006), consistent with a model where U1 promotes RNA lengthening by suppressing proximal PAS (Supplementary Fig. 7b). To test whether the encoded bias in U1 and PAS signal distribution explains the cleavage bias observed from our 3′-end sequencing analysis, we performed a cleavage site simulation using predicted strong U1 sites and canonical PAS (AATAAA) sequences. Specifically, we defined a protection zone of 1 kb downstream of a strong U1 site and used the first unprotected PAS as the cleavage site. The metagene plot of simulated cleavage events (Fig. 2d) recapitulate the major features of the observed distribution (Fig. 1a), including an antisense peak around 700 bases upstream and a ~2-fold difference between sense and antisense strands. Similar patterns were robustly observed when varying the size of the protection zone (Supplementary Fig. 8). Thus, we identified a U1-PAS axis flanking gene promoters that may explain why uaRNAs undergo early termination.
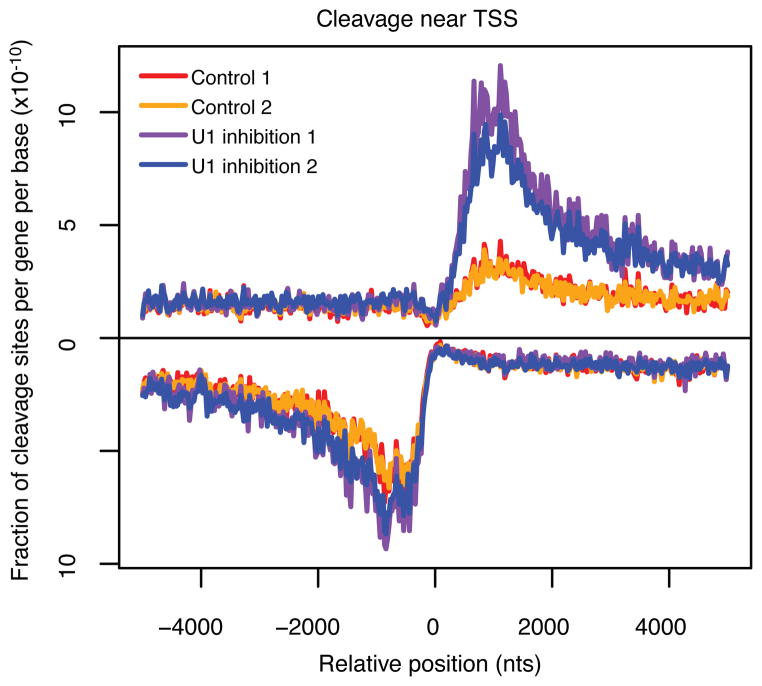
To validate the U1-PAS axis model, we functionally inhibited U1 in mESCs. Specifically, we transfected mESCs with either an antisense morpholino oligonucleotide (AMO) complementary to the 5′ end of U1 snRNA to block its binding to 5′ splice sites (or similar sequences) or a control AMO with scrambled sequences followed by 3′-end RNA sequencing21,22. Interestingly, we observe in two biological replicates a dramatic increase in promoter-proximal cleavage events in coding genes but only a slight increase in upstream antisense regions, which eliminates the asymmetric bias in promoter-proximal cleavage we observed in either the wild-type cells or cells treated with scrambled control AMOs (Fig. 3). These observations confirm that U1 protects sense RNA in protein-coding genes from premature cleavage and polyadenylation in promoter proximal regions, thus, reinforcing transcriptional directionality of genes. However, in the antisense direction, the activity of U1 is much less and there is little enhancement in cleavage sites upon inhibition of U1 recognition.
Figure 3.
Promoter-proximal cleavage sites are altered upon functional U1 inhibition. Y-axis is the number of cleavage sites per gene per base divided by the total number of cleavage sites identified in each 3′ end-sequencing library in a 5 kb region flanking coding gene TSS. Signal for the antisense strand is set as negative. U1 inhibition 1 (purple) and U1 inhibition 2 (blue) represent 3′-end sequencing libraries generated from mESCs treated with a U1-targeting AMO. Control 1 (red) and Control 2 (orange) represent 3′-end sequencing libraries generated from mESCs treated with a scrambled control AMO. Mean cleavage density of first 2 kb: sense/antisense = 2.5/4.4 (Control 1), 2.4/4.3 (Control 2), 7.0/5.8 (U1 inhibition 1), 5.9/5.5 (U1 inhibition 2).
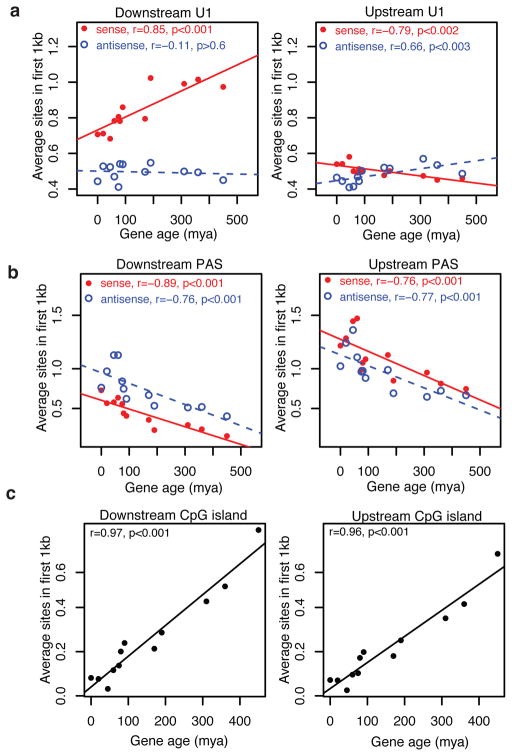
The conservation of the asymmetric cleavage pattern across human and mouse (Supplementary Fig. 2) led us to examine if there is evolutionary selection on the U1-PAS axis. Previously, mouse protein-coding genes have been assigned to 12 evolutionary branches and dated by analyzing the presence or absence of orthologs in the vertebrate phylogeny24. We find strong trends of progressive gain of U1 sites depending on the age of a gene (Fig. 4a) and loss of PAS sites (Fig. 4b) over time at the 5′ end (the first 1 kb) of protein-coding genes, suggesting that suppression of promoter-proximal transcription termination is important for maintaining gene function. Interestingly, the same trends, although weaker, are observed in upstream antisense regions, suggesting at least a subset of uaRNAs may be functionally important in that over time they gain U1 sites and lose PAS sites to become more extensively transcribed. In addition to the coding strand of genes (downstream sense region), PAS sites were also progressively lost on the other three strands flanking TSS (Fig. 4b). This observation probably reflects on the increases in CpG-rich sequences within 1 kb of gene TSS and suggests that coding genes acquire CpG islands as they age (Fig. 4c). However, the bias of low PAS site density in the sense direction extends across the total transcription unit (Supplementary Fig. 9) and is distinct from the CpG density near the promoter.
Figure 4.
Evolutionary gain and loss of U1 and PAS sites. a, Average number of strong U1 sites in the first 1 kb of protein-coding genes and upstream regions. b, Average number of PAS sites in the first 1 kb downstream and upstream of coding gene TSS, respectively. c, Average number of CpG islands overlapping the first 1 kb of protein-coding genes and upstream regions. Genes are divided into 12 ordered groups by gene age. X-axis indicates the age (myr, million years) of gene groups. The number of genes in each group (from old to young): 11934, 1239, 914, 597, 876, 1195, 279, 175, 198, 315, 926, and 1143. Solid red dots and blue circles indicate sites on the sense and antisense strands, respectively.
We also propose that some long noncoding RNAs (lncRNAs) generated from bidirectional promoters might represent an evolutionary intermediate between uaRNAs and protein-coding genes. Consistent with this, annotated head-to-head mRNA-lncRNA pairs as a whole showed a bias (in terms of promoter-proximal cleavage site, U1 site, and PAS site distributions flanking coding gene TSS) weaker than head-to-head mRNA-uaRNA pairs but stronger than mRNA-mRNA pairs (Supplementary Fig. 10). This is also consistent with recent results suggesting that de novo protein-coding genes originate from lncRNAs at bidirectional promoters25.
The U1-PAS axis likely has a broader role in limiting pervasive transcription throughout the genome. The enrichment of U1 sites and depletion of PAS sites are confined to the sense strand within the gene body, whereas intergenic and antisense regions show relatively high PAS but low U1 density (Supplementary Fig. 9), indicating the U1-PAS axis may serve as a mechanism for terminating transcription in both antisense and intergenic regions.
Together, we propose that a U1-PAS axis is important in defining the directionality for transcription elongation at divergent promoters (Supplementary Fig. 11). Although the U1-PAS axis may explain the observed cleavage bias at promoters surprisingly well, it seems likely that additional cis-elements may influence PAS usage26 and will need to be integrated into this model. There may also be other PAS-independent mechanisms that contribute to termination of transcription in upstream antisense regions and across the genome27–29. However, evidence for the U1-PAS axis is found in several different tissues of mouse and human, indicating its wide utilization as a general mechanism to regulate transcription elongation in mammals. Like protein-coding transcripts, lncRNAs must also contend with the U1-PAS axis. These RNAs and short non-coding RNAs from divergent transcription of gene promoters may be considered part of a continuum that varies in the degree of the activity of the U1-PAS axis.
Methods (Full length)
Cell Cuture
V6.5 (C57BL/6–129) mouse embryonic stem cells (mESCs) (Koch Institute Transgenic Facility) were grown under standard ES cell culture conditions2.
Poly(A) 3′-End sequencing
Total RNA was extracted from V6.5 mESCs using Ambion’s Ribopure kit (AM1924M). Poly (A) selected RNA was fragmented using RNase T1 (AM2283). Reverse transcription was performed with an RT oligo (Table S1) at 0.25 uM final concentration using Invitrogen’s Superscript III Reverse Transcriptase (18080–44) according to the manufacturer’s protocol. The resulting cDNA was run on a 6% TBE-Urea polyacrylamide gel (National Diagnostics) and the 100–300 size range of products were gel extracted and eluted overnight. The gel-purified cDNA products were circularized using CircLigase II (CL9025K) according to the manufacturer’s protocol. Circularized cDNA was PCR-amplified using the Phusion High-Fidelity DNA Polymerase (MO530L) for 15–18 cycles using the primers described in Table S1. Amplified products were run on a 1.5 % agarose gel and the 200–400 size range was extracted using Qiagen’s MinElute Gel Extraction Kit (28604). The 3′-end library was then submitted for Illumina sequencing on the HI-Seq 2000 platform.
U1 inhibition with antisense morpholino oligonucleotides (AMO)
V6.5 mESCs were transfected using the Amaxa Nucleofector II with program A-23 (mESC-specific) according to the manufacturers protocol. Specifically, 2.5 million V6.5 mESCs were transfected with 7.5 uM of U1-targeting or a scrambled AMO for 8 hrs,21,22 prior to RNA sequencing analysis.
3′-RACE
Total RNA was extracted using Ambion’s Ribopure kit and DNase-treated using Ambion’s DNA Free-Turbo. 3′-RACE was performed using Ambion’s Gene Racer Kit according to the manufacturer’s instructions. 3′-end PCR products were run on a 1.5% agarose gel, gel extracted using Qiagen’s gel extraction kit, and Sanger sequenced. All primers are described in Table S1.
Reads mapping
Raw reads were processed with the program cutadapt 30 to trim the adaptor sequence (TGGAATTCTCGGGTGCCAAGGAACTCCAGTCACATCAC) from the 3′ end. Reads longer than 15 nts after adaptor trimming were mapped to the mouse genome (mm9) with bowtie31 requiring unique mapping with at most two mismatches (options: -n 2 -m 1 --best --strata). Mapped reads were collapsed by unique 3′ end positions.
Internal priming filter
To remove reads whose A-tail is encoded in the genome rather than added post-transcriptionally, we filtered reads that have 1) more than 10 As in the first 20 nt window or 2) more than 6 As in the first 10 nt window downstream from the detected cleavage site of the 3′-end. The threshold used is based on the bimodal distribution of the number of As downstream of annotated TES.
PAS filter
In addition to a set of 12 hexamers identified previously in mouse and human EST analysis13,14, we analyzed the annotated TES in the mouse genome to identify additional potential PAS variants. All hexamers with at most two mismatches to the canonical AATAAA motif were used to search in the sequence up to 100 nts upstream of annotated TES. The distribution of the position of each hexamer relative to the TES (a histogram) is compared to that of AATAAA. Hexamers with a position profile similar to AATAAA will have a peak around position 20–24. We quantified the similarity by Pearson correlation coefficient and used a cut-off of 0.5 after manual inspection. In total, 24 new hexamers were identified as potential PAS and a hierarchy was assigned for the 36 hexamers (PAS36): first, the 12 known variants are ranked by their frequency of usage in the mouse genome, and then the newly identified PAS ranked by their correlation with AATAAA in terms of the positional profile defined above. To define a window where most PAS or variants are located, we searched for each of the 36 PAS variants within 100 nts of annotated gene 3′ ends and chose the best one according to the designated hierarchy. We summarized the distance of the best PAS to the annotated TES and defined a window of (0–41) around the position 22 peak such that 80% of the annotated TES have their best-matched PAS within that window. Using these criteria, we searched for PAS36 variants within the 0–41 window upstream of our experimentally sequenced 3′-ends. If there were multiple PAS hexamers identified within this window for a given 3′-end, we chose the best one defined by the hierarchy described above. Reads without any of the 36 PAS variants within the 0–41 window were discarded.
Remove potential false positive cleavage sites
Due to sequencing error, abundant transcripts such as ribosomal gene mRNAs can produce error-containing 3′ end reads that mapped to other locations in the genome, leading to false positive cleavage sites. To remove such potential false positive sites, we defined a set of 71674 (7.5%) abundant cleavage sites that are supported with more than 100 reads from the pooled library. A bowtie reference index was built using sequences within 50 nts upstream of those abundant sites. Non-abundant sites within these 50 nts reference regions were not used to search for false positives. Reads initially mapped to sites outside these reference regions were re-mapped against the new index allowing up to two mismatches. Reads mapped to any of the reference regions in this analysis were treated as potential false positive reads. Cleavage sites containing only potential false positive reads are defined as potential false positive sites and were removed from subsequent analysis. In total, 7.2% (389185) of initially mapped reads are outside the reference regions. 0.34% of all mapped reads were classified as potential false positive reads and 9.1% (86425) of all cleavage sites were identified as potential false positive sites.
Remove B2 SINE RNA associated cleavage sites
We further removed cleavage sites associated with B2_Mm1a and B2_Mm1t SINE RNAs. These B2 SINE RNAs are transcribed by RNA Pol III but contain AAUAAA sequences near the 3′ end. In total, 3.5% (33696) of all cleavage sites passing the internal priming filter and the PAS filter were mapped within B2 regions or within 100 nts downstream of B2 3′ end. These sites were removed.
Prediction of U1 sites/putative 5′ splice sites
A nucleotide frequency matrix of 5′ splice sites (3 nt in exon and 6 nt in intron) was compiled using all annotated constitutive 5′ splice sites in the mouse genome. The motif was then used by FIMO32 to search significant matches (p<0.05) on both strands of the genome. Matches were then scored by a Maximum Entropy model33. Maximum entropy scores for all annotated 5′ splice sites were also calculated to define thresholds used to classify the predicted sites into strong, medium and weak. Sites with scores larger than the median of annotated 5′ splice sites (8.77) were classified as ‘strong’. Sites with scores lower than 8.77 but higher than the threshold dividing the first and second quarter of annotated 5′ splice sites (7.39) were classified as ‘medium’, and the rest of the predicted sites with scores higher than 4 were classified as ‘weak’. Sites with scores lower than 4 were discarded.
Define a set of divergent promoters
GRO-seq data from mESCs11 were used to define a set of active and divergent promoters. Active promoters were defined as promoters with GRO-seq signal detected within the first 1 kb downstream sense strand. A promoter was considered divergent if it contained GRO-Seq signal in the first 1 kb downstream the sense strand and within the first 2 kb of the upstream antisense strand. A minimum number of two reads within the defined window (downstream 1 kb or upstream 2 kb) were used as a cut-off for background signals.
Define RNAPII Ser5P bound TSS
ChIP-seq data for ser5p RNA Pol II and corresponding input was downloaded from GEO database (accession number GSE2053034) and peaks called using MACS35 with default settings. TSS less than 500bps away from a peak summit are defined as bound.
Discriminative hexamer analysis
An unbiased exhaustive enumeration of all 4096 hexamers was performed to find hexamers that are discriminative of downstream sense and upstream antisense strands of protein-coding gene promoters. Specifically, the first 1000 nucleotides downstream sense and upstream antisense of all protein-coding gene TSS were extracted from repeat masked genome (from UCSC genome browser, non-masked genome sequence gave similar results). For each hexamer, the total number of occurrences on each side was counted and then the log2 ratio of the occurrences on sense versus antisense strand was calculated as a measure of enrichment on the sense but depletion on the antisense strand.
Cleavage site simulation
Protein-coding genes and 10 kb upstream antisense regions were scanned for strong U1 sites and PAS sites (AATAAA). Starting from protein coding gene TSS, the first unprotected PAS was predicted to be the cleavage site. A PAS is protected only if it is within a designated protection window (in nucleotides) downstream (+) of a strong U1 site.
Binding of 3′ end processing factors in uaRNA regions
RNA 3′ end cleavage and polyadenylation sites and CLIP-seq read density of ten 3′ end processing factors in wild type HEK293 cells were downloaded from Gene Expression Omnibus (GEO) dataset GSE37401. A cleavage site is defined as a uaRNA cleavage site if it is outside any protein-coding gene but locates within 5 kb upstream antisense of a protein-coding gene. mRNA cleavage sites are defined as cleavage sites within 100 bases of annotated protein-coding gene ends. For each 3′ end-processing factor, CLIP read density within 200 bases of all cleavage sites are added up every 5bp bin and then normalized such that the max value is 1.
Evolutionary analysis of U1 sites, PAS sites, and CpG islands
Mouse protein-coding gene branch/age assignment was obtained from a previous analysis24. The number of strong U1 sites, PAS (AATAAA) sites, and CpG islands (UCSC mm9 annotations) in the first 1 kb region flanking TSS on each strand were calculated, and the average number of sites in each branch/age group was plotted against gene age. Pearson correlation coefficient and linear regression fitting were done using R. Significance of the correlation was assessed by comparing to a null distribution of correlation coefficients calculated by shuffling gene branch/age assignments 1000 times.
Bidirectional promoter analysis
For each annotated TSS the closest upstream antisense TSS was identified and those TSS pairs within 1 kb were defined as head-to-head pairs. LncRNAs were defined as noncoding RNAs longer than 200 bps. UCSC mm9 gene annotations were used in this analysis.
Supplementary Material
Acknowledgments
The authors wish to dedicate this paper to the memory of Officer Sean Collier, for his caring service to the MIT community and for his sacrifice. We would like to thank Noah Spies for generously sharing his optimized 3′-end sequencing protocol, Charles Lin for providing computational assistance, Mary Lindstrom for assistance on constructing Supplementary Figure 11, and Sidi Chen, Anthony Chiu, Mohini Jangi, Qifang Liu, Jeremy Wilusz, and Jesse Zamudio for critical reading of the manuscript. We also thank the Core Facility in the Swanson Biotechnology Center at the David H. Koch Institute for Integrative Cancer Research at M.I.T for their assistance with high-throughput sequencing. This work was supported by United States Public Health Service grants RO1-GM34277 and R01-CA133404 from the National Institutes of Health (P.A.S.), partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute, and by Public Health Service research grant (GM-085319) from the National Institute of General Medical Sciences (C.B.B.). X.W. is a Howard Hughes Medical Institute International Student Research fellow.
Footnotes
Full Methods and Supplementary Information is available in the online version of the paper at www.nature.com/nature
Author Contributions. A.E.A, X.W. and P.A.S. conceived and designed the research. A.E.A. performed experiments. X.W. and A.J.K. performed computational analysis. A.E.A., X.W., C.B.B, and P.A.S. analyzed the data and wrote the manuscript.
Author information. 3′-end sequencing data is deposited in the Gene Expression Omnibus under accession number GSE46433. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to P.A.S (sharppa@mit.edu).
References
- 1.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 5.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci U S A. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preker P, et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spies N, Burge CB, Bartel DP. Global analysis of how 3′UTR-isoform choice influences mRNA stability and translational efficiency. (In Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sigova A, et al. Divergent transcription of lncRNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3′end processing reveals a decisive role of human cleavage factor I in the regulation of 3′UTR length. Cell Rep. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 18.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 19.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 20.Vanacova S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaida D, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen PK, Lykke-Andersen S, Jensen TH. Promoter-proximal polyadenylation sites reduce transcription activity. Genes Dev. 2012;26:2169–2179. doi: 10.1101/gad.189126.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010;8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie C, et al. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 2012;8:e1002942. doi: 10.1371/journal.pgen.1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connelly S, Manley JL. A CCAAT box sequence in the adenovirus major late promoter functions as part of an RNA polymerase II termination signal. Cell. 1989;57:561–571. doi: 10.1016/0092-8674(89)90126-8. [DOI] [PubMed] [Google Scholar]
- 28.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Ding Q, Wang P, Wang Z. An upstream promoter element blocks the reverse transcription of the mouse insulin-degrading enzyme gene. Biochem Biophys Res Commun. 2013;430:26–31. doi: 10.1016/j.bbrc.2012.11.052. [DOI] [PubMed] [Google Scholar]
- 30.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EmbBnet.journal. 2011;17:10–12. [Google Scholar]
- 31.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 34.Rahl PB, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.