Abstract
The naked mole-rat displays exceptional longevity, with a maximum lifespan exceeding 30 years1–3. This is the longest reported lifespan for a rodent species and is especially striking considering the small body mass of the naked mole-rat. In comparison, a similarly sized house mouse has a maximum lifespan of 4 years4,5. In addition to their longevity, naked mole-rats show an unusual resistance to cancer. Multi-year observations of large naked mole-rat colonies did not detect a single incidence of cancer2,6. Here we identify a mechanism responsible for the naked mole-rat’s cancer resistance. We found that naked mole-rat fibroblasts secrete extremely high molecular weight hyaluronan (HA), which is over five times larger than human or mouse HA. This high molecular weight HA accumulates abundantly in naked mole rat tissues due to the decreased activity of HA-degrading enzymes and a unique sequence of hyaluronan synthase 2 (HAS2). Furthermore, the naked mole-rat cells are more sensitive to HA signaling, as the naked mole rat cells have a higher affinity to HA than the mouse or human cells. Perturbation of the signaling pathways sufficient for malignant transformation of mouse fibroblasts fails to transform naked mole-rat cells. However, once high molecular weight HA is removed by either knocking down HAS2 or overexpressing the HA-degrading enzyme, Hyal2, naked mole-rat cells become susceptible to malignant transformation and readily form tumors in mice. We speculate that naked mole-rats have evolved a higher concentration of HA in the skin to provide skin elasticity needed for life in underground tunnels. This trait may have then been co-opted to provide cancer resistance and longevity to this species.
Mice and rats are standard animal models for cancer research due in part to their short lifespan and high incidence of cancer. However, these traits imply that mice and rats have fewer anticancer mechanisms, and novel tumor resistance mechanisms are less likely to be discovered using these models. Here we focused our research on a small rodent, the naked mole-rat, which in contrast to mice and rats, is long-lived and cancer resistant.
Our previous studies identified a novel anticancer mechanism in the naked mole-rat, termed early contact inhibition (ECI)7. Contact inhibition is a process of arresting cell growth when cells come in contact with each other or the extracellular matrix. Contact inhibition is a powerful anticancer mechanism that is lost in cancer cells8. Naked mole-rat cells arrest at a much lower density than mouse cells, and the loss of the ECI makes cells more susceptible to malignant transformation7. However, the signals triggering ECI in naked mole-rats remained unknown.
While culturing multiple lines of naked mole-rat fibroblasts we noticed that the culture media became very viscous after a few days. Viscosity measurements confirmed that the media conditioned by the naked mole-rat cells was more viscous than the media conditioned by human, guinea pig, or mouse cells (Figure 1a). We included the guinea pig because it is phylogenetically closer to the naked mole-rat than the mouse. We identified the viscous “substance” secreted by the naked mole-rat fibroblasts as high molecular weight HA (HMW-HA). Treatment with hyaluronidase (HAase) that specifically digests HA reduced the media viscosity to background levels (Figure 1a). Naked mole-rat embryonic fibroblasts, which do not display ECI, did not increase viscosity of the culture media (Figure 1a, and Supplementary Figure 1).
Figure 1. Naked mole-rat cells secrete HA of exceptionally high molecular weight.
(a) Naked mole-rat cells make the culture media viscous. The viscosity of water, media or media conditioned with human skin (HSF), guinea pig skin (GP SF), mouse skin (MSF) or naked mole-rat skin (NMR SF) fibroblasts for 20 days. The NMR SF + HAase bar shows naked mole-rat conditioned media digested with HAase to specifically digest HA. The NMR SF Mut are naked mole-rat skin fibroblasts that spontaneously lost the ECI phenotype7. The NMR EF are naked mole-rat embryonic fibroblasts that do not show ECI. The experiment was repeated three times; error bars show s.d. (b) Purified HA separated on pulse-field gel. Each sample was either run intact or pre-digested with HAase. The experiment was repeated five times, using both skin and lung fibroblasts (Supplementary Figure 9), and a representative gel is shown. (c) Western blot showing the levels of HA synthases in naked mole-rat adult skin fibroblasts, naked mole-rat embryonic fibroblasts, human skin fibroblasts or mouse skin fibroblasts. (d) Conserved catalytic domain of mammalian HAS2 proteins. The top sequence is the naked mole-rat HAS2. Dots indicate amino acids identical to the naked mole-rat sequence. The two amino acid changes unique to the naked mole-rat are indicated by red boxes.
HA is an unbranched disaccharide glucuronic acid/N-acetylglucosamine polymer and is one of the major components of the extracellular matrix9. Biological responses triggered by HA depend on the HA polymer length. HMW-HA represses mitogenic signaling and has anti-inflammatory properties10, while low molecular weight HA promotes proliferation and inflammation11.
Analysis of HA from tissue culture media using pulse field electrophoresis showed that the HA secreted by naked mole-rat cells has a molecular weight of 6–12 MDa, while mouse and guinea pig HA ranges from 0.5–3 MDa (Figure 1b); human HA has a molecular weight of 0.5–2 MDa12. Naked mole-rat embryonic fibroblasts did not secrete HMW-HA (Figure 1b).
Importantly, a mutated clone NMR SF Mut, which spontaneously lost the ECI phenotype and p16INK4a expression7, still produced HMW-HA (Figure 1a, b), indicating that the physical presence of HMW-HA is not sufficient for the ECI phenotype; rather the intact signaling pathway leading from HMW-HA to induction of p16 is required. These experiments establish HMW-HA as the extracellular signal that triggers ECI.
In vertebrate cells HA is produced by HA synthases HAS1, 2, and 3, that differ in tissue distribution and the size of HA produced13. Naked mole-rat skin fibroblasts overexpressed HAS2, the enzyme responsible for the synthesis of HMW-HA in comparison with mouse and human fibroblasts (Figure 1c). Naked mole-rat embryonic fibroblasts, which do not secrete HMW-HA, did not show increased levels of HAS2. The levels of HAS1 and HAS3 were similar between mouse, human, and naked mole-rat cells (Figure 1c). Collectively, these results show that naked mole-rat cells, which display ECI, secrete HA of exceptionally high molecular weight.
Hyaluronan synthases are highly conserved in vertebrates. The HAS2 protein has 98.7% identity and 100% similarity between human and mouse. We cloned and sequenced HAS2 cDNA from the naked mole-rat and compared it to other mammalian HAS2 genes (Figure 1d). Two Asparagines that are 100% conserved among mammals were replaced with Serines in the naked mole-rat HAS2. This change occurs in no other mammalian HAS2 genes deposited in GenBank, including the naked mole-rat’s close relative, the guinea pig. HAS2 contains seven putative transmembrane domains and a cytoplasmic loop14. The conserved regions carrying Asparagine to Serine substitutions correspond to the cytoplasmic loop containing the enzyme’s active site. These unique amino acid changes may be responsible for the high processivity of the naked mole-rat HAS2. Indeed, when the cDNA for the naked mole-rat HAS2 was overexpressed in human HEK293 cells, they began secreting HMW-HA (Figure 2a).
Figure 2. Naked mole-rat tissues contain high levels of HA.
(a) Naked mole-rat HAS2 overexpressed in human HEK293 cells secretes HMW-HA. Small panel on the right shows immunoblot with anti HAS2 antibodies on whole cell extracts from the control and HAS2-transfected cells. (b) Tissues from the naked mole-rat, mouse, and guinea pig stained with Alcian Blue. The control samples treated with HAase do not show blue staining, demonstrating that the staining is specific to HA. Staining was performed on three different animals and representative skin and heart images are shown. Brain and kidney are shown in Supplementary Figure 2. (c) Naked mole-rat fibroblasts have low HAase activity. Naked mole-rat skin fibroblasts (NMR SF), guinea pig skin fibroblasts (GP SF), human skin fibroblasts (HSF), mouse skin fibroblasts (MSF) or HeLa cells were incubated with the media containing HMW-HA for four days. The levels of HA were then analyzed by pulse-field gel. Control samples were incubated in the absence of cells. The experiments were repeated four times (all samples except GP), and three times for GF, error bars show s.d.; asterisk indicates P<0.01 by t-test. (d) Naked mole-rat tissues have low HAase activity. Media containing HMW-HA was incubated with corresponding tissue fragments from naked mole-rats (NMR) or mice for six hours and HA levels analyzed by pulse-field gel. The experiments were repeated three times and error bars show s.d.; asterisk indicates P<0.05 by t-test.
We then examined whether naked mole-rat tissues contain high levels of HA in comparison to mouse and guinea pig. Tissue sections were stained with alcian blue, and control samples were treated with HAase prior to staining to show that the staining is specific to HA. Naked mole-rat skin, heart, brain and kidney were highly enriched for HA (Figure 2b, Supplementary Figure 2). Furthermore, the HA extracted from naked mole-rat tissues had a higher molecular weight than HA from mouse tissues (Supplementary Figure 3). These results indicate that production of HMW-HA by naked mole-rat cells is not an artifact of tissue culture, but a unique in vivo property of this species.
HA levels are regulated by HA-degrading enzymes, HAases15. We measured HAase activity in naked mole-rat, mouse and human cells by quantifying HA degradation after incubation with these cells. HAase activity of the naked mole-rat cells was much lower than that of human, mouse or guinea pig cells (Figure 2c). Similarly, HAase activity was lower in the naked mole-rat tissues than in the mouse tissues (Figure 2d). These results indicate that two mechanisms contribute to accumulation of HMW-HA in the naked mole-rat: more robust synthesis and slower degradation.
We previously demonstrated that ECI contributes to cancer resistance of the naked mole-rat by arresting cell cycle via the induction of p16INK4a 7. To determine the role of HMW-HA in ECI we cultured naked mole-rat fibroblasts in the presence of bacterial HAase. Enzymatic digestion of HMW-HA abrogated the ECI phenotype and caused naked mole-rat cells to grow to complete confluence (Figure 3a, b). Upon subsequent removal of HAase from the culture media, a fraction of cells detached from the plate and died by apoptosis (Supplementary Figure 4), while the remaining cells re-acquired the ECI phenotype (Figure 3c).
Figure 3. HMW-HA is required for ECI.
(a) Naked mole rat cells (NMR SF) grown in the presence of HAase do not display ECI and proliferate to high cell density. (b) Quantification of cell growth, showing the maximum cell number per plate achieved under indicated growth conditions. The last bar shows naked mole-rat cells grown in the presence of CD44-blocking antibody. The experiments were repeated four times (except the last bar, which was repeated three times) and error bars show s.d.; asterisk indicates P<0.001 by t-test. (c) Naked mole-rat cells were grown in the presence of HAase for 12 days, then HAase was removed. (d) Naked mole-rat cells have higher affinity to HA. Cells were incubated with fluorescein-labeled HA, and the average fluorescence was plotted. Experiment was repeated four times; error bars are s.e.m.; asterisk indicates P<0.001 by t-test.
CD44 is a major HA receptor in human and mouse cells9,11,16. To confirm that HA signaling triggers ECI via the CD44 receptor we cultured naked mole-rat cells in the presence of a CD44-blocking antibody. Naked mole-rat cells grown with CD44 antibodies reached a higher cell density (Figure 3b) indicating that the ECI signal from HMW-HA is in part transmitted via the CD44 receptor.
We then used a Flow cytometric assay to measure the affinity of the naked mole-rat cells to fluorescently labeled HA. Naked mole-rat cells displayed a two-fold higher affinity to HA than mouse or human cells (Figure 3d), which can contribute to higher sensitivity of naked mole rat cells to HA signaling.
On the cytoplasmic face, the CD44 receptor interacts with NF2 (merlin), which mediates contact inhibition17. The phosphorylated, growth promoting, form of NF2 appeared in naked mole-rat cells grown in the presence of HAase, while cells cultured without HAase contained mainly the unphosphorylated growth-inhibitory form of NF2 (Supplementary Figure 5). We previously showed that ECI is associated with induction of p16INK4a, while the NMR SF Mut cells that do not display ECI have lost p16INK4a expression7. Accordingly, naked mole-rat cells grown in the presence of HAase displayed reduced levels of p16INK4a (Supplementary Figure 5). Collectively these results establish that ECI is controlled by the HA/CD44/NF2 pathway.
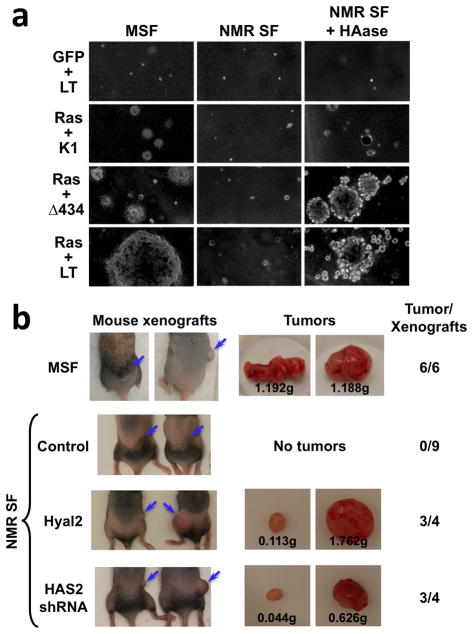
We then tested the role of HMW-HA in the resistance of naked mole-rat cells to malignant transformation in a soft agar assay. SV40 Large T antigen (SV40 LT) is a viral oncoprotein that binds and inactivates p53 and pRb. The mutant derivative LTK1 (K1) inactivates only p53, while LTΔ434–444 (Δ434) inactivates only pRb and its family members18. A combination of H-Ras V12 and SV40 LT is sufficient to transform mouse fibroblasts19, but, as we showed earlier, is not sufficient to confer anchorage independent growth to naked mole-rat cells7. To test the role of HMW-HA in the naked mole-rat’s resistance to transformation, we transfected naked mole-rat fibroblasts with H-Ras V12 combined with SV40 LT or its mutants K1 or Δ434 and cultured them in soft agar in the presence of HAase. Under these conditions, cells transfected with H-Ras V12 and SV40 LT, or H-Ras V12 and Δ434 formed robust colonies (Figure 4a). Similarly, naked mole rat cells cultured in the presence of CD44 blocking antibody formed colonies in soft agar (Supplementary Figure 6). These results demonstrate that if HMW-HA is degraded by HAase or HA signaling is blocked by a CD44 antibody, naked mole-rat cells become susceptible to anchorage-independent growth triggered by H-Ras V12 and SV40 LT. In embryonic naked mole-rat fibroblasts, which do not secrete HMW-HA, H-Ras V12 and SV40 LT or Δ434 were sufficient to trigger anchorage-independent growth (Supplementary Figure 6).
Figure 4. Removal of HMW-HA makes naked mole-rat cells susceptible to malignant transformation.
(a) Soft agar assays of anchorage-independent growth. Mouse (MSF) or naked mole-rat (NMR SF) cells were transfected with vectors encoding SV40 LT (LT) or its mutant derivatives K1 or Δ434 and H-Ras V12 (Ras), and plated in soft agar. Cells were cultured with or without HAase. The image shows colonies after 3 weeks of growth at 200× magnification. The experiment was repeated three times. (b) Mouse xenograft experiment with naked mole-rat cells in which HMW-HA was abolished. NIH-III immunodeficient mice were injected with mouse (MSF) cells expressing SV40LT and H-Ras V12 as a positive control, or naked mole rat (NMR SF) cells expressing SV40LT and H-Ras V12 and either control shRNA, Hyal2 cDNA, or shRNA to HAS2. All xenografts with mouse cells formed large tumors. Xenografts with naked mole-rat cells expressing control shRNA did not form tumors, while naked mole-rat cells overexpressing Hyal2 or HAS2 shRNA formed tumors in mice. The images show xenograft sites (blue arrow) and representative tumors. The number of xenografts resulting in tumor formation per the total number of xenografts with each cell type is shown on the right.
We then generated H-Ras V12 and SV40 LT expressing naked mole-rat cells, in which HMW-HA was abolished by either integrating shRNA targeting HAS2 (Supplementary Figure 7a) or overexpressing an HA-degrading enzyme Hyal2 (Supplementary Figure 7b). These cells no longer increased the viscosity of their culture media (Supplementary Figure 7c) and readily formed colonies in soft agar (Supplementary Figure 8). To confirm that HMW-HA inhibits tumor formation in vivo, we performed xenograft experiments with naked mole-rat cells containing a knockdown of HAS2, or overexpressing Hyal2 (Figure 4b). In the positive control, mouse cells expressing H-Ras V12 and SV40 LT readily formed tumors in mice. Naked mole-rat cells expressing H-Ras V12 and SV40 LT did not form tumors, consistent with an earlier report20. Remarkably, naked mole-rat cells expressing H-Ras V12 and SV40 LT and shRNA to HAS2 or overexpressing Hyal2 formed tumors in mice. This experiment establishes HMW-HA, produced by HAS2, as a key component responsible for the elevated cancer resistance of the naked mole-rat.
The HMW-HA in the naked mole-rat could have evolved as an adaptation to subterranean lifestyle to provide flexible skin needed to squeeze through underground tunnels. Interestingly, we found that cells of a different subterranean rodent, the blind mole-rat, which is phylogenetically closer to mice and rats than to the naked mole-rat also secreted HMW-HA (Supplementary Figure 9). In summary, our results demonstrate that extremely HMW-HA, its binding to the CD44 receptor, and lower HAase activity play a key role in mediating the cancer resistance of the naked mole-rat. Using naked mole-rat HMW-HA in the clinic or targeting Hyal2, or the HA-CD44 signaling pathway opens new avenues for cancer prevention and life extension.
Methods
Animals
All animal experiments were approved and performed in accordance with guidelines set up by the University of Rochester Committee on Animal Resources. Naked mole-rats were from the University of Rochester colonies. C57BL/6 mice and NIH III nude mice (NIH-Lyst bg-JFoxn1nu Btk xid) were purchased from Charles River Labs. Non-albino guinea pigs were obtained from Elm Hill Labs. Cells and tissues were obtained from at least three different animals.
Cell culture
Primary mouse, guinea pig, blind mole-rat and naked mole-rat cells were isolated from lung and underarm skin. Cells were obtained from five naked mole-rats, three mice, three guinea pigs, and three blind mole-rats. The growth characteristics, and HA secretion did not differ between the cell lines from different animals therefore we performed the experiments on three skin cell lines from three animals. All cell lines were used at early passage (<12–15 PDs). Human primary skin fibroblasts HCA2 were a kind gift from Olivia Pereira-Smith. Embryonic naked mole rat fibroblasts were isolated from eight mid-gestation embryos.
Mouse, human, guinea pig and blind mole-rat cells were cultured at 37°C, 5% CO2, 3% O2; naked mole-rat cells were cultured at 32°C, 5% CO2, 3% O2 on treated polystyrene culture dishes (Corning) in EMEM media (ATCC) supplemented with 15% fetal bovine serum (Gibco), nonessential amino acids, sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin (Gibco).
Viscosity Assay
To determine relative kinematic viscosity, 3 ml of distilled H2O, unused complete EMEM media, or media conditioned with naked mole-rat, mouse, or human cells were run through a 0.6 mm capillary Ostwald viscometer (Barnstead International) at 22°C and timed for the passage of the media or distilled H2O through the capillary. For HAase control, naked mole-rat media was treated with hyaluronidase 1 U/ml HAase from Streptomyces hyalurolyticus (Sigma-Aldrich). The relative viscosity of unused and conditioned media was determined by comparing times required to pass through the capillary to that of distilled H2O. Samples were run three times to determine an average relative viscosity.
Cell growth analysis
To measure cell proliferation and the confluent density, cells were seeded on 60 mm gridded plates (Corning). Three 2×2 mm squares were marked on each plate and the number of cells in those squares was counted each day for 20 days. For cell growth in the presence of HAase, 24 hours post plating the media was changed to media containing 3 U/ml HAase (Sigma). Media was then replaced with fresh media containing the enzyme every 48 hours. Images of the squares were taken using SPOT Advanced (Diagnostic Instruments) and analyzed using the colony counting program on ImageQuant TL (GE). The average count of the three squares for each day was multiplied by 458.33 to give the total cell count per 10 cm plate. Cell count data was analyzed using Microsoft Excel.
To calculate maximum cell number cells were harvested from the confluent plates and counted using Z2 Coulter counter (Beckman Coulter). Every sample was counted three times and the averages were used to calculate the maximum cell number from at least three independent experiments.
HA analysis by pulse field gel electrophoresis
HA was purified from conditioned media (typically at day 20) by first treating 2 ml of conditioned media with 500 μg of Proteinase K (Roche) at 50 °C for 45 minutes to remove proteins. Samples were then precipitated by adding 2 ml of 100% ethanol. The pellet was dissolved in 500 μl TE Buffer and incubated overnight at 4 °C. The following day, aliquots were removed and control samples treated with 1 U/ml of HAase from Streptomyces hyalurolyticus(Sigma-Aldrich). Twenty-five μl of each sample was mixed with 5 μl 4M sucrose loading solution and loaded to a 0.4% pulse field SeaKem Gold agarose gel (Cambrex). Ten μl of HA molecular size markers; HiLadder (~500 kDA to ~1,500 kDa) and Mega-HA Ladder (1,500 kDa to 6,000 kDa) (from Hyalose) were run to determine the size of HA from each sample. Samples were run overnight at 4 °C at 75 volts with a 1 to 10 running ratio in TBE buffer using CHEF-DRII system (Bio-Rad). The gel was next stained in a method adapted from21. Briefly, the gel was placed in a 0.005% (w/v) Stains-All (Sigma-Aldrich) in 50% ethanol solution overnight. To de-stain, the gel was placed in distilled H2O for 18-hours in the dark and then placed under ambient light in distilled H2O for 1 hour to complete the final de-staining stages and then photographed under white light. The amount of HA was quantified by counting pixels using Image J software.
HA extraction from tissues
Tissues were excised immediately after sacrificing the animals and weighed. Tissues were chopped and the same amount of tissues were digested at 50°C overnight in the digestion buffer containing 10 mM Tris-Cl, 25 mM EDTA, 100 mM NaCl, 0.5% SDS and 0.1 mg/ml proteinase K (Roche). Then 27 mM MgCl2 was added to chelate EDTA and Pefabloc SC was added to inhibit Proteinase K. 500-unit Benzonase® endonuclease (Sigma-Aldrich) was added to remove nucleic acid. The complete digestion of DNA and RNA was confirmed by running the agarose gel and staining with ethidium bromide (Bio-Rad). Total polysaccharide extraction was performed by phenol/chloroform extraction followed by ethanol precipitation. Finally, corresponding volume (100ul/100mg) of 10 mM Tris-Cl (pH 8.5) was added to dissolve the pellet.
Expression of naked mole rat HAS2 gene in human cells
HEK293 cells were transfected with an expression vector containing HAS2 under the CMV promoter and allowed to express HAS2 for 2 days, after which HA secreted into the media was analyzed by pulse-field gel. Control cells were transfected with a GFP expression vector.
HAase activity assay
Culture media containing HMW-HA secreted by naked mole-rat cells was mixed 1:1 with fresh media and incubated with 2 × 105 naked mole-rat fibroblasts, human diploid fibroblasts, mouse fibroblasts, or HeLa cells for four days. Then the media was harvested, HA was extracted and analyzed on a pulse-field gel as described above. HA levels before and after incubation were compared as a measure of HAase activity. For analysis of tissues, corresponding tissues were chopped into 1 mm cubes and washed twice with PBS. HMW-HA containing media was incubated with equal amounts (by weight) of tissue fragments of each tissue type for six hours and HA levels were analyzed as above.
Antibodies
The following antibodies were used: HAS1 (ab104864 Abcam), HAS2 (sc-66916 Santa Cruz), HAS3 (sc-66917 Santa Cruz), α-tubulin (ab4074 Abcam), CD44 (Monoclonal Mouse IgG2A Clone #2C5, Catalog Number BBA10, R&D Systems), NF2/Merlin (ab30329 Abcam), p16 (ab14244 Abcam), Hyal2 (ab68608bAbcam).
Tissue staining
HA detection in tissues was done as follows. Tissue samples from young animals (3 year old naked mole-rats, 3–5 months old mice, 1 year old guinea pigs) were fixed in 10% buffered neutral formalin, embedded in paraffin and quadruple sections cut at 5 μm were mounted on glass slides. Slides were deparaffinized and hydrated in distilled H2O. They were then placed in Coplin jars containing 40 ml of hyaluronidase digestion solution (40 U hyaluronidase from Streptomyces hyalurolyticus (Sigma) in 40 mL of PBS) for the samples to be digested or in 40 ml of PBS for the non-digested samples. The jars were microwaved for one minute at 60 W and then transferred to a 37 °C oven for 1 hour. The slides were rinsed four times in distilled H2O, followed by three rinses with 3% acetic acid. Next, slides were placed in new Coplin jars containing 1.0% filtered alcian blue solution at pH 2.5 (Alcian Blue 8GX, C.I. 74240 (Leica) in 3% acetic acid and microwaved at 60 W for 3 minutes followed by an additional 5 minute incubation in the hot 1.0% alcian blue solution. Slides were rinsed three times in distilled H2O, dehydrated in graded alcohols and rinsed three times in xylene. Images were taken by light microscopy.
Naked mole-rat cell growth assays with CD44 antibody
Naked mole-rat cells were seeded 50 cells/square onto cell culture treated 6 cm polystyrene gridded tissue culture plates (Corning). Twenty-four hours post plating, the media was changed to contain 5 μg/ml of CD44 specific antibody (Monoclonal Mouse IgG2A Clone #2C5, Catalog Number BBA10, R&D Systems) or no antibody control. Media was changed every 24-hours and images were taken daily using SPOT Advanced imaging software (Diagnostic Instruments). Images from three different squares from two independent plates were counted from both the CD44 antibody treated or control groups.
HA affinity assay
Naked mole-rat and mouse cells were harvested at subconfluent exponential phase. One hundred thousand (105) cells of each type were incubated for 45 min on ice in 210 μl PBS containing 1.5% Fetal Calf Serum and 35 μg/ml fluorescein-labeled HA (Fluorescein-Labeled HA from Bovine Trachea, sc-221733, Santa Cruz Biotech, Santa Cruz, CA). Five thousand cells from each replicate were analyzed by FACS. The experiment was repeated four times.
Transfections
Naked mole-rat skin fibroblasts were seeded at 2×105 cells/100 mm plate seven days prior to transfection. Mouse skin fibroblasts were seeded at 5×105 cells/100 mm plate two days prior to transfection. For transfection, cells were harvested, counted and 106 cells were transfected with 5 μg of plasmid DNA using Amaxa Nucleofector II on program U-020 and solution NHDF (Amaxa). After transfection, cells were seeded at 2×105 live cells per 10 cm plate for apoptosis analysis and 7×104 live cells per 6 cm grid plates (Corning) for cell growth analysis in the same media as stated above. Media was replaced 24 hours post transfection to remove dead cells due to electroporation.
Anchorage-independent soft agar growth assay
One million mouse, naked mole-rat wild type and naked mole-rat mutant exponentially growing skin fibroblast cells were transfected by Amaxa with the following plasmid DNA mixtures: 5 μg pEGFP-N1 (Clontech) and 5 μg pSG5 Large T (Addgene 9053), 5 μg pRas-V12 (Clontech) and 5μg pSG5 Large T, 5 μg pRas-V12 and 5μg pSG5 Large T K1 (Addgene 9055), or 5μg pRas-V12 and 5μg pSG5 Large T Δ434–444 (Addgene 9054). After transfection, cells were seeded and allowed to recover for 24 hours on 10 cm treated polystyrene plates (Corning) in 1X Minimum Essential Medium, Eagle with Earle’s Balances salt Solution supplemented with 15% fetal calf serum and antibiotics (Gibco). The following day, a 2 ml final solution of 0.5% Difco Agar Noble (BD Bioscience) and 1X media mixture was poured into 6 cm treated polystyrene plates (Corning) and allowed to solidify in incubators at 37°C. After harvesting and counting cells transfected 24 hours previous, 50 to 50,000 cells were serially diluted and resuspended in 1 ml of 2X media. This cell suspension was then quickly mixed with 1 ml 0.7% liquid Difco Agar Noble, making a final 0.35% agar/1X media solution, and seeded on top of the solidified 0.5%/1X media. Plates were incubated at 32 °C, 5% CO2, ad 3% O2 for twenty-four hours before the addition of 1ml of EMEM media with or without HAase from Streptomyces hyalurolyticus (Sigma) at 3 U/ml. To test the effect of CD44 antibody, 5 μg/ml of CD44 antibody (BBA10, R&D Systems) was added and changed daily. Cells were grown for six weeks, with the removal of old liquid EMEM media and the addition of 1 ml of new EMEM with or without 3 U/ml HAase every 48 hours to ensure efficient digestion of HMW-HA. Plates were monitored every 48 hours and photographed at week 3 (mouse cells) and week 6 (naked mole-rat cells) after plating at 200x on a Nikon TS100 phase contract microscope using SPOT software (Diagnostic Instruments).
shRNA-mediated HAS knockdown and Hyal2 overexpression
shRNAs were designed by Integrated DNA Technology (IDT) with shRNA Design Tool: 5′-GATCCGCCAGCTGCCTCAGAGGAATTCAAGAGATTCCTCTGAGGCAGCTGGCTTTTT TGGAAA-3′; 5′-AGCTTTTCCAAAAAAGCCAGCTGCCTCAGAGGAATCTCTTGAATTCCTCTGAGGCAG CTGGCG-3′. The corresponding 63bp DNA oligonucleotides harboring the 19-mer hairpin sequence, loop sequence, polythymidine tract (U6 terminator), BamHI and HindIII restriction site overhangs were designed according to the user manual of pSilencer™ 2.1-U6 neo kit (Life Technologies) and chemically synthesized by IDT. The complementary oligonucleotides were annealed and ligated to the pre-cut pSilencer2.1-U6 neo vector using the rapid DNA ligation kit (Roche). Following transformation into Top10 competent cells (Life Technologies), successful ligation was confirmed using restriction digestion and DNA sequencing with M13F primer (5′-GTAAAACGACGGCCAGT-3′).
Human Hyal2 cDNA was amplified from pCMV6-HYAL2 (sc117754 OriGene) using the primers 5′-CCGGAATTCGCCACCATGCGGGCAGGCCCAGGCCCCACCG-3′ and 5′-ATAAGAATGCGGCCGCCTACAAGGTCCAGGTAAAGGCCAGGGC-3′ and cloned into pEGFP-N1-neo plasmid to replace EGFP fragment using EcoR1 and NotI restriction enzymes.
Transfection grade plasmids were prepared with EndoFree plasmid maxi kit (Qiagen) and linearized with ScaI. One μg linearized plasmid was transfected into ~1×106 cells by Nucleofector (Amaxa) with U20 program, followed by G418 selection at 1 mg/ml for 2 weeks. Clones that stably expressed the shRNA were picked and expanded to characterize the knockdown efficiency. Clones with highest levels of HAS knockdown efficiency or the best Hyal2 expression were used for the in vivo xenograft assay.
Quantitative RT-PCR
Total RNA was extracted from cells at 80% confluence (2 days after splitting) using RNeasy Mini Kit (Qiagen). cDNA was generated using SuperScript®III reverse transcriptase (Life Technologies) with Oligo(dT)18 primer. First-strand cDNA was amplified using FastStart Universal SYBR Green Master (Roche; 04913850001) with corresponding primers in which QuantumRNA™ beta-actin internal standards (Life Technologies) were used as reference. Quantitative PCR was conducted with Applied Biosystems 7300 real-time PCR systems at 95°C for 10 min, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The standard curves for the quantitative PCR were set using 2, 1, 1/2, 1/4, 1/8, 1/16, 1/32, 1/64, 1/128 and 1/256 μl cDNA generated with RNA from NSF-LT-RAS cells. qPCR Primers: HAS2-Forward: 5′-GAAAAGGGTCCTGGTGAGACGGATGAG-3′; HAS2-Reverse: 5′-TTCACCATCTCCACAGATGAGGCAGG-3′
Tumor xenograft assay
NIH-III nude mice (Crl:NIH-Lystbg-J Foxn1nuBtkxid) were purchased from Charles river Laboratories Inc. (Wilmington, MA, USA). Seven-week-old female mice were used to establish xenografts. For each injection, 4 × 106 cells were harvested and resuspended in 100 μl of ice-cold 20% matrigel (BD Bioscience, Franklin Lakes, NJ) in PBS (Gibco). This 100 μl solution was injected subcutaneously close to the base of the external ear or into the flank just in front of the hind legs with 22 gauge needle. Transplantations of MSF-LT-Ras cells were allowed to grow for 2–3 weeks, while xenografts with NMR cells were allowed to grow for 65 days before sacrifice. Tumors were excised and size and weight were recorded. The mice were dissected and tumor metastasis was examined for each organ.
Supplementary Material
Acknowledgments
This work was supported by the grants from the US National Institutes of Health and Ellison Medical Foundation to V.G. We thank Michael Van Meter for critically reading the manuscript.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: X.T. performed HA analysis, and HAase assays, soft agar assays, and generated cells for xenograft experiments; J.A. performed immunoblots and cloning and analysis of HAS2; C.H. identified HA, performed tissue staining, and soft agar assays; A.V. performed xenografts, M.-M.R. performed HA affinity assays; J.A. purified HA; Z.M. performed experiments with HAS2 expression; E.N. provided essential materials; X.T., J.A., C.H., A.S., and V.G. designed the study and analyzed data; A.S. and V.G. wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints
Authors declare no competing financial interests.
References
- 1.Buffenstein R, Jarvis JU. The naked mole rat--a new record for the oldest living rodent. Sci Aging Knowledge Environ. 2002:pe7. doi: 10.1126/sageke.2002.21.pe7. [DOI] [PubMed] [Google Scholar]
- 2.Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol [B] 2008;178:439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- 3.Kim EB, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011 doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turturro A, et al. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- 5.de Magalhaes JP, Costa J, Toussaint O. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 2005;33:D537–543. doi: 10.1093/nar/gki017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaney MA, Nagy L, Kinsel MJ, Treuting PM. Spontaneous Histologic Lesions of the Adult Naked Mole Rat (Heterocephalus glaber): A Retrospective Survey of Lesions in a Zoo Population. Vet Pathol. 2013 doi: 10.1177/0300985812471543. [DOI] [PubMed] [Google Scholar]
- 7.Seluanov A, et al. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc Natl Acad Sci USA. 2009;106:19207–19208. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abercrombie M. Contact inhibition and malignancy. Nature. 1979;281:259–262. doi: 10.1038/281259a0. [DOI] [PubMed] [Google Scholar]
- 9.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat Rev Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 10.Kothapalli D, et al. Hyaluronan and CD44 antagonize mitogen-dependent cyclin D1 expression in mesenchymal cells. J Cell Biol. 2007;176:535–544. doi: 10.1083/jcb.200611058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pure E, Assoian RK. Rheostatic signaling by CD44 and hyaluronan. Cell Signal. 2009;21:651–655. doi: 10.1016/j.cellsig.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes MW, Bayliss MT, Muir H. Hyaluronic acid in human articular cartilage. Age-related changes in content and size. Biochem J. 1988;250:435–441. doi: 10.1042/bj2500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang D, Liang J, Noble PW. Hyaluronan in tissue injury and repair. Annu Rev Cell Dev Biol. 2007;23:435–461. doi: 10.1146/annurev.cellbio.23.090506.123337. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe K, Yamaguchi Y. Molecular identification of a putative human hyaluronan synthase. J Biol Chem. 1996;271:22945–22948. doi: 10.1074/jbc.271.38.22945. [DOI] [PubMed] [Google Scholar]
- 15.Stern R, Jedrzejas MJ. Hyaluronidases: their genomics, structures, and mechanisms of action. Chemical reviews. 2006;106:818–839. doi: 10.1021/cr050247k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 17.Morrison H, et al. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn WC, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Liang S, Mele J, Wu Y, Buffenstein R, Hornsby PJ. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 2010;9:626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HG, Cowman MK. An agarose gel electrophoretic method for analysis of hyaluronan molecular weight distribution. Anal Biochem. 1994;219:278–287. doi: 10.1006/abio.1994.1267. S0003-2697(84)71267-X [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.