Abstract
Evolutionary advances are often fueled by unanticipated innovation. Directed evolution of a computationally designed enzyme suggests that dramatic molecular changes can also drive the optimization of primitive protein active sites. The specific activity of an artificial retro-aldolase was boosted >4,400 fold by random mutagenesis and screening, affording catalytic efficiencies approaching those of natural enzymes. However, structural and mechanistic studies reveal that the engineered catalytic apparatus, consisting of a reactive lysine and an ordered water molecule, was unexpectedly abandoned in favor of a new lysine residue in a substrate binding pocket created during the optimization process. Structures of the initial in silico design, a mechanistically promiscuous intermediate, and one of the most evolved variants highlight the importance of loop mobility and supporting functional groups in the emergence of the new catalytic center. Such internal competition between alternative reactive sites may have characterized the early evolution of many natural enzymes.
Computational design has emerged in recent years as a promising tool for creating protein catalysts with tailored activities and specificities. Using this approach, mechanistically diverse chemical reactions, including transformations without biological counterparts, have been catalyzed1–6. Although rate accelerations are still low compared to those of natural enzymes, efficiency can be enhanced substantially by directed evolution7,8.
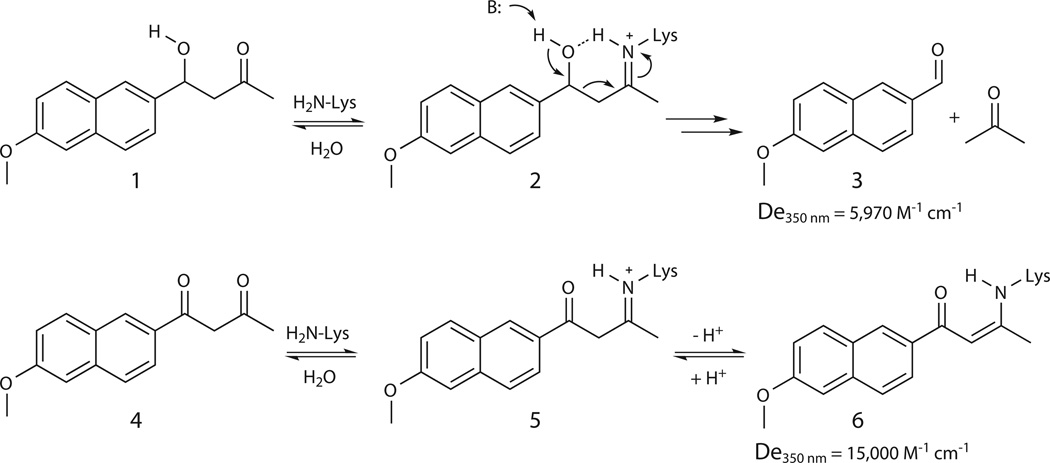
Retro-aldolases that promote the cleavage of 4-hydroxy-4-(6-methoxy-2-naphthyl)-2-butanone (1) to 6-methoxy-2-naphthaldehyde (3) and acetone (Fig. 1a) are, mechanistically, the most complex enzymes designed to date2,7. Like natural class I aldolases9, they exploit a multistep pathway involving amine catalysis and enzyme-bound Schiff base intermediates (e.g., 2) to convert substrate to product. Successful designs employed a catalytic motif comprising a reactive lysine residue, a hydrophobic pocket for substrate binding, and an explicit water molecule to promote proton transfers. While mechanistic and structural characterization of two representative designs has confirmed the importance of the lysine and the apolar substrate binding pocket for catalysis, other design features, such as the catalytic water, appear to be less effectively realized10,11. Moreover, ready optimization of the low starting activities of these catalysts (kcat/Km = 0.02 to 0.30 M−1 s−1) by random mutation, both at the active site and at more distant locations7, raises fundamental questions about the suitability of the original design model and/or the accuracy of its implementation.
Figure 1. Amine catalysis of the retro-aldol reaction.
(a) Amines accelerate the cleavage of 4-hydroxy-4-(6-methoxy-2-naphthyl)-2-butanone (usually called methodol, 1) by forming Schiff base intermediates (e.g., 2). 6-Methoxy-2-naphthaldehyde (3) and acetone are the final products. (b) The 1,3-diketone 4 is a mechanism-based inhibitor that traps the catalytic lysine upon deprotonation of the initially formed Schiff base 5 to give the stable vinylogous amide 6.
To gain deeper insight into the structure, function and optimization of such systems, we evolved a weakly active, computationally designed retro-aldolase to high efficiency and characterized key intermediates along the evolutionary trajectory structurally and biochemically. Complexes of the variants with a mechanism-based inhibitor (Fig. 1b) allow direct inferences regarding catalytic mechanism to be made in each case. In addition to providing valuable feedback for the design process, our results illuminate how mechanistic innovation can lead to large increases in catalytic activity.
RESULTS
In silico model versus experiment
As a starting point for this study we used the previously designed retro-aldolase RA95.07. This enzyme was created in a (βα)8 barrel scaffold by introducing eleven active site mutations predicted by the Rosetta software suite12. It contains a reactive lysine at position 210 to facilitate carbon-carbon bond cleavage via Schiff base formation, a large hydrophobic pocket between the L1 (β1α1) and L6 (β6α6) loops to accommodate the substrate naphthyl ring, and a glutamate at position 53 to orient the catalytic water molecule.
RA95.0 promotes the cleavage of 1 with a 15,000-fold rate acceleration over background (Table 1). As expected for a mechanism involving a lysine with a perturbed pKa, it exhibits a sigmoidal pH-rate profile with an inflection point at 8.1 (Supplementary Results, Supplementary Fig. 1) and a significant advantage over free lysine in solution (Table 1). The enzyme, which was designed to cleave (S)-methodol, is also enantioselective, but the preference for the (S) over the (R)-configured substrate is only 2.3-fold (Supplementary Table 1, Supplementary Fig. 1).
Table 1.
Cleavage of (±)-methodol by RA95 variantsa
| Variant | 103 × kcat(s−1) | KM(µM) | kcat/KM(M−1 s−1) | pKa | ||
|---|---|---|---|---|---|---|
| RA95.0 | 0.10 ± 0.02 | 530 ± 85 | 0.19 | 1.5 × 104 | 7.8 × 104 | 8.1 |
| RA95.5 | 3.0 ± 0.5 | 210 ± 12 | 14 | 4.6 × 105 | 6.1 × 106 | 7.6 |
| RA95.5-5 | 73 ± 4 | 230 ± 27 | 320 | 1.1 × 107 | 1.4 × 108 | 7.7 |
| RA95.5-8 | 170 ± 20 | 200 ± 50 | 850 | 2.6 × 107 | 3.7 × 108 | 7.6 |
| Ab 38C2 | 12 ± 1 | 25 ± 4 | 480 | 1.8 × 106 | 2.0 × 108 | 6.0b |
All measurements were carried out at 29 °C in 25 mM HEPES, 100 mM NaCl (pH 7.5) containing 2.7% acetonitrile. The estimated errors reflect the standard deviations of two to three independent measurements. For comparison: kuncat = 6.5 × 10−9 s−1; klysine = 2.3 × 10−6 M−1 s−1 at 25 °C10. Sequences of the variants are listed in Supplementary Fig. 5.
Ref. 21.
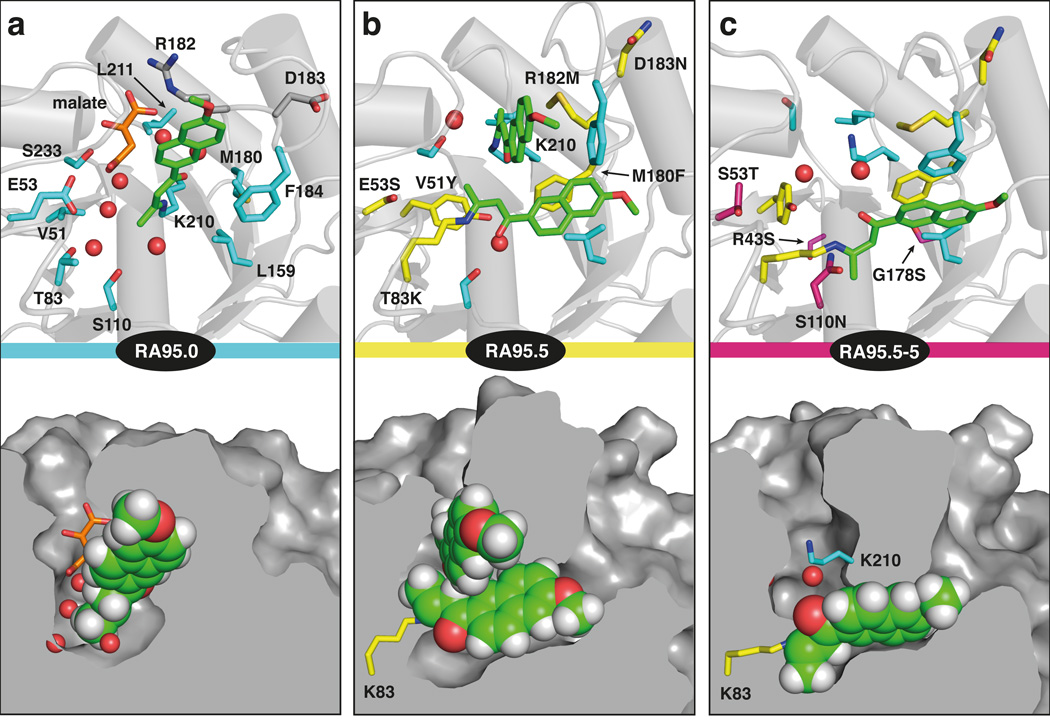
To investigate the structural basis of binding and catalysis by RA95.0, we determined a high-resolution X-ray structure of the enzyme in complex with the mechanism-based inhibitor 4 (Fig. 1b). The structure at 1.10 Å confirmed the overall features of the design model (Fig. 2a) with the Cα atom root mean square deviation (rmsd) being 0.69 Å for the entire protein and 1.0 Å for active site residues. The L1, L6, and L7 loops, which bracket the active site and contain many of the Rosetta mutations, showed the largest deviations from the model, with rmsd values between 2 and 4 Å. Despite these structural discrepancies, Lys210 adopts a conformation as in the design, which effectively lowers its pKa by placing the ε-amino group in a relatively hydrophobic environment.
Figure 2. Computationally designed retro-aldolase RA95.0.
(a) Overlay of the in silico design (blue) and the X-ray crystal structure of the enzyme (grey) complexed with 4 (green). The Lys210/Glu53 catalytic motif and the composite transition state (including water) used for design are shown (yellow carbons). The corresponding residues in the X-ray structure are gray. The L1, L6 and L7 loops bracketing the active site show significant deviations from the design model. A malate molecule from the crystallization buffer that binds at the active site proximal to the inhibitor but has no effect on catalytic activity (Supplementary Table 1) was omitted for clarity. (b) Electron density for the inhibitor covalently bound to Lys210. The Fo-Fc omit map is contoured at 3.0σ.
Lys210 reacts covalently with the 1,3-diketone inhibitor 4, as intended, to form a vinylogous amide (6 in Fig. 1b). This adduct, which was well ordered in the electron density map (Fig. 2b), is linked via its C2 carbon to the ε-amine of the catalytic lysine. The naphthyl ring binds snugly within the designed hydrophobic pocket but is rotated by 114° around its long axis relative to the model (Fig. 2a). This binding mode may reflect structural differences between the inhibitor, which has three consecutive sp2 centers, and the composite transition state used for modeling, which has three sp3 centers.
The altered orientation causes the carbonyl group of the inhibitor to point away from Glu53 and the water molecule designed to interact with the β-hydroxyl group of the substrate (Fig. 3a). Since the enzyme retained full activity when Glu53 was replaced by alanine (Supplementary Table 1), this residue either does not contribute to catalysis, contrary to the original design, or its function can be assumed by water in its absence. Several other ordered water molecules were observed at the active site, including a hydrogen bond donor to the enaminone β-carbonyl group, positioned by Ser181 and several backbone amides (Fig. 3a & Supplementary Fig. 2). The ability of active site waters to mediate mechanistically relevant proton transfers, in combination with the well-defined substrate binding site, may explain why RA95.0 is more efficient than simpler amine-containing aldolase mimics13–16
Figure 3. Structural optimization of the retro-aldolase.
Active sites of the de novo retro-aldolase RA95.0 (a), the promiscuous intermediate RA95.5 (b), and the evolved variant RA95.5-5 (c). The upper panels are close-up views of the respective ligand binding pockets. Residues introduced by design are shown in cyan, substitutions identified by cassette mutagenesis in yellow, and mutations found during full-scale directed evolution in magenta; all other residues are colored gray. The mechanism-based inhibitor and a malate molecule from the crystallization buffer are shown in green and orange, respectively. The lower panels show cross-sections of the binding pockets (not applied to bound inhibitors). They illustrate the dramatic remodeling of the active site and the increase in ligand shape complementarity of the binding pocket that arises in the course of evolution.
Active site optimization
Although the computationally designed retro-aldolase utilizes programmed hydrophobic interactions to activate the catalytic lysine and position the substrate for reaction, it is many orders of magnitude less efficient than natural aldolases. To augment its activity, it was previously optimized by iterative cassette mutagenesis of active site residues7. A robust medium-throughput assay, based on detection of the fluorescent naphthaldehyde product 3, facilitated identification of more active variants.
The best variant from that study, RA95.5 (Table 1), contains five active site mutations (V51Y, E53S, T83K, M180F, R182M), and one second-shell substitution (D183N)7. The appearance of a second lysine at position 83 is reminiscent of many natural class I aldolases, which exploit an additional cationic group at the active site to activate the Schiff base-forming lysine by decreasing its pKa through electrostatic interactions9. Together, these changes shift the apparent pKa of the catalytic lysine by half a unit to 7.6 (Supplementary Fig. 1) and increase kcat for cleavage of (±)-methodol 30-fold and kcat/Km 78-fold (Table 1). Remarkably, they also invert the enantioselectivity of the catalyst to favor cleavage of (R)-methodol by 3:1 (Supplementary Table 1).
To determine the origin of these effects, RA95.5 was co-crystallized with the 1,3-diketone 4 and its structure determined at 1.4 Å resolution (Fig. 3b). Unexpectedly, this variant is modified twice by the inhibitor, once at Lys210, as in the precursor, and once at the newly introduced Lys83. The other mutations substantially remodel the active site. For example, M180F, R182M, and D183N induce a major conformational change in the L6 loop that creates a deep hydrophobic binding pocket not present in the original design (Fig. 3b), making simultaneous labeling of both active site lysines possible. Active site plasticity may also have facilitated generation of the new reaction center: the L1, L6 and L7 loops of RA95.5, which are not involved in crystal contacts, exhibited significantly higher B-factors than the rest of the structure (Supplementary Fig. 3), consistent with enhanced mobility.
The Lys210 side chain is displaced from its position in RA95.0, as is the adjacent L7 loop, due to a steric clash with the bulky aromatic residues introduced at positions 51 and 180. Although the naphthyl ring of the appended inhibitor still docks in the same hydrophobic pocket its orientation is almost perpendicular to the original binding mode. In the case of Lys83, two distinct conformations are detected for the side chain and the ligand, which are populated to similar extents (Supplementary Fig. 4). Despite this heterogeneity, mutagenesis studies showed that this residue is more effective than Lys210 as a catalytic group under normal turnover conditions. Replacement of Lys83 with methionine reduced kcat and kcat/Km for the cleavage of (±)-methodol 3- and 14-fold, respectively, whereas replacement of Lys210 with methionine left kcat unchanged and increased kcat/Km 2-fold (Supplementary Table 2). Although both sites effectively compete for substrate at high concentrations, the more buried Lys83 has a ~30-fold advantage over Lys210 under sub-saturating conditions, perhaps because it is better shielded from bulk solvent.
As previously reported7, introducing just the T83K mutation into the RA95.0 scaffold increases catalytic activity by less than 5-fold. An engineered double mutant T83K/K210M of RA95.0, in which the lysine originally introduced by design is replaced by methionine, was also roughly 4-fold more active than RA95.0 (Supplementary Table 2). Comparison of the double mutant with K210M RA95.5, which also lacks the original lysine, suggests that the other favorable mutations identified during the active site optimization procedure preferentially enhanced the small intrinsic advantage exhibited by Lys83 over Lys210, presumably by helping to establish the alternative substrate binding pocket.
The emergence of the second reaction center can plausibly explain both the shift in apparent pKa value of the catalytic residue and the inversion in stereochemistry observed for RA95.5. Interestingly, mutagenesis studies suggest that progenitors of natural aldolases may have exhibited similar mechanistic ambiguity17,18. When the Schiff base-forming lysine in deoxyribose 5-phosphate aldolase (DERA) is mutated to arginine, for instance, its role can apparently be assumed by a supporting active site lysine on the neighboring β-strand with only minor losses in activity18.
Mechanistic promiscuity as a springboard to high activity
The promiscuous RA95.5 variant was subjected to multiple rounds of fully random gene diversification using error-prone PCR19 and DNA shuffling20. The most active clone from the fifth round of directed evolution, RA95.5-5 (Table 1), contained six additional mutations, three of which are in or near the active site (E53T, S110N, G178S) and three (R23H, R43S, T95M) on the protein surface. It was >20-fold more efficient than RA95.5 and >1,700-fold more active than the original in silico design (Table 1). Moreover, the (R) selectivity of the catalyst increased to >5:1 (Supplementary Table 1).
The structure of RA95.5-5 in complex with inhibitor 4, solved at 1.3 Å resolution, showed further modulation of the L1, L6 and L7 loops. In addition, this variant is modified exclusively at Lys83 (Fig. 3c), implying that the new catalytic pocket was more evolvable than the original computationally designed site. Site-directed mutagenesis confirmed that Lys83 is the key catalytic residue (Supplementary Table 2). When it was substituted by methionine, activity decreased >200-fold. In contrast, the K210M mutation had no effect on catalytic efficiency. Introduction of the S110N mutation, which provided a 5-fold boost in activity in the first round of directed evolution of RA95.5, was probably decisive in favoring this particular evolutionary trajectory. The asparagine side chain is only 3.8 Å away from the ε-amino group of Lys83 and may assist formation and breakdown of the initial carbinolamine intermediate through H-bonding interactions.
A reduction in conformational heterogeneity may have further enhanced the activity of the new catalytic site. In fact, only a single rotamer was detected for the Lys83 side chain (Supplementary Fig. 4), due to reorientation of Tyr51 in response to the E53T mutation (Fig. 3b and 3c). Although two roughly perpendicular orientations of the naphthyl moiety were still observed (Supplementary Fig. 4), one is populated to a greater extent than the other (70:30). The preferred binding mode is bolstered by more extensive hydrophobic interactions, including π-stacking of the naphthyl ring with the side chain of Phe184 (Fig. 3c).
Approaching the activity of natural aldolases
Additional mutagenesis and screening showed that the RA95.5-5 scaffold still possesses considerable evolutionary potential. The best clone from the eighth round of directed evolution, RA95.5-8, contains three additional active site mutations (K135N, S178V, G212D) and two more distant substitutions (S43R, F72Y). This variant is a remarkably effective retro-aldolase with a >3 × 108-fold rate acceleration relative to free lysine in solution (Table 1), conversion of >103 substrate molecules per active site, and improved (14:1) enantioselectivity towards (R)-methodol (Supplementary Table 1).
To place these results into context, RA95.5-8 has a 14-fold higher turnover number than the commercially available aldolase antibody 38C2 (Table 1), one of the most efficient artificial enzymes in the literature21. Moreover, the efficiency of the evolved retro-aldolase approaches that of human fructose-1,6-diphosphate (FDP) aldolase, a central enzyme in glucose metabolism. The kcat and kcat/Km values for cleavage of (R)-methodol (Supplementary Table 1) and FDP22 by the two enzymes differ by less than factors of 2 (0.36 vs. 0.61 s−1) and 170 (1.6 × 103 vs. 2.7 × 105 M−1 s−1), respectively. Though more efficient natural catalysts are known, this level of activity is notable for an artificial enzyme.
DISCUSSION
Although computation does not yet deliver true enzyme-like activities, in silico design combined with laboratory evolution has afforded a remarkably efficient catalyst for a complex, multistep retro-aldol reaction. Notably, improvements in RA95 were manifest primarily in increased kcat values, implying substantial fine-tuning of active site chemistry and not simply increased substrate affinity.
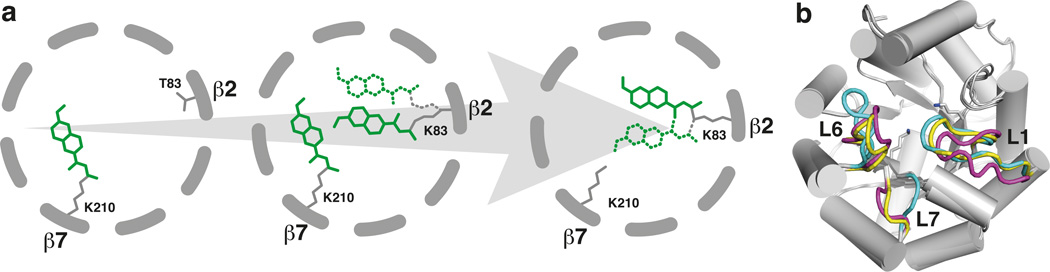
Characterizing the increasingly active retro-aldolases, as opposed to working backward from an already optimized natural enzyme, has provided direct insight into the molecular events responsible for enhanced efficiency. For RA95, optimization entailed unexpectedly extensive structural and mechanistic remodeling (Fig. 4a). In this instance, the shift from low to high activity arose via serendipitous emergence of a mechanistically promiscuous intermediate containing a new catalytic lysine and an alternative substrate binding pocket.
Figure 4. Evolutionary transformation of the computationally designed retro-aldolase.
(a) Schematic representation of the migration of the catalytic center within the core 8-stranded β-barrel. The mechanism-based inhibitor 4 is shown in green; minor conformations of the bound inhibitor at Lys83 are denoted as dotted lines. (b) Superimposition of the RA95.0 (cyan loops), RA95.5 (yellow) and RA95.5-5 (magenta) crystal structures highlighting the shifts in the L1, L6 and L7 loops that occurred during evolutionary optimization.
The dramatic changes observed during RA95 evolution naturally prompt the question whether generation of a highly active retro-aldolase required a computational design step. Although productive evolutionary trajectories might have been initiated from random libraries, recent experiments with the same scaffold demonstrate that chemical instruction conferred by computation greatly increases the probability of identifying catalysts23. While the programmed mechanisms of other computationally designed enzymes have been generally reinforced and refined by directed evolution7,8, the molecular acrobatics observed with RA95 attest to the functional leaps that unanticipated, innovative mutations — here, replacement of Thr83 by lysine — can initiate.
The remarkable transformation of RA95 was no doubt aided by the simplicity of the original design model. Although RA95.0 functions largely as intended, utilizing Lys210 for amine catalysis and an apolar pocket for substrate binding, it is much less sophisticated than natural aldolases24. The absence of supporting functional groups in the vicinity of the catalytic lysine, for instance, account for its comparatively low activity. Introduction of Tyr51 and Asn110 proximal to Lys83 probably facilitated the catalytic residue switch as the polar side chains of these residues can mediate critical proton transfers during catalysis, either directly or via bound water molecules.
Extensive remodeling of active site loops observed in the course of RA95 evolution (Fig. 4b) is reminiscent of the structural changes that occur in antibody proteins during affinity maturation as a result of somatic mutation25,26. As the new and ultimately superior substrate binding pocket was not apparent in the original design, conformational plasticity was undoubtedly crucial for the emergence and improvement of the new catalytic center. Indeed, the RosettaMatch searches in the original protein design calculations did not identify the more potent active site with a lysine nucleophile at position 83, indicating that the original scaffold backbone was not compatible with the theozymes (ideal active site geometries) used in the calculations. Changes in the loops may also have facilitated dynamic fluctuations allowing the active site to adapt to structurally distinct species on the multistep reaction path27.
Additional research will be needed to determine the factors that make some designed sites more evolvable than others. Nevertheless, increasing starting design accuracy and activity would be advantageous for generating better leads for laboratory evolution and for improving control over specific evolutionary trajectories. Our results suggest two approaches for achieving these goals. The initial design calculations were carried out assuming a rigid scaffold, which is clearly incorrect. Proper sampling of plausible loop conformations and better control of loop structure through specific designed sequences should significantly improve placement of the idealized active site within the target scaffold; considering backbone flexibility in the design cycle would also enable wider-ranging exploration of potential sites for introducing catalytic residues. A second important lesson from our study is that more sophisticated arrays of functional groups than those in our starting retroaldolase will be crucial to accessing higher activities. To that end, more complex theozymes can be employed as design templates, but accurate modeling of long-range electrostatics and inclusion of explicit water molecules in the calculations will also be necessary to fine-tune the reactivity of these groups11. Although advances in these and other directions can be anticipated, the challenges of handling multiple transition states along a reaction coordinate make it likely that laboratory evolution will remain an essential tool for tailoring the properties of computationally designed aldolases for the foreseeable future.
Conformational and functional promiscuity are believed to foster the evolution of enzyme function28. Structural snapshots of the evolving aldolase active site studied here support this view and provide a mechanistic description of the role catalytically ambiguous intermediates might play in such a process. Loop mobility and active site remodeling may have analogously shaped the evolution of many natural enzymes29.
METHODS
Starting point for in vitro evolution
Retro-aldolase RA95.0 was designed into the indole-3-glycerol phosphate synthase scaffold (PDB ID: 1LBL) using the RosettaMatch30 and RosettaDesign31 programs, as previously described2,7. In addition to 11 active site mutations (E51V, K53E, L83T, K110S, E159L, N180M, L184F, L187G, E210K, S211L, G233S), it contains five surface substitutions (K10E, F22V, S70A, F246L, and L248E) that enhance protein production. The active site was subsequently optimized by iterative cassette mutagenesis of active site residues; six mutations that individually increased activity (V51Y, E53S, T83K, M180F, R182M, and D183N) were combined to give variant RA95.57. This catalyst served as the starting point for multiple rounds of whole-gene random mutagenesis and screening.
Library construction
The gene encoding RA95.5 was subcloned into the commercial pET-29b(+) vector (Novagen). Primers for library cloning (forward primer: 5’-TGA TAA CAT ATG CCG CGT TAT CTG AAA GG and reverse primer: 5’-TTA TCA CTC GAG TGA TCC CTC GAT TAA TTC TTT) were purchased from Microsynth AG (Balgach, Switzerland). A DNA library was constructed by error-prone PCR (epPCR) using the GenemorphTM PCR mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) as recommended by the manufacturer32. A low mutation rate was chosen to achieve one to two amino acid substitutions on average per protein. The linear epPCR library fragments and the pET-29b(+) vector containing a stuffer fragment were digested using NdeI and XhoI endonucleases (New England Biolabs, Ipswich, MA, USA), gel purified, and subsequently ligated using T4 DNA ligase (New England Biolabs). The resulting plasmid library was purified by phenol/chloroform extraction and used to transform Escherichia coli BL21 Gold(DE3) cells (Invitrogen, Carlsbad, CA, USA). The transformation mixture was plated on Luria broth (LB) agar containing 30 µg mL−1 kanamycin. Library quality and mutation rate were assessed by sequence analysis of plasmids from single colonies before screening (see below).
The most active clone from the first round of mutagenesis and screening, RA95.5-S110N, served as the template for a second round of diversification by epPCR. In all subsequent rounds (three to eight), plasmids from the most active clones emerging from the previous round, typically 0.4% to 1% of the screened population, were isolated, pooled, and further diversified by a combination of epPCR and DNA shuffling20. Specifically, following epPCR, the gene pool was digested using 60 µg mL−1 bovine pancreatic DNase I (Roche, Basel, Switzerland) to give 50–100 bp fragments, which were gel-purified and re-assembled in a standard PCR reaction using the cloning primers. The full-length DNA library genes were digested and ligated into the pET-29b(+) vector, as described above. For the eighth mutagenesis round, two clones from a focused library that targeted position 178 (S178T and S178V) in RA95.5 were mixed with the seven best clones from the seventh round, and the resulting pool was diversified by epPCR and shuffled as already described.
Focused libraries
Based on their proximity to the new catalytic site, residues 57, 89, 110, and 178 in RA95.5-5 were individually targeted for a round of cassette mutagenesis. The DNA libraries were constructed by standard overlap extension PCR33 using the following degenerate primer pairs: RA95.5-5bs-P57f (5’-ATC GCG TAT TAT ACA CGC AAA TCG NNS TCA GGT CTG GAT GTT GAG CGT G) and RA95.5-5bs-P57r (5’-CGA TTT GCG TGT ATA ATA CGC GAT); RA95.5-5bs-F89f (5’-TT GGC CTG TCT ATT AAG ACA GAA GAG AAA TAC NDT AAT GGC AGC TAT GAA ATG CTG C) and RA95.5-5bs-F89r (5’-GTA TTT CTC TTC TGT CTT AAT AGA CAG GCC AA); RA95.5-5bs-N110f (5’-GCT AGC AGT GTC AGT ATT CCA ATT CTA ATG NDT GAT TTT ATC GTG AAA GAA AGC CAG ATT G) and RA95.5-5bs-N110r (5’-CAT TAG AAT TGG AAT ACT GAC ACT GCT AGC); and RA95.5-5bs-S178f (5’-CGT ATC GGT GCG CGT TTT ATT NNS ATC TTC TCA ATG AAT TTT GAA ACG GG) and RA95.5-5bs-S178r (5’-AAT AAA ACG CGC ACC GAT ACG). The randomized codons are shown in bold, where N is an equimolar mixture of all four nucleotides; D is an equimolar mixture of A, G, and T; and S is an equimolar mixture of G and C. The two most active variants isolated from these libraries, RA95.5-5-S178T and RA95.5-5-S178V, were included in the construction of the eighth round library as noted above.
Site-directed mutagenesis
The role of selected active site residues was assessed by site-directed mutagenesis. The glutamate at position 53 in RA95.0, which was designed to coordinate a catalytic water molecule, was mutated to alanine (forward primer 5’-ACA GCA ATC ATC GCG GTT TAT GCG CGC AAA TCG CCG TCA GG and reverse primer 5’-CAT AAA CCG CGA TGA TTG CTG T). The lysines at positions 210 (RA95.0, RA95.5, and RA95.5-5) and 83 (RA95.5 and RA95.5-5) were individually replaced by methionine (K210, forward primer 5’-CGT GGT GAA AGT CGC GAT GCT GGG CAT TAG TGA GCG and reverse primer 5’-CGC GAC TTT CAC CAC G; and K83, forward primer 5’-TTA CGC GGT TGG CCT GTC TAT TAT GAC AGA AGA GA ATA CTT TAA TGG CAG CTA TG and reverse primer 5’-TAA TAG ACA GGC CAA CCG CGT AA). In addition, for RA95.5 and RA95.5-5, K83M/K210M double knock-outs were generated. To assess the influence of lysine at position 83 in the starting scaffold, an additional mutation T83K was inserted into the RA95.0-K210M mutant (T83, forward primer 5’-GCG GTT GGC CTG TCT ATT AAA ACA GAA GAA AAA TAC and reverse primer 5’-AAT AGA CAG GCC AAC CGC). Full-length genes were assembled using the standard library cloning primers. Ligation into the pET-29b(+)-library vector and subsequent transformation was performed as described above.
Screening
Approximately 800 clones were screened per evolutionary round as previously described7. Single colony transformants were grown in LB medium containing 30 µg mL−1 kanamycin overnight in 96-well microtiter plates at 25 °C. These pre-cultures were used to inoculate 1 mL LB medium containing 30 µg mL−1 kanamycin (rounds one to six) in 2 mL 96-deep-well plates. The cell culture volume for protein production was reduced to 0.5 ml in the seventh round and to 0.3 mL in the eighth round. After 4 h at 37 °C (OD600 ≈ 0.4) gene expression from the T7 promoter was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and the cultures were further incubated for 5 h. Cells were then harvested and stored at −20 °C prior to lysis. Cell lysis was accomplished by addition of 300 µL lysis buffer (25 mM HEPES, 100 mM NaCl, pH 7.5, containing 1 mg mL−1 egg white lysozyme, Sigma) and subsequent incubation of the resuspended cells at 23 °C for one hour. The lysates were cleared by centrifugation and the supernatants transferred to black fluorescence microtiter plates (FluoroNunc, Nunc, Penfield, NY, USA). The substrate, (±)-methodol, was added and its conversion to 6-methoxy-2-naphthaldehyde and acetone was monitored in a Varioskan flush multimode reader (Thermo Scientific, Waltham, MA, USA) (excitation wavelength, 330 nm; emission wavelength, 452 nm; excitation band width, 12 nm) at 29 °C. The substrate concentration was either 80 µM (first round) or 30 µM (all other libraries). The most active clone from the preceding round was included on each plate in triplicate as a positive control and reference. The clones with the largest increase in activity relative to the standard, typically up to 1% of the total population, were picked from the replica plate for plasmid isolation, sequencing, and further diversification. For detailed characterization, the best variant in each round was identified and characterized as described below.
Protein production and purification
Individual variants were produced as C-terminally His-tagged proteins in E. coli BL21 Gold(DE3) and purified by affinity chromatography for biochemical characterization2,7. To ensure monoclonality, single-colony streakouts of the most active clones were prepared from the library master plates. Single colonies were used to prepare pre-cultures, of which 0.5 mL were inoculated in 500 mL of LB medium containing 30 µg mL−1 kanamycin. The bacterial cultures were incubated at 37 °C and 220 rpm until an OD600 of 0.4 was reached. Following induction of enzyme production with 0.1 mM IPTG, the cells were incubated for an additional 5 h at 37 °C, at which point they were harvested. Cell pellets were stored at -20 °C prior to lysis. Upon thawing, the pellets were resuspended in 25 mM HEPES, 300 mM NaCl, pH 7.5, containing 1 mg mL−1 egg white lysozyme and incubated for 1 h at 4 °C. The cells were then lysed by sonication, and cell debris was removed by centrifugation. Ni-NTA beads (Qiagen, Venlo, Netherlands) were equilibrated with 25 mM HEPES, 300 mM NaCl, pH 7.5, and the soluble protein fraction was loaded onto the column. The samples were washed once with the same buffer without imidazole and subsequently with buffer containing 20 mM and 40 mM imidazole. The protein was finally eluted using 200 mM imidazole. The sample was dialyzed at 4 °C against 25 mM HEPES, 100 mM NaCl, pH 7.5. Protein concentration was determined from the absorbance at 280 nm using calculated extinction coefficients (http://expasy.org/tools/protparam.html).
Substrate and product solubility
The solubility of (±)-methodol in aqueous buffer (25 mM HEPES, 100 mM NaCl, pH 7.5, containing 2.7% acetonitrile) was assessed by UV/Vis spectroscopy. Based on the experimentally determined extinction coefficient of 1,390 ± 10 M−1 cm−1 at 330 nm, racemic methodol is fully soluble in this buffer up to a concentration of 1000 µM at 29 °C. In contrast, the solubility of the retro-aldol product, 6-methoxy-2-naphthaldehyde (ε310 = 16,100 ± 370 M−1 cm−1) is much lower under these conditions. Because deviations from Beer’s law were observed above 50 µM 6-methoxy-2-naphthaldehyde, initial rates were only calculated from data in which no more than 25 µM product had accumulated.
Fluorescence assay
Kinetic assays were carried out at 29 °C in aqueous buffer (25 mM HEPES, 100 mM NaCl, pH 7.5) in 2 mm cuvettes. Acetonitrile at a final concentration of 2.7% was used as co-solvent to facilitate substrate solubility. Initial rates for the conversion of methodol to the fluorescent product 6-methoxy-2-naphthaldehyde were determined in a Photon Technology International spectrofluorometer (excitation wavelength, 330 nm; emission wavelength, 452 nm; band width, 2 nm) using a predetermined standard curve to convert fluorescence units into concentrations of product. The data were corrected for the buffer-catalyzed background reaction measured under the same conditions. Although the fluorescence assay is highly sensitive, photobleaching and side reactions of the photoactive species lead to complications7,10. Consequently, this assay was primarily used for preliminary characterization or for comparative purposes. In general, steady-state kinetic parameters were determined with the more robust UV-vis assay.
UV-vis spectroscopic assay
Reactions were carried out at 29 °C in aqueous buffer (25 mM HEPES, 100 mM NaCl, pH 7.5) in 1 mL sealed quartz cuvettes using either an RA95 variant or the catalytic antibody 38C2 (Sigma-Aldrich) as catalyst. Acetonitrile at a final concentration of 2.7% was included as co-solvent to facilitate substrate solubility. The retro-aldol cleavage of methodol to give 6-methoxy-2-naphthaldehyde and acetone was monitored spectroscopically at 350 nm (Δε = 5,970 M−1 cm−1) using a Perkin Elmer Lambda 35 UV-vis spectrometer equipped with a Peltier system for temperature control. The data were corrected for the buffer-catalyzed background reaction measured under the same conditions. Steady-state kinetic parameters were derived by fitting the experimental data to the Michaelis-Menten equation: v0/[E] = kcat · [S] / (KM + [S]), where v0 is the initial rate, [E] is the enzyme concentration, and [S] is the substrate concentration. The pH dependence was determined under sub-saturating conditions ([S] less than one-fifth of KM) using the buffers: 25 mM MES, 100 mM NaCl from pH 5.2 to 6; 25 mM HEPES, 100 mM NaCl from pH 7 to 8.5, and 25 mM AMP, 100 mM NaCl above pH 8.5. Data were fitted to the following equation: (kcat/KM)obs = (kcat/KM)max / (1 + 10pKa - pH).
Crystallization of RA95-inhibitor complexes
The RA95 retro-aldolases were mixed with the 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione (4) in a 1:50 to 1:100 molar ratio and incubated at 400 rpm overnight (RA95.5 and RA95.5-5) or for 3 days (RA95.0) at 30 °C. After incubation, the sample was concentrated to 10–20 mg mL−1 and purified by size-exclusion chromatography (S75 16/60, GE Healthcare). Peak fractions were analyzed by SDS-PAGE and pure fractions were pooled. The samples were concentrated to approximately 12 mg mL−1 using a 10 kDa molecular weight cutoff ultrafiltration membrane (Amicon, Millipore) and crystallized by the vapor diffusion method. Initial crystallization hits were obtained by using commercial sparse matrix screens PEG/Ion (Hampton Research) and PACT Premiere (Molecular Dimension). Successful crystallization conditions were improved by increasing the incubation temperature from 20 °C to 28 °C and by varying the pH, precipitant concentration and additives around the initial hits. In the case of RA95.5-5, microseeding afforded larger crystals. The optimized crystallization conditions were 0.2 M D,L-malic acid, pH 7.0, and 20% w/v PEG 3350 for RA95.0; 0.1 M HEPES-KOH, pH 7.6, containing 23% w/v PEG 3350 for RA95.5; and 0.1 M Tris-HCl, pH 8.5, 0.1 M Na2HPO4 and 23% w/v PEG 3350 for RA95.5-5. Crystals were cryoprotected by plunging into mother liquor complemented with 20% w/v glycerol prior to flash freezing in liquid nitrogen.
Crystallographic methods
Crystallographic data were collected at 100 K using a wavelength of 1.00 Å at the Swiss Light Source (SLS) X06SA PX beamline (Paul Scherrer Institute, Villigen, Switzerland). The diffraction data obtained were indexed and integrated with the program XDS34. All structures were solved by molecular replacement using the program PHASER35 with the deposited structure 1LBL36 as a starting model. The model was refined by using PHENIX37 and the obtained model was subsequently optimized by iterative model building with the program COOT38. Resolution estimation and data truncation were performed by using an I/sigma(I) criterion of 2 for the highest resolution shell. Additionally, we used a split half correlation CC(1/2) criterion of 50% to avoid discarding highly significant data39. The backbone dihedral angles for all three proteins were distributed within the most favored (92.9 to 95.6%) and additionally allowed regions (7.1 to 4.4%) of the Ramachandran map. Details of the refinement statistics are summarized in Supplementary Table 3.
Methodol synthesis
Racemic 4-hydroxy-4-(6-methoxy-2-naphthyl)-2-butanone, also referred to as (±)-methodol, was synthesized following a literature procedure40. The individual (R) and (S) enantiomers were separated by preparative normal phase HPLC using a chiral silica sorbent column (Reprosil Chiral-NR, 8 µm, 250 × 20 mm; Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) and 30% ethyl acetate in hexane at a flow rate of 20 mL min−1 for elution. The fractions of the well-separated isomers (retention times: 8.5 and 10.9 min) were collected and the solvent removed on a rotary evaporator. 1H NMR spectra and [α]D22 values were in agreement with the literature41.
1-(6-Methoxy-2-naphthalenyl)-1,3-butanedione synthesis
100 mg 4-hydroxy-4-(6-methoxy-2-naphthyl)-2-butanone were incubated with 1.2 equivalents of Dess-Martin periodinane in chloroform for 1 hour at room temperature7. The oxidized product was purified by silica gel chromatography, eluting with hexanes/ethyl acetate. The identity of the compound was confirmed by matrix-assisted laser desorption ionization mass spectrometry ([M+H]+, 242.09 calculated and 242.09 measured); its 1H-NMR spectrum was identical to that reported in the literature7.
Accession codes
Protein Data Bank: the crystal structures of RA95.0, RA95.5, and RA95.5-5 have been deposited under the accession numbers 4a29, 4a2s, and 4a2r, respectively.
Supplementary Material
Acknowledgments
We thank T. Tomizaki, M. Müller, V. Olieric, G. Pompidor, and A. Pauluhn at the Swiss Light Source for their outstanding support and all members of the Ban laboratory for suggestions and discussions. We are also grateful to E. Althoff for providing the genes for RA95.0 and RA95.5 and sharing data prior to publication and D. Gillingham and S. Tonazzi for inhibitor synthesis and resolution. The authors acknowledge support by the Swiss National Science Foundation (SNSF) (N.B. and D.H.), the National Center of Excellence in Research (NCCR) Structural Biology program of the SNSF (N.B.), the ETH Zurich (P.K., N.B., and D.H.), the Defense Advanced Research Projects Agency (DARPA) (D.B. and D.H.), and the Howard Hughes Medical Institute (HHMI) (D.B.). LG was funded by the Stipendienfonds der Schweizerischen Chemischen Industrie.
Footnotes
Author contributions
D.H., N.B., D.B., P.K., L.G., and S.C. designed the experiments. L.G. and R.O. evolved and biochemically characterized the variants; S.C. crystallized the proteins and solved their structures. The manuscript and figures were prepared by L.G., S.C., P.K., N.B., and D.H.
Competing financial interests
The authors declare no competing financial interests.
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Bolon DN, Mayo SL. Enzyme-like proteins by computational design. Proc. Natl. Acad. Sci. USA. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang L, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Röthlisberger D, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 4.Privett HK, et al. Iterative approach to computational enzyme design. Proc. Natl. Acad. Sci. USA. 2012;109:3790–3795. doi: 10.1073/pnas.1118082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel JB, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kries H, Blomberg R, Hilvert D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 2013;17:221–228. doi: 10.1016/j.cbpa.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Althoff EA, et al. Robust design and optimization of retroaldol enzymes. Protein Sci. 2012;21:717–726. doi: 10.1002/pro.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khersonsky O, et al. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc. Natl. Acad. Sci. USA. 2012;109:10358–10363. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gefflaut T, Blonski C, Perie J, Willson Ml. Class I aldolases: Substrate specificity, mechanism, inhibitors and structural aspects. Prog. Biophys. Mol. Biol. 1995;63:301–340. doi: 10.1016/0079-6107(95)00008-9. [DOI] [PubMed] [Google Scholar]
- 10.Lassila JK, Baker D, Herschlag D. Origins of catalysis by computationally designed retroaldolase enzymes. Proc. Natl. Acad. Sci. USA. 2010;107:4937–4942. doi: 10.1073/pnas.0913638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, et al. Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design. J. Mol. Biol. 2012;415:615–625. doi: 10.1016/j.jmb.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. De novo enzyme design using Rosetta3. PLoS One. 2011;6:e19230. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tagaki W, Yamamoto H. Polyamino-β-cyclodextrin as a model of aldolase. Tetrahedron Lett. 1991;32:1207–1208. [Google Scholar]
- 14.Tanaka F, Fuller R, Barbas CF., 3rd Development of small designer aldolase enzymes: catalytic activity, folding, and substrate specificity. Biochemistry. 2005;44:7583–7592. doi: 10.1021/bi050216j. [DOI] [PubMed] [Google Scholar]
- 15.Müller MM, Windsor MA, Pomerantz WC, Gellman SH, Hilvert D. A rationally designed aldolase foldamer. Angew. Chem. Int. Ed. 2009;48:922–925. doi: 10.1002/anie.200804996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wörsdörfer B, Henning L, Obexer R, Hilvert D. Harnessing protein symmetry for enzyme design. ACS Catal. 2012;2:982–985. [Google Scholar]
- 17.Wymer N, et al. Directed evolution of a new catalytic site in 2-keto-3-deoxy-6-phosphogluconate aldolase from Escherichia coli. Structure. 2001;9:1–9. doi: 10.1016/s0969-2126(00)00555-4. [DOI] [PubMed] [Google Scholar]
- 18.Heine A, et al. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science. 2001;294:369–374. doi: 10.1126/science.1063601. [DOI] [PubMed] [Google Scholar]
- 19.Leung DW, Chen E, Goeddel DV. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 20.Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbas CF, et al. Immune versus natural selection: Antibody aldolases with enzymic rates but broader scope. Science. 1997;278:2085–2092. doi: 10.1126/science.278.5346.2085. [DOI] [PubMed] [Google Scholar]
- 22.Esposito G, et al. Structural and functional analysis of aldolase B mutants related to hereditary fructose intolerance. FEBS Lett. 2002;531:152–156. doi: 10.1016/s0014-5793(02)03451-8. [DOI] [PubMed] [Google Scholar]
- 23.Kipnis Y, Baker D. Comparison of designed and randomly generated catalysts for simple chemical reactions. Protein Sci. 2012;21:1388–1395. doi: 10.1002/pro.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St-Jean M, Blonski C, Sygusch J. Charge stabilization and entropy reduction of central lysine residues in fructose-bisphosphate aldolase. Biochemistry. 2009;48:4528–4537. doi: 10.1021/bi8021558. [DOI] [PubMed] [Google Scholar]
- 25.Wilson IA, Stanfield RL. Antibody-antigen interactions: new structures and new conformational changes. Curr. Opin. Struct. Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 26.Cauerhff A, Goldbaum FA, Braden BC. Structural mechanism for affinity maturation of an anti-lysozyme antibody. Proc. Natl. Acad. Sci. USA. 2004;101:3539–3544. doi: 10.1073/pnas.0400060101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruscio JZ, Kohn JE, Ball KA, Head-Gordon T. The influence of protein dynamics on the success of computational enzyme design. J. Am. Chem. Soc. 2009;131:14111–14115. doi: 10.1021/ja905396s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 29.Babbitt PC, Gerlt JA. Understanding enzyme superfamilies. Chemistry as the fundamental determinant in the evolution of new catalytic activities. J. Biol. Chem. 1997;272:30591–30594. doi: 10.1074/jbc.272.49.30591. [DOI] [PubMed] [Google Scholar]
- 30.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 31.Zanghellini A, et al. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neylon C. Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution. Nucleic Acids Res. 2004;32:1448–1459. doi: 10.1093/nar/gkh315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain-reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hennig M, Darimont BD, Jansonius JN, Kirschner K. The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product. J. Mol. Biol. 2002;319:757–766. doi: 10.1016/S0022-2836(02)00378-9. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synch. Rad. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.List B, Barbas CF, 3rd, Lerner RA. Aldol sensors for the rapid generation of tunable fluorescence by antibody catalysis. Proc. Natl. Acad. Sci. USA. 1998;95:15351–15355. doi: 10.1073/pnas.95.26.15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner JM, Bui T, Lerner RA, Barbas CF, 3rd, List B. An efficient benchtop system for multigram-scale kinetic resolutions using aldolase antibodies. Chemistry. 2000;6:2772–2774. doi: 10.1002/1521-3765(20000804)6:15<2772::aid-chem2772>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.