SUMMARY
As the cellular power plant, mitochondria play a significant role in homeostasis. To maintain the proper quality and quantity of mitochondria requires both mitochondrial degradation and division. A selective type of autophagy, mitophagy, drives the degradation of excess or damaged mitochondria, whereas division is controlled by a specific fission complex; however, the relationship between these two processes, especially the role of mitochondrial fission during mitophagy, remains unclear. In this study, we report that mitochondrial fission is important for the progression of mitophagy When mitophagy is induced, the fission complex is recruited to the degrading mitochondria through an interaction between Atg11 and Dnm1; interfering with this interaction severely blocks mitophagy. These data establish a paradigm for selective organelle degradation.
Keywords: lysosome, mitophagy, phagophore, stress, vacuole, yeast
INTRODUCTION
Mitochondria are double-membrane-bound organelles that play significant roles in a variety of cellular metabolic reactions. This organelle is central to cellular physiology, supplying energy and certain metabolites, but it also generates harmful reactive oxygen species. Thus, mitochondrial homeostasis must be maintained, which can be a costly process. As a result, cells degrade superfluous, or extensively damaged, mitochondria; however, this is a considerable structural challenge considering the extended, reticular nature of this organelle. Constitutive mitochondrial fusion and fission, as well as biogenesis and degradation, make the mitochondria highly dynamic. In the budding yeast Saccharomyces cerevisiae, a complex containing Fis1, Dnm1, Mdv1 and Caf4 controls the fission of mitochondria, whereas fusion is regulated by the action of Fzo1, Ugo1 and Mgm1 (Okamoto and Shaw, 2005). The degradation of mitochondria is mediated by mitophagy, a selective type of macroautophagy (hereafter autophagy). The study of mitophagy has attracted increasing attention in recent years; this process plays significant roles in various aspects of normal physiology such as the removal of mitochondria during the maturation of erythroid cells, whereas its dysfunction is associated with certain pathophysiologies such as Wolfram Syndrome 2 and Parkinson disease (Kanki and Klionsky, 2009; Kanki et al., 2009b; Matsuda et al., 2010; Narendra et al., 2008; Narendra et al., 2010; Okamoto et al., 2009; Schweers et al., 2007; Vives-Bauza et al., 2010).
In eukaryotic cells, autophagy functions as a lysosome/vacuole-dependent mechanism for the degradation of damaged or obsolete proteins and organelles, and can occur in either nonselective or selective modes. Nonselective autophagy functions to sequester bulk cytoplasm into double-membrane vesicles, termed autophagosomes, which are then transported to the lysosome/vacuole where the cargo is degraded (Xie and Klionsky, 2007). In contrast, selective autophagy targets specific proteins or organelles as cargos, such as peroxisomes (pexophagy) and mitochondria. In the case of selective autophagy, a general model has been established in which a ligand on the target interacts with a specific receptor; the receptor in turn binds a scaffold protein, which links the cargo-receptor complex with the autophagy machinery (Mijaljica et al., 2012). For example, in yeast mitophagy, Atg32 is a mitochondrial protein that serves as a selective receptor (a ligand, if one exists, has not been identified), which binds the Atg11 scaffold. Atg11 is needed for subsequent engagement of the mitochondria with Atg8–PE, which lines the initial sequestering compartment, the phagophore. Due to its potentially large size, one key question with regard to mitophagy concerns the role of mitochondrial fission. Work in mammalian cells suggests that mitochondrial fission facilitates the process of mitophagy (Tanaka et al., 2010; Twig et al., 2008); however, it is not clear whether these two processes occur independently, or whether they function in a coordinated manner.
Here, we show that the Atg32-Atg11 interaction marks degrading mitochondria. Furthermore, Dnm1 is recruited to these mitochondria through an interaction with Atg11. When Dnm1 loses its interaction with Atg11, the degradation of mitochondria is severely blocked. These results support the hypothesis that mitochondrial fission machinery participates in, and facilitates, mitochondrial division in an early step of mitophagy, and indicate that Atg11 plays a unique role as a scaffold that recruits the fission components in addition to its role in connecting the target with the autophagic machinery.
RESULTS
The Mitochondrial Fission Complex Is Required for Mitophagy
In our recent genome-wide yeast mutant screen for mitophagy-defective strains, we found that DNM1, a gene encoding a dynamin-related GTPase required for mitochondrial fission, is required for efficient mitophagy (Kanki et al., 2009a). In Saccharomyces cerevisiae, the mitochondrial fission complex consists of four components, Fis1, Dnm1, Mdv1 and Caf4. Fis1 is a conserved integral membrane protein and is required for the proper localization of Dnm1 and Mdv1 on mitochondria (Karren et al., 2005; Mozdy et al., 2000; Tieu and Nunnari, 2000). Dnm1 assembles specifically at the sites where mitochondrial fission occurs (Bleazard et al., 1999). Mdv1 and Caf4 redundantly bridge the interaction between Fis1 and Dnm1 (Griffin et al., 2005; Tieu and Nunnari, 2000). Although Fis1 is distributed evenly on mitochondria, Dnm1 and Mdv1 show colocalized puncta on mitochondrial tubules, and it is thought that those puncta are the sites where mitochondrial division takes place. We first sought to determine whether all four of the corresponding gene products are involved in mitophagy.
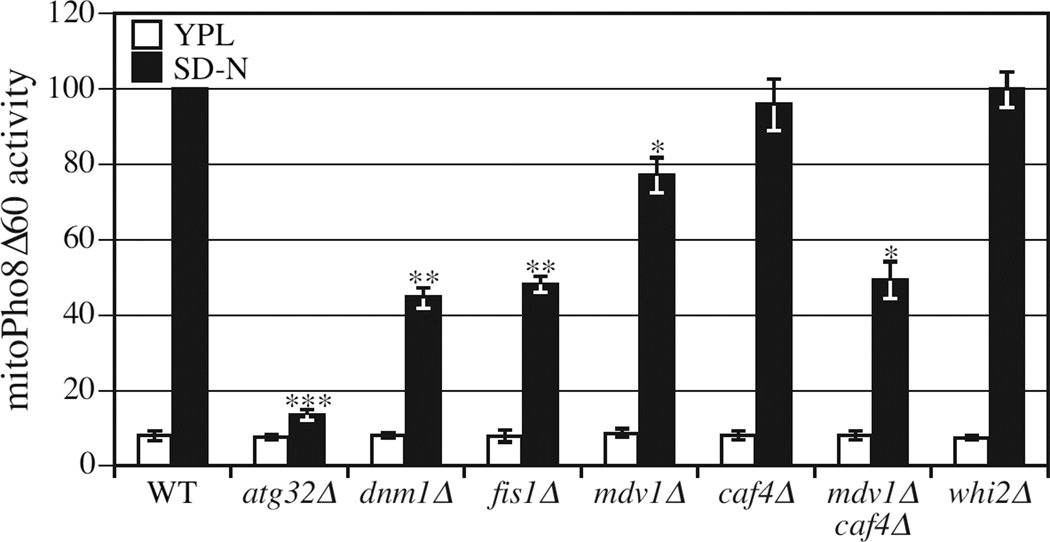
In Saccharomyces cerevisiae, mitochondria proliferate when cells are cultured in a non-fermentable carbon source, such as lactic acid or glycerol. When these cells are subjected to conditions of nitrogen starvation in the presence of a fermentable carbon source, such as glucose, mitophagy is induced to degrade the excess mitochondria (Kanki and Klionsky, 2008). We examined the activity of mitophagy in the absence of DNM1, FIS1, MDV1 or CAF4, using an enzymatic assay in which a mitochondrially-targeted zymogen, mitoPho8Δ60, is activated following mitophagic delivery to the vacuole (Kanki et al., 2009a). PHO8 encodes an alkaline phosphatase that is transported to the vacuole via the ALP pathway (Klionsky and Emr, 1989). Pho8Δ60, a mutant form of Pho8 in which the N-terminal 60 amino acids including the transmembrane domain have been removed, localizes to the cytoplasm and is unable to be transported to the vacuole through the ALP pathway. We fused cytochrome c oxidase subunit IV (Cox4) with Pho8Δ60 and named the fusion protein mitoPho8Δ60. This fusion protein is localized on mitochondria and its transport to the vacuole, which results in enzyme activation, is dependent on mitophagy (Kanki et al., 2009a). Therefore, calculating the phosphatase activity of mitoPho8Δ60 can be used to monitor mitophagy activity. We found that mitophagy was severely blocked in dnm1Δ and fis1Δ cells, partially blocked in mdv1Δ cells, and essentially normal in caf4Δ cells (Figure 1). Previous work implied a redundant function for Mdv1 and Caf4, with Mdv1 being more important than Caf4 in mitochondrial fission (Naylor et al., 2006). Accordingly, we examined the mdv1Δ caf4Δ double-deletion mutant, which showed a strong defect in mitophagy, similar to dnm1Δ and fis1Δ (Figure 1). These results suggest that the intact mitochondrial fission machinery is required for mitophagy.
Figure 1. Mitochondrial Fission Is Required for Mitophagy.
MitoPho8Δ60 activity is reduced in strains with deletions of genes encoding mitochondrial fission proteins. Wild-type (KWY20), atg32Δ (KWY22), dnm1Δ (KDM1013), fis1Δ (KDM1002), mdv1Δ (KDM1006), caf4Δ (KDM1011), mdv1Δ caf4Δ (KDM1012) and whi2Δ (KDM1010) cells in the mitoPho8Δ60 background were cultured in YPL to mid-log phase, then shifted to SD-N for 6 h. The mitoPho8Δ60 assay was performed as described in Experimental Procedures. Error bars correspond to the standard error, and were obtained from three independent repeats. * p<0.01; ** p<0.001; *** p<0.0001.
Previously, it was reported that mitochondrial fission is not required for rapamycin-induced mitophagy (Mendl et al., 2011). In this study, the diminished mitophagy activity in fis1Δ cells is reported to be due to a secondary mutation in the WHI2 gene. Therefore, we tested mitophagy activity in whi2Δ cells using the mitoPho8Δ60 assay. Mitophagy activity in whi2Δ cells was essentially identical to that of the isogenic wild-type cells (Figure 1). The discrepancy between our results and those of Mendl et al. may be due to the different methods used to induce mitophagy. In the previous study, yeast cells were cultured in medium containing glycerol, and mitophagy was induced by the addition of rapamycin; however, these conditions require 24 h of drug treatment to induce a high level of mitophagy. In our study, mitochondrial proliferation was achieved by growth in lactic acid, and mitophagy was induced by nitrogen starvation in the presence of glucose; a similar level of mitoPho8Δ60 activity was detected in the wild-type and whi2Δ strains within 6 h. In general, nitrogen starvation induces a substantially stronger response than treatment with rapamycin.
The Atg32-Atg11 Interaction Marks Degrading Mitochondria During Mitophagy
Mitochondrial fission is proposed to occur on mitochondrial termini (Griffin et al., 2005), which may therefore represent a very early stage of mitophagy. Thus, the first requirement was to identify a marker that would enable us to specifically identify mitochondria destined for degradation. Previously, others and we showed that Atg32 and Atg11 directly participate in an early event of mitophagy (Aoki et al., 2011; Kanki et al., 2009b; Kondo-Okamoto et al., 2012; Okamoto et al., 2009). When mitophagy is induced, Atg32 recruits Atg11 to the mitochondria, and their interaction is required for the delivery of mitochondria to the phagophore assembly site (PAS), the location of autophagosome formation, and eventually into the vacuole for degradation. In mammalian cells, PARK2/Parkin is recruited to, and thereby selectively marks, depolarized mitochondria (Narendra et al., 2008); in yeast, however, Atg32 is evenly distributed on the mitochondrial tubules, even though only a relatively small portion of the total mitochondrial population will ultimately be degraded (Figure S1A; Mao et al., 2011). Therefore, Atg32 alone cannot serve as the sole marker of the degrading mitochondria.
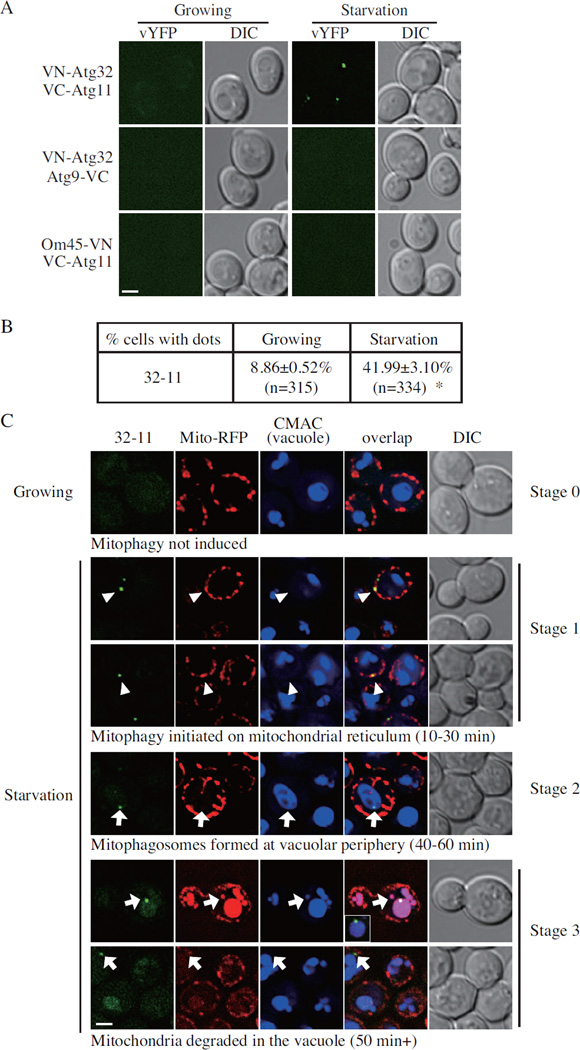
We took advantage of the bimolecular fluorescence complementation (BiFC) assay (Sung and Huh, 2007), in which the Venus yellow fluorescent protein (vYFP) is split into two fragments, VN (N terminus of vYFP) and VC (C terminus of vYFP); we fused VN to Atg32, and VC to Atg11 by integrating the corresponding constructs at the chromosomal ATG32 and ATG11 loci. Fluorescence from these chimeras can only be observed when the two proteins interact and bring the two fluorophore fragments into close proximity. We tested the VN-Atg32-VC-Atg11 interaction in both growing (YPL) and mitophagy-inducing (SD-N) conditions. When expressing both VN-Atg32 and VC-Atg11, ~9% of the cells showed vYFP dots in growing conditions; however, when mitophagy was induced, ~42% of the cells displayed vYFP dots, indicating the colocalization of these two proteins (Figure 2A and B). In contrast, neither VN-Atg32 in combination with another autophagy-related protein fusion construct, Atg9-VC, nor a different chimera containing the OM45 mitochondrial outer membrane protein fused to VN (OM45-VN) in combination with VC-Atg11, generated fluorescent puncta in either growing or mitophagy-inducing conditions (Figure 2A). These results were consistent with our previous report that the interaction of Atg32 and Atg11 is enhanced when mitophagy is induced (Kanki et al., 2009b). The chimeric constructs were stable under mitophagy-inducing conditions and did not show an appreciable change in protein level, indicating that the appearance of the fluorescent dots reflected colocalization, and was not due to a change in protein concentration (Figure S1B).
Figure 2. BiFC 32-11 Dots Mark Degrading Mitochondria During Mitophagy.
(A) A BiFC assay was performed for Atg32, Atg1 1, Om45 and Atg9. Cells containing BiFC pairs (Atg32-Atg1 1 in KDM1501, Atg32-Atg9 in KDM1519 and Atg11-Om45 in KDM1520) were cultured in YPL and shifted to SD-N for 1 h, followed by analysis by fluorescence microscopy; images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar, 2 µm.
(B) Quantification of (A). 12 Z-section images were projected and the percentage of cells that contained 32-11 dots was determined. Standard error was calculated from three independent experiment. * p<0.01.
(C) VN-ATG32 VC-ATG11 (KDM1501) cells, transformed with pMito-RFP, were cultured in SML and shifted to SD-N from 10 min to 1 h, and the cell samples were observed by fluorescence microscopy. CellTracker Blue CMAC was used to stain the vacuolar lumen. Arrowheads indicate the 32-11 dots that localized on the mitochondrial reticulum; and arrows indicate the 32-11 dots that localized on the vacuolar periphery. All of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar, 2 µm. The inset in row 5, panel 4 corresponds to the large vacuole, reducing the red intensity to demonstrate that the intravacuolar punctum corresponds to a green 32-11 dot that has not yet been degraded. Also see Fig. S1.
We next asked whether the VN-Atg32-VC-Atg11 BiFC fluorescent puncta (hereafter, 32-11 dots) represented degrading mitochondria. We first examined the activity of mitophagy in the presence of the fusion proteins VN-Atg32 and VC-Atg11. Wild-typeand VN-Atg32-VC-Atg11 cells were grown in a non-fermentable carbon source, then shifted to SD-N to induce mitophagy. The mitoPho8Δ60 activity of VN-Atg32-VC-Atg11 cells was similar to that of wild-type cells, which indicated that mitophagy occurred normally when Atg32 and Atg11 were fused with VN and VC, respectively (Figure S1C). The atg32Δ strain served as a negative control, and displayed only a background level of activity. Second, we stained the mitochondria with MitoTracker Red dye and examined the localization of the 32-11 dots relative to the organelle. When mitophagy was induced following a 30 min shift to SD-N, most of the 32-11 dots were localized on the mitochondrial reticulum (Figure S1D). Subsequently, we used the FM 4–64 dye to mark the vacuole. Immediately after the shift to SD-N, no 32-11 dots were detected in proximity to the vacuole (data not shown). Within 1 h after mitophagy induction, however, we detected 32-11 dots localized close to the vacuole limiting membrane (Figure S1E); this perivacuolar location likely corresponds to the PAS. Finally, by 6 h after shifting to SD-N, very few 32-11 dots could be detected in wild-type cells; however, atg1Δ mutant cells that are defective in mitophagy continued to accumulate 32-11 dots, which indicated that the degradation of mitochondria marked by the VN-Atg32-VC-Atg11 BiFC interaction was dependent on an intact autophagy pathway (Figure S1F and G). Therefore, based on the observations that 1) VN-Atg32 and VC-Atg11 are functional fusion proteins; 2) 32-11 dots localized on mitochondria shortly after inducing mitophagy; 3) at a later time point, the 32-11 dots subsequently showed a vacuole-peripheral PAS localization; and 4) the loss of the 32-11 signal was dependent on autophagy, we concluded that the 32-11 dots were an appropriate marker to monitor degrading mitochondria.
In order to clarify the sequential steps of mitophagy, we used a plasmid containing Mito-RFP and CellTracker Blue CMAC dye to mark the mitochondria and vacuole lumen, respectively, which enabled us to observe both organelles and determine the localization of the 32-11 signal. In growing conditions, when mitophagy was not induced, very few 32-11 dots were detected, which was defined as Stage 0 (Figure 2C, row 1). After a short time of starvation (10 to 30 min), when mitophagy was initiated, 32-11 puncta were formed on the mitochondrial reticulum, which was defined as Stage 1 (Figure 2C, rows 2 and 3). After longer times of starvation (approximately 40 to 60 min), even though most of the 32-11 puncta were still localized on the mitochondrial reticulum, some of the puncta were detected in proximity to the vacuole, which corresponded to the presence of mitophagosomes. We defined this step as Stage 2 (Figure 2C, row 4). The earliest we were able to detect the vYFP signal in the vacuole was after approximately 50 min starvation, and the RFP signal was also seen in the vacuole at this time, which suggested the degradation of mitochondria in the vacuole lumen. We defined this step as Stage 3 (Figure 2C, rows 5 and 6).
Dnm1 Is Recruited to the Degrading Mitochondria through its Interaction with Atg11
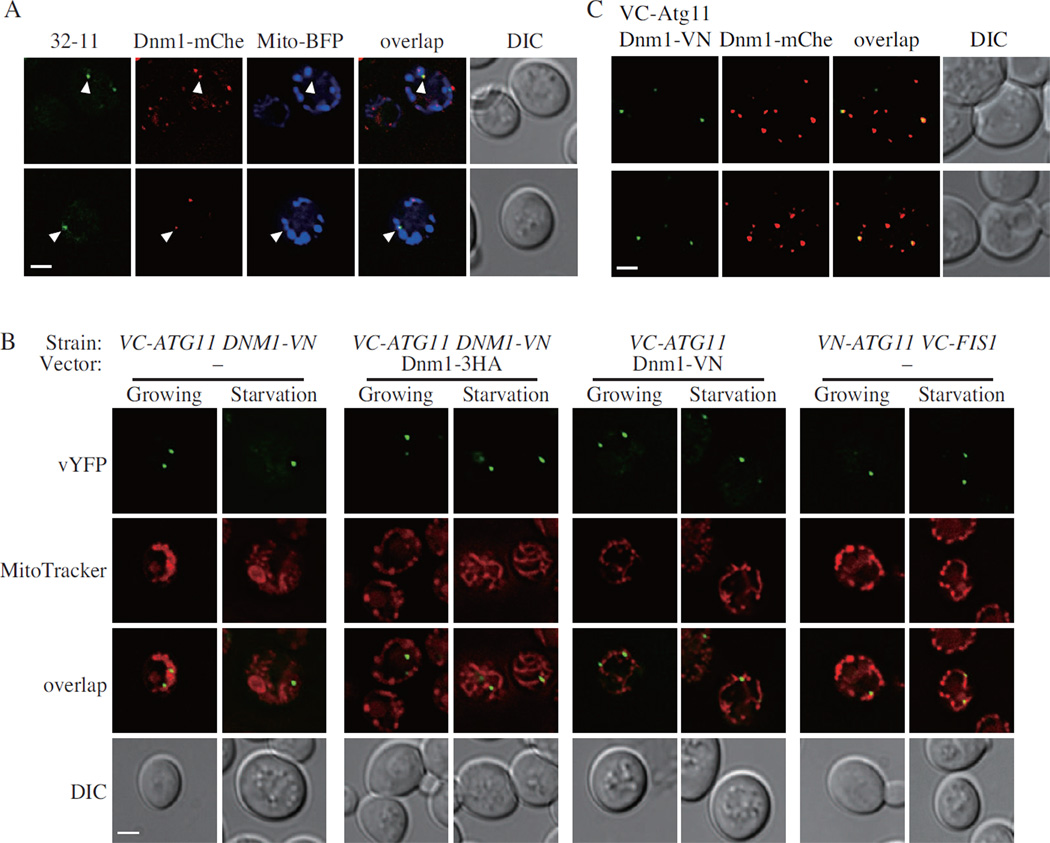
Dnm1 assembles specifically at the sites where mitochondrial fission occurs. Therefore, we next asked whether Dnm1 accumulated on the degrading mitochondria, which were marked by 32-11 dots. Accordingly, we chromosomally tagged Dnm1 with mCherry and examined its localization together with the 32-11 dots. Several Dnm1-mCherry puncta that colocalized with 32-11 dots were seen when mitophagy was induced by nitrogen starvation, and this colocalization occurred on the mitochondrial reticulum, which was marked by Mito-BFP (Figure 3A). We also chromosomally tagged Mdv1 and Caf4 with mCherry. Similar to Dnm1-mCherry, both Mdv1-mCherry and Caf4-mCherry were colocalized with 32-11 dots (Figure S2A and B).
Figure 3. Atg11 Recruits Dnm1 to the Degrading Mitochondria.
(A) VN-ATG32 VC-ATG11 DNM1-mCherry cells, transformed with pMito-BFP, were cultured in SML and shifted to SD-N for 1 h, and samples were observed by fluorescence microscopy. Arrowheads indicate the colocalized 32-11 dots with Dnm1-mCherry on the mitochondrial reticulum. All of the images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bar, 2 µm.
(B) VC-ATG11 DNM1-VN cells transformed with empty vector or pDnm1-3HA, VC-ATG11 cells transformed with pDnm1-VN, and VN-ATG11 VC-FIS1 cells transformed with empty vector were cultured in SML and shifted to SD-N for 1 h. Samples were observed by fluorescence microscopy as in (A). Scale bar, 2 µm.
(C) VC-ATG11 DNM1-mCherry cells, transformed with pDnm1-VN, were cultured in SML and shifted to SD-N for 30 min. Samples were observed by fluorescence microscopy as in (A). Scale bar, 2 µm.
Also see Fig. S2.
Based on these observations, we hypothesized that there were two groups of Dnm1, one of them assembling on the mitochondria destined for degradation in order to promote mitophagy-specific fission, with the other functioning in the normal process of constitutive mitochondrial division. A key question with regard to this first population would be the mechanism through which Dnm1 can be specifically recruited to sites that correspond to mitochondria that are destined for degradation. We hypothesized that Atg32 or Atg11 might be able to interact with Dnm1, Fis1, Mdv1 and/or Caf4. To determine whether Atg32 or Atg11—which we just showed mark this population of mitochondria—played a role in Dnm1 localization, we constructed eight strains expressing different combinations of Atg11, Atg32 and the fission machinery components as BiFC chimeras: VC-Atg11 co-expressed with Dnm1-VN, VN-Atg11 with VC-Fis1, VC-Atg11 with Mdv1-VN, VC-Atg11 with Caf4-VN, VN-Atg32 with Dnm1-VC, VN-Atg32 with VC-Fis1, VN-Atg32 with Mdv1-VC, and VN-Atg32 with Caf4-VC; we used the BiFC assay to test the interactions of these chimeric pairs. All eight strains were tested in both growing (YPL) and mitophagy-inducing (SD-N) conditions. The fluorescent signal could only be observed in cells expressing VC-Atg11 and Dnm1-VN or VN-Atg11 and VC-Fis1 (Figure S2C). The chimeric constructs were stable and did not change in expression level during the course of the analysis, indicating that the fluorescent signals reflected protein localization rather than changes in protein concentration (Figure S2D).
To extend our analysis, we then examined where the interactions occurred. Therefore, we stained the mitochondria with MitoTracker Red dye and examined the localization of the Atg11-Dnm1 and Atg11-Fis1 dots relative to the organelle. Atg11-Fis1 dots were localized on the mitochondrial reticulum in both growing and starvation conditions (Figure 3B). However, in VC-Atg11 Dnm1-VN cells, although Atg11-Dnm1 dots were observed on the mitochondrial reticulum, the mitochondrial morphology was abnormal, which resembled that in dnm1Δ cells. It was reported that the BiFC assay allows the detection of weak and transient interactions, but that these interactions are stabilized by the formation of an intact YFP. Thus, we considered that the stabilized binding of VC-Atg11 and Dnm1-VN prevented the free Dnm1, which functions as an oligomer, from participating in the normal process of constitutive mitochondrial fission. Therefore, in order to provide additional free Dnm1 in the cells, we either transformed a plasmid harboring Dnm1-3HA into the VC-Atg11 VN-Dnm1 cells or a plasmid harboring Dnm1-VN into the chromosomally integrated VC-Atg11 cells. In both cases, the mitochondrial morphology was normal and the Atg11-Dnm1 dots were localized on the mitochondrial reticulum (Figure 3B).
To further demonstrate the existence of two different populations of Dnm1, one of which interacted with Atg11, we asked whether the localization of Dnm1 was affected when Atg11 or Atg32 was absent. Accordingly, we chromosomally tagged Dnm1 with GFP in wild-type, atg11Δ and atg32Δ cells, and tracked the mitochondria with a plasmid-driven Mito-RFP. In both growing and nitrogen starvation conditions, the cellular pattern of Dnm1-GFP and mitochondrial morphology was normal in either atg11Δ or atg32Δ cells, which suggested that neither Atg11 nor Atg32 would affect the function of the normal mitochondrial fission machinery (Figure S2E). We also noticed that even though many Dnm1-GFP puncta were detected in each cell (Figure S2E), only a few Dnm1-Atg11 interacting dots were formed (Figure 3B), which also supported our hypothesis that there were two different population of Dnm1. To directly observe these two groups of Dnm1, we generated a yeast strain with VC-Atg11 and Dnm1-mCherry on the genome and transformed these cells with a plasmid harboring Dnm1-VN. The Dnm1 puncta corresponded to an oligomeric mixture of both Dnm1-mCherry and Dnm1-VN, and as a result all of the Dnm1 puncta showed a mCherry signal; however, only the small population associated with VC-Atg11 also showed a vYFP signal (Figure 3C).
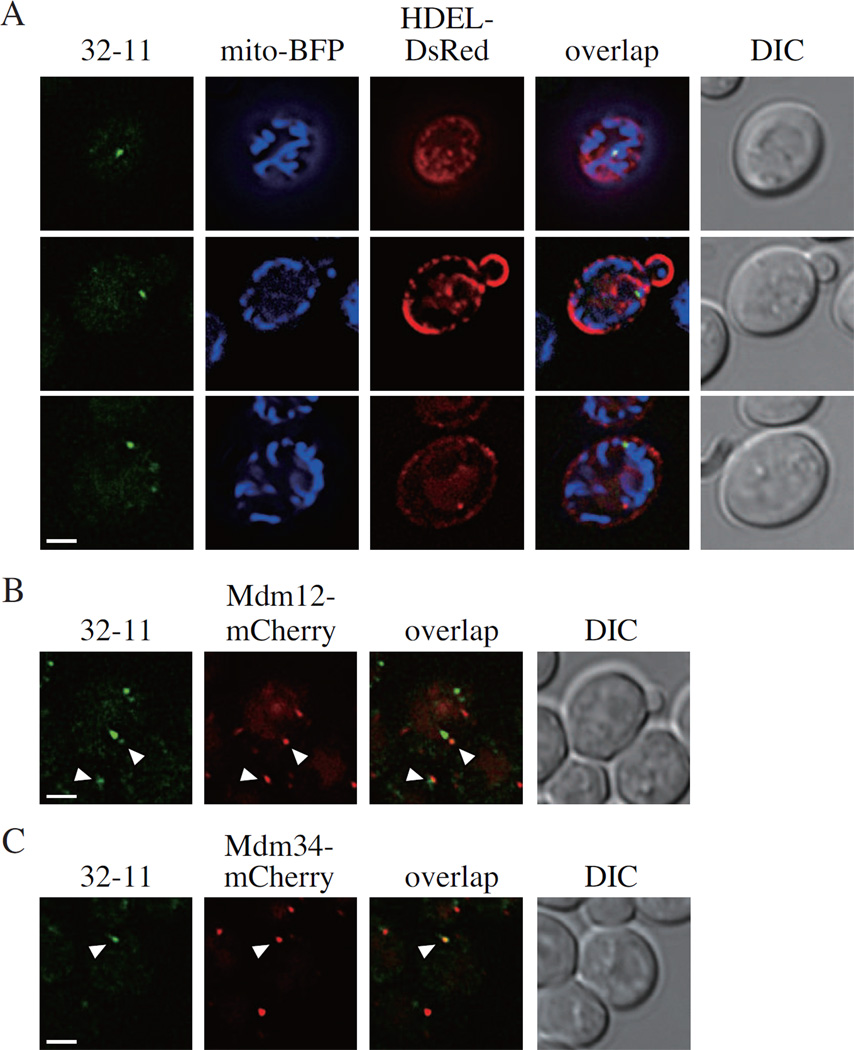
A previous report demonstrated that mitochondrial fission happens at the ER-mitochondria contact sites (Friedman et al., 2011). We wondered whether mitophagy-specific fission also occurred at these sites. Accordingly, we co-transformed plasmids containing Mito-BFP and HDEL-DsRed to track the mitochondria and ER, respectively. We found that some of the 32-11 dots were indeed localized at the ER-mitochondria contact sites (Figure 4A). Previous work also reported an ERMES (ER-mitochondria encounter structure) complex localized at the ER-mitochondria contact sites (Kornmann et al., 2009). This complex includes four proteins: Mmm1 and Mdm12, which are on the ER side, and Mdm10 and Mdm34, which are on the mitochondria side of the complex. We fused mCherry at the C terminus of Mdm12 or Mdm34 in the yeast genome, and observed the localization of these proteins together with the 32-11 dots. We observed the colocalization of 32-11 dots with some of the Mdm12-mCherry and Mdm34-mCherry puncta (Figure 4B and C). These observations suggested that the ER, in particular at the ER-mitochondria contact sites, might also participate in mitophagy-specific fission.
Figure 4. The ER Participates in Mitophagy-Specific Fission.
(A) VN-ATG32 VC-ATG11 (KDM1501) cells, transformed with pMito-BFP and pHDEL-DsRed, were cultured in SML and shifted to SD-N for 30 min.
(B, C) VN-ATG32 VC-ATG11 MDM12-mCherry (KDM1561) and VN-ATG32 VC-ATG11 MDM34-mCherry (KDM1562) cells were cultured in YPL and shifted to SD-N for 30 min.
The cells in (A, B, and C) were analyzed by fluorescence microscopy. The images are representative pictures from single Z-sections. DIC, differential interference contrast. Scale bars, 2 µm.
The Dnm1-Atg11 Interaction Is Required for Mitophagy
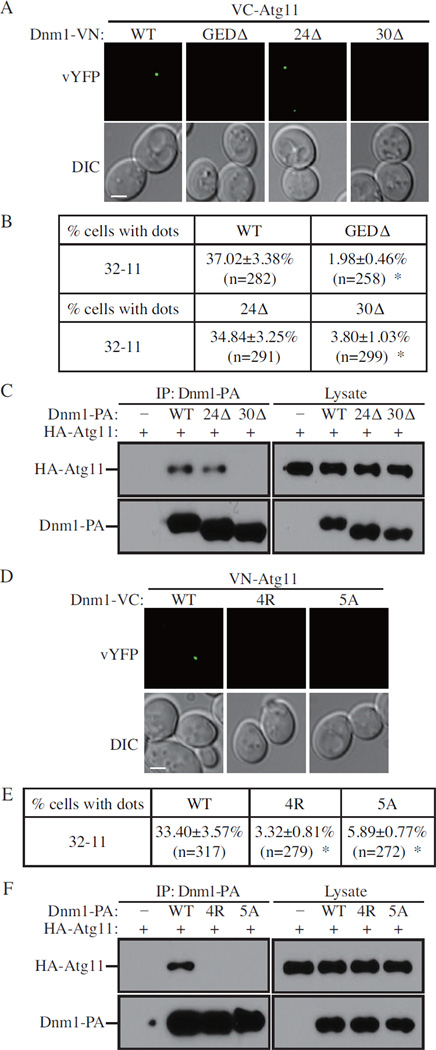
Dnm1 contains four domains: GTPase, middle, insert B, and GED (GTPase effector domain) (Fukushima et al., 2001; Figure 5). In order to find a Dnm1 mutant that lost its binding to Atg11, we examined the interaction of Dnm1 mutants with Atg1 1 by the BiFC assay. Almost no interacting dots were observed when the GED (~87 amino acids at the C terminus) was deleted (Figure 6A and B), which implied that the GED is required for binding to Atg11. We then made truncations of Dnm1 from the C terminus. The interacting dots were detected when we deleted the last 24 amino acids (Dnm1-VN 24Δ); however, very few fluorescent dots were seen when the last 30 amino acids were absent (Dnm1-VN 30Δ) (Figure 6A and B). To verify the observation from the BiFC assay, we carried out protein A affinity isolation with IgG-Sepharose. Protein A-tagged wild-type Dnm1 or the mutant without the last 24 amino acids co-precipitated HA-Atg11; however, the Dnm1 mutant lacking the last 30 amino acids was not able to precipitate this protein (Figure 6C).
Figure 5. Domain Structure of Dnm1 and GED Mutations.
The domains of Dnm1 are depicted in the top diagram, and the position of the GED domain (amino acids 670–757) are indicated. The C-terminal truncations, GEDΔ, 24Δ and 30Δ are depicted in the middle diagrams. The sequence of the six amino acid residues comprising E728 through A733 in the GED that are required for the interaction with Atg11 and the mutations 5A and 4R are shown at the bottom.
Figure 6. Mutation of the Dnm1 C Terminus Blocks Mitophagy.
(A) Cells containing BiFC pairs (Atg11-Dnm1 in KDM1523, Atg11-Dnm1 (GEDΔ) in KDM1528, Atg11-Dnm1 (24Δ) in KDM1532, and Atg11-Dnm1 (30Δ) in KDM1533) were cultured in YPL and shifted to SD-N for 1 h. Samples were observed by fluorescence microscopy, and all the images are representative pictures from single Z-sections. DIC, differential interference contrast.
(B) Quantification of (A). 12 Z-section images were projected and the percentage of cells that contained BiFC Dnm1-Atg11 dots was determined. Standard error was calculated from three independent experiment. * p<0.01.
(C) The plasmid pCuHA-Atg11 was transformed into atg11Δ (YTS147), atg11Δ DNM1-PA (KDM1247), atg11Δ DNM1(24Δ)-PA (KDM1248), and atg11Δ DNM1(30Δ) (KDM1249) cells. Cells were cultured in SML and shifted to SD-N for 1.5 h. Cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation as described in Experimental Procedures. The eluted proteins were separated by SDS-PAGE and detected with monoclonal anti-HA antibody and an antibody that binds to PA.
(D) VN-ATG11 (KDM1535) cells, transformed with pDnm1-VC, pDnm1(4R)-VC, or pDnm1(5A)-VC, were cultured in SML and shifted to SD-N for 1 h. Samples were observed by fluorescence microscopy as in (A).
(E) Quantification of (D). 12 Z-section images were projected and the percentage of cells that contained BiFC Dnm1-Atg11 dots was determined. Standard error was calculated from three independent experiment. * p<0.01.
(F) The plasmid pCuHA-Atg11 together with pDnm1-VC, pDnm1(4R)-VC, or pDnm1(5A)-VC were transformed into atg11Δ dnm1Δ (KDM1251) cells. The cells were cultured in SML and shifted to SD-N for 1.5 h. Cell lysates were prepared and analyzed as in (C).
Also see Fig. S3.
We hypothesized that the six amino acids (EDQTLA) that exist in Dnm1 24Δ, but not Dnm1 30Δ, might be important for Dnm1 binding to Atg11. Therefore, we made two Dnm1 mutants: Dnm1 4R in which the first four amino acids, EDQT, were substituted with arginine, and Dnm1 5A in which the first five amino acids, EDQTL, were substituted with alanine. We then examined the interaction of these two Dnm1 mutants with Atg11 by the BiFC assay. Interacting dots were detected with wild-type Dnm1, but almost none were seen with the Dnm1 4R or 5A mutants (Figure 6D and E). Consistent with this result, protein A-tagged Dnm1 4R or 5A were not able to co-precipitate HA-Atg11 by affinity isolation using IgG-Sepharose (Figure 6F). Furthermore, these mutants resulted in a block in mitophagy activity similar to that seen with the complete absence of Dnm1 (Figure S3A).
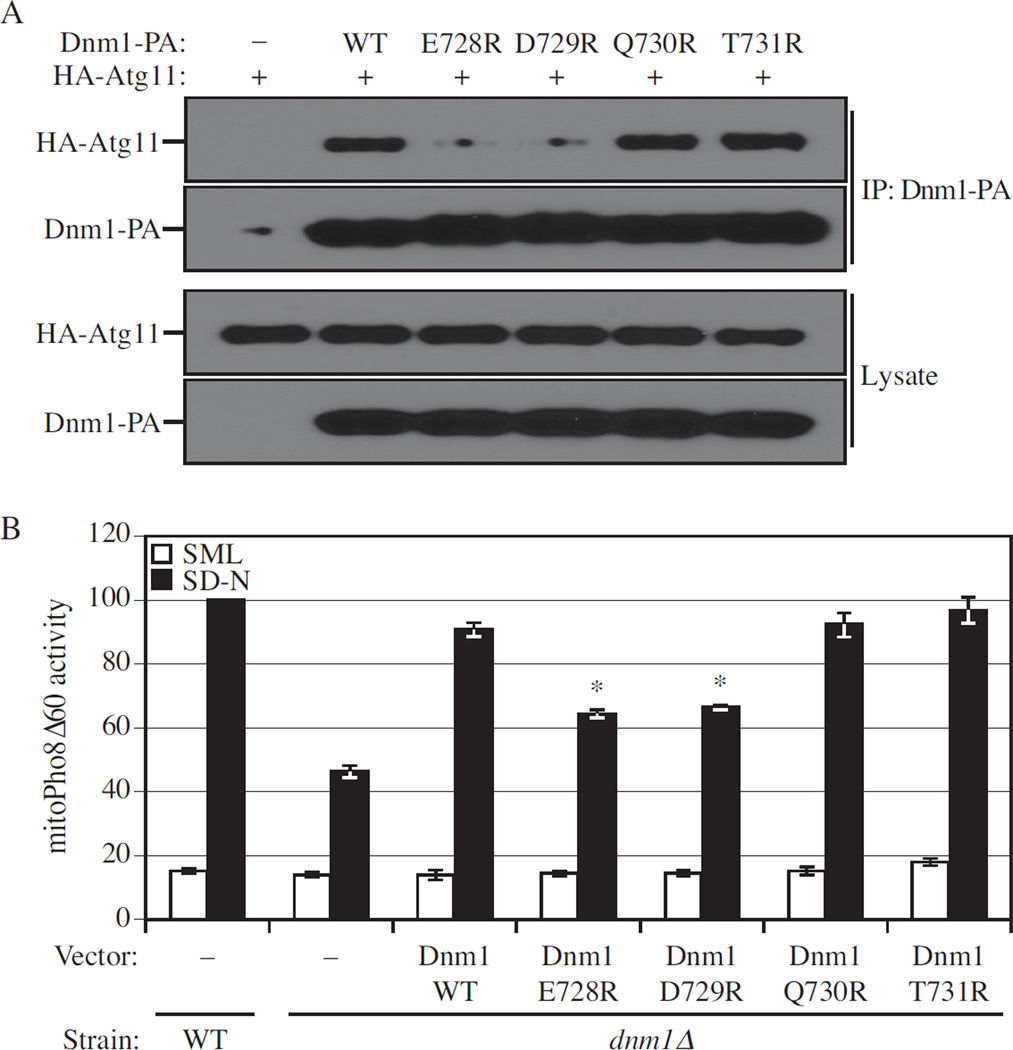
Even though Dnm1 4R or 5A lost the ability to interact with Atg11, we asked whether these two mutants still retained the normal function of Dnm1. Therefore, we used the plasmid-driven Mito-RFP to determine the mitochondrial morphology in the presence of the Dnm1 4R or 5A mutant. Mitochondrial morphology was normal in wild-type cells, whereas enlarged mitochondria were seen in dnm1Δ cells (Figure S3B). The expression of proteinA-tagged wild-type Dnm1 from the plasmid was able to rescue the defect of mitochondrial fission in dnm1Δ cells, whereas neither Dnm1 4R nor 5A could rescue the defect (Figure S3B). We suspected the loss of function of Dnm1 4R or 5A was due to the loss of interaction with Mdv1, Caf4 and/or Fis1. Therefore, we carried out affinity isolation with IgG-Sepharose and found that proteinA-tagged Dnm1 4R or 5A were unable to co-precipitate HA-Mdv1, HA-Caf4, GFP-Fis1 or GFP-Atg32 (Figure S4). Accordingly, we generated additional Dnm1 mutants with single amino acid changes in the interaction domain that we had identified. We found that Dnm1 E728R and D729R, but not Q730R or T731R, had a reduced interaction with Atg11 (Figure 7A), but maintained the normal function of Dnm1 with regard to mitochondrial fission, and interaction with Mdv1, Caf4, and Fis1 (Figure S4A, B, and C). In contrast, Dnm1 E728R and D729R were not able to co-precipitate GFP-Atg32, in contrast to Dnm1 Q730R or T731R, in agreement with a role for Atg11 as a scaffold that bridges the interaction between Dnm1 and Atg32 (Figure S4D).
Figure 7. Dnm1 Mutants that Lose Binding to Atg11 are Mitophagy Defective.
(A) The plasmid pCuHA-Atg11 together with pDnm1-PA, pDnm1(E728R)-PA, pDnm1(D729R)-PA, pDnm1(Q730R)-PA, or pDnm1(T731R)-PA was co-transformed into atg11 Δ dnm1 Δ (KDM1251) cells. Cells were cultured in SML and shifted to SD-N for 1.5 h. Cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation as described in Experimental Procedures. The eluted proteins were separated by SDS-PAGE and detected with monoclonal anti-HA antibody and an antibody that binds to PA.
(B) MitoPho8Δ60 wild-type (KDM1009) cells were transformed with empty vector; mitoPho8Δ60 dnm1 Δ (KDM1014) cells were transformed with empty vector, pDnm1-PA, pDnm1(E728R)-PA, pDnm1(D729R)-PA, pDnm1(Q730R)-PA, or pDnm1(T731R)-PA. Cells were cultured in SML to mid-log phase, then shifted to SD-N for 6 h. The mitoPho8Δ60 assay was performed as described in Experimental Procedures. Error bars correspond to the standard error, and were obtained from three independent repeats. * p<0.01.
Also see Fig. S4.
Finally, we asked whether the Dnm1 mutants that lost binding to Atg11 were competent for mitophagy. Therefore, we determined the mitophagy activity of the dnm1Δ strain containing empty vector, or vector harboring either wild-type Dnm1, or the single amino acid mutants using the mitoPho8Δ60 assay. The presence of wild-type Dnm1 largely suppressed the mitophagy defect in the dnm1Δ mutant strain, and similar results were obtained with the Dnm1 Q730R or T731R mutants. In contrast, the Dnm1 E728R or D729R mutants displayed decreased mitoPho8Δ60 activity (Figure 7B). Based on these results, we suggest that the interaction of Dnm1 and Atg11 is required for efficient mitophagy, although we do not yet know if this interaction is direct or is mediated by another protein(s).
DISCUSSION
Autophagy regulates the degradation of cytoplasmic components and organelles. In nonselective autophagy, the amount of Atg8 controls the size of the autophagosomes (Xie et al., 2008). In contrast, during selective types of autophagy the phagophore membrane is in close apposition to the cargo, excluding bulk cytoplasm. Thus, partly different mechanisms may be involved in determining curvature of the phagophore, and the ultimate size of the autophagosome. In the cytoplasm-to-vacuole targeting (Cvt) pathway, a biosynthetic route that delivers resident hydrolases to the vacuole, the size of the cargo is relatively small; however, when the primary cargo of the Cvt pathway is overexpressed, it forms a larger complex that is no longer efficiently sequestered (Baba et al., 1997), indicating that there is a size limit for Cvt vesicle formation. Similarly, size may be an issue for organelles that are destined for degradation, especially for organelles such as mitochondria, which exist largely as an extended, reticular structure. Therefore, it is possible that large organelles need to be divided into smaller pieces in order to be effectively engulfed by phagophores.
Here, we showed that mitochondrial division is important for mitophagy, which leads to the question of how mitochondrial fission participates in this process. If fission and mitophagy occur independently, we could imagine that mitochondrial fission happens constitutively and only the resulting small mitochondrial fragments would be chosen for degradation. In this case, there would be no requirement for a direct connection between the fission complex and the mitophagy machinery. In contrast, if fission and mitophagy occur in a coordinated manner, the mitochondria destined for degradation should be selected first, and then the fission complex would be recruited to drive the separation of these mitochondria from the mitochondrial reticulum. The direct interaction between Atg11 and Dnm1 supports the second model. Therefore, we propose that when mitophagy is induced, Atg32 recruits Atg11 to the degrading mitochondria. Atg11 in turn brings Dnm1 and other mitochondrial fission proteins to these “marked” mitochondria, and promotes their division. These small fragments of mitochondria are subsequently transported to the PAS where other Atg proteins accumulate, initiating the formation of mitochondria-specific autophagosomes (mitophagosomes).
As a scaffold protein and selective autophagy adaptor, Atg11 binds to a variety of cargo receptors to mediate different types of selective autophagy. For example, Atg11 binds to the Cvt pathway receptor, Atg19, to Atg32 and to Atg36 for cargo selection during the Cvt pathway, mitophagy and pexophagy, respectively (Kanki et al., 2009b; Kim et al., 2001; Motley et al., 2012; Shintani et al., 2002; Yorimitsu and Klionsky, 2005). Atg11 also interacts with Atg1 and Atg17, which connects the step of cargo selection to the initiation of autophagosome formation (Yorimitsu and Klionsky, 2005). Here, we unveil a role for Atg11: recruiting mitochondrial fission machinery to facilitate mitophagy.
Previous results indicated that Atg32 is evenly distributed on the mitochondria, even in mitophagy-inducing conditions (Mao et al., 2011). In contrast, the BiFC Atg32-Atg11 interacting pair displayed a punctate pattern on the mitochondrial tubules. We suggest that these dots represent the degrading mitochondria. However, it remains unknown as to how Atg11 recognizes and binds only the Atg32 that marks the organelles, or segments of the organelles, that are to be degraded. Thus, some other factor(s) might determine this very early event in mitophagy.
The degradation of peroxisomes and chloroplasts through autophagy pathways in Pichia pastoris, S. cerevisiae, Arabidopsis thaliana and other organisms have been reported (Farre et al., 2008; Hutchins et al., 1999; Motley et al., 2012; Wada et al., 2009). It was also reported that the mitochondrial fission complex (Dnm1, Fis1, Mdv1 and Caf4) controls the fission of peroxisomes in yeast, and peroxisomes and chloroplasts in plants (Kuravi et al., 2006; Motley et al., 2008; Zhang and Hu, 2010). Thus, we propose that the mechanism we show in this study (receptor-adaptor-fission complex) may also be relevant in selective pexophagy and chloroplast autophagy.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
See supplementary experimental procedures for the yeast strains used in this paper. Yeast cells were grown in rich (YPD; 1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal (SMD; 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as needed) media. For mitochondria proliferation, cells were grown in lactate medium (YPL; 1% yeast extract, 2% peptone, and 2% lactic acid) or synthetic minimal medium with lactic acid (SML; 0.67% yeast nitrogen base, 2% lactic acid, and auxotrophic amino acids and vitamins as needed). Mitophagy was induced by shifting the cells to nitrogen starvation medium with glucose (SD-N; 0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% glucose).
Fluorescence Microscopy
For fluorescence microscopy, yeast cells were grown to OD600 ~0.6 in YPL or SML media and shifted to SD-N for nitrogen starvation. Samples were then examined by microscopy (Delta Vision, Applied Precision) using a 100x objective and pictures were captured with a CCD camera (CoolSnap HQ; Photometrics). For each microscopy picture, 12 Z-section images were captured with a 0.3-µm distance between two neighboring sections. MitoTracker Red (Invitrogen/Molecular Probes) was used to stain the mitochondria, CellTracker Blue CMAC (Invitrogen/Molecular Probes) to stain the vacuolar lumen, and FM 4–64 (Invitrogen) to stain the vacuolar membrane.
Additional Assays
The mitoPho8Δ60 assay and immunoprecipitation were performed as described previously (Kanki et al., 2009a; Kanki et al., 2009b).
Supplementary Material
HIGHLIGHTS.
The Atg32-Atg11 interaction marks the degrading mitochondria.
Mitochondrial fission is required for efficient mitochondrial degradation.
The fission machinery is recruited to mitochondria by the Atg11 scaffold.
The Atg11-Dnm1 interaction coordinates fission with mitophagy.
ACKNOWLEDGMENTS
This work was supported by grant GM053396 to DJK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, Kang D. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell. 2011;22:3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima NH, Brisch E, Keegan BR, Bleazard W, Shaw JM. The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol. Biol. Cell. 2001;12:2756–2766. doi: 10.1091/mbc.12.9.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitochondrial abnormalities drive cell death in Wolfram syndrome 2. Cell Res. 2009;19:922–923. doi: 10.1038/cr.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen W-L, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell. 2009a;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009b;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karren MA, Coonrod EM, Anderson TK, Shaw JM. The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J. Cell Biol. 2005;171:291–301. doi: 10.1083/jcb.200506158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr., Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Noda NN, Suzuki SW, Nakatogawa H, Takahashi I, Matsunami M, Hashimoto A, Inagaki F, Ohsumi Y, Okamoto K. Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J. Biol. Chem. 2012;287:10631–10638. doi: 10.1074/jbc.M111.299917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae . J. Cell Sci. 2006;119:3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- Labbé S, Thiele DJ. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae . J. Cell Biol. 2011;193:755–767. doi: 10.1083/jcb.201102092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendl N, Occhipinti A, Muller M, Wild P, Dikic I, Reichert AS. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2 . J. Cell Sci. 2011;124:1339–1350. doi: 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- Mijaljica D, Nazarko TY, Brumell JH, Huang WP, Komatsu M, Prescott M, Simonsen A, Yamamoto A, Zhang H, Klionsky DJ, et al. Receptor protein complexes are in control of autophagy. Autophagy. 2012;8:1701–1705. doi: 10.4161/auto.21332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae . EMBO J. 2012;31:2852–2868. doi: 10.1038/emboj.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley AM, Ward GP, Hettema EH. Dnm1p–dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 2008;121:1633–1640. doi: 10.1242/jcs.026344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae . Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009;149:885–893. doi: 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hu J. The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell. 2010;22:431–442. doi: 10.1105/tpc.109.071324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.