Abstract
A major challenge of modern human biology is to understand how a differentiated somatic cell integrates the response to external signals in the complex context of basic metabolic and tissue-specific gene expression programs. This requires exploring two interconnected basic processes: the signaling network and the global function of the key transcription factors on which signaling acts to modulate gene expression. An apparently simple model to study these questions has been steroid hormones action, since their intracellular receptors both initiate signaling and are the key transcription factors orchestrating the cellular response. We have used progesterone action in breast cancer cells to elucidate the intricacies of progesterone receptor (PR) signaling crosstalk with protein kinases, histone modifying enzymes and ATP-dependent chromatin remodeling complexes.1 Recently we have described the cistrome of PR in these cells at different times after addition of hormone and its relationship to chromatin structure.2 The role of chromatin in transcription factor binding to the genome is still debated, but the dominant view is that factors bind preferentially to nucleosome-depleted regions, usually identified as DNaseI-hypersensitive sites (DHS). In contrast with this vision, we have shown that PR requires nucleosomes for optimal binding and function. In breast cancer cells treated with progestins we identified 25,000 PR binding sites (PRbs), the majority encompassing several copies of the hexanucleotide TGTYCY, highly abundant in the genome. We found that strong functional PRbs accumulate around progesterone-induced genes mainly in enhancers, are enriched in DHS but exhibit high nucleosome occupancy. Progestin stimulation results in remodeling of these nucleosomes with displacement of histones H1 and H2A/H2B dimers. Our results strongly suggest that nucleosomes play crucial role in PR binding and hormonal gene regulation.
Keywords: chromatin, progesterone receptor, nucleosome, breast cancer, gene regulation, DNase hypersensitivity, chromatin remodeling, steroid hormone
Introduction
Gene expression in a differentiated eukaryotic cell type is determined by its specific set of transcription factors and by the particular organization of its genome in chromatin that are both establish during development. Understanding how sequence-specific transcription factors control gene regulatory networks requires knowledge of their intracellular interactions and in particular of their genomic binding sites, which are influenced by the cell-specific chromatin structure. But how the nucleosomal organization and the higher order chromatin structure of these genomic regions influences access of transcription factors to its target DNA sequences is still a matter of debate.
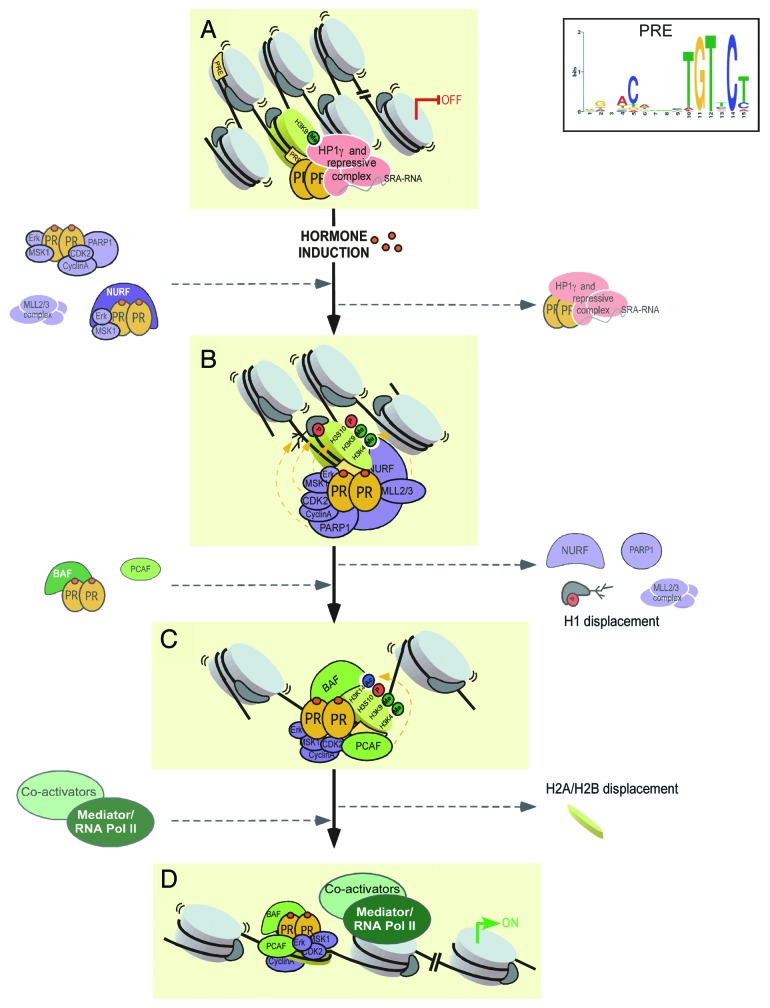
Steroid hormones exert their effects by binding to intracellular receptors, which regulate gene expression mainly by interacting with specific DNA sequences in chromatin and recruiting chromatin remodeling complexes and transcriptional co-regulators. In addition, hormone receptors can signal via crosstalk with kinase signaling. In breast cancer cells a small fraction of the progesterone and estrogen receptors (PR and ER, respectively) is attached to the cytoplasmic side of the cell membrane via a palmitoylated residues.3,4 When activated upon hormone binding these membrane receptors interact with members of the c-SRC family of protein tyrosine kinases leading to activation of various downstream kinase cascades, including ERK1/2.5 We have previously shown that progesterone-activated ERK1/2 phosphorylates PR and the downstream kinase MSK1 in the cell nucleus. The resulting ternary complex pPR/pERK/pMSK1 is the active form of the receptor that interacts productively with chromatin in a subset of target genes, where pMSK1 phosphorylates histone H3 at S10.6 This modification contributes to the displacement of a repressive complex containing HP1γ, LSD1, HDACs and the non-coding RNA SRA, which is anchored to the DNA via non-liganded PR (Fig. 1).7 Already one minute after hormone treatment pPR interacts with and activates the CyclinA/CDK2 complex, which is recruited to regulated promoters and phosphorylates histone H1.8 Hormone activated CDK2 also phosphorylates poly (ADP-ribose) polymerase 1 (PARP1), which converts NAD+ to poly(ADP-ribose) (PAR) attached to itself and to linker and core histones, facilitating displacement of histone H1.9 This initial remodeling step requires also NURF, an ATP-dependent chromatin remodeling complex that is recruited to target sites via interaction with PR and is stabilized by trimethylation of histone H3 at lysine 4 catalyzed by MLL2/3 of the ASCOM complex.8 In a subsequent remodeling cycle PR recruits the BAF complex that is stabilized by PCAF-mediated acetylation of histone H3 at K14 and catalyzes displacement of histones H2A/H2B dimers (Fig. 1).10 We have previously demonstrated that local displacement of histones H1 and H2A/H2B are needed for optimal induction of the MMTV promoter and other progestin-target genes. Thus, crosstalk of PR with kinase signaling pathways, histone modifying enzymes and ATP-dependent complexes impinges on chromatin that is remodeled in two consecutive cycles as a requisite for gene regulation.1
Figure 1. Model for PR binding to PREs organized in nucleosomes. (A) Functional PREs are exposed in well-positioned nucleosomes. A subset of these PREs is occupied by unliganded PR associated to a repressive complex containing HP1γ and SRA-RNA.7 On poorly positioned nucleosomes (upper left corner), PREs are not efficiently recognized by PR. (B) Upon hormone binding, the activated PR reaches these PREs in association with kinases, histone tails modifiers and NURF that stabilize PR binding via contact with histones and initiate chromatin remodeling (H1 displacement).8,9(C) In a subsequent step BAF catalyzes histone H2A/H2B displacement.10(D) After chromatin remodeling, PR interacts with other TFs, co-regulators and components of the transcriptional machinery, to promote formation of the transcription initiation complex.24
But, how the initial chromatin organization of potential PR binding sites affects the access of activated PR to the hormone responsive element (HRE) is not clear. The generally accepted view is that hormone receptors bind preferentially to HREs located in nucleosome-depleted “open” chromatin regions that are marked by so-called pioneer factors, such as FOXA1 and AP1.11 We wanted to test whether this assumption holds for PR. To this end we determined genome-wide the PR binding sites at different times after progestin treatment and performed global gene expression analysis, DNaseI sensitivity assays and nucleosome mapping experiments in breast cancer cells. Intriguingly, we found that prior to hormone addition functional HREs are enriched in stable well-positioned nucleosomes, which are remodeled but not evicted upon hormone induction. Our results strongly suggest that the nucleosomal organization of HREs is required for optimal PR binding and hormonal induction.2
PR binding Sites are Marked by High Nucleosomes Occupancy
Chromatin immunoprecipitation and next generation sequencing (ChIP-seq) with an antibody to PR, identifies 25,876 PR binding sites (PRbs) in T47D-MTVL breast cancer cells12 treated for 30 min with the synthetic progestin R5020 (Pg). Whole genome expression microarrays with cells stimulated for different times with Pg reveals around 4,000 hormone target genes, half of them upregulated and the other half downregulated.2 The nature of the regulated genes is concordant with previous reports demonstrating a proliferative burst induced by progesterone in breast cancer cells.13 Comparison of gene expression results with PR occupancy in response to hormone shows a significant enrichment of PRbs around Pg upregulated genes, which correlates with the magnitude of hormone response. Sequence analysis of the 10% most significant PRbs defines a new consensus Progesterone Responsive Element (PRE) motif (Fig. 1, right upper panel), present in up to 89% of all PRbs. However, there are 8.17 million such PRE matches in the human haploid genome, indicating that a PRE sequence is not sufficient to recruit PR. Although cooperation with other transcription factors, such as STAT, FKHD, AP1 and SP1, may be required for significant PR binding, it is also possible that the chromatin structure limits the fraction of accessible PREs. Indeed, the PRbs show an enrichment of binding sites for members of several TF families, but none of them was significant enough to explain the large discrepancy between potential PRbs and actually bound sites.2
Previous reports have shown that receptors for other steroid hormone bind preferentially to sites within regions of “open” chromatin, detected as DNaseI hypersensitive sites (DHS),14 which are usually represented as depleted of nucleosome. We used DNaseI-sequencing (DNaseI-Seq) to determine genome wide the location of DHS in T47D-MTVL cells before and after hormone treatment.2 A large proportion of the genomic regions that bind PR in presence of progestin already exhibit a high sensitivity to DNaseI before hormone stimulation, which increases after progestin treatment. We used MNase digestion and deep sequencing to test whether these sites are indeed depleted of nucleosomes. We found high nucleosome occupancy before hormone treatment over the regions where PR will bind after hormone addition. Thus, in contrast to current views high sensitivity to DNaseI does not necessary reflect nucleosome depletion and PR binding seems to be favored on sequences well organized in nucleosomes. The high level of nucleosome occupancy over a narrow region where PR will bind indicates that the nucleosomes containing functional PREs are well positioned compared with flanking nucleosomes.
Nucleosomes Containing PRbs are Remodeled upon Hormone Stimulation and are Associated with Hormone Regulation
One hour after progestin stimulation a significant decrease in nucleosome occupancy takes place over PRbs, suggesting nucleosome displacement or remodeling. To measure the magnitude of this change we calculated the nucleosome-remodeling index (NRI = ratio between the nucleosome occupancy before and after hormone administration) and ranked all PRbs according to their NRI. PRbs with higher NRI exhibit higher density of well-positioned nucleosomes before hormone, with a dramatic decrease in signal upon Pg stimulation.2 Also, the PRbs with high nucleosome density exhibit high sensitivity to DNaseI before hormone addition and become even more sensitive after stimulation. In contrast, low NRI PRbs show low nucleosome occupancy, reflecting poor enrichment in positioned nucleosomes. These PRbs are also less sensitive to DNaseI before and after hormone treatment.2 Therefore, high DNaseI sensitivity correlates with high occupancy of stable nucleosomes, in contrast with the generally accepted occluding function of nucleosomes.
Analysis of gene expression revealed that the top NRI PRbs accumulate in the vicinity of progestin-regulated genes and are preferentially associated with enhancers and promoters as defined by epigenetic marks. Together, our results suggest a functional role of high NRI PRbs in progestin-mediated gene regulation.
Chromatin Remodeling at PRbs is Accompanied by Displacement of Histones H1 and H2A/H2B Dimers
We have previously reported that the initial chromatin remodeling on the MMTV promoter in response to hormone leads to displacement of linker histone H1 catalyzed by the chromatin remodeling complex NURF, which is recruited to hormone target genes by the activated PR.8 ChIP-seq experiments show that hormone treatment causes genome-wide enrichment of BPTF (the large subunit of NURF) and displacement of histone H1 over PRbs with high NRI but no over low NRI PRbs.2
Following H1 displacement, hormone treatment leads to recruitment of the ATP-dependent remodeling complex BAF, which catalyzes displacement of core histones H2A and H2B from the MMTV promoter.8,10 Consistently we find a significant recruitment of the Brg1 subunit of BAF as well as H2A/H2B displacement in PRbs with high NRI but not in low NRI PRbs. Eviction of H1 and/or H2A/H2B dimers destabilizes the nucleosome particle leading to internal cleavage of nucleosomal DNA by MNase and consequent reduction of nucleosomal DNA fragments.
Taken together our results support the hypothesis that stable well-positioned nucleosomes over PREs are necessary for efficient and sustained PR recruitment, as required to trigger chromatin remodeling and gene regulation. Chromatin remodeling involves displacement of histones H1 and of H2A and H2B dimers, leaving behind the DNA wrapped around a histone H3/H4 tetramer, which is an optimal template for hormonal gene regulation.15 Thus, we postulate a positive role of nucleosomes both in PR recognition of functional PREs and stabilization of PR binding, as well as in chromatin remodeling and hormonal gene induction.
Discussion
The large discrepancy between the number of potential PR binding sequences (millions) and the number of actually detected PRbs (25,000) cannot be explained by the need for cooperation of PR with other transcription factors (TF), but is likely due to the chromatin organization of the PREs. However, while estrogen receptor cannot bind to its genomic targets unless they are first “open” by the pioneer factor FoxA1,16-18 PR can bind to the PREs on the MMTV promoter assembled in reconstituted nucleosomes, as well as in minichromosomes, integrated in the genome of the yeast Saccharomyces cerevisiae or in chromosomes of breast cancer cells in culture.12,19-21 We believe that efficient PR binding is facilitated by well-positioned stable nucleosomes that expose a PRE in their surface for a sufficiently long period of time to be recognized by the receptor. In contrast, PREs covered by poorly positioned nucleosomes are accessible only in a tiny fraction of the cells and for brief periods of time, reducing the probability of efficient PR binding (Fig. 1).
It is assumed that the open chromatin regions, pre-established by pioneer factors, are depleted of nucleosomes.11 However, our results show that prior to hormone treatment strong functional PRbs exhibit high nucleosome occupancy and partly overlap with pre-established DHS, clearly showing that high nucleosome occupancy can occur in DNase I accessible regions. The DNase I sensitivity may be due to capacity of non-liganded PR to bind to a subset of PREs in a complex with HP1γ, LSD1, HDACs and the non-coding RNA SRA (Fig. 1, top panel).7 At many other regions, PR binding to nucleosomal PREs upon hormone treatment occurs de novo and causes the appearance of novel DHS, confirming that PR can initiate chromatin binding and remodeling. Therefore, PR qualifies as a bona fide “pioneer factor.”
We have previously shown that at the MMTV promoter a well-positioned nucleosome not only does not interfere with PR binding but is also essential for proper hormonal induction.20,21 Based on our recent results, we conclude that a similar situation applies genome-wide. Nucleosomes containing functional PRbs are required for optimal hormonal induction and become more sensitive to cleavage by DNaseI and particularly by MNase after hormone treatment. Remodeling is at least in part mediated by the ATP-dependent complexes NURF and BAF, which remove histones H1 and histone H2A/H2B dimers, respectively. Thus, mechanisms of remodeling previously observed in the MMTV promoter6,8,10,22 are representative of a genome-wide behavior. We hypothesize that removal of histone H1 may be the main mechanism for initiating cooperative decondensation of the chromatin fiber, though we cannot exclude that different histone H1 isoforms are also involved and the extent of histones or DNA methylation play also a role.
What could be the mechanism by which well-positioned nucleosomes favor the function of PREs? We know that PR binds PREs contacting only one side of the DNA double helix and with relatively low affinity compared with other transcription factors that fully embrace the DNA, like NF1.23 It is possible that PR binding to the DNA of exposed PREs in the surface of well-positioned nucleosomes is not sufficiently stable to orchestrate transcriptional regulation. On the other side, activated PR reaches the target PREs in association with kinases and chromatin remodeling complexes that contact the core and linker histones.1,24 We know that CDK2 and PARP1 interact with histone H1, MSK1 with H3S10, MLL2/3 and NURF with H3K4me3, PCAF and BAF with H3K14 (Table 1). Therefore, one possibility is that binding of PR to PREs organized in stable nucleosomes is stabilized by interaction of associated factors with histone tails, thus increasing the residence time on chromatin and favoring remodeling and recruitment of co-activators. After chromatin remodeling cooperation between different PR molecules bound to adjacent or more distant PREs, or between PR and other TFs, could be facilitated in the surface of a H3/H4 tetramer particle.15 This may be the reason for the frequent occurrence of clustered PREs and for the preferential association of PREs with binding sites for other sequence-specific TFs in functional hormone regulatory regions.
Table 1. PR-associated chromatin-interacting factors.
| PR complexes | Modify | Recognize |
|---|---|---|
| HP1γ |
- |
H3K9me3 |
| ERK/MSK1 |
H3S10phos |
- |
| CDK2/CyclinA/PARP1 |
H1phos/H1parylation |
- |
| MLL2/3 |
H3K4me3 |
- |
| NURF |
- |
H3K4me3 |
| PCAF |
H3K14ac |
- |
| BAF | - | H3K14ac |
Finally, it is also possible that nucleosomes fulfill an unspecific repressor function in the absence of hormone by making regulatory information unavailable for binding of other TFs binding or by offering histone marks for anchoring of repressive complexes recruited by the unliganded hormone receptor.7 The unspecific repressive function of nucleosomes will make these promoter/enhancers dependent on hormone-activated PR, which act as a pioneer factor recognizing target sequences on nucleosomes. Binding of PR with associated remodeling complexes and histone-modifying enzymes will overcome nucleosome repression and modulate binding of either other TFs/co-activators to mediate activation or co-repressors to mediate repression. Thus, in contrast with the currently established view, nucleosomes may play a double role, functioning as repressor for general TFs and as facilitators for PR binding and function, which requires ATP-dependent nucleosome remodeling to initiate the opening of chromatin needed for gene regulation.
Acknowledgments
The experimental work summarized in this review was supported by grants from the Spanish government (BMC 2003–02902 and 2010–15313; CSD2006–00049), the European Union (IP HEROIC) and the Catalan government (AGAUR).
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/25108
References
- 1.Beato M, Vicent GP. Impact of chromatin structure and dynamics on PR signaling. The initial steps in hormonal gene regulation. Mol Cell Endocrinol. 2012;357:37–42. doi: 10.1016/j.mce.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Ballaré C, Castellano G, Gaveglia L, Althammer S, González-Vallinas J, Eyras E, et al. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell. 2013;49:67–79. doi: 10.1016/j.molcel.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 3.Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, Levin ER. A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem. 2007;282:22278–88. doi: 10.1074/jbc.M611877200. [DOI] [PubMed] [Google Scholar]
- 4.Ballaré C, Uhrig M, Bechtold T, Sancho E, Di Domenico M, Migliaccio A, et al. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol Cell Biol. 2003;23:1994–2008. doi: 10.1128/MCB.23.6.1994-2008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Migliaccio A, Piccolo D, Castoria G, Di Domenico M, Bilancio A, Lombardi M, et al. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. EMBO J. 1998;17:2008–18. doi: 10.1093/emboj/17.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vicent GP, Ballaré C, Nacht AS, Clausell J, Subtil-Rodríguez A, Quiles I, et al. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell. 2006;24:367–81. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Vicent GP, Nacht AS, Zaurin R, Font-Mateu J, Soronellas D, Le Dily F, et al. Unliganded progesterone receptor-mediated targeting of an RNA-containing repressive complex silences a subset of hormone-inducible genes. Genes Dev. 2013;27:1179–97. doi: 10.1101/gad.215293.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicent GP, Nacht AS, Font-Mateu J, Castellano G, Gaveglia L, Ballaré C, et al. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 2011;25:845–62. doi: 10.1101/gad.621811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright RH, Castellano G, Bonet J, Le Dily F, Font-Mateu J, Ballaré C, et al. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012;26:1972–83. doi: 10.1101/gad.193193.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vicent GP, Zaurin R, Nacht AS, Li A, Font-Mateu J, Le Dily F, et al. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet. 2009;5:e1000567. doi: 10.1371/journal.pgen.1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truss M, Bartsch J, Schelbert A, Haché RJ, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995;14:1737–51. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groshong SD, Owen GI, Grimison B, Schauer IE, Todd MC, Langan TA, et al. Biphasic regulation of breast cancer cell growth by progesterone: role of the cyclin-dependent kinase inhibitors, p21 and p27(Kip1) Mol Endocrinol. 1997;11:1593–607. doi: 10.1210/me.11.11.1593. [DOI] [PubMed] [Google Scholar]
- 14.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–24. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Vicent GP, Zaurin R, Nacht AS, Font-Mateu J, Le Dily F, Beato M. Nuclear factor 1 synergizes with progesterone receptor on the mouse mammary tumor virus promoter wrapped around a histone H3/H4 tetramer by facilitating access to the central hormone-responsive elements. J Biol Chem. 2010;285:2622–31. doi: 10.1074/jbc.M109.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piña B, Brüggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–31. doi: 10.1016/0092-8674(90)90087-U. [DOI] [PubMed] [Google Scholar]
- 20.Di Croce L, Koop R, Venditti P, Westphal HM, Nightingale KP, Corona DF, et al. Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol Cell. 1999;4:45–54. doi: 10.1016/S1097-2765(00)80186-0. [DOI] [PubMed] [Google Scholar]
- 21.Chávez S, Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci U S A. 1997;94:2885–90. doi: 10.1073/pnas.94.7.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vicent GP, Nacht AS, Smith CL, Peterson CL, Dimitrov S, Beato M. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol Cell. 2004;16:439–52. doi: 10.1016/j.molcel.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Eisfeld K, Candau R, Truss M, Beato M. Binding of NF1 to the MMTV promoter in nucleosomes: influence of rotational phasing, translational positioning and histone H1. Nucleic Acids Res. 1997;25:3733–42. doi: 10.1093/nar/25.18.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beato M, Vicent GP. When every minute counts: the enzymatic complexity associated with the activation of hormone-dependent genes. Cell Cycle. 2011;10:2407–9. doi: 10.4161/cc.10.15.16200. [DOI] [PubMed] [Google Scholar]