Abstract
Linkers of the nucleoskeleton to the cytoskeleton (LINC) complexes formed by SUN and KASH proteins are conserved eukaryotic protein complexes that bridge the nuclear envelope (NE) via protein-protein interactions in the NE lumen. Revealed by opisthokont studies, LINC complexes are key players in multiple cellular processes, such as nuclear and chromosomal positioning and nuclear shape determination, which in turn influence the generation of gametes and several aspects of development. Although comparable processes have long been known in plants, the first plant nuclear envelope bridging complexes were only recently identified. WPP domain-interacting proteins at the outer NE have little homology to known opisthokont KASH proteins, but form complexes with SUN proteins at the inner NE that have plant-specific properties and functions. In this review, we will address the importance of LINC complex-regulated processes, describe the plant NE bridging complexes and compare them to opisthokont LINC complexes.
Keywords: nuclear envelope, LINC complex, SUN, KASH, plant, Arabidopsis
The nuclear envelope (NE) is a double membrane system consisting of an inner nuclear membrane (INM) and an outer nuclear membrane (ONM). Studies in opisthokonts revealed that the two membranes are bridged by protein complexes formed by the INM Sad1/UNC-84 (SUN) proteins and the ONM Klarsicht/ANC-1/Syne homology (KASH) proteins.1-3 SUN-KASH complexes have been found from yeast to humans, and they serve as linkers of the nucleoskeleton or chromosomes to the cytoskeleton (LINC) complexes.4-7 LINC complexes regulate nuclear shape, rigidity and position of the nucleus as well as chromatin organization. The recent identification of a plant NE-bridging complex showed that it has plant-specific properties and functions.8 Here we will briefly review opisthokont LINC complexes and then focus on novel insights on plant NE-bridging proteins.
Opisthokont SUN and KASH Proteins
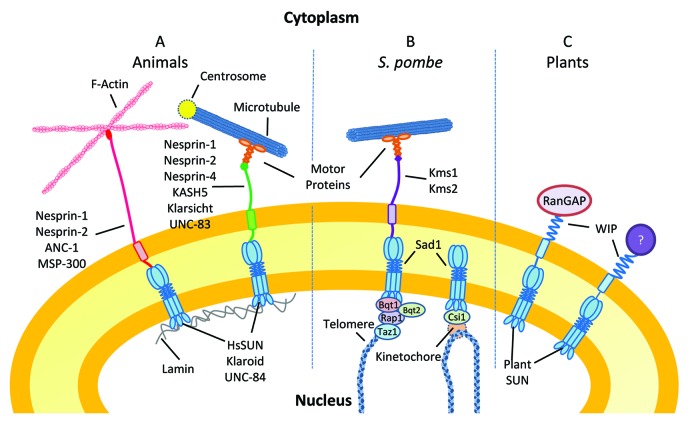
The founding member of SUN proteins is Caenorhabditis elegans UNC-84. The C-terminus of UNC-84 has a domain sharing homology with two human proteins and Schizosaccharomyces pombe Sad1, a component of the spindle pole body.9 This domain was subsequently found to interact with a C-terminal domain conserved in a series of ONM proteins including Klarsicht, ANC-1, and Syne (also known as Nesprin).10 Therefore, the two interacting domains were named Sad1/UNC-84 (SUN) domain9 and Klarsicht/ANC-1/Syne homology (KASH) domain,10 respectively. SUN proteins typically locate at the INM with their C-terminal SUN domain positioned in the perinuclear space (PNS) (Fig. One and see below). KASH proteins are tail-anchored proteins at the ONM harboring a C-terminal KASH domain, which contains a short PNS tail (~30 amino acids) typically ending with a PPPX (X represents any amino acid) motif that is essential for interacting with the SUN domain. The SUN-KASH complexes bridge their binding partners across the NE (Fig. 1). In the nucleoplasm, the N-termini of many SUN proteins interact directly or indirectly with nuclear lamins, which are intermediate filament proteins located underneath the INM and are considered components of the nucleoskeleton.11 At the cytoplasmic side, KASH proteins are linked to motor proteins, intermediate filaments, microtubules, or F-actin.4-7,12
Figure 1. Nuclear envelope bridging complexes in different organisms. Since SUN domains are relatively conserved across species, all SUN proteins are drawn as trimers according to the evidence provided for HsSUN2.39,40 For simplicity, all KASH proteins are drawn as monomers. See text for details.
Mammalian SUN1 and SUN2 interact with Lamin A, and the NE localization of SUN2 depends on Lamin A.13,14 SUN1 and SUN2 interact with the mammalian KASH proteins Nesprin-1 and Nesprin-2 at the NE.4,15,16 These two KASH proteins consist of N-terminal F-actin-binding calponin homology domains, a long stalk domain composed of spectrin repeats, and a C-terminal KASH domain.4,16 The spectrin repeats each assemble into a three-helix bundle, which makes the protein flexible in length and may assist in buffering against mechanical stress.17 Nesprin-1 and -2 interact with F-actin and connect the INM lamins through SUN1 and SUN2 (Fig. 1A).4,18 These LINC complexes are responsible for anchoring the synaptic nuclei at the mouse neuromuscular junction.18 Nesprin-1 and -2 also link the centrosome to the nucleus through interactions with the dynein/dynactin complex (Fig. 1A).19 This connection is essential for interkinetic nuclear migration and nucleokinesis in mice.19
In Drosophila, the localization of Klarsicht at the nuclear periphery depends on a type B lamin and a SUN protein, Klaroid.20 Klaroid also forms nuclear aggregates in transgenic flies expressing a mutated Lamin C that lacks the first 42 amino acids,21 suggesting that Klaroid might be associated with Lamin C. Klarsicht is connected to microtubules and is responsible for the apical nuclear migration during photoreceptor formation.20,22 A similar nuclear migration process in mouse cone photoreceptor development is regulated by SUN1, but the responsible KASH proteins and the involved elements of the cytoskeleton are unknown.23 The localization at the nuclear periphery of another KASH protein, MSP-300 also depends on Klaroid.24 MSP-300 is an ortholog of mammalian Nesprin-1 and -2. Although its role in nuclear anchorage was previously unclear,24-26 a recent study revealed that MSP-300 interacts with D-Titin/Sallimus and anchors mitochondria and endoplasmic reticulum (ER) to the striated muscle Z-discs.27 It also cooperatively functions with Klarsicht to promote even nuclear spacing in striated muscle.27 In C. elegans, UNC-84 co-localizes with Ce-lamin at the NE and its localization depends on Ce-lamin.28 UNC-84 recruits the KASH proteins UNC-83 and ANC-1 to the NE (Fig. 1A).10,29 UNC-83 in turn targets kinesin-1 and/or dynein to the nuclear periphery, and the force provided by the motor proteins drives the nuclear migration in C. elegans hypodermal P cells and embryonic hypodermal cells.30-32 ANC-1 interacts with F-actin and is required for nuclear anchorage in the adult C. elegans syncytial hypodermis.10
SUN-KASH complexes can also link chromosomes to the cytoskeleton. In Schizosaccharomyces pombe, centromeres are tethered to the SUN protein Sad1 through Csi1 (Fig. 1B).33 Loss of Sad1 or Csi1 leads to high-frequency centromere clustering defects.33 At meiotic prophase, telomeres are tethered to Sad1 through Bqt1, Bqt2 and telomere-associated proteins Taz1 and Rap1.6 Telomeres are further linked to dynein motors by the KASH protein Kms1 and Kms2 (Fig. 1B).34-37 This results in telomere clustering and later in nuclear oscillation between the cell poles, which facilitates homologous paring and recombination.38 In mammals, a recent study revealed the formation of a similar chromosomal bouquet mediated by a SUN1-KASH5 complex.7Table 1 provides an overview of known SUN-KASH pairs from different organisms and their known or proposed functions.
Table 1. Known SUN-KASH pairs from different organisms, their cytoplasmic partners, and their known or proposed functions.
| SUN |
KASH |
Cytoplasmic partner: function Reference |
|---|---|---|
| Mammalian | ||
| SUN1/2 |
Nesprin-1 |
F-actin: anchoring the synaptic nuclei under the mouse neuromuscular junction.18 Dynein/dynactin complex: connecting the nucleus to centrosome for interkinetic nuclear migration and nucleokinesis.19 |
| SUN1/2 |
Nesprin-2 |
F-actin: anchoring the synaptic nuclei under the mouse neuromuscular junction.18 Dynein/dynactin complex and kinesin: connecting the nucleus to centrosome for interkinetic nuclear migration and nucleokinesis.19 |
| SUN1/2 |
Nesprin-3 |
Plectin, BPAG1, or MACF: connecting the nucleus to intermediate filaments or microtubules which stabilizes the anchorage of the nucleus and maintains the structure/shape of the nucleus (reviewed in ref. 12). |
| Maybe SUN1/2 |
Nesprin-4 |
Kinesin-1: predicted to promote nuclear migration toward the base of the secretory epithelial cells.82 |
| SUN1/2 |
KASH5 |
Dynein/dynactin complex: telomere movement during meiosis.7 |
| SUN3 |
Nesprin-1 |
Proposed to be kinesin II, dynein/dynactin, or F-actin: probably links the nucleus to posterior manchette during sperm head formation.83 |
| SUN1η |
Nesprin-3 |
Proposed to be plectin: proposed to be a non-NE complex anchoring acrosome to the anterior actin filaments during sperm head formation.83 |
| SPAG4 |
Unknown |
Unknown: Testis-specific, non-NE localized, function unknown.84 |
| SPAG4L/4L-2 | Unknown | Unknown: restricted to the apical nuclear region of round spermatids facing the acrosomic vesicle, and probably involved in linkage of the acrosomic vesicle to the spermatid nucleus and in acrosome biogenesis.85 |
| Drosophila | ||
|---|---|---|
| Klaroid |
MSP-300 |
F-actin: nuclear anchoring during Drosophila oogenesis.26 |
| Unknown |
MSP-300 |
D-Titin/Sallimus: anchoring mitochondria and endoplasmic reticulum to the striated muscle Z-discs.27 Unknown: anchoring microtubules to the NE in striated muscle.27 Unknown: Anchoring the nuclei to myofibril compartment in striated muscle.27 |
| Klaroid |
Klarsicht |
Proposed to be microtubule motors: nuclear migration during eye development.20,22 |
| Unknown |
Klarsicht |
Unknown: anchoring microtubules to the NE in striated muscle.27 Unknown: promoting even myonuclear spacing in both striated muscle and nonstriated myotubes.27 |
| SPAG4/Giacomo | Unknown | Yuri and dynein/dynactin: involved in centriolar-nuclear attachment during spermatogenesis.86 |
| C. elegans | ||
|---|---|---|
| UNC-84 |
ANC-1 |
F-actin: nuclear anchorage in the adult C. elegans syncytial hypodermis.10 |
| UNC-84 |
UNC-83 |
Kinesin-1 and dynein: nuclear migration in embryonic C. elegans hypodermal cells.30,31 |
| SUN-1/matefin |
ZYG-12 |
Dynein and ZYG-12A: linkage between the centrosome and nucleus,87 meiotic chromosome paring and synapsis,88,89 and nuclear positioning within the syncytial gonad.90 |
| SUN-1/matefin | KDP-1 | Unknown: cell-cycle progression.67 |
| S. cerevisiae | ||
|---|---|---|
| Mps3 |
Unknown |
Unknown: Mps3 is involved in spindle pole body insertion into the NE and NE homeostasis91; it interacts with Mps2 to connect the spindle pole body to the NE and functions in spindle pole body duplication.92 |
| Mps3 |
Csm4 |
Probably F-actin: meiotic telomeres are tethered to Mps3 at the NE by Ndj1 and further connected to the cytoskeleton (perhaps actins) by Csm4.93-95 |
| Mps3 | Unknown | Unknown: Mitotic telomeres are tethered to Mps3 at the NE by Sir4 and the telomere clustering is mediated by two Mps3 associated proteins,96 Ebp2 and Rrs1.97 |
| Dictyostelium | ||
|---|---|---|
| SUN-1 |
Unknown |
Unknown: SUN-1 connects the centrosome to chromatin and ensures genome stability98 |
| Unknown | Interaptin | F-actin: Function unknown99 |
The structure of the mammalian SUN-KASH complex was recently resolved.39,40 In this complex, three KASH domains are anchored in one cloverleaf-like trimer of SUN domains. The SUN domain protomer has several functional domains: an α-helical stalk, a compact β-sandwich core, a cation-loop, and a protruding anti-parallel β-sheet named the KASH-lid (Fig. 2). The α-helix serves as an extension of the trimeric coiled-coil domain of SUN proteins, which facilitates the trimerization. The trimer is further stabilized by a large interacting surface on the β-sandwich core and the hydrogen bonds between the α-helix of one protomer to the β-sandwich of the adjacent protomer. The KASH domain is clamped between the KASH lid of one protomer and the β-sandwich core of the adjacent protomer. Without binding the KASH domain, the KASH-lid conformation is rather random.40 The very C-terminal “PPPX” motif is positioned in a KASH pocket formed by S641, Y703, Y707, H628 and the cation-loop (Q593-C601), which explains why these four amino acids are critical for SUN-KASH interactions.40
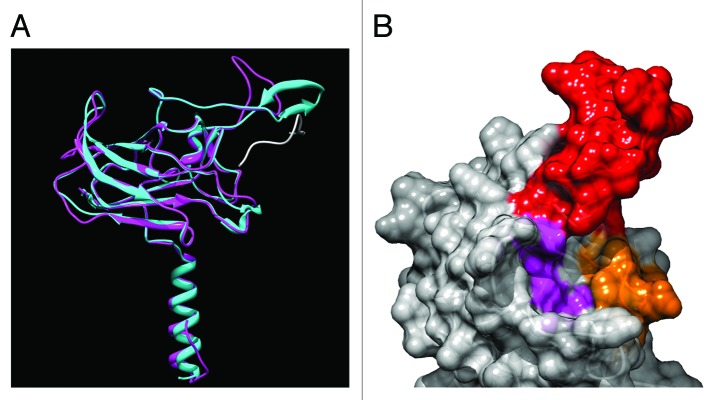
Figure 2. Computed three-dimensional model of the SUN domain of AtSUN1. (A) The S251-D453 fragment of AtSUN1 was modeled using MODELER.79 The SUN domain of HsSUN2 was used as a template (PDB: 4FI9). Three models were computed and the one with the lowest zDOPE score is shown. Magenta, model of the AtSUN1 SUN domain. Cyan, SUN domain of HsSUN2. Gray, Nesprin-2 KASH domain in the HsSUN2-KASH complex. (B) Computed surface of the binding pocket for the KASH C-terminus in AtSUN1. Red, V301-N318 fragment of AtSUN1 corresponding to the KASH lid of HsSUN2 (Y567-S587). Orange, S324-C333 fragment of AtSUN1, corresponding to the cation-loop of HsSUN2 (Q593-C601). Purple, residue H360, S371, H439, and Y443 of AtSUN1, corresponding to H628, S641, Y703, Y707 of HsSUN2, respectively. Images were generated using UCSF Chimera package80 and POV-Ray (http://www.povray.org/).
Nuclear Positioning in Plants
Nuclear positioning events are involved in considerable aspects of the plant life cycle. The most obvious example of nuclear movement is pollen tube growth, during which sperm cells and the vegetative nucleus migrate over long distances toward the tip of the growing pollen tube (for a recent review see ref. 41). In growing root hairs the nucleus is positioned at a relatively fixed distance from the apex. Interrupting this nuclear positioning by depolymerizing F-actin or trapping the nucleus with a laser beam prohibits root hair growth.42,43 Similarly, in Arabidopsis trichomes—branched, single-cell leaf hairs—the nucleus migrates to a position close to the first branch point.44
Asymmetric cell divisions play an important role in plant development. The nucleus has to be positioned at the future division site during this process. During lateral root formation, the nuclei of two neighboring lateral root founder cells migrate toward the common cell boundary. This is followed by asymmetric cell divisions leading to two small adjacent cells and two larger peripheral cells.45,46 During pollen mitosis I, the nucleus in microspores migrates to allow asymmetric division to produce a large vegetative cell and a small generative cell. Arabidopsis microtubule organization 1 (MOR1; also known as GEM1) and its tobacco ortholog tobacco microtubule bundling polypeptide of 200 kDa (TMBP200) are required for this process, and mutations in these proteins lead to defects in pollen production.47-49 Guard cell formation also involves several steps of asymmetric cell division (reviewed in refs. 50 and 51). In monocots, the guard mother cells arise from one asymmetric division and then divide symmetrically to produce guard cell pairs. Subsequently, the adjacent epidermal cells divide asymmetrically to generate subsidiary cells. In dicots, a protodermal cell divides asymmetrically to produce a meristemoid, which is capable of generating either epidermal pavement cells or guard mother cells.
Nuclear movement is also actively involved in plant-microbe interactions. When arbuscular mycorrhizal fungi penetrate host root cells, the host cell nucleus rapidly positions itself beneath the appressorium contact site and promotes cytoskeleton and ER rearrangements.52,53 The nucleus then migrates away from the contact site, accompanied by a formation of a column comprised of cytoskeleton and ER.52,53 This newly formed structure strictly defines the future intracellular path of hyphal penetration and is called the pre-penetration apparatus.52,53 However, in case of a pathogenic fungus, the host cell nucleus stays at the appressorium contact site accompanied by cell wall thickening and papilla formation that blocks the fungal penetration.54
Plant Meiotic Telomere Movement
Similar to mammalian telomere organization, plant telomeres are also attached to the NE during meiosis.55,56 During maize (Zea mays L.) meiotic prophase I, starting at the end of leptotene, telomeres begin to cluster at the nuclear periphery to form a bouquet.57 This process persists through zygotene and ends at early pachytene when telomeres start to disperse at the nuclear periphery.57 In Arabidopsis, telomeres move to the nuclear periphery and form a loose cluster, perhaps a temporary bouquet.58
Despite the importance of nuclear positioning and chromosome movement during the plant life cycle, little is known about the molecular players involved. The recent identification of NE-bridging complexes in plants has now provided tools to begin investigating the underlying molecular mechanisms.
Plant SUN Proteins
Two types of SUN protein have been identified in plant genomes: the canonical C-terminal SUN domain (CCSD) type and the plant-prevalent mid-SUN 3 transmembrane (PM3) type.59,60 The CCSD type has a SUN domain at the C-terminus, while the signature of the PM3 type is a centrally positioned SUN domain followed by a highly conserved domain of unknown function.60 The PM3 type was first discovered in maize, but is also present in other plant species, as well as in opisthokonts.60 PM3-type SUN proteins have not yet been functionally investigated. To date, all well-studied SUN proteins belong to the CCSD type, and therefore, only plant CCSD-type SUN proteins will be discussed in detail here.
The Arabidopsis genome encodes two CCSD-type SUN proteins—AtSUN1 and AtSUN2.59,61 They share a similar domain organization with non-plant SUN proteins: an N-terminal domain with a nuclear localization signal (NLS), a transmembrane domain, a coiled-coil domain, and a SUN domain. Deleting the N-terminal domain, the NLS, or the coiled-coil domain of AtSUN1 or AtSUN2 affects their NE localization, suggesting that these domains are involved in NE targeting and integration. Both genes are expressed ubiquitously in various tissues, including roots, hypocotyl, cotyledons, and leaves. As analyzed by fluorescence resonance energy transfer assays, AtSUN1 and AtSUN2 can form homo- or hetero-complexes,59 perhaps similar to the mammalian SUN protein trimers.
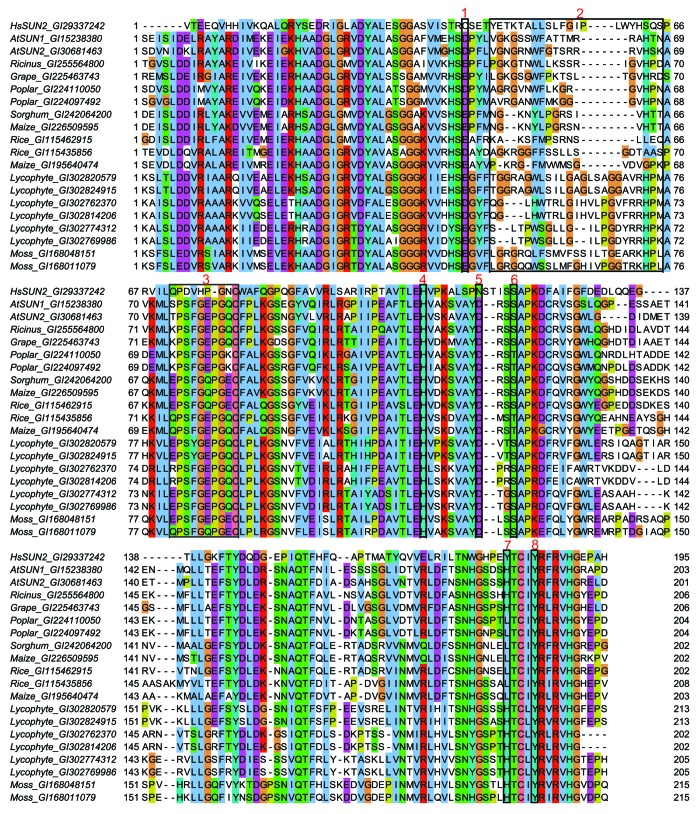
This is supported by the predicted 3D structure of the AtSUN1 SUN domain derived from comparative modeling with the SUN domain of Homo sapiens SUN2 (HsSUN2) as a template (Fig. 2A). According to the HsSUN2 model, the SUN2 trimer harbors three binding site for the KASH domain, such that the SUN-KASH complex is a hexamer.40 AtSUN1 likely harbors the same binding configuration, because the essential structures and amino acids involved in KASH binding are mostly conserved (Fig. 2B and Fig. 3): (1) the KASH-binding pocket is very well-conserved; (2) the cation loop is conserved, especially within plant SUN domains; (3) although the residues of the KASH lid are not conserved, a corresponding fragment is present in all plant SUN domains (Fig. 3). In contrast, the residues that are dispensable for SUN-KASH interaction—residue C563 of HsSUN2 that forms a disulfide bond with a cysteine in the KASH domain and residue N636 of HsSUN2, the N-glycosylation site—are not conserved in plant SUN domains (Fig. 3).
Figure 3. Amino acid sequence alignment of plant SUN domains with the SUN domain of HsSUN2. Alignment was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default settings, except that the output sequences were kept as input order. Image was generated using JalView81 and ClustalX color. GI, NCBI GenInfo identifier. Black frames in the alignment are numbered at the top. Frame 1 indicates C563 in HsSUN2, which forms a disulfide bond with Homo sapiens Nesprin-2 C6862. This disulfide bond is dispensable for SUN-KASH interaction, and in plant SUN domains this position is instead a conserved D or E. Frame 2 indicates the KASH-lid in HsSUN2, however, the sequences have low similarity between HsSUN2 and plant SUN proteins. Frame 3 represents the cation loop in HsSUN2. This cation loop and residues indicated by frame 4, 6, 7, 8 (correspond to H628, S641, Y703, and Y707 of HsSUN2, respectively) form the pocket holding the KASH C-terminus and are well conserved in plant SUN domains. Frame 5 represents N636 of HsSUN2, the N-glycosylation site. N-glycosylation of HsSUN2 is dispensable for KASH binding, and this position is a conserved D in plant SUN domains.
Plant ONM SUN-Binding Proteins
Although the sequences and the KASH binding features are conserved between mammalian and plant SUN domains, plant genomes do not encode any homologs of opisthokont KASH proteins.
WPP domain-interacting proteins (WIPs) are plant tail-anchored ONM proteins that contain a cytoplasmic coiled-coil domain, a transmembrane domain, and a PNS tail terminated in a conserved “VPT” motif. WIPs are plant-specific proteins and Arabidopsis has three homologs—AtWIP1, AtWIP2, and AtWIP3. They are redundantly involved in anchoring the Ran GTPase activating protein (RanGAP) to the NE. This anchorage is achieved by the interaction between the N-terminal plant-specific WPP domain of RanGAP and the coiled-coil domain of WIPs. A combination of immunofluorescence and immunogold-labeling experiments showed that WIPs are associated with the ONM.62,63
A combination of co-immunoprecipitation and fluorescence recovery after photobleaching experiments demonstrated that AtWIP1, AtWIP2, and AtWIP3 interact with AtSUN1 and AtSUN2 at the plant NE.8 The PNS tail of AtWIP1, especially the “VPT” motif, is required for the interaction, similar to the interactions between mammalian Nesprins and SUNs.4,16,64 Surprisingly, the PNS tail of WIP1 is only 9 amino acids long. The PNS tail of Homo sapiens Nesprin-2 is 30 amino acids long, and its C-terminal fragment of 14 amino acids is the shortest one able to bind the SUN domain.40 The ability of Arabidopsis SUN proteins to bind a very short PNS tail might be connected to the presence of a stretch of additional conserved residues in plant SUN domains,8 however, further work is required to understand the exact biochemical nature of the unusual plant NE-bridging complex.
Function of the AtSUN-AtWIP Complex in RanGAP NE Anchoring
The NE localization of AtWIP1 is reduced in a sun1-knockout sun2-knockdown (sun1-KO sun2-KD) mutant, suggesting that the localization of AtWIPs depends on AtSUNs, analogous to the animal KASH proteins.8 Consistent with these findings, the NE localization of AtRanGAP1 is also reduced in undifferentiated root cells of the sun1-KO sun2-KD mutant, indicating that plant SUN proteins play a role in RanGAP-NE association by forming a RanGAP-WIP-SUN complex. The existence of this complex is supported by co-immunoprecipitation data.8
AtRanGAP1 is currently the only confirmed cytoplasmic partner of an NE-bridging complex that does not appear to be associated with elements of the cytoskeleton. RanGAP is the GTPase activating protein for the small GTPase Ran. RanGTP hydrolysis is an important step in nucleocytoplasmic trafficking and implied in the release of cargo from export receptors after exiting from the nuclear pore at the cytoplasmic surface.65 Both metazoan and plant RanGAP is associated with the outer NE, but the mechanisms differ. The RanGAP-WIP-SUN complex appears to be specific for plants. Mammalian nuclear pore-associated RanGAP1 is in a complex containing the SUMO E3-ligase nucleoporin RanBP2, SUMOylated RanGAP1 and the E2 SUMO-conjugating enzyme UBC9.66 The finding that plants have recruited an NE-bridging complex to anchor RanGAP to the NE suggests that there may be additional functions for such complexes beyond linking the nucleus and the cytoskeleton. Thus, novel, cytoskeleton-unrelated binding partners might also exist for opisthokont KASH proteins. One such KASH protein candidate might be C. elegans KDP-1, which functions in cell cycle progression from late S to M phase, and for which a cytoplasmic partner is currently unknown.67
Function of the AtSUN-AtWIP Complex in Nuclear Shape Determination
It is possible that WIPs additionally interact with motor proteins or the cytoskeleton. However, neither the wip1–1 wip2–1 wip3–1 nor the sun1-KO sun2-KD mutant has obvious defects in nuclear positioning or plant development.8,61 The only phenotype observed in these mutants was a reduced nuclear polarity in root hairs, leaf epidermal cells, and trichomes.8,61 In wild-type Arabidopsis root hair cells, trichomes, and some of the leaf epidermal cells, the nucleus is spindle- shaped.8 Especially in root hair cells, the super-elongated nucleus resembles a “pod” with two thin tails attached to its poles.8 In the wip1-1 wip2-1 wip3-1 mutant or the sun1-KO sun2-KD mutant, this spindle shape is lost, indicating that both WIP and SUN are required for maintaining an elongated nuclear structure in these cell types.8 It is possible that an unknown factor anchored by the SUN-WIP complex regulates the nuclear shape in the Arabidopsis epidermis (Fig. 1C, indicated by the oval with a question mark).
Intriguingly, loss-of-function mutants of two Arabidopsis nuclear long coiled-coil proteins, LITTLE NUCLEI 1 and 2 (LINC1 and LINC2; recently renamed to CROWDED NUCLEI 1[CRWN1] and 2 [CRWN2] to avoid confusion with the LINC complex), also lead to the loss of nuclear elongation, in addition to a reduced nuclear size.68 Plant genomes do not encode homologs of animal Lamin genes. The Daucus carota L. (carrot) Nuclear Matrix Constituent Protein 1 (NMCP1) and its Arabidopsis homologs—CRWN1, CRWN2, CRWN3 and CRWN4—have been proposed as putative plant counterparts of animal lamins. They are conserved in many plant species and contain long coiled-coil domains similar to lamins, though at almost twice the size, and NMCP1 and CRWN1 are located at the nuclear periphery.68,69 In addition to the nuclear shape change, the crwn1-1 crwn2-1 double mutant has a significantly reduced nuclear volume, fewer chromocenters, and a dwarf plant phenotype.68
Nuclear shape change caused by mutations in Lamin genes was observed in a series of human diseases called laminopathies.70 One such disease is Hutchinson–Gilford Progeria Syndrome (HGPS), which is linked to point mutations in the human lamin A gene (Lmna). Patients suffer from a series of premature-aging symptoms—among others, loss of hair, restrictive joint mobility, and cardiovascular disease. One of the hallmarks of HGPS cells is the occurrence of blebbed and lobulated nuclei.71 This defect of nuclear shape is reversely proportional to the amount of Nesprin-2 at the NE,72 and mutation or silencing of Nesprin-2 also causes blebbed nuclei in both mouse and human cells.73 Unexpectedly, the mutation of SUN1 in Lmna−/− mice did not accelerate the pathological phenotypes, but instead ameliorated them and corrected the nuclear aberrancies in fibroblasts.74 In light of the SUN1 overaccumulation observed in Lmna−/− cells, this implies SUN1 overabundance as a pathogenic event in HGPS and a trigger of nuclear shape aberrancies.
The involvement of both lamins and lamin-like proteins and SUN-KASH complexes in nuclear shape in both plants and mammals and their connection to human disease make it worthwhile to further investigate the biological relevance of nuclear morphology at the cellular and organismal level.
Possible Function of Plant SUN Proteins in Mitosis and Meiosis
Mammalian SUN1 is associated with NPCs and interacts with lamins.75 It is suggested that this kind of INM protein links mitotic ER to chromatin in telophase and might mediate NE reassembly during mitosis.76 A similar mechanism might also exist in plants. In Arabidopsis, AtSUN1 diffuses to the ER after NE breakdown and is mainly located at the distal side of the separated chromosomes throughout anaphase.61 At telophase, an enriched AtSUN1 signal starts to enclose the chromosomes from the distal surface to the proximal surface. At the same time, the signal at the ER becomes gradually reduced, indicating the translocation of AtSUN1 to the newly formed NE.61 Studies of AtSUN1 and AtSUN2 in BY-2 cells showed similar results.77 Additional work using BY-2 cells also showed that AtSUN1 and AtSUN2 are associated with membranes around the spindle and close to the chromosomes.77
During plant meiotic prophase I, the telomere bouquet associated with the NE has roles in interhomolog pairing, synapsis, and homologous recombination. Evidence for a possible involvement of plant SUN proteins in this process comes from the maize desynaptic (dy) mutant.78 This mutant is defective in chromosome synapsis, recombination, telomere-NE tethering, and chromosome segregation. Linkage mapping combined with a candidate-gene approach make it likely that a splice variant of maize SUN3 is responsible for this phenotype.78
Perspectives
Our understanding of the nature and role of plant NE proteins has just begun. Compared with the nuclear pore proteins, NE proteins appear to be even less conserved between plants and animals. This suggests that the hunt for plant NE proteins largely will be by de novo identification, rather than homology searches. It is exciting to learn that NE-bridging complexes are conserved in plants and that they perform plant-specific functions through plant-specific ONM partners. A cornucopia of new questions follows from these findings: How is the SUN-WIP complex involved in the developmental changes in nuclear morphology? Are there additional plant NE-bridging complexes and are they connected to the cytoskeleton? Are they involved in nuclear migration? Do plant SUN proteins interact with other IMN proteins and do they interact with a plant lamina? How are plant SUN proteins involved in chromosomal positioning? Perhaps the answers to these questions will help resolve a more general enigma: why is the complement of NE proteins so different in plants? And what does this tell us about the separate evolution of NE functions in plants and opisthokonts? A rapid resolution of these questions is unlikely, but their answers will certainly lead to a broader, more comparative understanding of the physical interaction of the nucleus with its cellular environment.
Glossary
Abbreviations:
- CCSD
canonical C-terminal SUN domain
- ER
endoplasmic reticulum
- INM
inner nuclear membrane
- KASH
Klarsicht/ANC-1/Syne Homology
- LINC
linkers of the nucleoskeleton to the cytoskeleton
- NE
nuclear envelope
- ONM
outer nuclear membrane
- PM3
plant-prevalent mid-SUN 3 transmembrane
- SUN
Sad1/UNC-84
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/24088
References
- 1.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–72. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graumann K, Runions J, Evans DE. Nuclear envelope proteins and their role in nuclear positioning and replication. Biochem Soc Trans. 2010;38:741–6. doi: 10.1042/BST0380741. [DOI] [PubMed] [Google Scholar]
- 3.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–44. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–38. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 7.Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, et al. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol. 2012;198:165–72. doi: 10.1083/jcb.201204085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Graumann K, Evans DE, Meier I. Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol. 2012;196:203–11. doi: 10.1083/jcb.201108098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–81. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 10.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–9. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 11.Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 12.Ketema M, Sonnenberg A. Nesprin-3: a versatile connector between the nucleus and the cytoskeleton. Biochem Soc Trans. 2011;39:1719–24. doi: 10.1042/BST20110669. [DOI] [PubMed] [Google Scholar]
- 13.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–51. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, et al. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–98. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei K, Zhang XC, Ding X, Guo X, Chen MY, Zhu BG, et al. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci U S A. 2009;106:10207–12. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–30. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 17.Djinovic-Carugo K, Gautel M, Ylänne J, Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–23. doi: 10.1016/S0014-5793(01)03304-X. [DOI] [PubMed] [Google Scholar]
- 18.Zhang XC, Xu RN, Zhu BG, Yang XJ, Ding X, Duan SM, et al. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134:901–8. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–87. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15:600–10. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dialynas G, Speese S, Budnik V, Geyer PK, Wallrath LL. The role of Drosophila Lamin C in muscle function and gene expression. Development. 2010;137:3067–77. doi: 10.1242/dev.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kracklauer MP, Banks SML, Xie XH, Wu YN, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- 23.Razafsky D, Blecher N, Markov A, Stewart-Hutchinson PJ, Hodzic D. LINC complexes mediate the positioning of cone photoreceptor nuclei in mouse retina. PLoS One. 2012;7:e47180. doi: 10.1371/journal.pone.0047180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Technau M, Roth S. The Drosophila KASH domain proteins Msp-300 and Klarsicht and the SUN domain protein Klaroid have no essential function during oogenesis. Fly (Austin) 2008;2:82–91. doi: 10.4161/fly.6288. [DOI] [PubMed] [Google Scholar]
- 25.Xie XH, Fischer JA. On the roles of the Drosophila KASH domain proteins Msp-300 and Klarsicht. Fly (Austin) 2008;2:74–81. doi: 10.4161/fly.6108. [DOI] [PubMed] [Google Scholar]
- 26.Yu JH, Starr DA, Wu XH, Parkhurst SM, Zhuang Y, Xu T, et al. The KASH domain protein MSP-300 plays an essential role in nuclear anchoring during Drosophila oogenesis. Dev Biol. 2006;289:336–45. doi: 10.1016/j.ydbio.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 27.Elhanany-Tamir H, Yu YV, Shnayder M, Jain A, Welte M, Volk T. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J Cell Biol. 2012;198:833–46. doi: 10.1083/jcb.201204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee KK, Starr D, Cohen M, Liu J, Han M, Wilson KL, et al. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136:2725–33. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, et al. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–50. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- 32.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–28. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, et al. Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol. 2012;199:735–44. doi: 10.1083/jcb.201208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci. 1998;111:701–12. doi: 10.1242/jcs.111.6.701. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145:1233–49. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol Genet Genomics. 2004;270:449–61. doi: 10.1007/s00438-003-0938-8. [DOI] [PubMed] [Google Scholar]
- 37.Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, et al. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet. 1997;254:238–49. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- 38.Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell. 2004;6:329–41. doi: 10.1016/S1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhou ZC, Du XL, Cai Z, Song XM, Zhang HT, Mizuno T, et al. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Biol Chem. 2012;287:5317–26. doi: 10.1074/jbc.M111.304543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–47. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCue AD, Cresti M, Feijó JA, Slotkin RK. Cytoplasmic connection of sperm cells to the pollen vegetative cell nucleus: potential roles of the male germ unit revisited. J Exp Bot. 2011;62:1621–31. doi: 10.1093/jxb/err032. [DOI] [PubMed] [Google Scholar]
- 42.Ketelaar T, Faivre-Moskalenko C, Esseling JJ, de Ruijter NC, Grierson CS, Dogterom M, et al. Positioning of nuclei in Arabidopsis root hairs: an actin-regulated process of tip growth. Plant Cell. 2002;14:2941–55. doi: 10.1105/tpc.005892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chytilova E, Macas J, Sliwinska E, Rafelski SM, Lambert GM, Galbraith DW. Nuclear dynamics in Arabidopsis thaliana. Mol Biol Cell. 2000;11:2733–41. doi: 10.1091/mbc.11.8.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Folkers U, Berger J, Hülskamp M. Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development. 1997;124:3779–86. doi: 10.1242/dev.124.19.3779. [DOI] [PubMed] [Google Scholar]
- 45.De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol. 2010;20:1697–706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 46.De Smet I, Tetsumura T, De Rybel B, Frey NFD, Laplaze L, Casimiro I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–90. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 47.Park SK, Howden R, Twell D. The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development. 1998;125:3789–99. doi: 10.1242/dev.125.19.3789. [DOI] [PubMed] [Google Scholar]
- 48.Twell D, Park SK, Hawkins TJ, Schubert D, Schmidt R, Smertenko A, et al. MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat Cell Biol. 2002;4:711–4. doi: 10.1038/ncb844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh SA, Pal MD, Park SK, Johnson JA, Twell D. The tobacco MAP215/Dis1-family protein TMBP200 is required for the functional organization of microtubule arrays during male germline establishment. J Exp Bot. 2010;61:969–81. doi: 10.1093/jxb/erp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Smet I, Beeckman T. Asymmetric cell division in land plants and algae: the driving force for differentiation. Nat Rev Mol Cell Biol. 2011;12:177–88. doi: 10.1038/nrm3064. [DOI] [PubMed] [Google Scholar]
- 51.Scheres B, Benfey PN. Asymmetric cell division in plants. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:505–37. doi: 10.1146/annurev.arplant.50.1.505. [DOI] [PubMed] [Google Scholar]
- 52.Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell. 2005;17:3489–99. doi: 10.1105/tpc.105.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P. Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell. 2008;20:1407–20. doi: 10.1105/tpc.108.059014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmelzer E. Cell polarization, a crucial process in fungal defence. Trends Plant Sci. 2002;7:411–5. doi: 10.1016/S1360-1385(02)02307-5. [DOI] [PubMed] [Google Scholar]
- 55.Bozza CG, Pawlowski WP. The cytogenetics of homologous chromosome pairing in meiosis in plants. Cytogenet Genome Res. 2008;120:313–9. doi: 10.1159/000121080. [DOI] [PubMed] [Google Scholar]
- 56.Bass HW. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell Mol Life Sci. 2003;60:2319–24. doi: 10.1007/s00018-003-3312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roberts NY, Osman K, Armstrong SJ. Telomere distribution and dynamics in somatic and meiotic nuclei of Arabidopsis thaliana. Cytogenet Genome Res. 2009;124:193–201. doi: 10.1159/000218125. [DOI] [PubMed] [Google Scholar]
- 59.Graumann K, Runions J, Evans DE. Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 2010;61:134–44. doi: 10.1111/j.1365-313X.2009.04038.x. [DOI] [PubMed] [Google Scholar]
- 60.Murphy SP, Simmons CR, Bass HW. Structure and expression of the maize (Zea mays L.) SUN-domain protein gene family: evidence for the existence of two divergent classes of SUN proteins in plants. BMC Plant Biol. 2010;10:269. doi: 10.1186/1471-2229-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oda Y, Fukuda H. Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J. 2011;66:629–41. doi: 10.1111/j.1365-313X.2011.04523.x. [DOI] [PubMed] [Google Scholar]
- 62.Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol. 2007;17:1157–63. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- 63.Jeong SY, Rose A, Joseph J, Dasso M, Meier I. Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J. 2005;42:270–82. doi: 10.1111/j.1365-313X.2005.02368.x. [DOI] [PubMed] [Google Scholar]
- 64.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Güttler T, Görlich D. Ran-dependent nuclear export mediators: a structural perspective. EMBO J. 2011;30:3457–74. doi: 10.1038/emboj.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGee MD, Stagljar I, Starr DA. KDP-1 is a nuclear envelope KASH protein required for cell-cycle progression. J Cell Sci. 2009;122:2895–905. doi: 10.1242/jcs.051607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–803. doi: 10.1105/tpc.107.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masuda K, Xu ZJ, Takahashi S, Ito A, Ono M, Nomura K, et al. Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long α-helical domain. Exp Cell Res. 1997;232:173–81. doi: 10.1006/excr.1997.3531. [DOI] [PubMed] [Google Scholar]
- 70.Burke B, Stewart CL. The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu Rev Genomics Hum Genet. 2006;7:369–405. doi: 10.1146/annurev.genom.7.080505.115732. [DOI] [PubMed] [Google Scholar]
- 71.Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am J Med Genet A. 2006;140:2603–24. doi: 10.1002/ajmg.a.31346. [DOI] [PubMed] [Google Scholar]
- 72.Kandert S, Lüke Y, Kleinhenz T, Neumann S, Lu W, Jaeger VM, et al. Nesprin-2 giant safeguards nuclear envelope architecture in LMNA S143F progeria cells. Hum Mol Genet. 2007;16:2944–59. doi: 10.1093/hmg/ddm255. [DOI] [PubMed] [Google Scholar]
- 73.Lüke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu WS, et al. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci. 2008;121:1887–98. doi: 10.1242/jcs.019075. [DOI] [PubMed] [Google Scholar]
- 74.Chen CY, Chi YH, Mutalif RA, Starost MF, Myers TG, Anderson SA, et al. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–77. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, et al. Functional association of Sun1 with nuclear pore complexes. J Cell Biol. 2007;178:785–98. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–91. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 77.Graumann K, Evans DE. Nuclear envelope dynamics during plant cell division suggest common mechanisms between kingdoms. Biochem J. 2011;435:661–7. doi: 10.1042/BJ20101769. [DOI] [PubMed] [Google Scholar]
- 78.Murphy SP, Bass HW. The maize (Zea mays) desynaptic (dy) mutation defines a pathway for meiotic chromosome segregation, linking nuclear morphology, telomere distribution and synapsis. J Cell Sci. 2012;125:3681–90. doi: 10.1242/jcs.108290. [DOI] [PubMed] [Google Scholar]
- 79.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 80.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 81.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–9. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Göb E, Schmitt J, Benavente R, Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS One. 2010;5:e12072. doi: 10.1371/journal.pone.0012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shao XP, Tarnasky HA, Lee JP, Oko R, van der Hoorn FA. Spag4, a novel sperm protein, binds outer dense-fiber protein Odf1 and localizes to microtubules of manchette and axoneme. Dev Biol. 1999;211:109–23. doi: 10.1006/dbio.1999.9297. [DOI] [PubMed] [Google Scholar]
- 85.Frohnert C, Schweizer S, Hoyer-Fender S. SPAG4L/SPAG4L-2 are testis-specific SUN domain proteins restricted to the apical nuclear envelope of round spermatids facing the acrosome. Mol Hum Reprod. 2011;17:207–18. doi: 10.1093/molehr/gaq099. [DOI] [PubMed] [Google Scholar]
- 86.Kracklauer MP, Wiora HM, Deery WJ, Chen X, Bolival B, Jr., Romanowicz D, et al. The Drosophila SUN protein Spag4 cooperates with the coiled-coil protein Yuri Gagarin to maintain association of the basal body and spermatid nucleus. J Cell Sci. 2010;123:2763–72. doi: 10.1242/jcs.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Malone CJ, Misner L, Le Bot N, Tsai M-C, Campbell JM, Ahringer J, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–36. doi: 10.1016/S0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 88.Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, Woglar A, et al. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009;139:920–33. doi: 10.1016/j.cell.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 89.Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, et al. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–19. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose WA. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol. 2009;186:229–41. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Friederichs JM, Ghosh S, Smoyer CJ, McCroskey S, Miller BD, Weaver KJ, et al. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 2011;7:e1002365. doi: 10.1371/journal.pgen.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jaspersen SL, Martin AE, Glazko G, Giddings TH, Jr., Morgan G, Mushegian A, et al. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J Cell Biol. 2006;174:665–75. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Conrad MN, Lee C-Y, Chao G, Shinohara M, Kosaka H, Shinohara A, et al. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–87. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 94.Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wanat JJ, Kim KP, Koszul R, Zanders S, Weiner B, Kleckner N, et al. Csm4, in collaboration with Ndj1, mediates telomere-led chromosome dynamics and recombination during yeast meiosis. PLoS Genet. 2008;4:e1000188. doi: 10.1371/journal.pgen.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol. 2007;179:845–54. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Horigome C, Okada T, Shimazu K, Gasser SM, Mizuta K. Ribosome biogenesis factors bind a nuclear envelope SUN domain protein to cluster yeast telomeres. EMBO J. 2011;30:3799–811. doi: 10.1038/emboj.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, et al. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–24. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- 99.Rivero F, Kuspa A, Brokamp R, Matzner M, Noegel AA. Interaptin, an actin-binding protein of the α-actinin superfamily in Dictyostelium discoideum, is developmentally and cAMP-regulated and associates with intracellular membrane compartments. J Cell Biol. 1998;142:735–50. doi: 10.1083/jcb.142.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]