Introduction
The exponential rise in genomics research over the past decade has yielded a growing number of sequence variants associated with medication response that may have clinical utility. Despite existing barriers, attention is turning to strategies that integrate these data into clinical care. The CLIPMERGE PGx Program is establishing a best-practices infrastructure for implementation of genome-informed prescribing using a biobank-derived clinical cohort, pre-emptive genetic testing, and real-time clinical decision support deployed through the electronic health record.
The Road to Personalized Medicine
Personalized medicine aims to optimize clinical decisions about a patient’s care by utilizing all available data, including genomic data. The first wave of genomic variants to emerge with potential clinical utility are those associated with medication response; so-called pharmacogenomic variants. To date, more than 100 medications have had their U.S. Food and Drug Administration (FDA) label modified to include pharmacogenomic information (1). Despite this, the clinical implementation and evaluation of pharmacogenomics lags behind the increasing knowledge base (2). There are a number of factors that may account for this, including an unwillingness to separate pharmacogenomics from the relatively embryonic concept of genomic medicine. However, the risk of pharmacogenomic interventions is generally low, and some suggest that they need only reach evidence levels of non-inferiority compared with current prescribing practices to merit use (3). Additionally, the rapid ascent of genomics from research to clinical translation has left many in the provider workforce under-prepared. The recent, widespread implementation of electronic health records (EHRs) may offer part of the solution to these obstacles. EHRs can help facilitate the implementation and evaluation of pharmacogenomics through point-of-care clinical decision support (CDS), which provides clinicians with knowledge presented at appropriate times, usually through computerized alerts (4). CDS has the potential to translate and integrate current genomic knowledge into existing clinical workflows, allowing clinicians to make genome-informed decisions at the point of care.
The Biobank as a Model for Implementation
Large-scale DNA biobanks, repositories that collect, store and manage samples and associated data, have allowed for the efficient and in-depth study of genotype-phenotype associations. Just as biobanks have been an efficient tool for genomic discovery, they may also form the basis of cohorts used to study strategies for the implementation and evaluation of genomic medicine in clinical care. To achieve this goal, a biobank must 1) be able to identify and re-contact participants, 2) have access to their clinical data on an ongoing basis, and 3) have opportunities to universally or selectively implement an intervention that may be studied.
Since 2007, almost 25,000 Mount Sinai Medical Center (MSMC) patients have enrolled in the EHR-linked, BioMe Biobank program. Patients provide opt-in consent for de-identified research on their EHR-derived clinical data and allow it to be linked to their genomic data. Importantly, patients also consent to recontacting for further research. Over 70% of enrolled patients reside in the local communities of upper Manhattan, one of the most ethnically and socio-economically diverse areas in the U.S. with broad health disparities (Table 1). This representative, urban clinical cohort fulfills the requirements for clinical implementation and forms the foundation of the CLIPMERGE PGx Program.
Table 1.
BioMe Participant Characteristics
| All Ethnicities* |
Hispanic/ Latino |
European Ancestry |
African Ancestry |
||
|---|---|---|---|---|---|
| N | 23,435 | 8,372 | 7,147 | 5,767 | |
| % Female | 59.3 | 63.2 | 53.0 | 63.7 | |
| Mean | Mean | Mean | Mean | ||
| Age (yrs) | 54.0 | 54.9 | 54.9 | 52.8 | |
| BMI (kg/m2) | 28.5 | 29.5 | 26.4 | 30.5 | |
| % | % | % | % | ||
| Hard Coronary Artery | |||||
| Disease** | Diagnosis | 16.0 | 17.9 | 15.7 | 12.4 |
| Diabetes | Diagnosis *** | 28.8 | 37.7 | 14.8 | 34.9 |
| Medications‡ | 16.8 | 22.7 | 16.8 | 19.1 | |
| Hypertension | Diagnosis | 53.0 | 60.6 | 38.6 | 63.5 |
| Medications | 55.6 | 61.0 | 45.4 | 63.9 | |
| Hyperlipidemia | Diagnosis | 26.9 | 32.8 | 18.3 | 31.3 |
| Medications | 37.5 | 42.4 | 34.6 | 35.4 | |
| Depression | Diagnosis | 20.7 | 26.6 | 17.0 | 20.2 |
| Medications | 23.5 | 28.8 | 23.0 | 20.5 |
BioMe is representative of Mount Sinai’s local populations. More than 90% of participants self-identify as being Hispanic/Latino, Caucasian of European ancestry or African American
Myocardial Infarction, Percutaneous Coronary Intervention or Coronary Artery Bypass Graft
Diagnosis is by relevant ICD-9 code(s)
Medications are grouped by drug class commonly prescribed for the treatment of the indicated diagnosis
Project Overview
CLIPMERGE PGx is a Mount Sinai Institutional Review Board (IRB)-approved (HS#: 12-00501, GCO#: 12-0931) program that aims to establish an infrastructure for the use and evaluation of genomic information in clinical care. To achieve these goals, a pilot cohort of 1,500 BioMe patients are being subjected to pre-emptive genotyping for known variants associated with medication response. All physicians who provide primary care through Mount Sinai’s Internal Medicine Associates (IMA) are being invited to participate. To facilitate clinical translation, a novel CDS engine has been developed that delivers guidance on actionable genomic variants in a manner that integrates with existing physician work processes.
A Platform for Genome-Informed CDS
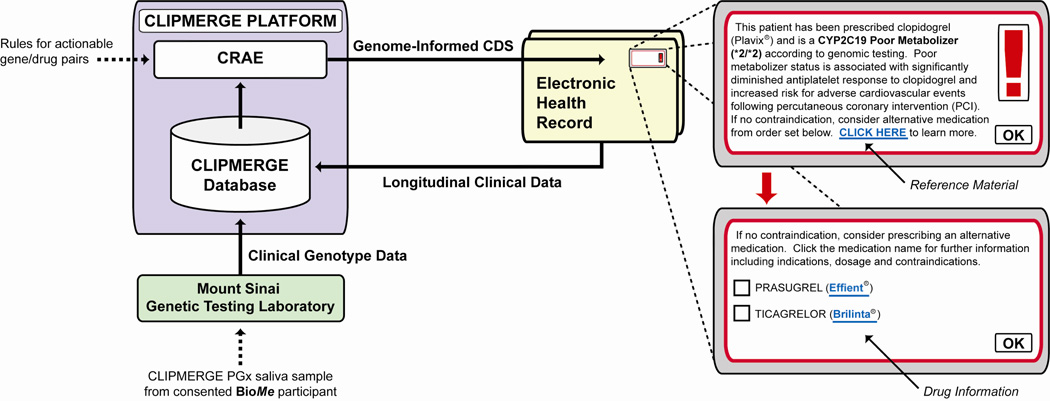
CLIPMERGE PGx utilizes an advanced data management system that is external to, but communicates with, Mount Sinai’s Epic EHR (Figure 1). An external system was developed to allow maximal flexibility when adding and modifying rules in response to new developments in the pertinent literature, which may be less efficient if there is a dependency on a commercial vendor. CLIPMERGE PGx decision support rules are based on actionable variants distilled from each patient’s genotype data and are combined with relevant phenotypic data in the CLIPMERGE database, which includes longitudinal clinical data extracted from the EHR. A Clinical Risk Assessment Engine (CRAE) includes this database and a rules engine that relates actionable genotype-phenotype pairs to genome-informed advice messages. If predefined rules are met, decision support is delivered in real-time at the point-of-care through the EHR. Decision support messages consist of a short text segment, a link to reference material and an order set with suggested medications/doses, if appropriate. CDS content is viewable only when there is a match of both an enrolled patient and enrolled provider. Transactional data regarding CDS metrics, usage and clinical parameters are automatically collected on an ongoing basis from participant’s EHRs.
Figure 1.
A platform for implementation of genome-informed clinical decision support (CDS). Saliva samples from BioMe patients sent to the Mount Sinai Genetic Testing Laboratory are subjected to clinical pharmacogenomic testing. Valid genotypes are released to the CLIPMERGE database, which also contains longitudinal clinical data extracted from the Electronic Health Record (EHR). These data are assessed by the Clinical Risk Assessment Engine (CRAE), which contains pre-specified rules relating actionable genotype-drug pairs to genome-informed advice messages. If a rule is fulfilled, decision support is delivered in real time via the EHR. A mockup of CDS for a clopidogrel (Plavix®) poor metabolizer is shown, consisting of a text segment, a reference link and an order set with suggested alternative medications.
Patient Selection and Recruitment
Although the CLIPMERGE PGx Program is initially consenting 1,500 BioMe participants, the eventual aim is to recruit all participants. The pilot cohort was selected for having regularly attended Mount Sinai IMA over the past 2 years for their primary care. This criterion was applied primarily to ensure that BioMe patients who are cared for by CLIPMERGE-enrolled IMA physicians are selected. Additionally, in order to enrich the program with patients likely to have pharmacogenomically-relevant interactions (and therefore likely to receive CLIPMERGE PGx CDS) certain inclusion criteria were applied, including targeting patients who were known to be taking medications with pharmacogenomic interactions such as clopidogrel, warfarin, simvastatin, tricyclic antidepressants (TCAs), and selective serotonin reuptake inhibitors (SSRIs). Patients were also identified for being at increased risk for receiving these medications in the near future. Since February 2013, interested BioMe patients have been meeting with CLIPMERGE PGx research coordinators for enrollment, consent and donation of a saliva sample for DNA isolation and genetic testing in a CLIA-certified laboratory.
Clinical Pharmacogenomic Testing
Saliva specimens are subjected to clinical pharmacogenomic testing by the Clinical Laboratory Improvement Amendments (CLIA)-certified Mount Sinai Genetic Testing Laboratory (MGTL). The MGTL is currently approved for targeted genotyping of several pharmacogenomic variants using commercially available and laboratory-developed tests. The goal of CLIPMERGE PGx is not to validate or use a single testing methodology, but rather to create an infrastructure that will allow the clinical implementation of useful genomic information, regardless of how it is generated. As new pharmacogenomic practice guidelines become available for ‘actionable’ gene/drug pairs, stored genotype data will be released for use in CLIPMERGE PGx pending regulatory approval.
Provider Recruitment, Education and Feedback
Following consultation with institutional leadership, it was decided that the program would initially be limited to a group of consented providers, by practice, in order to minimize potential disruption to the institution-at-large whilst the infrastructure is established and its’ impact evaluated. The eventual aim as the program develops is to include all Mount Sinai providers, which will allow for greater generalizability of the outcome data generated. Before participating in CLIPMERGE PGx, providers are required to attend a 1-hour recruitment session. Sessions are held regularly and on an ongoing basis to ensure a high level of provider enrollment. In addition to stand-alone sessions, scheduled teaching slots for trainees and divisional meetings have been utilized for recruitment. All sessions are advertised and communicated to relevant providers through existing channels. During the sessions, providers first complete a ‘pre-training questionnaire’ about their current knowledge of genomics, personalized medicine, and CDS (see Supplementary Information). After completing the questionnaire, they watch a 30 minute presentation outlining the scientific justification and content of the CDS. They then complete a ‘post-training questionnaire’ about their background and attitudes toward prescribing decision aids and personal genome testing. Those who complete the sessions are invited to consent and their credentials are added to a list of participating users. In addition, each time providers encounter CLIPMERGE CDS in the course of patient care, they will receive a survey via email to gauge their opinions and the appropriateness of the CDS they encountered.
Development and Evaluation of CDS Content
The Clinical Pharmacogenetics Implementation Consortium (CPIC) of the Pharmacogenomic Research Network (PGRN) develops practice guidelines for implementing pharmacogenomics (5). These guidelines include recommendations regarding medication selection or dosing based on combinations of genotype and phenotype, which are an ideal resource for CLIPMERGE PGx. For example, initial CDS content was developed for clopidogrel (CYP2C19), warfarin (CYP2C9 and VKORC1), simvastatin (SLCO1B1), TCAs (CYP2D6 and CYP2C19) and SSRIs (CYP2D6) as these have CPIC guidelines published or in development and/or modifications to the FDA label, with clinically-approved genotyping assays available for implicated alleles. The creation of CDS content was undertaken by a multidisciplinary working group of geneticists, pharmacologists, physicians and informaticists that formed CDS content by consensus. This group will continue to review existing CDS and to extend new CDS in response to developments in the field, FDA label revisions, and publication of new guidelines. CDS content was also evaluated as part of user acceptance testing by a group of CLIPMERGE-enrolled providers.
Outcome and Process Measures
CLIPMERGE PGx is concerned predominantly with the process of genomic medicine implementation. This program will contribute to the emerging body of pilot data needed for forthcoming larger studies that will assess the utility of genomic information in optimizing medication efficacy and safety. In addition to quantitative transactional data (e.g., genotype results, CDS type and frequency) and questionnaire data (e.g., appropriateness of CDS deployment); qualitative data are being collected to provide a deeper understanding of the barriers and facilitators to genome-informed CDS adoption. The program is designed such that the influences of moderators like prior physician perceptions about the usefulness of, experience with, and awareness of genome-informed CDS can be assessed.
Conclusions
In order for personalized medicine to materialize into reality, the utility of genomic information in clinical care must be demonstrated on a large scale. But before this can be done, or at least in parallel, tools and best-practices, both technical and human, to facilitate the delivery of this information must be developed and evaluated so that the question of utility can be answered without the burden of questionable process. The CLIPMERGE PGx Program will provide valuable insight into the mechanisms and processes that will best support the use of genomic information in clinical care and provide an infrastructure for the ongoing evaluation of genomic information in the context of clinical utility.
Acknowledgements
Funding for this program was provided by the Andrea and Charles Bronfman Philanthropies and The Icahn School of Medicine at Mount Sinai. O.G, S.A.S, J.L.K and E.P.B are supported in part by the National Human Genome Research Institute (NHGRI) through Grant U01HG006380 (eMERGE Network). S.A.S. is supported in part by the National Center for Research Resources (NCRR) and the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) grant KL2TR000069. E.P.B is supported in part by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant 5U01DK085688 (Chronic Kidney Disease Biomarker Consortium). J.S.H. is supported in part by National Heart, Lung, and Blood Institute (NHLBI) grant 5R01HL113497. C.L.O. is supported by Columbia Training in Biomedical Informatics (NIH NLM #T15 LM007079).
The authors would like to thank Yolanda Keppel, Tanisha Brown, Stacy Paris, Ana Mejia, Barbara Murphy, Alex Federman, Juan Wisnivesky, Eva Waite, Scott Lorin, Jenny Lin, George Hripsak, Emilia Bagiella, Miriam Udler, Noura Abul-Husn, Naresh Hinduja, Deepesh Chandra, Bill Fultz, Vaneet Lotay, Rajiv Nadukuru, Quingbin Song, Bernadette Liggayu, Patrick Shanley, Yumin Li, Marie Teil, The Mount Sinai Epic team, Mount Sinai Information Technology and Epic Corporation for their contributions to the CLIPMERGE PGx Program.
O.G, S.B.E and E.P.B are co-inventors on patent applications related to personalized clinical decision support. S.A.S has received consulting fees from United States Diagnostic Standards (USDS), Inc. J.S.H has received research grant support (to the institution) from Biotronik, Medco Research Institute; and consulting fees from Biotronik, Medco Health Solutions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication. NPG is providing this early version of the manuscript as a service to our customers. The manuscript will undergo copyediting, typesetting and a proof review before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers apply.
Conflict of Interest
The other authors declared no conflicts of interest.
References
- 1. [Accessed February 2013];FDA - Table of Pharmacogenomic Biomarkers in Drug Labels. 2013 http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm.
- 2.Scott SA. Personalizing medicine with clinical pharmacogenetics. Genet Med. 2011;13:987–995. doi: 10.1097/GIM.0b013e318238b38c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman RB. Pharmacogenomics: "noninferiority" is sufficient for initial implementation. Clin Pharmacol Ther. 2011;89:348–350. doi: 10.1038/clpt.2010.310. [DOI] [PubMed] [Google Scholar]
- 4.Osheroff JA, Teich JM, Middleton B, Steen EB, Wright A, Detmer DE. A roadmap for national action on clinical decision support. J Am Med Inform Assoc. 2007;14:141–145. doi: 10.1197/jamia.M2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]