SUMMARY
Genomic instability at repetitive DNA regions in cells of the nervous system leads to a number of neurodegenerative and neuromuscular diseases, including those with an expanded trinucleotide repeat (TNR) tract at or nearby an expressed gene. Expansion causes disease when a particular base sequence is repeated beyond the normal range, interfering with the expression or properties of a gene product. Disease severity and onset depend on the number of repeats. As the length of the repeat tract grows, so does the size of the successive expansions and the likelihood of another unstable event. In fragile X syndrome, for example, CGG repeat instability and pathogenesis are not typically observed below tracts of roughly 50 repeats, but occur frequently at or above 55 repeats, and are virtually certain above 100–300 repeats.
Recent evidence points to bidirectional transcription as a new aspect of TNR instability and pathophysiology. Bidirectional transcription of TNR genes produces novel proteins and/or regulatory RNAs that influence both toxicity and epigenetic changes in TNR promoters. Bidirectional transcription of the TNR tract appears to influence aspects of its stability, gene processing, splicing, gene silencing, and chemical modification of DNAs. Paradoxically, however, some of the same effects are observed on both the expanded TNR gene and on its normal gene counterpart. In this review, we discuss the possible normal and abnormal effects of bidirectional transcription on trinucleotide repeat instability, the role of DNA repair in causing, preventing, or maintaining methylation, and chromatin environment of TNR genes.
INTRODUCTION
More than 20 human neurodegenerative diseases are caused by amplification of simple DNA repeat tracts [1–3]. In this review, we will focus primarily on expansion of trinucleotide repeats (TNR). In the past, the position of the TNR in the coding or noncoding region of the gene was identified as a parameter of significant impact on both the mechanism of toxicity and the size of the expansion. TNRs in the coding sequence supported expression of long and “sticky” homopolymeric proteins that disrupted cellular machinery, interfering with cellular processes including trafficking, lipid homeostasis and transcription [4–6]. In a non-coding region of a gene, abnormal heterochromatin formation, gene silencing and hypermethylation frequently accompanied expansion. With the recent discovery of bidirectional transcription of TNR repeat tracts, however, these lines have been blurred.
Historically it was believed that, in eukaryotic genomes, each protein-coding gene has a unique promoter in the 5'-flanking region, and the direction of the promoter is usually unidirectional. Advances in transcriptome analysis, however, have revealed that bidirectional transcription of coding genes is common [7]. Bidirectional transcription across repeat tracts occurs in most, if not all TNR genes [8], and provides previously unknown aspects of disease pathophysiology encompassing both protein- and RNA-mediated toxicity [8–11]. Furthermore, TNR coding transcripts can be alternatively translated in the absence of a starting ATG site, in a process coined as Repeat Associated Non-ATG translation (RAN-translation) [12]. RAN-translation occurs in all reading frames. At CAG tracts, for example, translation of CAG, AGC, and GCA produces toxic homopolymeric proteins of polyglutamine, polyserine, and polyalanine tracts [12, 13], respectively. These emerging discoveries provide entirely new disease mechanisms arising from a single gene defect.
The impact of bidirectional transcription on the DNA expansion mutation is unknown. The occurrence of bidirectional transcription, itself, does not require an expanded TNR tract, nor does it depend on the TNR sequence of the disease gene. To name just a few, bidirectional transcription occurs in myotonic dystrophy type 1 (DM1) (CTG) [14], in Huntington's disease-like 2 (HDL2) (CTG) [15], in the FMR1 gene of fragile X syndrome (FXS) (CGG) [16], in Huntington's disease (HD) (CAG) [17], and in Spinocerebellar ataxia type 7 (SCA7) (CAG) [18], in which CAG, CTG, and CGG repeats are contained within the sense strand. The occurrence of bidirectional transcription also does not depend on the position of the TNR tracts in the gene. The CAG and CTG repeats in HD, HDL2 and SCAs reside in coding regions of the gene, while DM1, FXS and SCA8 contain repeats in the untranslated regions of the gene.
Since TNR sequences potentially encode multiple repeat-containing RNAs, bidirectional transcription increases the repertoire of RNA-mediated regulatory mechanisms that are likely to influence toxicity. However, a major question is how and whether processing of TNR expansion-containing RNA influences TNR instability. Many of the RNA-mediated processing events that arise from TNR expansion genes also occur in their normal counterparts. Thus, a key question is whether TNR expansion has an influence on bidirectional transcription or vice versa. TNR expansion leads to aberrant phenotypes such as abnormal heterochromatin spreading. Yet, heterochromatin changes are used in nature to silence and regulate gene expression for specific uses during development. Hypermethylation often accompanies unscheduled gene silencing of TNR genes, yet methylation is required for normal or reversible heterochromatin formation. How a cell knows to regulate the extent of methylation is unknown. Hypermethylation of some TNR genes results in faulty suppression of gene expression, yet there are DNA repair activities present in the cell to maintain or regulate the normal methylated state. Do they fail to work at TNR genes? For example, deamination of 5-methylcytosine to thymine results in formation of mismatch pairs that are removed by either DNA glycosylases or mismatch repair (MMR) [19, 20]. The growth arrest and DNA damage 45α protein (Gadd45α) participates with NER factors to maintain active DNA demethylation and maintain the promoter of active genes under a hypomethylated state [21]. While some RNAs arise from loci destined to encode regulatory RNAs, bidirectional transcription of TNR genes gives rise to RNAs that exert abnormal activity. Thus, TNR expansion, genome locus, DNA and histone methylation, and DNA repair are connected, but the links among them are poorly understood. Distinguishing normal from abnormal events is difficult.
In this review, we address whether and how the regulatory RNAs that arise from bidirectional transcription of TNR genes influence TNR instability. Specifically, we will review examples of bidirectional transcription of TNR genes, and discuss models for (1) how sense and antisense regulatory RNA mechanisms that operate at TNR genes are altered for an abnormal outcome, (2) the impact of chromatin modulation on methylation patterns in TNR expansion, and (3) the role of excision repair in regulating expansion, hypermethylation, and unscheduled silencing.
DUAL SOURCES OF TOXICITY FROM BIDIRECTIONALLY TRANSCRIBED TNR EXPANSION GENES
In order to understand possible mechanisms for expansion, we must first consider the consequences of bidirectionally transcribed aberrant RNA processing and its effects on toxicity (Fig. 1). Bidirectional transcription across an expanded TNR gene leads to three distinct scenarios for RNA regulation of TNR genes (Table 1): (1) RNAs that code for protein in both the sense and antisense directions, (2) RNAs that code for protein in only one direction, (3) some non-coding RNAs induce heterochromatin spreading and hypermethylation. Thus, the sense and antisense transcripts serve as the basis for protein-mediated toxicity, RNA-mediated toxicity, or both [9–11].
Figure 1. Possible fates of bidirectional RNA transcripts at trinucleotide repeats.
Coding and non-coding RNAs from TNR repeats form secondary structure with several possible fates. (Homopolymeric proteins) The RNA could serve as a coding template for homopolymeric toxic proteins (red squiggle). (Splicing defects) TNR RNA binds to RNA splicing factors Muscleblind (MBLD) (blue circle) and CUG binding proteins (CUG-BP) (orange circle) to alter RNA splicing and binding of other splicing factors (yellow and red balls). (SiRNA) A non-coding RNA hairpin is cleaved by DICER into short 21nt double stranded RNA. The siRNA can induce silencing in cis, by formation of heterochromatin formation, or in trans by targeting complementary sequences of other genes for degradation via an RNA-I silencing complex (RISC) and Argonaute (AGO) complex.
Table 1.
Antisense Transcription and CTCF Genomic Insulators in Trinucleotide Repeat Expansion Disease Loci
| Disease | Gene Product |
Repeat | Antisense transcripta |
Sense (S)/ Antisense (AS) Expressionb |
Antisense Function | TXN Silencing in Expanded State |
Flanking CTCF sites (5’/ 3’)c |
Refs (PMID) |
|---|---|---|---|---|---|---|---|---|
| Dentorubral pallidoluysian atrophy (DRPLA) |
DRPLA | CAG | * | S: C | no | 5’ and 3’ (flanking the CAG repeats) |
22342974 11479593 |
|
| Fragile X syndrome (FXS) |
FMR1 | CGG | ASFMR1 | S: C AS: NC |
Silenced in full-mutation, upregulated in carriers of premutation alleles. May be involved in pathogenesis of FXTAS and FXS |
yes | CTCF-binding sites flank the CGG repeat and the promoter of ASFMR1 |
17921506 |
| Fragile X tremor/ataxia syndrome (FXTAS) |
FMR1 | CGG | ASFMR1, FMR4 | S: C AS: NC |
Silenced in full-mutation, upregulated in carriers of premutation alleles. May be involved in pathogenesis of FXTAS and FXS |
no | CTCF-binding sites flank the CGG repeat and the promoter of ASFMR1 |
17921506 |
| Friedrich's ataxia (FRDA) |
FXN | GAA | FAST-1 | S: C AS: ORF (154 aa) |
Increased expression of FAST-1 causes heterochromatin formation and transcriptional silencing of FXN |
yes | 5’ (5’ UTR) and 3’ |
19956589 19956589 |
| Huntington's disease (HD) |
HTT | CAG | HTTAS | S: C AS: C |
Functions to regulate HTT expression. Regulation is Dicer dependent |
no | 5’ |
21672921 11479593 |
| Huntington's Disease Like 2 (HDL2) |
JPH3 | CTG | HDL2-CAG | S: C AS: C |
Leads to polyglutamine pathogenesis and neuronal dysfunction |
yes | 5’ and 3’ |
21555070 22367996 |
| Myotonic dystrophy type 1 (DM1) |
DMPK | CTG | DMPKAS | S: C AS: NC |
Converted into 21 nt siRNA that induces heterochromatin formation, favoring transcription of the flanking gene SIX5. Repeat expansion causes loss of binding to adjacent CTCF sites, leading to abnormal spreading of heterochromatin and consequent repression of SIX5. |
Decreased DMPK transcripts in adult myotonic dystroph; TXN repression of SIX5 |
5’ and 3’ (flanking the CTG repeats) |
16285929 11479593 |
| Myotonic dystrophy type 2 (DM2) |
ZNF9 | CCTG | none | S: C | no | nd | 16876389 | |
| Oculopharyngeal muscular dystrophy (OPMD) |
PABPN1 | GCG | * | S: C | no | 5’ and 3’ | 9462747 | |
| Spinal and bulbar muscular atrophy (SBMA) |
AR | CAG | * | S: C | no | 5’ | 22609045 | |
| Spinocerebellar ataxia type 1 (SCA1) |
ATXN1 | CAG | * | S: C | no | 5’ and 3’ | 18957430 | |
| Spinocerebellar ataxia type 2 (SCA2) |
ATXN2 | CAG | * | S: C | yes | 5’ to the CAG repeats, and 3’ to the gene |
22037902 11479593 |
|
| Spinocerebellar ataxia type 3 (SCA3) |
ATXN3 | CAG | ATXN3OS | S: C | no | 5’ and 3’ | 21827905 | |
| Spinocerebellar ataxia type 6 (SCA6) |
CACNA1A | CAG | * | S: C | no | 5’ and 3’ | 21827907 | |
| Spinocerebellar ataxia type 7 (SCA7) |
ATXN7 | CAG | SCAANT1 | S: C AS: NC |
Loss of SCAANT1 derepresses ataxin-7 sense transcription in a cis-dependent fashion and is accompanied by chromatin remodeling. |
no | 5’ and 3’ (flanking the CAG repeats) |
21689595 19008940 |
| Spinocerebellar ataxia type 8 (SCA8) |
ATXN8 | CAG | ATXN8OS | S: C AS: NC |
RNA gain-of-function effects. Expanded CUG transcripts dysregulate MBNL/CELF regulated pathways in the brain. |
no | nd |
16804541 19680539 |
| Spinocerebellar ataxia type 10 (SCA10) |
ATXN10 | ATTCT | * | S: C | no | 5’ and 3’ | 21531163 | |
| Spinocerebellar ataxia type 12 (SCA12) |
PPP2R2B | CAG | * | S: C | no | 5’ and 3’ | 21827912 | |
| Spinocerebellar ataxia type 17 (SCA17) |
TBP | CAG | * | S: C | no | nd | 18418687 | |
| Spinocerebellar ataxia type 31 (SCA31) |
TK2 | TGGAA | * | S: C AS: C | no | 5’ and 3’ | 19878914 |
* antisense transcripts reported in Dion and Wilson 19540013, compiled from 19056939.
S = Sense strand, AS = Antisense strand, C = Coding, NC = Non-coding.
CTCF site search performed using CTCFBSDB: a CTCF binding site database for characterization of vertebrate genomic insulators (ref. 17981843). Sites searched by gene. Experimentally identified CTCF sites listed and published CTCF sites listed. nd is not determined
RNAs containing CAG repeats in coding transcripts express polyglutamine in the sense strand (Fig. 1), but by RAN-translation can also produce polyalanine and polyserine, exacerbating the potential toxicity [12, 13]. At the same time, expanded CUG non-coding RNAs form ribonuclear foci and mislocalize or misregulate RNA binding proteins, notably splicing factors (Fig. 1) [9–12]. Two key splicing factors exhibit altered affinities for RNA as a consequence of CUG repeats [22, 23]. CUG-BP1 (CUG binding protein 1) is a member of the CUG-BP/ETR-3-like (CELF) family of proteins involved in RNA splicing, RNA editing, and translation [22]. The second splicing regulatory protein is muscleblindlike (MBNL) [24]. Although discovered first in conjunction with DM1, MBNL is implicated in multiple TNR expansion diseases [24]. Both CUG-BP1 and MBNL bind to CUG repeats, but the two proteins antagonistically regulate alternative splicing [23]. In the case of the skeletal muscle chloride channel CLCN1/CIC-1, for example, it is alternatively spliced in DM1 to include exon 7A [25]. Elevation of this splice form leads to a reduction in CLCN1 function, which contributes to myotonia [25]. CUG RNA binding to MBNL represses inclusion of exon 7A, while binding of CELF4 proteins promotes inclusion [25]. The two opposing effects occur through binding of the respective proteins to distinct regions of the RNA. The length of the repeat affects only MBNL binding [26]. Thus, as the CTG region expands, there is a gain-of-function in CELF4 and a loss-of-function in MBNL, conditions that tend to favor generation of the alternatively spliced forms of DM1 [27, 28].
In general, the RNA-dependent mechanisms for toxicity generate multiple toxic sources. Expression of misfolded proteins, generation of siRNA, and aberrant splicing appear to be common features (Fig. 1). To illustrate them, we discuss a few examples of the bidirectional transcription of TNR disease genes, and consider how RNA processing plays a role in the toxicity of TNR genes.
RNAs that code for protein in both the sense and antisense directions
A natural antisense transcript exists at the HD repeat locus that contains the repeat tract [17]. The sense strand of the normal Huntington’s disease gene (htt) harbors a CAG tract that codes for a polyglutamine region, the antisense HD transcript (httas) is 5′ capped, poly (A) tailed and contains three exons, alternatively spliced into two products: htt AS-1 (exons 1 and 3) and htt AS-2 (exons 2 and 3). Exon 1 includes the TNR repeat. The first splice product htt AS-1 has a weak promoter, and is expressed at low levels in multiple tissue types and throughout the brain [17]. In cell systems, overexpression of htt AS-1 specifically reduces endogenous htt transcript levels, while siRNA knockdown of htt AS-1 increases htt transcript levels. Thus, bidirectional expression of htt AS-1 regulates the amount of htt in the cell. CAG expansion in the mutant Huntington’s disease gene (mhtt) reduces promoter efficiency and the level of htt AS-1 decreases in human HD frontal cortex. The effect is dependent on repeat length and is at least partially Dicer-dependent, implying that some sort of processing of the antisense RNA to small RNAs also occurs.
Huntington's disease-like-2 transcripts in the sense and the antisense direction also code for two proteins [15]. HDL2 is a phenocopy of Huntington's disease caused by CTG/CAG repeat expansion at the Junctophilin-3 (JPH3) locus [29]. The neuropathology of postmortem HDL2 brains is strikingly similar to that of HD [30, 31]. Both exhibit robust and selective striatal and cortical atrophy, with neurodegeneration primarily targeting the striatal medium spiny neurons and a subset of cortical pyramidal neurons. The CTG repeat expansion is located within the variably spliced exon 2A of JPH3, which is not part of the main transcript encoding JPH3 protein [32]. On the sense strand, three splice variants that include exon 2A have been described, placing the expanded CTG repeat in polyleucine or polyalanine open reading frames (ORFs), or in the 3′ untranslated region (UTR) [13]. The mechanisms underlying HDL2 pathogenesis remain unclear, but expression of an expanded CAG repeat-containing transcript emanating from the strand antisense to JPH3 has been suggested as a possible mechanism [15]. This mutant HDL2-CAG transcript results in polyglutamine protein aggregation providing a molecular pathogenic link between HD and HDL2.
RNAs that code for protein in only one direction
At the spinocerebellar ataxia type 8 (SCA8) locus on chromosome 13q21 [33], a transcript from an untranslated gene in one direction (ATXN8OS) contains a CUG repeat expansion, while the gene in the opposite direction expresses a transcript in which a CAG repeat appears to encode polyglutamine (ataxin 8, ATXN8) [34]. Evidence from flies [35], cells [36], mouse [37] models and human brain tissue suggests that both transcripts contribute to neurotoxicity [9, 34, 36]. Expression of two transcripts leads to novel pathogenesis with the generation of dcr-2 and ago2-dependent 21-nt triplet repeatderived siRNAs [38]. These small RNAs targeted the expression of CAG-containing genes, such as Ataxin-2 and TATA binding protein (TBP), which bear long CAG repeats in both fly and man [39]. The alternative splicing abnormalities lead to accumulation of RNA foci, and sequestration of the RNA binding protein MBNL1 [40]. These findings indicate that the generation of triplet repeat-derived siRNAs enhances toxicity and suppresses the expression of other TNR genes.
RNAs that cause heterochromatin formation and hypermethylation
The Dystrophia Myotonica protein kinase (DMPK) gene (DM1) harbors a binding site for the zinc finger protein CCCTC-binding factor (CTCF). CTCF binding sites serve as a functional insulator in establishing a heterochromatin environment around the gene [41]. DMPK is located on chromosome 19q13.2–q13.3 spanning 13 kbp and comprising at least 15 exons [42–44]. The encoded protein is 624 amino acids in length and contains an N-terminal domain that shares strong homology to the cAMP-dependent serine-threonine protein kinase family. The CTG repeats in DMPK are a component of a CTCF-dependent insulator element and repeat expansion results in conversion of the region to heterochromatin [41]. The DM1 insulator is maintained in a local heterochromatin context: an antisense transcript emanating from the adjacent SIX5 regulatory region extends into the insulator element [14]. The antisense transcript at this locus is converted into 21 nt small interfering RNA (siRNA) that induces heterochromatin formation [14], associated regional histone H3 lysine 9 (H3-K9) methylation and heterochromatin protein 1-gamma (HP1γ) recruitment [14]. The antisense transcript originates in a 124 bp region previously characterized as a positive regulator and hypersensitive-site enhancer (HSE) of the sense transcript for the SIX5 gene [14]. The expansion of the CUG repeat in DMPK RNA leads to misregulation of alternative splicing events, which occurs in trans. The CTG expansion results in loss of the CTCF zinc finger and DM1 insulator function. As a consequence, heterochromatin spreads to the SIX5 gene and suppresses its expression.
Collectively, bidirectional transcription of expanded repeats reveals common features that provide potential mechanisms for toxicity: production of long, toxic homopolymers, generation of siRNA and gene suppression, and aberrant splicing. Each expansion gene has abnormal features that distinguish them from their normal counterparts, yet the three candidate RNA-mediated mechanisms themselves are observed throughout nature as normal modes of genome regulation. How, why and even if these normal processes go awry to create instability in TNR genes is unknown.
For example, siRNA processing occurs at many normal genes and mediates regulated silencing of genes. Are the toxic effects of TNR expansions a consequence of Dicer activity? A second question is why are TNR diseases not more alike? The expanded genes themselves are obviously distinct and are expressed differently among brain regions. However, the newly discovered bidirectional transcription draws them mechanistically closer, and provides the potential for more similar consequences of the expansion. For example, HD is ubiquitously expressed. If RAN-translation of CAG/CTG tracts is capable of expressing polyglutamine, polyserine, or polyalanine, the coding HD CAG transcript would produce the same homopolymers, as would SCA8 (for example), with at least some overlap in their regional expression. Alternative splicing and sequestration of MBNL is a common consequence of expanded non-coding RNAs. Do all RNA foci contain the same proteins? All TNR genes expand, suggesting common basic mechanisms, yet only some expanded genes form heterochromatin, with differential effects on the level and types of DNA modifications in the tract. How is chromatin folding related to instability? In the following sections, we address (1) possible differences among normal and abnormal steps arising from bidirectional transcription, (2) at what steps does the normal process disconnect to cause mutation, and (3) the effects of bidirectional transcription on DNA repair and expansion.
NORMAL AND ABNORMAL MECHANISMS REGULATING CHROMATIN CONTEXT, TOXICITY AND EXPANSION
Although heterochomatin is abnormally formed in some expanded genes, heterochomatin formation at repetitive elements is normally observed in many prokaryotic and eukaryotic systems. This process underlies the old concept of “facultative” heterochromatin, which is localized to promoters and is established either in a developmentally regulated manner or in response to environmental triggers [45]. Facultative heterochromatin ensures the epigenetic silencing of genes in a certain cell type or tissue, and is an important mechanism for developmental programming and cell fate [46]. By definition, facultative heterochromatin is, “optional” or “discretionary”. It can form anywhere in the nucleus and its formation is required for specialized functions in which gene silencing regulates reversibility or timed expression. X inactivation is a classic example of the formation of facultative heterochromatin that occurs in mammals (Fig. 2) [46]. In mammals, the inactive X chromosome (Xi) appears within interphase nuclei as a heteropycnotic Barr body usually positioned at the nuclear or nucleolar periphery [47–49]. The inactive X is silenced early in development to ensure gene dosage between the male and female species. Inactivation of the X-chromosome occurs due to production of a non-coding untranslated gene, X-inactive specific transcript gene (Xist gene) (Fig. 2A) [50]. Expression of Xist RNA induces heterochromatin spreading from its position to the end of the X chromosome (Fig. 2B), resulting in its inactivation [51, 52].
Figure 2. Inactivation of the X-chromosome occurs after formation of heterochromatin in a regulated and reversible process induced by methylation.
(A) X-inactivation is triggered by the expression of the interference RNA (red lines) coded for by the X-inactive specific transcript gene (Xist gene). Xist triggers DNA methylation (small blue balls), and chromatin remodeling stably inactivates the chromosome (blue). The RNAi promotes methylation, and loss of accessibility to nucleases of the inactive X chromosome implies that it has formed a condensed heterochromatic state. (B) The Xist transcript (blue dashed lines) is expressed exclusively in the inactive X in the Barr body. Xist resides at the X inactivation center (XIC), and spreading of heterochromatin radiates outward from the XIC. The inactive heterochromatin state can be reversed to an active state by treatment with 5-deazacytidine (black arrow).
Use of siRNA to regulate chromatin folding is also a normal process. Untranslated regions (UTRs) throughout the genome contain tri- or tetranucleotide repetitive regions capable of being transcribed and processed into repeat-associated small interfering RNAs (siRNAs) [53–55]. In vertebrates, the biogenesis of intronic siRNA occurs in a series of five steps [56, 57] (Fig. 3). The micro RNA (miRNA) is transcribed as a long primary precursor miRNA. Transcription occurs by a type II RNA polymerase (Pol-II) from the intron or UTR of a primary gene transcript [58]. MiRNAs can be located within the noncoding DNA or protein-coding region of DNA [59]. After transcription, instead of being translated, the primary miRNA is processed by Drosha-like RNaseIII endonucleases/microprocessors to a short hairpin-like stem-loop precursor (shRNA). During the third step, the shRNA is exported out of the cell nucleus into the cytoplasm, by Ran-GTP and exportin receptors [60, 61]. Once in the cytoplasm, a Dicer-like endoribonuclease cleaves the shRNA to form a single-stranded mature siRNA [57, 62]. Finally, the mature siRNA is assembled into a ribonuclear particle (RNP) to form an RNA-induced silencing complex (RISC) with argonaute proteins [63] or RNA-induced transcriptional silencing (RITS) complex [64]. The siRNA binds to matched sequences of one or more messenger RNAs (mRNAs) to execute RNAi-related gene silencing, through either direct mRNA degradation or translational suppression [57, 62, 65, 66].
Figure 3. Normal siRNA processing.
Two types of small ribonucleic acid (RNA) molecules, microRNA (miRNA) and small interfering RNA (siRNA) are central to normal RNA interference processes. The RNAi pathway is initiated by processing of long double-stranded RNA precursor miRNA into short hairpins (shRNA) by a Drosha-like RNaseIII endonuclease/microprocessor. The shRNA are exported out of the nucleus via the RAN-GTP exportin. In the cytoplasm, the shRNA form a complex with Dicer, which cleaves shRNA into short fragments of ∼20 nucleotides siRNA. Each siRNA is unwound into two single-stranded RNAs, the passenger strand and the guide strand. The passenger strand is degraded, and the guide strand is incorporated into a ribonuclear particle to form a RNA-induced silencing complex (RISC). The most well-studied outcome is post-transcriptional gene silencing, which occurs when the guide strand base pairs with a complementary sequence in a messenger RNA molecule and induces cleavage by Argonaute (AGO), the catalytic component of the RISC complex.
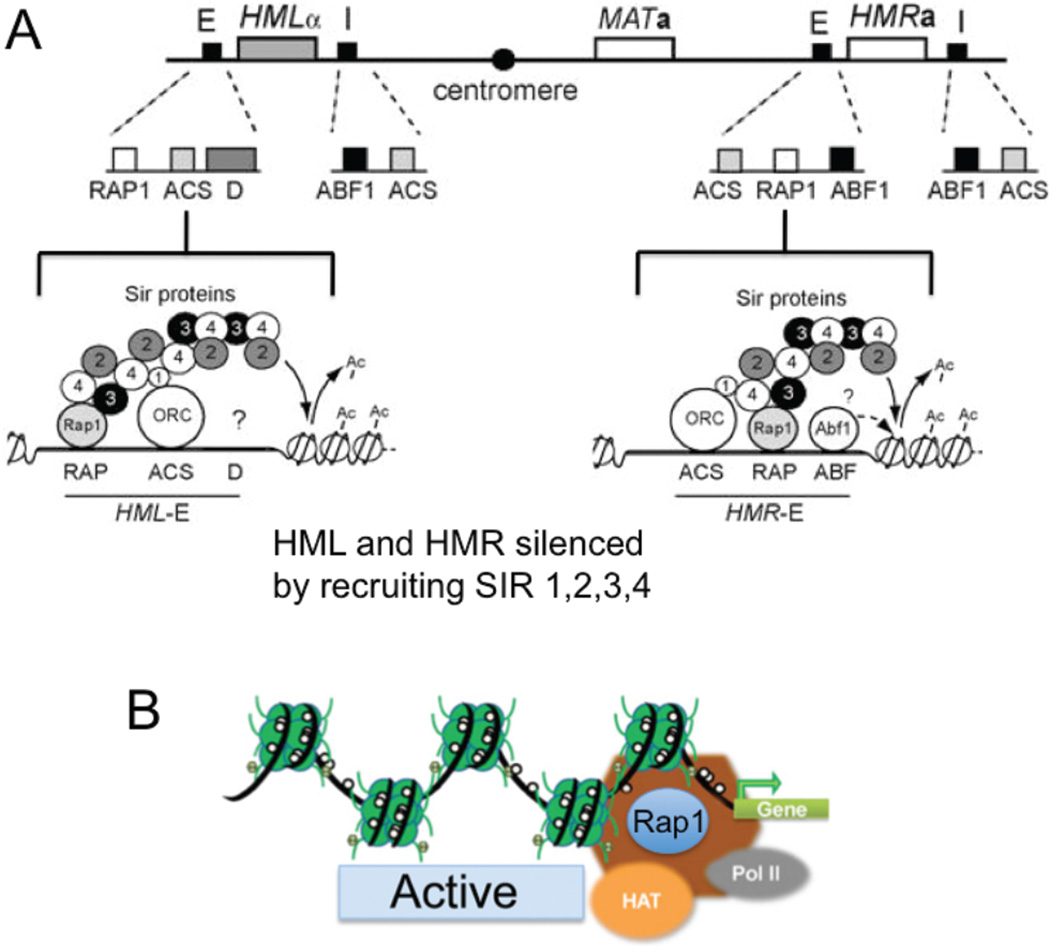
Thus, antisense RNA transcripts and the use of siRNAs to induce local chromatin modifications and gene silencing are normal and conserved in most organisms [67–69]. Indeed, a well-known example is position-effect variegation (PEV) [70] that is a reversible hallmark of yeast mating (Fig. 4). Yeast haploid cells can exist in one of two mating types, either a or alpha [71]. The two types can mate with each other to form an a/alpha diploid that can undergo meiosis (and DNA recombination) resulting in four meiotic products or haploid spores. The Mata and Matalpha genes are located at HMLalpha and HMRa on the left and right arms of chromosome III, respectively (Fig. 4). The MAT locus serves as a recipient of either an a-specific sequence or an alphaspecific sequence, which are placed via gene conversion into the active MAT locus site after HO endonuclease cleavage [71]. Arrangement of the Mata and Matalpha genes proximal to the “E-box” and “I-box” in the HMLalpha and HMRa sites results in heterochromatic silencing at the specific locus, and only the gene that is copied into the active MAT locus is expressed (Fig. 4). Initiation of facultative heterochromatin depends on site-specific transcriptional repressors, which, in turn, recruit a silencing complex, in this case, the silence information regulator (SIR) proteins [71]. Thus, genes are on or off depending on whether they reside near an insulator. Insulators act as barriers in rare cases where constitutive heterochromatin and highly active genes are juxtaposed in mammalian cells. In a similar process, PEV is used by mammalian insulators such as that present upstream of the 5' HS4 (5’ hypersensitive site 4) in the chicken β -globin locus [72]. Heterochromatin encroaches on adjacent genes or recedes from genes at the extremes of domains, and PEV represses them by being positioned (in cis) at these boundary domains.
Figure 4. Mating type locus and heterochromatin regulation.
Genome expression is active at the MAT locus (A) The donor sequences for gene conversion are located at HMLalpha and HMRa on the left and right arms of chromosome III respectively. Each donor has binding sites for key transcription factors repressor/activator protein 1 (Rap1), autonomously replicating sequence-binding factor 1 (ABF1), and an ARS consensus sequence (ACS), which binds the origin recognition complex. ABF1 and RAP1 contribute to transcriptional activation of genes involved in carbon source regulation, sporulation, amino acid biosynthesis, and ribosomal functions. The transcription factors (RAP1 and ABF-1) at HMLalpha and HMRa recruit the silencing information proteins (SIR), which keep these loci in a silent, heterochromatic state where genes at the two loci cannot be expressed, nor can they be cleaved by the HO endonuclease. The MATa cells or HMLalpha loci also contain an autonomously replicating sequence bound with the origin recognition complex. During mating, either the MATa cells or HMLalpha locus is copied into the active MAT locus for expression (Active). A phenomenon of “donor preference” refers to the ability of MATa cells to choose the HMLalpha locus as a donor about 90% of the time. Likewise MATalpha cells choose HMRa about 90% of the time. (B) The MAT locus is an active site with open chromatin suitable for transcription of MATa cells or HMLalpha genes.
So, is siRNA generation at TNR genes abnormal? Normal unexpanded TNR genes retain the same features of normal silent loci. Indeed, in the case of the wild-type DM1 insulator, the antisense transcript emanating from the adjacent SIX5 regulatory region extends into the insulator element and is converted into 21 nucleotide (nt) fragments accompanied by transcriptional silencing [14, 73]. CTCF and HP1γ are enriched in the region of the wt CTG repeats, implying that the siRNA processing step does not distinguish the normal and expanded genes. CTCF is bound to the insulator, and appears to prevent the propagation of heterochromatin to the surrounding regions [73]. Gene silencing by siRNA, generation of riboparticles [74], as well as heterochromatin formation and insulator activity associated with DM1 mimic normal processes.
Abnormal mechanisms
Clearly, the consequences of TNR expansion at disease loci are abnormal. So what is different in an expanded gene? At the wt DMPK locus, the region is in a phased nucleosome array with a nucleosome positioned over the repeats [41], whereas CTG expansion is sufficient to reduce or exclude CTCF binding and to nucleate the spreading of heterochromatin to surrounding regions. Spreading results in a region more resistant to nuclease access [73] and suppression of the SIX5 gene. The spread of H3-K9 dimethylation to a broader region also occurs if the CTCF site is mutated [14].
Why CTCF is lost by expansion-mediated heterochromatin structural changes in the expanded but not in the wt gene is unknown. The silenced genes in model organisms contain transcription factors, histone- and DNA-modifying enzymes, and other structural proteins that maintain a silent state (Fig. 5). The level of 21 nt fragments in normal wt fibroblasts is similar to that of adult onset and congenital DM cells implying that siRNA is not overproduced in disease cells [14]. It seems likely that if siRNA processing is normal, and the repressive histone marks are present on the wt gene, then exclusion of CTCF is unlikely to result from gaining histone marks, the heterochromatin spreading, or the hypermethylation that appears to follow CTCF loss. Much more needs to be learned to sort out the steps between expansion and chromatin spreading, but several mechanisms for loss of CTCF might be envisioned. One possibility is abnormal splicing. Since alternative splicing is also perturbed in DM1 cells, it might be speculated that the expanded CUG RNA generates alternatively spliced fragments that compete for binding of CTCF, resulting in failure to establish normal heterochromatin status. Another possibility is abnormal siRNA. Although the level of siRNA production does not appear to be distinct between wt and disease transcripts, it is still possible that the siRNAs from the DM1 loci have longer half-lives and lead to more severe suppression of target genes. Indeed, CUG-BP regulates alternative splicing of the human cardiac troponin T (cTNT) pre-messenger RNA, which is disrupted in DM striated muscle [74]. In SCA8, splicing of the CAG tract leads to siRNAs that suppress normal counterparts of expanded genes [36]. Whether suppression of this array of targeted genes is sufficient to cause toxicity is an issue of intense interest.
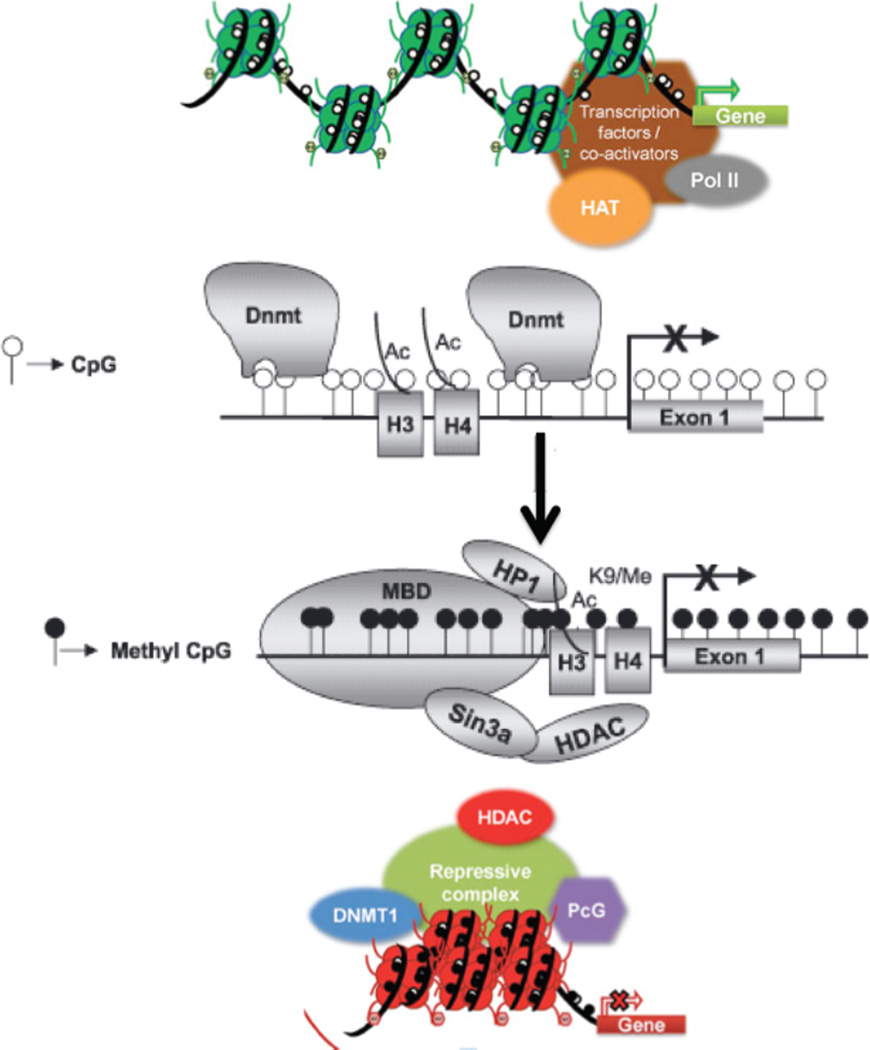
Figure 5. Schematic representation of heterochromatin formation.
A model for heterochromatin formation at repetitive elements has emerged from studies in lower eukaryotes (reviewed in [67]). In this model, bidirectional RNA transcripts are converted to siRNA, which recruits histone methyltransferases, HP1, and DNA methyltransferases with associated conversion of the region to heterochromatin. The DNA methyltransferase (DNMT) transfers a methyl group to DNA at CpG sites (white balls) resulting in 5-Me-cytosine at these positions (black balls). Histone deacetylases (HDACs) remove the acetyl groups on the histone tails.
The physical relationship between altered protein splicing and heterochromatin spreading is unknown. A caveat in assessing the effects on heterochromatin, however, is our poor understanding of its definition. In DM1 and FXS, hypermethylation results in a reduction of nuclease sensitivity, but whether this corresponds to actual condensation of interphase chromatin versus changes in protein coating of DNA is not formally known [14, 75]. Dense packaging and staining at the nuclear periphery remains the classic cytochemical signature of heterochromatin. However, the distinction between chromatin packaging and gene expression has become blurred. Interphase chromatin is typically thought to be de-condensed at locations in which genes are transcribed and condensed where genes are silent [76]. However, this generalization is contradicted by observations that inactive genes reside in domains of open chromatin whereas active genes reside in regions of low gene density and can be embedded within compact chromatin fibers [77, 78]. This might be relevant to the DM1 gene, which is embedded within a region of euchromatin-associated H3 lysine 4 (H3-K4) methylation [14]. A diversity of theoretical models for the nuclear organization of active and silent chromatin stem from our limited understanding of higher-order chromatin structure beyond the 30-nm fiber. The technical limitations in imaging of chromatin by light microscopy inhibits a more detailed look, but with the generation of super resolution microscopes, some of these processes may be better resolved at the cytogenetic level.
DNA INSTABILITY AT BIDIRECTIONALLY TRANSCRIBED TNR TRACTS
The involvement of the CTCF insulator in DM1 pathology raises another puzzling aspect of disease pathology. Many TNR genes lie close to a CTCF site boundary element (Table 1) and might be subject to heterochromatin-driven repression, yet the chromatin spreading of repressed chromatin is restricted to only a few (Table 1). Is chromatin context and antisense processing directly connected to instability? The results from ATX7 suggest that it is. Indeed, the CAG repeat in the spinocerebellar ataxia type 7 (SCA7) gene is the most unstable of all the CAG repeat loci [79]. Transgenic mice carrying SCA7 genomic fragments with CTCF binding-site mutations promote triplet repeat instability both in the germ line and in somatic tissues [80], and CpG methylation or mutation of CTCF binding sites further destabilizes triplet repeat expansions. SCA7 is bidirectionally transcribed. CTCF is required for the SCA7 antisense (SCAANT1) expression, and loss of SCAANT1 de-repressed ataxin-7 sense transcription [18]. Although SCAANT1 expression in trans does not reduce ataxin-7 alternative sense promoter activity in vitro or in vivo, convergent transcription of SCAANT1 in cis leads to repression that is accompanied by post-translational modification of histones and DNA. Thus, CTCF transactivation of antisense non-coding RNA, histone modification and the ensuing DNA hypermethylation state imply that TNR instability occurs through an epigenetic mechanism. Below, we discuss possible mechanisms by which expansion might occur in the context of bidirectional transcription arising from both coding and non-coding RNAs.
DNA DAMAGE AND REPAIR OF TNRs ARISING FROM BIDIRECTIONAL TRANSCRIPTION
In neurons, DNA expansion occurs in the process of excising chemically modified bases from DNA, providing a direct means to instability [1, 81]. Progressive somatic expansion throughout the lifetime of an individual results from the process of removing base lesions and may determine onset of disease via a critical threshold mechanism of repeat length. If RNA processing influences genomic instability at the DNA level, then RNA-mediated toxicity either increases oxidative stress or methylation, or alters the chromatin context and accessibility of the TNR repeat to damage. The two distinct classes of RNA-bidirectional genes, while they cause the problem, perhaps provide at least some insight as to mechanistic possibilities.
THE ROLE FOR DNA REPAIR OF CODING RNAs
Oxidative damage is abundant in non-dividing neuronal cells and DNA expansion of TNRs arises in the process of correcting oxidized and chemically modified DNA bases through use of the excision repair machinery [81, 82]. A hallmark of neurodegenerative diseases is the accumulation and aggregation of misfolded pathogenic proteins. Considering RAN-translation, CAG/CTG coding tracts have the possibility to generate homopolymeric proteins of polyglutamine, polyserine, and polyalanine tracts [13]. Oxidative stress is a common feature of these expansion diseases and is well documented and extensively reviewed [83, 84]. Inflammation [85], ER stress [86], and MT respiration [87] provide sources for reactive oxygen species (ROS), which affect modification of both the nuclear and the mitochondrial genomes.
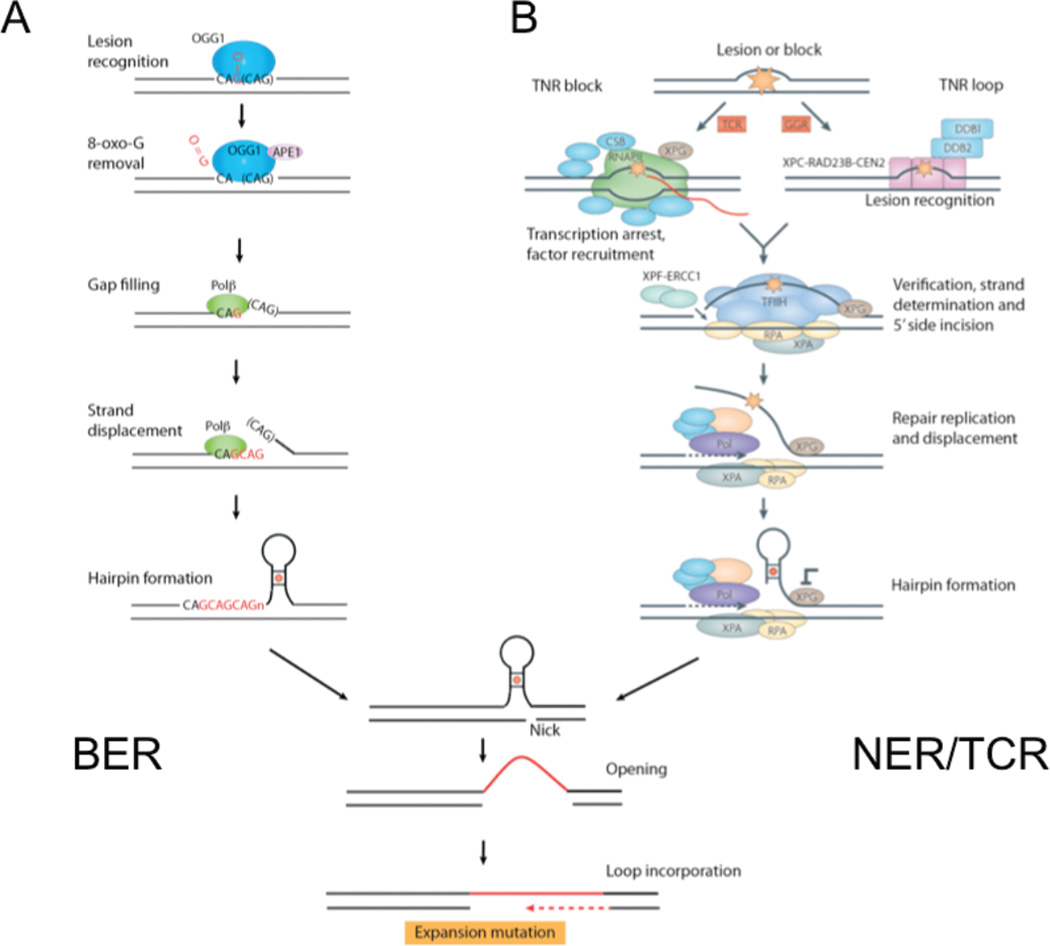
Two pathways are implicated in TNR expansion through removal of oxidized bases: the base excision repair (BER) pathway and the transcription coupled repair pathway (TCR) [1, 81, 88–91] (Fig. 6). In mammalian cells, BER of oxidized bases is initiated by the action of a lesion-specific DNA N-glycosylase, which catalyses the hydrolysis of the C1 N-glycosyl bond that links the base to the sugar-phosphate backbone [92]. Release of the damaged base generates an apurinic (AP) site, where the base on one strand has no partner on the other [93–95]. A current model for expansion arising from BER posits that a polymerase, during gap filling synthesis, displaces the TNR repeat, allowing for formation of hairpin loops [1, 90]. After its incorporation into genomic DNA, the loop becomes the expansion. To date, more than 10 mammalian glycosylases have been identified, with different substrate specificities for removal of oxidized bases [96]. However, to date, only OGG1 [81], and more recently, NEIL1 [88] glycoslyases have been identified as positive regulators of expansion.
Figure 6. BER and TCR.
(A) Schematic diagram for BER-mediated expansion. Loops formed during base excision repair by strand displacement: the toxic oxidation cycle. In base excision repair (BER), 7,8-dihydro-8-oxoguanine DNA glycosylase (OGG1) (blue oval) recognizes and removes oxidized guanines (O = G) (red) in the DNA template. OGG1 has two enzymatic functions: glycosylase activity and lyase activity. The glycosylase activity cleaves the C1 glycosidic bond between the ribose sugar and the base. Removal of the oxidized guanine creates an apurinic site in which the widowed cytosine has no partner. OGG1 can nick the phosphodiester backbone between the C3 bond of the ribose ring and the phosphate on the 3′ side of the apurinic site. Apurinic/apyrimidinic endonuclease 1 (APE1, also known as APEX1) (pink oval) processes the AP site. This produces a 3′ hydroxyl group (OH) suitable for extension by a DNA polymerase (Pol) (green oval). The trinucleotide repeat (TNR) strand is displaced during gap-filling synthesis and TNRs from the displaced ‘flap’ can fold back into a hairpin (loop formation). The hairpin DNA is ligated and expansion occurs after the DNA hairpin loop is incorporated into duplex DNA. (B) Schematic diagram for TCR-mediated expansion. Nucleotide excision repair has two branches: transcription-coupled repair (TCR) (left) and global genome repair (GGR) (right). In a model for the involvement of TCR in trinucleotide repeat (TNR) loop formation, the GC-rich TNR sequences block (orange star) progression of RNA polymerase II (RNAPII) (the red line represents mRNA). TCR rescues the stalled polymerase through the combined action of a group of accessory proteins (blue), including Cockayne syndrome protein CSB (also known as ERCC6). Xeroderma pigmentosum complementation group A (XPA) stabilizes the transcription bubble. The TNR tract should eventually be removed from DNA by the action of two nucleases: the XPF–ERCC1 complex (composed of XPF and excision cross complementing repair 1 (ERCC1)) and the endonuclease XPG. However, the incision by XPF–ERCC1 is made first. XPG has been implicated in TNR instability; it both binds and stabilizes the open transcription ‘bubble’. XPG is recruited to the opened DNA together with TFIIH (a transcription factor complex associated with RNAPII). Helicase and ATPase activities within TFIIH stimulate further opening of the bubble. Replication protein A (RPA) and XPA protect the ssDNA in the denatured bubble and stabilize the complex. Gap-filling repair (involving DNA polymerase (Pol) and associated proteins) and strand displacement from the 5′ side incision begin before the 3′ side incision is made by XPG. Thus, the transient flap created during strand displacement repair can fold back to form a stable hairpin loop at TNRs (mismatched bases in the stem of the hairpin are indicated by a red circle).
Oxidized bases are also removed by transcription-coupled repair (TCR) [97–100]. In TCR, a complex of xeroderma pigmentosum complementation group F (XPF) and excision repair cross complementing 1 (ERCC1) makes an incision 5′ to the lesion, and the endonuclease XPG makes an incision 3′ to the lesion, reviewed in [1, 101]. This excises a patch consisting of 10–20 nucleotides. In vivo, the 5′ incision by XPF–ERCC1 is made before the 3′ incision by XPG [102], so ssDNA containing the TNR has the potential to form a looped intermediate, at least transiently (Fig. 6B). A TCR mediated expansion model is similar to the BER model: the TNR flap folds back to allow formation of a hairpin loop [1, 89, 102]. After its incorporation into genomic DNA, the loop becomes the expansion [103]. In mice, loss of XPC, which functions specifically to recognize aberrant bases in the whole genome by global genome repair (GGR) (Fig. 6B), has little effect on CAG expansion in HD, suggesting that global removal of oxidized bases does not occur frequently enough to substantially increase TNR expansion [104]. In contrast, loss of Cockayne syndrome protein CSB (also known as ERCC6), which is specific to TCR (Fig. 6), has effects on instability [90]. The way in which TCR influences expansion is, as yet, poorly understood. In the mouse, loss of CSB actively promotes deletion of TNRs in the germ cells, with weak effects in somatic cells, implying the CSB protects against expansion [90]. However, these results are not necessarily observed in other models systems. In addition to roles in repair, CSB has been shown to play a role in chromatin maintenance and remodeling, and transcriptional regulation [105]. Effects of CSB on TNR expansion may be influenced both by its role in repair and its effects on chromatin structure.
In Drosophila melanogaster, transcriptional induction of a transgene (under the control of regulatory protein GAL4) that contains exon 10 of ataxin 3 (ATXN3, also known as SCA3) results in expansion and contraction of the TNR tract within this exon [106]. Loss of XPG in this line reduces instability [106]. Similarly, in human cell lines, small interfering RNA knockdown of ERCC1, XPG and CSB reduces the CAG tract within an intron of an active hypoxanthine guanine phosphoribosyl transferase (HPRT) minigene [107, 108]. The different effects of TCR loss among mice [90], flies [106], and human cells may perhaps be explained by crosstalk between BER and TCR [109]. TCR can cooperate with BER to correct oxidative DNA damage [110]. In the mouse, CSB in vivo protects against CAG expansion, by either active reduction of the CAG tracts, or by antagonizing the action of OGG1 [90], which tends to expand them. However, CSB also interacts with BER machinery such as AP endonuclease 1 (APE1) [111] and poly(ADP-ribose) polymerase (PARP1) [112]. Thus, CSB may act through two distinct pathways. CSB and OGG1 may compete for binding to the same oxidized base in somatic cells, with opposite effects. In this model, engagement with the TCR machinery results in tract deletion, while engagement with the BER machinery results in expansion. The varying degrees of expansion or contraction may depend on which complex is bound more frequently. While a detailed mechanism awaits further investigation, the genetic results obtained in mice suggest that different repair pathways for removal of oxidative lesions can have distinct outcomes in vivo at CAG tracts. The genetic results will need to be coupled to biochemical analyses to sort out the mechanisms. Oxidation of DNA occurs frequently and has the potential to promote expansion in disease in which RNAs are coding into proteins, however, chromatin context is likely to regulate the frequency of these modifications.
THE ROLE FOR DNA REPAIR IN GENE SILENCING OF NON-CODING TNRs
Methylation and Demethylation
Methylated bases can also be removed by BER, but whether removal of methylation damage generates expansion is not clear. Recent evidence suggests that hypermethylation is not the primary force for expansion: (1) Rare individuals having full mutations but normal intelligence lack hypermethylation and maintain expression of FMR1 mRNA [113–115]. (2) In FXS, carriers with 55–200 repeats of CGGs are present without methylation, and there is excess transcription of FMR1 mRNA. (3) Pharmacologic treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine (azadC) reactivates transcription and FMRP expression but does not alter the repeat tract [116–119]. Collectively, it appears that either the expansion mutation does not require methylation, or expansion by removal of methylated bases is rapid. Under the latter conditions, a BER model for expansion is possible. Indeed removal of methylated bases is accomplished by several DNA glycosylases with different specificities [120], although none are known to promote TNR expansion.
TDG and MBD4
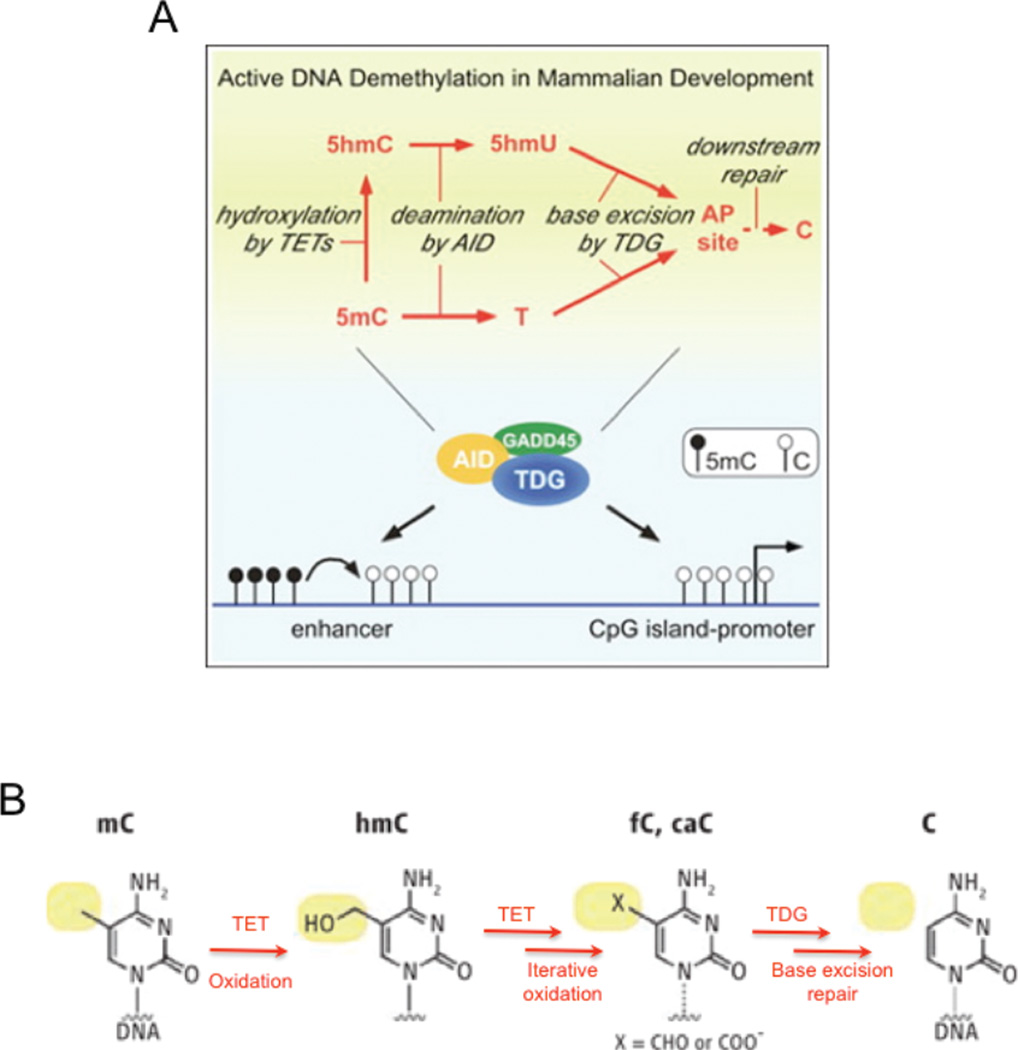
The relationship between methylation and expansion, however, is far from clear. Current evidence suggests that BER has a complex role, which is used to establish an epigenetic balance between DNA demethylation and protection from hypermethylation (Fig. 7A). This type of epigenetic modulation is likely to be key since the degree of methylation establishes genetic “homeostasis”: too much methylation leads to gene shut-down while too little leads to genome instability. A class of DNA glycosylases in vertebrates plays a key role in this type of epigenetic regulation [121–123]. Thymine DNA glycosylase (TDG) is a multi-functional protein that is coupled to demethylation [124–128]. TDG overexpression in human cells correlates with demethylation and activation of a reporter gene promoter [129], and down-regulation of TDG in mouse myoblasts causes a genome-wide methylation [130]. The process of demethylation has been extensively analyzed in germ cells, which undergo genetic reprogramming during development. Active demethylation in the paternal genome during early embryogenesis involves DNA repair via cytosine deaminases of the activation-induced cytosine deaminase (Aid) and apolipoprotein B editing complex Apobec1 family [131]. Interestingly, the Apobec1 family is the same complex that binds to CUG repeats for alternative splicing [132]. Aid and Apobec1 can deaminate 5-methylcytosine to thymine, creating T–G mismatches [133]. This Aid-Apobec1 complex works in conjunction with the glycosylase methyl-CpG-binding domain protein 4 (MBD4) and the growth arrest and DNA-damage-inducible α (Gadd45α) [123] (Fig. 7), a nucleotide excision repair (NER) component to remove T replace it with C. This process prevents hypermethylation of CpG residues (Fig. 7). The same chemical reaction could occur via mismatch repair (MMR) [134], as has been suggested in somatic hypermutability, or even via nucleotide excision repair (NER) [135]. However, both TDG and MBD4 have strong preference for removal of methyl groups in CpG sequences [136, 137]. TDG knockouts result in neonatal lethality, indicating that it is essential for differentiation and development [19, 20], and may play a role in establishing gene expression patterns during differentiation.
Figure 7. TDG keeps CpG islands unmethylated and actively demethylates promoters and enhancers.
(A) Model of the dual role of TDG in DNA demethylation and protection from hypermethylation. TDG interacts with AID and Gadd45α and regulates the levels of AID. TDG removes 5-hydromethyluracil originated by deamination of 5-hydroxymethylcytosine. (B) The TET enzymes are proposed to oxidize 5-methylcytosine (mC) to 5 hydroxy-methylcytosine (hmC) and subsequently to generate the higher oxidation substituents 5-formylcytosine (fC) and 5-carboxylcytosine (caC) (shown as the structure with the 5-X substituent). Unmodified cytosine (C) is on the far right. Base excision repair, initiated by thymine-DNA glycosylase (TDG), releases and replaces the entire modified oxidized base with unmodified C.
If TDG is involved in prevention of hypermethylation, for instance at the hypermethylated FMR1 promoter of fragile X, a key question is how TDG is directed to the local site. Candidate proteins that act as loading factors are those that interact physically at the FMR1 promoter region. Several transcription factors (TFs) and co-activators that target these sites in vivo have been identified by chromatin immunoprecipitation (ChIP) analysis [138–141]. Some of them, such as Sp1 and CBP/ p300, are bound to active but not to the fully mutated FMR1 promoter [138, 140] (Fig. 7). Therefore, possible ways for TDG localization to the FMR1 promoter include (1) association with CBP/p300 proteins that can be recruited to the promoter by CREB; (2) two retinoic acid-responsive factors, Sp1 and AP-2, and (3) CGG repeats may form a G-quartet structure, which may be bound by FMRP. For example, CREB binding protein (CBP), known to integrate multiple regulatory pathways within chromatin, can physically interact with and acetylate TDG in vitro and in vivo [142, 143]. As a complex, CBP binding to TDG does not compromise CBP function as a transcriptional activator nor does complex formation alter the glycosylase function of TDG, but the functional relationship between TDG and CBP is as yet poorly understood.
There are no data to address whether CBP recruitment by TDG to T-G pairs facilitates repair by chromatin remodeling, or whether TDG is brought to promoter regions by the transcription machinery to repair the lesion. TDG acetylation by CBP leads to its release from the TDG/CBP complex, and prevents recruitment of APE to the abasic lesion [142]. Thus, it is possible that the TDG acetylation state regulates switching from its function in transcription to its function in repair. Recent data suggest that it may be the non-catalytic interactions of TDG that provide the context with which it can effectively carry out its catalytic activities. For example, a complex containing TDG acts as a co-activator for estrogen receptor (ER)-regulated transcription, but stimulation does not depend on its glycosylase domain [144]. It is likely that proper silencing requires a balance (or antagonism) between methylation and active demethylation, which is disrupted by expansion [145, 146].
TET PROTEINS
The human TET1 protein is another relevant DNA repair protein that catalyzes the conversion of 5-methylcytosine (5mC) of DNA to 5-hydroxymethylcytosine (5hmC), raising the possibility that DNA demethylation may be regulated by a Tet1-mediated process [147] (Fig. 7C). All Tet-family proteins (Tet1, Tet2 and Tet3) catalyze a similar reaction [148–150], which has roles in epigenetic regulation and demethylation [151–155]. Both TET1 and TET3 proteins contain a DNA-binding motif that is believed to target CpG sites [156–158]. Due to the activity of DNA methyltransferases (DNMTs) in maintenance of methylation patterns, levels of 5-meC remain relatively constant, however, levels of 5-hmC tend to vary significantly between tissue types. The highest levels of 5-hmC are in neuronal cells of the central nervous system [159, 160]. The TET proteins themselves are also differentially expressed; TET1 is expressed early in embryonic development and in only selected tissues in adulthood, including the post-mitotic cells of the brain. TET2 and TET3 are more ubiquitously expressed [147, 150]. An important new feature of Tet activity is the production of hydroxymethylation of DNA, which also provides a third source for oxidative damage that would be removed by excision repair. Thus, methylation/demethylation may not directly generate an expansion. However, the degree of methylated bases may in the end, indirectly cause expansion via BER removal of the oxidized bases generated by TET activity. Collectively, there are multiple excision repair processes that modulate the methylation status and oxidation status of TNRs. The balance among them is critical to maintain normal genomic stability and epigenetic balance, which is upset by TNR expansion.
CONCLUSION
Given that antisense transcription occurs widely in the human genome, defining the range of potential roles and impact of antisense repeat transcripts on trinucleotide repeat diseases and on the expansion mutation itself is of great importance. The number of possible regulatory mechanisms provides the next frontier of biology that is likely to not only shed light on the triplet repeat pathophysiology, but also on the DNA repair mechanisms that regulate and stabilize the human genome.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grants NS40738 (CTM), GM066359 (CTM), NS062384 (to CTM), and NS060115 (to CTM), and CA092584 (CTM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11(11):786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11(4):247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dion V, Wilson JH. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25(7):288–297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. The Journal of biological chemistry. 2009;284(12):7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imarisio S, et al. Huntington's disease: from pathology and genetics to potential therapies. The Biochemical journal. 2008;412(2):191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 6.Messaed C, Rouleau GA. Molecular mechanisms underlying polyalanine diseases. Neurobiol Dis. 2009;34(3):397–405. doi: 10.1016/j.nbd.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Trinklein ND, et al. An abundance of bidirectional promoters in the human genome. Genome research. 2004;14(1):62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y, et al. The antisense transcriptomes of human cells. Science. 2008;322(5909):1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda Y, Daughters RS, Ranum LP. Bidirectional expression of the SCA8 expansion mutation: one mutation, two genes. Cerebellum. 2008;7(2):150–158. doi: 10.1007/s12311-008-0010-7. [DOI] [PubMed] [Google Scholar]
- 10.Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum Mol Genet. 2010;19(R1):R77–R82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamori M, Pearson CE, Thornton CA. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats. Hum Mol Genet. 2011;20(3):580–8. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(1):260–5. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson CE. Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames , potentially nine toxic entities! PLoS genetics. 2011;7(3):e1002018. doi: 10.1371/journal.pgen.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho DH, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20(3):483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Wilburn B, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington's disease-like 2 mice. Neuron. 2011;70(3):427–440. doi: 10.1016/j.neuron.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladd PD, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16(24):3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 17.Chung DW, et al. A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Hum Mol Genet. 2011;20(17):3467–3477. doi: 10.1093/hmg/ddr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sopher BL, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70(6):1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortellino S, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146(1):67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortazar D, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470(7334):419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 21.Le May N, et al. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38(1):54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Timchenko NA, et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. The Journal of biological chemistry. 2001;276(11):7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 23.de Haro M, et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet. 2006;15(13):2138–2145. doi: 10.1093/hmg/ddl137. [DOI] [PubMed] [Google Scholar]
- 24.Pascual M, et al. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation; research in biological diversity. 2006;74(2–3):65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 25.Kino Y, et al. MBNL and CELF proteins regulate alternative splicing of the skeletal muscle chloride channel CLCN1. Nucleic acids research. 2009;37(19):6477–6490. doi: 10.1093/nar/gkp681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du H, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17(2):187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suenaga K, et al. Muscleblind-like 1 knockout mice reveal novel splicing defects in the myotonic dystrophy brain. PloS one. 2012;7(3):e33218. doi: 10.1371/journal.pone.0033218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llamusi B, Artero R. Molecular Effects of the CTG Repeats in Mutant Dystrophia Myotonica Protein Kinase Gene. Curr Genomics. 2008;9(8):509–516. doi: 10.2174/138920208786847944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margolis RL, Rudnicki DD, Holmes SE. Huntington's disease like-2: review and update. Acta neurologica Taiwanica. 2005;14(1):1–8. [PubMed] [Google Scholar]
- 30.Greenstein PE, et al. Huntington's disease like-2 neuropathology. Movement disorders : official journal of the Movement Disorder Society. 2007;22(10):1416–1423. doi: 10.1002/mds.21417. [DOI] [PubMed] [Google Scholar]
- 31.Rudnicki DD, et al. A comparison of huntington disease and huntington disease-like 2 neuropathology. Journal of neuropathology and experimental neurology. 2008;67(4):366–374. doi: 10.1097/NEN.0b013e31816b4aee. [DOI] [PubMed] [Google Scholar]
- 32.Holmes SE, et al. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nature genetics. 2001;29(4):377–378. doi: 10.1038/ng760. [DOI] [PubMed] [Google Scholar]
- 33.Koob MD, et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nature genetics. 1999;21(4):379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 34.Moseley ML, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nature genetics. 2006;38(7):758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 35.Mutsuddi M, et al. The spinocerebellar ataxia 8 noncoding RNA causes neurodegeneration and associates with staufen in Drosophila. Curr Biol. 2004;14(4):302–308. doi: 10.1016/j.cub.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 36.Daughters RS, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS genetics. 2009;5(8):e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Y, et al. Targeted deletion of a single Sca8 ataxia locus allele in mice causes abnormal gait, progressive loss of motor coordination, and Purkinje cell dendritic deficits. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26(39):9975–9982. doi: 10.1523/JNEUROSCI.2595-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cernilogar FM, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480(7377):391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Z, Teng X, Bonini NM. Triplet repeat-derived siRNAs enhance RNA-mediated toxicity in a Drosophila model for myotonic dystrophy. PLoS genetics. 2011;7(3):e1001340. doi: 10.1371/journal.pgen.1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. The EMBO journal. 2000;19(17):4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filippova GN, et al. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nature genetics. 2001;28(4):335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 42.Brook JD, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;68(4):799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 43.Jansen G, et al. Physical and genetic characterization of the distal segment of the myotonic dystrophy area on 19q. Genomics. 1992;13(3):509–517. doi: 10.1016/0888-7543(92)90118-c. [DOI] [PubMed] [Google Scholar]
- 44.Mahadevan M, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255(5049):1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 45.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28(1):1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 46.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12(8):542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 47.Barr ML, Carr DH. Correlations between sex chromatin and sex chromosomes. Acta Cytol. 1962;6:34–45. [PubMed] [Google Scholar]
- 48.Belmont AS, Bignone F, Ts'o PO. The relative intranuclear positions of Barr bodies in XXX non-transformed human fibroblasts. Exp Cell Res. 1986;165(1):165–179. doi: 10.1016/0014-4827(86)90541-0. [DOI] [PubMed] [Google Scholar]
- 49.Puck TT, Johnson R. DNA exposure and condensation in the X and 21 chromosomes. Stem cells. 1996;14(5):548–557. doi: 10.1002/stem.140548. [DOI] [PubMed] [Google Scholar]
- 50.Hall LL, Lawrence JB. XIST RNA and architecture of the inactive X chromosome: implications for the repeat genome. Cold Spring Harbor symposia on quantitative biology. 2010;75:345–356. doi: 10.1101/sqb.2010.75.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinter SF, et al. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome research. 2012 doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. 2011;130(2):223–236. doi: 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Handa V, Saha T, Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic acids research. 2003;31(21):6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin SL, Chang SJ, Ying SY. First in vivo evidence of microRNA-induced fragile X mental retardation syndrome. Molecular psychiatry. 2006;11(7):616–617. doi: 10.1038/sj.mp.4001828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krol J, et al. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Molecular cell. 2007;25(4):575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 56.Lin SL, Miller JD, Ying SY. Intronic microRNA (miRNA) Journal of biomedicine & biotechnology. 2006;2006(4):26818. doi: 10.1155/JBB/2006/26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin SL, Kim H, Ying SY. Intron-mediated RNA interference and microRNA (miRNA) Frontiers in bioscience : a journal and virtual library. 2008;13:2216–2230. doi: 10.2741/2836. [DOI] [PubMed] [Google Scholar]
- 58.Lin SL, et al. A novel RNA splicing-mediated gene silencing mechanism potential for genome evolution. Biochemical and biophysical research communications. 2003;310(3):754–760. doi: 10.1016/j.bbrc.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 59.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 60.Yi R, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & development. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lund E, et al. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 62.Lin SL, Chang D, Ying SY. Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene. 2005;356:32–38. doi: 10.1016/j.gene.2005.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12(1):19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Creamer KM, Partridge JF. RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast. Wiley Interdiscip Rev RNA. 2011;2(5):632–646. doi: 10.1002/wrna.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 66.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 67.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19(14):1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 68.Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431(7005):211–217. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- 69.Morris KV, et al. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305(5688):1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 70.Dernburg AF, et al. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996;85(5):745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 71.Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191(1):33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Burgess-Beusse B, et al. The insulation of genes from external enhancers and silencing chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(Suppl 4):16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Otten AD, Tapscott SJ. Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(12):5465–5469. doi: 10.1073/pnas.92.12.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280(5364):737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 75.Kumari D, Usdin K. The distribution of repressive histone modifications on silenced FMR1 alleles provides clues to the mechanism of gene silencing in fragile X syndrome. Hum Mol Genet. 2010;19(23):4634–4642. doi: 10.1093/hmg/ddq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wegel E, Shaw P. Gene activation and deactivation related changes in the three-dimensional structure of chromatin. Chromosoma. 2005;114(5):331–337. doi: 10.1007/s00412-005-0015-7. [DOI] [PubMed] [Google Scholar]
- 77.Gilbert N, Bickmore WA. The relationship between higher-order chromatin structure and transcription. Biochem Soc Symp. 2006;73:59–66. doi: 10.1042/bss0730059. [DOI] [PubMed] [Google Scholar]
- 78.Gilbert N, et al. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118(5):555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 79.Garden GA, La Spada AR. Molecular pathogenesis and cellular pathology of spinocerebellar ataxia type 7 neurodegeneration. Cerebellum. 2008;7(2):138–149. doi: 10.1007/s12311-008-0027-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Libby RT, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS genetics. 2008;4(11):e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447(7143):447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johri A, Beal MF. Antioxidants in Huntington's disease. Biochimica et biophysica acta. 2012;1822(5):664–674. doi: 10.1016/j.bbadis.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends in biochemical sciences. 2012;37(4):162–172. doi: 10.1016/j.tibs.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quintanilla RA, Johnson GV. Role of mitochondrial dysfunction in the pathogenesis of Huntington's disease. Brain Res Bull. 2009;80(4–5):242–247. doi: 10.1016/j.brainresbull.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferguson LR. Chronic inflammation and mutagenesis. Mutat Res. 2010;690(1–2):3–11. doi: 10.1016/j.mrfmmm.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 86.Doyle KM, et al. Unfolded proteins and endoplasmic reticulum stress in neurodegenerative disorders. J Cell Mol Med. 2011;15(10):2025–2039. doi: 10.1111/j.1582-4934.2011.01374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pan Y. Mitochondria, reactive oxygen species, and chronological aging: a message from yeast. Exp Gerontol. 2011;46(11):847–852. doi: 10.1016/j.exger.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 88.Mollersen L, et al. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salinas-Rios V, Belotserkovskii BP, Hanawalt PC. DNA slip-outs cause RNA polymerase II arrest in vitro: potential implications for genetic instability. Nucleic acids research. 2011;39(17):7444–7454. doi: 10.1093/nar/gkr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kovtun IV, Johnson KO, McMurray CT. Cockayne syndrome B protein antagonizes OGG1 in modulating CAG repeat length in vivo. Aging. 2011;3(5):509–514. doi: 10.18632/aging.100324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hubert L, Jr., et al. Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum Mol Genet. 2011;20(24):4822–4830. doi: 10.1093/hmg/ddr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell research. 2008;18(1):27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 94.Lindahl T. DNA glycosylases in DNA repair. Basic Life Sci. 1986;38:335–340. doi: 10.1007/978-1-4615-9462-8_36. [DOI] [PubMed] [Google Scholar]
- 95.Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 96.Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121(1):1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, et al. Transcriptional inhibition by an oxidized abasic site in DNA. Chem Res Toxicol. 2006;19(2):234–241. doi: 10.1021/tx050292n. [DOI] [PubMed] [Google Scholar]
- 98.Brooks PJ. The 8,5'-cyclopurine-2'-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA repair. 2008;7(7):1168–79. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.You C, et al. A quantitative assay for assessing the effects of DNA lesions on transcription. Nature chemical biology. 2012 doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim N, Jinks-Robertson S. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Molecular and cellular biology. 2010;30(13):3206–3215. doi: 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA repair. 2011;10(7):722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Staresincic L, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. The EMBO journal. 2009;28(8):1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Volker J, et al. Energy landscapes of dynamic ensembles of rolling triplet repeat bulge loops: implications for DNA expansion associated with disease states. Journal of the American Chemical Society. 2012;134(13):6033–6044. doi: 10.1021/ja3010896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dragileva E, et al. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009;33(1):37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Newman JC, Bailey AD, Weiner AM. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(25):9613–9618. doi: 10.1073/pnas.0510909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315(5820):1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 107.Lin Y, Hubert L, Jr., Wilson JH. Transcription destabilizes triplet repeats. Molecular carcinogenesis. 2009;48(4):350–361. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Molecular and cellular biology. 2007;27(17):6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kovtun IV, McMurray CT. Crosstalk of DNA glycosylases with pathways other than base excision repair. DNA repair. 2007;6(4):517–529. doi: 10.1016/j.dnarep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 110.Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutation research. 2003;531(1–2):231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 111.Wong HK, et al. Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic acids research. 2007;35(12):4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thorslund T, et al. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Molecular and cellular biology. 2005;25(17):7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hagerman RJ, et al. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. Am J Med Genet. 1994;51(4):298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- 114.Smeets HJ, et al. Normal phenotype in two brothers with a full FMR1 mutation. Hum Mol Genet. 1995;4(11):2103–2108. doi: 10.1093/hmg/4.11.2103. [DOI] [PubMed] [Google Scholar]
- 115.Pietrobono R, et al. Molecular dissection of the events leading to inactivation of the FMR1 gene. Hum Mol Genet. 2005;14(2):267–277. doi: 10.1093/hmg/ddi024. [DOI] [PubMed] [Google Scholar]
- 116.Chiurazzi P, et al. In vitro reactivation of the FMR1 gene involved in fragile X syndrome. Hum Mol Genet. 1998;7(1):109–113. doi: 10.1093/hmg/7.1.109. [DOI] [PubMed] [Google Scholar]
- 117.Coffee B, et al. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nature genetics. 1999;22(1):98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 118.Pietrobono R, et al. Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic Acids Res. 2002;30(14):3278–3285. doi: 10.1093/nar/gkf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tabolacci E, et al. Differential epigenetic modifications in the FMR1 gene of the fragile X syndrome after reactivating pharmacological treatments. Eur J Hum Genet. 2005;13(5):641–648. doi: 10.1038/sj.ejhg.5201393. [DOI] [PubMed] [Google Scholar]
- 120.Sedgwick B, et al. Repair of alkylated DNA: recent advances. DNA repair. 2007;6(4):429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 121.Maiti A, et al. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(26):8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133(7):1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 123.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu B, et al. Overexpression of 5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293 demethylates the promoter of a hormoneregulated reporter gene. Proc Natl Acad Sci U S A. 2001;98(9):5031–5036. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jost JP, et al. 5-Methylcytosine DNA glycosylase participates in the genomewide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic Acids Res. 2001;29(21):4452–4461. doi: 10.1093/nar/29.21.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Metivier R, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 127.Cortazar D, et al. The enigmatic thymine DNA glycosylase. DNA repair. 2007;6(4):489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 128.Hardeland U, et al. Thymine DNA glycosylase. Prog Nucleic Acid Res Mol Biol. 2001;68:235–253. doi: 10.1016/s0079-6603(01)68103-0. [DOI] [PubMed] [Google Scholar]
- 129.Zhu B, et al. Overexpression of 5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293 demethylates the promoter of a hormoneregulated reporter gene. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5031–5036. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jost JP, et al. 5-Methylcytosine DNA glycosylase participates in the genomewide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic acids research. 2001;29(21):4452–4461. doi: 10.1093/nar/29.21.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sanz LA, Kota SK, Feil R. Genome-wide DNA demethylation in mammals. Genome biology. 2010;11(3):110. doi: 10.1186/gb-2010-11-3-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Anant S, et al. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. The Journal of biological chemistry. 2001;276(50):47338–47351. doi: 10.1074/jbc.M104911200. [DOI] [PubMed] [Google Scholar]
- 133.Morgan HD, et al. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. The Journal of biological chemistry. 2004;279(50):52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 134.Pena-Diaz J, Jiricny J. Mammalian mismatch repair: error-free or errorprone? Trends in biochemical sciences. 2012;37(5):206–214. doi: 10.1016/j.tibs.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 135.Rechkunova NI, Lavrik OI. Nucleotide excision repair in higher eukaryotes: mechanism of primary damage recognition in global genome repair. Sub-cellular biochemistry. 2010;50:251–277. doi: 10.1007/978-90-481-3471-7_13. [DOI] [PubMed] [Google Scholar]
- 136.Waters TR, Swann PF. Thymine-DNA glycosylase and G to A transition mutations at CpG sites. Mutat Res. 2000;462(2–3):137–147. doi: 10.1016/s1383-5742(00)00031-4. [DOI] [PubMed] [Google Scholar]
- 137.Hang B, Guliaev AB. Substrate specificity of human thymine-DNA glycosylase on exocyclic cytosine adducts. Chem Biol Interact. 2007;165(3):230–238. doi: 10.1016/j.cbi.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 138.Smith KT, Coffee B, Reines D. Occupancy and synergistic activation of the FMR1 promoter by Nrf-1 and Sp1 in vivo. Hum Mol Genet. 2004;13(15):1611–1621. doi: 10.1093/hmg/ddh172. [DOI] [PubMed] [Google Scholar]
- 139.Lim JH, et al. AP-2alpha selectively regulates fragile X mental retardation-1 gene transcription during embryonic development. Hum Mol Genet. 2005;14(14):2027–2034. doi: 10.1093/hmg/ddi207. [DOI] [PubMed] [Google Scholar]
- 140.Smith KT, Nicholls RD, Reines D. The gene encoding the fragile X RNA-binding protein is controlled by nuclear respiratory factor 2 and the CREB family of transcription factors. Nucleic Acids Res. 2006;34(4):1205–1215. doi: 10.1093/nar/gkj521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carrillo C, Cisneros B, Montanez C. Sp1 and AP2 transcription factors are required for the human fragile mental retardation promoter activity in SK-N-SH neuronal cells. Neurosci Lett. 1999;276(3):149–152. doi: 10.1016/s0304-3940(99)00798-3. [DOI] [PubMed] [Google Scholar]
- 142.Tini M, et al. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9(2):265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 143.Mohan RD, et al. SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment. Molecular and cellular biology. 2007;27(1):229–243. doi: 10.1128/MCB.00323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen D, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. The Journal of biological chemistry. 2003;278(40):38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 145.Xiao W, et al. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell. 2003;5(6):891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
- 146.Penterman J, Uzawa R, Fischer RL. Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 2007;145(4):1549–1557. doi: 10.1104/pp.107.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell selfrenewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Williams K, Christensen J, Helin K. DNA methylation: TET proteinsguardians of CpG islands? EMBO reports. 2012;13(1):28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25(23):2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guo JU, et al. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell cycle. 2011;10(16):2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13(1):7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 156.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473(7347):389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42(4):451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Globisch D, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PloS one. 2010;5(12):e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]