Abstract
Cell-based therapies hold the potential to alleviate the growing burden of liver diseases. Such therapies require human hepatocytes, which, within the stromal context of the liver, are capable of many rounds of replication. However, this ability is lost ex vivo and human hepatocyte sourcing has been limiting many fields of research for decades. Here, we developed a high-throughput screening platform for primary human hepatocytes to identify small molecules in two different classes that can be used to generate renewable sources of functional human hepatocytes. One class induced functional proliferation of primary human hepatocytes in vitro. The second class enhanced hepatocyte functions and promoted differentiation of iPS-derived hepatocytes, toward a phenotype more mature than what was previously obtainable. The identification of these small molecules can help to address a major challenge impacting many facets of liver research and may lead to the development of novel therapeutics for liver diseases.
Chronic liver disease affects more than 500 million people worldwide1. Most treatments are palliative; the only therapy shown to directly alter outcome and prevent mortality is organ transplantation, but its utility is limited by a persistent shortage of donor organs2. Cell-based therapies, such as cell transplantation, engineered hepatocellular tissue constructs and bioartificial liver devices, have long held promise as alternatives to whole organ transplantation3. Such therapies require human hepatocytes due to substantial species-specific differences between animal and human hepatocellular functions including apolipoprotein expression, metabolic regulation of cholesterol and phase I detoxification enzymes4,5.
Human hepatocytes, within the stromal context of the liver in vivo, are capable of extensive expansion. Following 2/3 partial hepatectomy (PHx), the residual mature cell populations comprised mainly of hepatocytes are able to proliferate and replace lost liver mass. In vitro, hepatocytes, particularly human ones, lose this phenomenal proliferative ability6,7. Various attempts have been made in the last several decades to harness the innate replication potential of hepatocytes ex vivo. Investigations have yielded a number of different culture conditions that can support moderate expansion of rodent hepatocytes8–13, including a multi-factor media formulation that expands rat hepatocytes through a dedifferentiated bi-potential intermediate11. However, translation of these findings to human cultures has not been reported. To overcome the growth limitations of primary human cells, human hepatocyte cell lines have been developed, derived from tumor cells or generated through introduction of the SV40 immortalizing gene14,15. Although these cell lines are growth-competent, their use is limited by safety concerns and their abnormal levels and repertoire of hepatic functions, including notable divergences in cytochrome P450 activity15,16.
Human stem cells are an attractive alternative cell source to primary human hepatocytes and immortalized cell lines, holding great promise due to their ability to self-renew without limit and to differentiate along many lineages, including hepatocytes. Induced pluripotent stem cells (iPS cells) additionally create the possibility of establishing patient-specific cell types, thus facilitating in vitro modeling of rare diseases, and enabling personalized medicine. Human iPS cells are generated from somatic cells via forced expression of reprogramming factors, and can be differentiated towards hepatocyte-like cells (iHeps) in a step-wise manner, using defined factors17–19. However, the resulting iHeps exhibit an immature hepatic phenotype, which resembles fetal hepatocytes more than adult hepatocytes. Notably, iHeps persistently express fetal markers like alpha fetoprotein (AFP) and lack key mature hepatocyte functions, as reflected by drastically reduced activity (0.1%) of many detoxification enzymes (e.g. CYP2A6, CYP3A4)17–19. These key differences between iHeps and adult hepatocytes have limited the use of stem cells as a renewable source of functional human hepatocytes, especially for in vitro applications.
For decades, human hepatocyte sourcing has been a bottleneck for many fields of research and clinical therapies. To overcome this limitation, we utilized a small-molecule screening approach and identified factors that can either induce proliferation of mature primary human hepatocytes or induce maturation of human iPS-derived hepatocyte-like cells.
RESULTS
Human hepatocyte screening platform
To identify factors that permit renewable sourcing of functional human hepatocytes, we developed a high-throughput liver platform that enables unbiased chemical screening using primary human hepatocytes, in order to avoid species-specific differences and reliance on cell lines and their attendant mutations. To date, chemical screening on such cells has been prohibited by their limited availability in large quantities, and their precipitous decline in viability and liver-specific functions in vitro. Recent advances in cryopreservation technologies enabled the storage of sufficiently large batches of human hepatocytes from single donors. Based on extensive previous characterizations, we have demonstrated that some cryopreserved primary human hepatocytes exhibit phenotypes that approach fresh hepatocytes and are thus very useful for in vitro liver studies 20–22. To maintain these cells in culture, we co-cultivated them with murine embryonic J2-3T3 fibroblasts, which have been shown to transiently stabilize hepatocytes in vitro23,24. We designed the screening platform to contain a sparse population of hepatocytes on top of a confluent feeder layer of J2-3T3s within 384-well plates (Supplementary Results, Supplementary Fig. 1a). This design enabled fibroblast-mediated hepatocyte stabilization (Fig. 1a) without hepatocyte crowding, thus offering both time and space for hepatocyte expansion.
Fig. 1. High-throughput identification of small molecules that induce proliferation and enhance functions of primary human hepatocytes.
(a) High-throughput liver platform. Primary human hepatocytes (green) were seeded on a feeder layer of confluent J2-3T3 fibroblasts (red) in 384-well plates. Line graph shows representative rate of albumin secretion in screening co-cultures and hepatocyte-only cultures (green) over time. Bar graph displays albumin secretion as a function of hepatocyte density in screening cultures. Upper inset phase-contrast imaging shows morphology of feeder-layer co-cultures (scale bar = 100 µm). Far right, Hoechst staining of screening co-cultures shows that hepatocyte nuclei (green *) have uniform texture while fibroblast nuclei (red *) are punctate (scale bar = 50 µm). Automated high-content imaging assay identifies and classifies individual nuclei. (b) Chemical screening. Bar graph depicts categories of screened and hit compounds. Scatter plots display replicates of the screen, shown separately for the image-based proliferation and competitive-ELISA functional readouts. Blue and red data points represent DMSO and experimental small molecules, respectively. Boxed regions indicate hit zones. (c) Hit validation. All primary hits were retested in an 8-point dose-response curve. Active hits had increasing curves of hepatocyte nuclei counts and/or decreasing curves of competitive [Albumin] with flat curves of cell-free [Albumin]. (d) Chemical structures of hit compounds FPH1, FPH2 and FH1. All data presented as mean ± s.d.
To assess cell fates in this platform, we developed two separate high-throughput readouts. The functional readout evaluated the phenotype of treated cells via competitive ELISA. This biochemical assay measured the level of secreted albumin as a marker for protein synthesis functions of the cultured hepatocytes (Fig. 1a).
The proliferation readout detected hepatocyte proliferation via a customized, automated, high-content imaging protocol. In brief, the assay quantified the number of hepatocyte nuclei, using nuclear morphologies to distinguish the hepatocyte and fibroblast sub-populations that co-exist within the screening platform. When visualized with Hoechst stain, hepatocyte nuclei are more uniform in texture while fibroblast nuclei are punctate (Fig. 1a). We developed automated image analysis pipelines to identify every nucleus in every Hoechst-stained image of the screening cultures and to measure various characteristics (e.g. shape, size, intensity, texture) of each nucleus using the open-source CellProfiler software25. We used these characteristics to train machine-learning algorithms to identify and count the number of hepatocyte nuclei in each image using CellProfiler Analyst (Supplementary Fig. 2)26,27. Assay validation data showed that this image-based readout can confidently (Z’>0) detect doublings in hepatocyte nuclei numbers with low variance (CV<20%) and good reproducibility. In addition to quantifying hepatocyte nuclei that have completed mitosis, we also found and quantified the number of nuclei in the process of mitosis. Two additional analysis pipelines were built to detect nuclear morphologies consistent with cells undergoing metaphase and anaphase.
High-throughput identification
Using the high-throughput liver platform with both readouts, we screened 12,480 small molecules supplied by the Broad Institute. Screen details are provided in Supplementary Tables 1 and 2. Briefly, the collection included commercially available compounds, known bioactives and natural products (Fig. 1b). In order to avoid donor-to-donor variability, human primary hepatocytes from a single donor (donor a) were pre-conditioned for seven days by J2-3T3 fibroblasts in collagen-coated 384-well plates (Supplementary Fig. 1b). Upon stabilization in vitro, cultures were treated with individual molecules from the chemical library for 48 hours. All compounds were tested in duplicate, at a single concentration of ~15 µM. Following compound treatment, media supernatants were collected for functional analyses via competitive ELISA and cultures were fixed in 4% paraformaldehyde for proliferation analyses via imaging (Supplementary Tables 1 and 2).
To identify proliferation hits, we integrated the three image-based readouts (the number of nuclei in metaphase, in anaphase and hepatocyte nuclei in interphase). Functional hits were separately identified based on the ELISA readout. 93 compounds met all hit selection criteria, qualifying as functional proliferation hits (FPH); Figure 1b shows the types of compounds that constituted this set of hits. In addition to these 93 FPHs, we also identified proliferation-only and function-only hits. A total of 400 primary hits across these classes were retested in eight-point dose-response curves using a different donor of cryopreserved hepatocytes (donor b) for biological diversity (Supplementary Fig. 1c). Remaining hits were further tested using a cell-free counter assay to eliminate compounds that interfered with the ELISA assay chemically. Ultimately, we obtained 12 confirmed hits (Fig. 1c and Supplementary Fig. 3).
We divided these confirmed hits into three separate classes of compounds: proliferation hits, functional hits and functional proliferation hits (Supplementary Table 3). Each of the three classes may enable a distinct approach towards generating renewable sources of functional human hepatocytes and may synergize when used in combination. We chose to focus on candidates within the functional proliferation hit (FPH) and functional hit (FH) categories only (Supplementary Fig. 1d). FPHs induced functional proliferation of hepatocytes in vitro (FPH1 (1) and FPH2 (2), Fig. 1d), and thus may be useful for expanding mature human primary hepatocytes. FHs enhanced the functions of cultured hepatocytes (FH1 (3), Fig. 1d), prompting us to hypothesize that these molecules may promote the differentiation of iPS-derived hepatocytes toward a phenotype more mature than what has been obtainable to date.
We characterized top hits through LC/MS, 1H-NMR and 13C-NMR (Supplementary Note 2). In an effort to determine if any structure-activity relationships (SARs) were present for the three strongest hits (FH1, FPH1 and FPH2), we searched the remaining 12,477 compounds screened for analogs (similarity score ≥ 0.8). Using this criterion, FH1 and FPH2 were singletons, while 21 analogs of FPH1 were found. These analogs all contained the same N-phenyl-2-(N-phenylmethylsulfonamido) acetamide core as FPH1, but had varying substitutions around the sulfonamide and amide phenyl rings (Supplementary Fig. 4). The key driver for compound activity in the FPH1 series was the presence of a 5-chloro-2-methyl substitution on the sulfonamido phenyl ring and a small functional group at the para position of the phenylamide ring. This is depicted in Supplementary Figure 4, where, FPH1 and 5 were proliferation hits, 6 a weak hit, and 7 and 8 were inactive. 9 and 10 illustrate that low steric bulk at the para position of the phenylamide ring alone is not sufficient to drive compound activity of the FPH1 series. Ultimately, while the SAR around FPH1 makes it the most suitable for chemical optimization, the three strongest hits (FH1, FPH1 and FPH2) were each selected for follow-up in downstream assays based upon their ability to increase function and proliferation of primary human hepatocytes.
Expansion of human primary hepatocytes
We assayed the ability of our strongest FPHs to expand human primary hepatocytes in vitro. As a first step, we tested Prostaglandin E2 (PGE2) as a putative positive control as it has been shown to promote liver regeneration in zebrafish and mouse via Wnt signaling28,29. We tested PGE2 on human primary hepatocytes in our high-throughput liver platform and found it to indeed perform as a FPH in this system (Supplementary Fig. 3). Two other strong FPHs (FPH1 and FPH2), identified through unbiased screening, were also tested. Both induced an increase in hepatocyte nuclei count and/or elevated the number of nuclei undergoing mitosis during primary screening (Fig. 2a and Supplementary Fig. 5a), and these effects on hepatocytes were concentration dependent (Fig. 1c). Cells treated with these FPHs also maintained their liver-specific functions (Figs. 1c and 2a).
Fig. 2. Expansion of primary human hepatocytes.
(a) Primary screening data for FPH1 and FPH2. Data presented as mean ± s.d. (b) Ki67 (red) and albumin (green) immunofluorescent staining after 6 days of culture. Bar graphs show CellProfiler quantification of displayed images. (c) FACS (right) and Cellometer (left) Automated Cell Counter analysis. Fibroblasts were labeled with CM-DiI prior to initiation of culture in order to allow identification of hepatocytes via negative selection. FACS cell counting was further enabled by fluorescent counting beads. Control cultures were treated with empty vehicle (DMSO). Data presented as mean ± SEM.
To further characterize the effects of FPH1 and FPH2, we stained for proliferation marker Ki67, counted hepatocyte cell numbers and evaluated the morphology, gene expression and functional output of treated hepatocytes. Human primary hepatocytes were cultured in standard 12-well tissue culture plates on top of a feeder layer of growth-arrested J2-3T3 fibroblasts. A single FPH was supplemented into the culture media on days 1 and 5 at a concentration of 20 µM for FPH1 and 40 µM for FPH2. Treated hepatocyte colonies increased in area over time, with more hepatocytes populating each colony (Supplementary Fig. 5b).
Immunofluorescent staining showed that FPH treatment increased Ki67 staining, which co-localized with Hoechst stains for cell nuclei, and also with human albumin stains for hepatocytes. Quantitative image analysis showed an up to 6.6-fold increase in the area of albumin-positive colonies upon small-molecule treatment (Fig. 2b). The vast majority of Ki67-positive nuclei exhibited hepatocyte nuclear morphologies, which is consistent with the lack of proliferating cells in fibroblast-only cultures treated with FPHs (Fig. 2b, Supplementary Fig. 5c). These results strongly indicate that human primary hepatocytes can be induced to proliferate in vitro using FPHs.
To characterize the degree and kinetics of proliferation, we quantified the number of hepatocytes in culture, using both an automated cell counter and FACS analysis. Results showed a dramatic, up to 10-fold increase in the number of hepatocytes when treated with various FPHs (Fig. 2c and Supplementary Fig. 5d). The strongest proliferation inducer was FPH2. Over 7 days, FPH2 induced hepatocyte doublings at a rate that is consistent with reported liver regeneration kinetics in vivo30.
To generalize our findings across multiple donors, we obtained primary human hepatocytes from six additional cell sources and treated them with FPH1 and FPH2. Immunofluoresent staining for Ki67 and albumin, along with FACS-mediated cell counting revealed that hepatocytes from every source examined expanded upon small-molecule exposure (Supplementary Fig. 6). These results, together with the primary screening and validation data obtained using cells from donors a and b, suggest that the FPHs are active across a wide range of genetically diverse individuals.
To assess the phenotype of the treated hepatocytes, we performed phase-contrast imaging, gene expression profiling and biochemical analyses. Imaging showed that normal hepatocyte morphology was maintained throughout the treatment period (Supplementary Fig. 5b). Gene expression profiling demonstrated that there are no significant differences between FPH-treated and untreated primary human hepatocytes (Supplementary Fig. 7a). Urea synthesis, a surrogate marker of nitrogen metabolism, and albumin secretion were both stable throughout FPH treatment (Supplementary Fig. 7c). Metabolic functions were assessed via examinations of cytochrome P450 (CYP450) activity and canalicular transport. Results showed active MRP2 transport, which is consistent with intact bile canaliculi (Supplementary Fig. 7b), and that CYP450 activity was not compromised by small-molecule treatment (Supplementary Fig. 7c). These data agree with published findings of sustained liver functions throughout liver regeneration31.
Maturation of human iHeps
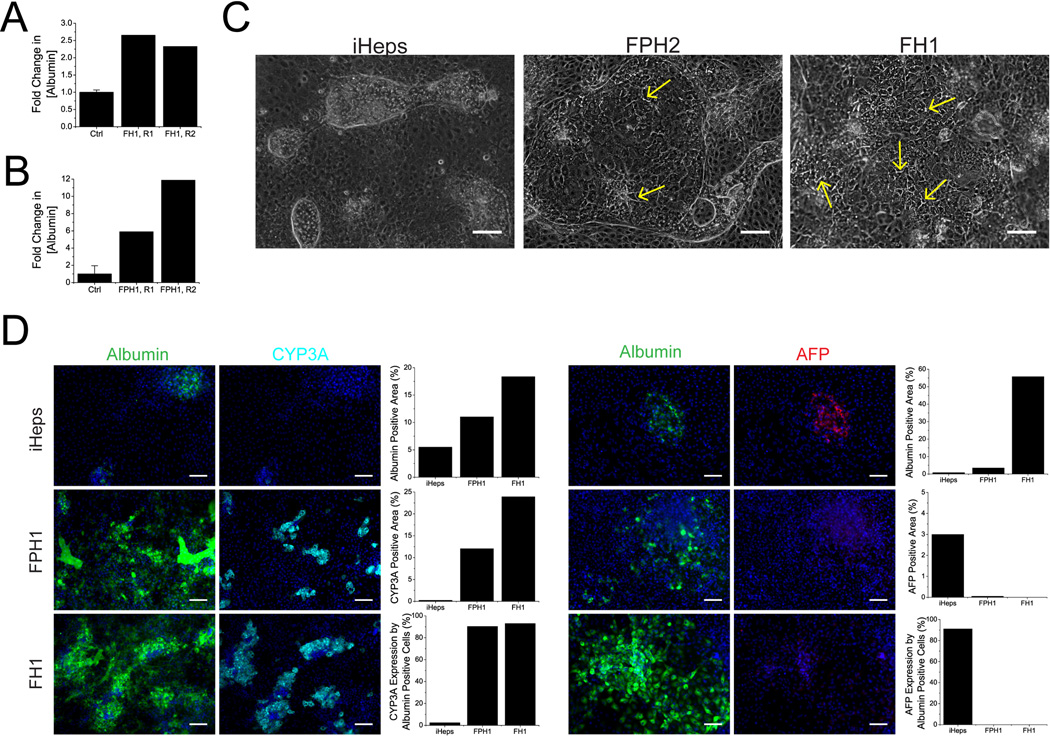
We tested the ability of hit molecules to promote the differentiation of iPS cells towards a hepatic lineage and to induce the maturation of iHeps towards more adult-like liver phenotype. Undifferentiated iPS cells were cultured on Matrigel, supported by conditioned media from primary mouse embryonic fibroblasts and differentiated into iHeps as previously described18. In brief, iPS cells were cultured in differentiation media, with sequential addition of growth factors (Activin A, BMP-4, bFGF, HGF and OSM) to guide differentiation, first into endoderm, then into hepatic specified endoderm, then into hepatic progenitor cells and finally into iHeps (Supplementary Fig. 8). Small molecules were added to basal media (Lonza, without OSM in Fig. 3, Fig. 4 and Supplementary Fig. 9; with OSM in Supplementary Fig. 10) on day 21 post initiation of differentiation and acted over a period of 9 days. One FH (FH1) and one FPH (FPH1) were used to treat iHeps. FH1 doubled albumin secretion during primary screening (Fig. 3a) and the effects were dose-responsive (Fig. 1c). FPH1 had a similar effect on hepatocyte functions (Fig. 3b), which interestingly, was much more pronounced in the younger donor (a, age 1 year).
Fig. 3. Functional enhancement of human primary hepatocytes and iHeps.
(a) Primary screening data of FH1. (b) Primary screening data of FPH1. Control cultures (Ctrl) were treated with empty vehicle (DMSO). All data presented as mean ± s.d. (c) Morphology and colony size of FPH1- and FH1-treated iHeps in 6-well plates 9 days post-initiation-of-treatment. Untreated iHeps are shown for comparison (scale bar = 100 µm). Yellow arrows point to select bile canaliculi. (d) Albumin, CYP3A and AFP staining of iHeps after 9 days of culture (scale bar = 100um). Bar graphs represent quantifications of displayed images.
Fig. 4. Maturation of human iHeps.
Gene expression profiling of FPH1-treated (a) and FH1-treated (b) iHeps. Heat map displays of Luminex analysis for 83 liver-specific genes, shown separately for independent experiments. Columns of the heatmap are averaged values of replicate (n≥3) loadings of mRNA extracted from various populations of iPS cells (250ng total RNA per replicate). mRNA expression was determined relative to the average of control gene transferrin, and heat maps are row-normalized. Bar graphs are select gene sets comparing the relative mRNA expression of small molecule-treated hepatocytes (colored bars) to iPS (light gray), untreated iHeps (gray bars) and fetal human hepatocytes (dark gray), normalized to control (black) for nuclear receptors, phase I, phase II and phase III drug metabolism genes. Control refers to adult cryopreserved human primary hepatocytes stabilized by micropatterned co-culture (Adult Control, more details in Supplementary Note 1). Data represent the mean ± SEM of Luminex-loaded replicates. (c) ELISA assays for secreted albumin and AFP, and quantitative CYP3A4 and CYP2A6 activity assays. For all analyses, iHeps were cultured for 9 days post-differentiation, in 6-well plates (n=3). All data presented as mean ± SEM.
Cultures treated with FH1 and FPH1 contained larger colonies of iHeps, which exhibited more pronounced hepatocyte morphologies, including polygonal cell shapes, visible nuclei, and more noticeable bile cannaliculi between hepatocytes (Fig. 3c). We subsequently examined the maturity of treated iHeps via gene expression profiling, immunofluorescent staining and various biochemical assays probing protein secretion and enzyme activity.
Gene expression profiles showed that treated iHeps more closely resemble mature hepatocytes than untreated cells (Fig. 4a,b). Euclidian clustering analyses grouped untreated cells with fetal hepatocytes, and treated cells with adult hepatocytes. Of particular interest are the expression levels of various ABC transporters and GSTP1. ABC transporters are known to mature after birth. ABCB11, also known as bile-salt export pump (BSEP), increased ~4-fold in expression levels with FH1 treatment, reaching 100% of adult hepatocyte levels. In contrast, GSTP1 expression, whose levels decrease with maturity, remained low upon small molecule treatment.
To examine the effects of FH1 and FPH1 at the protein level in iHeps, we visualized AFP, albumin and CYP3A levels via immunofluorescent staining (Fig. 3d and Supplementary Fig. 9). Images showed dramatic increases in albumin upon both FH1 and FPH1 treatment, although the effects of FH1 are more pronounced. This pattern is in agreement with morphological findings. Consistent with prior work17–19, untreated cultures contained islands that were strongly positive for both albumin and the fetal marker AFP, but showed minimal staining for the mature marker CYP3A. In contrast, treated islands double stained strongly for albumin and CYP3A, with AFP largely absent. This more mature phenotype was stable for at least 1 week after removal of small-molecule treatment (Supplementary Fig. 10).
To confirm the staining results, we measured secreted levels of albumin and AFP via ELISA, and CYP450 activities through isoenzyme-specific substrates with fluorescent or luminescent metabolites. ELISA results verified that small-molecule treatment both increased albumin secretion and decreased AFP secretion (Fig. 4c). CYP3A4 activity was found to increase by 16 and 45 times upon treatment with FH1 and FPH1, respectively. CYP2A6, another mature CYP450, was also found to increase significantly upon small-molecule treatment (Fig. 4c). We considered enzyme induction as a possible explanation for these elevations in CYP450 activity. However, this is unlikely. While human hepatocytes treated with specific inducers do exhibit elevated CYP450 activity, such elevations are typically reverted by 24 hrs after removal of the inducer (Supplementary Fig. 11). A period of at least 48 hrs separates small-molecule treatment and the measurement of CYP450 activity, thus the iHeps are expected to have recovered from any general elevations in CYP450 activity.
DISCUSSION
Developing novel cell-based therapies for liver disease requires human hepatocytes due to substantial species-specific differences4,5. Human hepatocytes, in their native environment, have phenomenal regenerative capabilities 30,32–35, which is lost ex vivo. Consequently, for decades, human hepatocyte sourcing has been a bottleneck for many fields of research and clinical therapies.
In traditional cultures of other cells, small molecules have shown promising roles in modulating a wide range of complex cell phenotypes. These outcomes include stem cell self-renewal and differentiation, and the proliferation of normally quiescent mature adult cells such as pancreatic β-cells and cardiomyocytes36–38. Compounds can act through a variety of mechanisms to induce cell division, including activation of developmental signaling pathways such as Wnt28 or recruitment of GEFs to the plasma membrane for RAS/MAPK pathway activation38. However, such systems have limited applicability towards primary human hepatocytes due to the rapid loss of hepatocyte phenotype and viability in vitro. To date, high-throughput screens related to the liver have been restricted to hepatocyte cell lines or hepatocyte extracts, neither of which offers the full repertoire of hepatocellular functions and responses exhibited by primary human hepatocytes. Such deviation from clinically-relevant biology has been a major cause of the high attrition rates currently troubling drug discovery. The high-throughput platform developed here leveraged stromal interactions, known to be important in vivo, to enable screening using primary human hepatocytes. Such a platform enables studies of many previously inaccessible aspects of liver biology and small-molecule bioactivity. In this work, we used the platform to address two long-standing issues: lack of a renewable human hepatocyte source and incomplete differentiation of human stem cells.
Early work on hepatocyte expansion focused on in vitro addition of growth factors, hormones, serum and vitamins8,10,11,39, and has led to a media formulation for the expansion of rodent hepatocytes11. Our study contributed to this progress through the identification of small molecules that can induce proliferation of human hepatocytes in vitro. With this advance, we foresee the potential to extend our findings to generate renewable materials (i.e. functional human hepatocytes) for various cell-based therapies, and in vitro liver models and assays, including tissue engineered liver constructs, cell transplantation, ADME or toxicity screening, and humanized mice for in vivo disease modeling. Our future work will determine the maximum expansion potential and functionality of treated hepatocytes using animal rescue experiments, and explore use of hepatocytes expanded from patient biopsies for cell transplantation therapy and other forms of personalized medicine. Such outcomes can contribute to our understanding of liver regeneration, and carry implications for regenerative medicine.
An alternative cell source to primary human hepatocytes is human iPS cells, which can be differentiated towards hepatocyte-like cells (iHeps)17,18,40. Among the factors used to drive differentiation is Oncostatin M. It has been observed that continued supplementation of Oncostatin M beyond the first 20 days of differentiation promotes survival of the resulting hepatocyte-like cells but suppresses the late stages of hepatocyte maturation. Thus, in order to isolate the effect of hit molecules on maturation (rather than differentiation), Oncostatin M was excluded beyond the 20-day point for the experiments depicted in Figures 3, 4 and Supplementary Figure 9. Under these conditions, a small percentage of hepatocyte-like cells were derived; however, the few that were obtained exhibited dramatic improvements in maturation. When Oncostatin M was maintained throughout the treatment period (Supplementary Fig. 10), high efficiencies of differentiation to hepatocyte-like cells were observed. Interestingly and contrary to our expectations, the hit molecules were also able to mature these hepatocyte-like cells; however, as expected, the levels of maturation as judged by CYP2A6 and 3A4 activity were not as robust as in the absence of Oncostatin M.
The efficiency of differentiation is generally stochastic, yielding some highly efficient differentiations and others far less efficient—even in the same laboratory. The variables that underlie this stochasticity are yet poorly understood but may include the variability in other cell types that emerge over time during differentiation, effectively creating a variable co-culture environment. Even the most robust cultures of differentiated iHeps persistently express fetal markers (e.g. AFP) and lack key mature hepatocyte functions (e.g. CYP2A6, CYP3A4)17–19, an outcome which presents a major hurdle to the application of these cells in both therapeutic and basic research assays. Our iPS results suggest that FH1 and FPH1 are able to promote the maturation of well-differentiated cultures of iHeps beyond what has been obtainable to date, thus potentially alleviating a major obstacle to the use of iPS cells as a renewable source of functional human hepatocytes. We found that this effect generalized across two pluripotent cell lines; nonetheless, we did observe stochasticity in the efficiency of maturation akin to the variability observed in response to existing directed differentiation protocols.
Based on tetraploid complementation experiments with mice, it is clear that murine iPS-derived progeny are capable of differentiating into mature, adult liver cells, when provided with the correct in vivo context and developmental cues41,42. Given that it is not possible to conduct the parallel experiment with human cells, it remains to be seen whether similar conditioning is sufficient to promote complete maturation to a comparable, fully mature hepatocyte. The evidence that human iHeps mature when xenografted in a 'gold standard' adult, in vivo environment is mixed43,44; the remaining donor cells still appear to exhibit low function, on a per cell basis18,40,45–47. This apparent defect could indicate that the cells are truly blocked from late stages of tissue-specific maturation, or that the pre-conditioning protocols provided in vitro are deficient in some way - perhaps lacking specific cues that the human embryo experiences in the third trimester of development, for example. Our data suggest that human iPS cells are indeed capable of further maturation in vitro, consistent with their potential observed in the mouse system. In light of similar challenges in other cell types48, our platform and its resulting compounds offer a roadmap for testing the generality of this finding in other tissues, and for identification of regulatory networks that are influenced by small-molecule exposure.
Through a high-throughput screen of 12,480 small molecules on primary human hepatocytes, we identified compounds in two separate classes that can be used to establish renewable sources of functional human hepatocytes. One class, FPH, induced proliferation of mature human primary hepatocytes. The second class, FH, helped differentiate human iPS cells towards more mature, adult hepatocytes. The identification of these small molecules may impact several areas of research including maturation of other iPS-derived cell types, expansion of other ‘terminally’ differentiated cell types and the translational potential of the resultant hepatocytes and other cell types.
Online methods
High-throughput screen
384-well screening plates (Corning) were incubated with a solution of type-I collagen in water (100 µg/ml, BD Biosciences) for 1 h at 37°C. A feeder layer of J2-3T3 fibroblasts (gift from Howard Green) were robotically plated onto the collagen at a density of 8,000 cells/well (designated as day -2), and allowed to reach confluency over 48 hours, when their growth became contact inhibited. Primary human hepatocytes were plated onto the fibroblasts on day 0 at a density of 2,000 cells/well and maintained under standard culture conditions with daily replacement of hepatocyte medium (detailed composition listed in supplementary note 1) for 7 days. Primary human hepatocytes were purchased in cryopreserved suspension from Celsis In vitro Technologies (donor a) and Invitrogen (donor b), and pelleted by centrifugation at 50g for 10 min. The supernatant was discarded before re-suspension of cells in hepatocyte medium. A library of 12,480 compounds was added on day 7 at a final concentration of ~15uM, and allowed to incubate for 48 hours. On day 9, culture supernatants were collected for automated ELISA analysis, and cells were fixed in 4% PFA for imaging analysis. Hepatocyte functions were determined via competitive ELISA (MP Biomedicals) using horseradish peroxidase detection and chemiluminescent luminol (Pierce) as a substrate. The cell-free counter assay involved ELISAs on fresh media incubated for 48 hrs with compounds of interest (without cells). Hepatocyte proliferation was assessed through customized, automated high-content imaging protocol (Supplementary Fig. 2). Fixed cells were permeabilized with 0.1% Triton-X, nuclei visualized with Hoechst stain (invitrogen) and robotically imaged (Thermo, Molecular Devices) at 21 dispersed sites per well. Images were digitized and analyzed using CellProfiler and CellProfiler Analyst (Broad Institute). To identify functional proliferation hits, we integrated all four readouts using p values. FPHs were selected using the criterion pprod < 1×10−6 and pmax < 0.25 and palb < −0.05, where pprod = pHepInterphase × pmetaphase × panaphase, pmax = max (pHepInterphase, pmetaphase, panaphase), and palb = ELISA p value. Additional proliferation-only hits were selected based on Z > 3 in image-based assays; function-only hits were selected by an ELISA Z < −4. Compounds with ELISA z > 3 were eliminated as toxic.
Cell Counting
J2-3T3 fibroblasts were covalently labeled with Vybrant CM-DiI (Invitrogen) before initiation of co-culture. For FACS analysis, cells were treated with trypsin, or collagenase then accutase, and suspended in PBS-0.2%FBS. Cell suspensions were supplemented with 50,000 fluorescent counting beads (CountBright, Invitrogen) per sample. Data were acquired with a 4-color flow cytometer (FACSCalibur, BD Biosciences) and analyzed with CellQuest (BD Biosciences). For Cellometer analysis, cells were trypsinized and placed on cell counting chambers (Nexcelom) for automated cell counting.
Luminex analysis
Cells were lysed using RLT buffer (Qiagen) or Trizol (Invitrogen) and purified using the Mini-RNeasy kit (Qiagen). Gene expression was determined using Luminex analysis, as previously described22. Briefly, total RNA was immobilized on a Qiagen turbo capture 384-well plate, and reverse-transcribed using oligodT priming. A biotinylated FlexMap tag sequence unique to each gene of interest and a phosphorylated downstream probe were then added to resulting cDNAs to generate biotinylated FlexMap-tagged amplicons. Universal PCR was then performed for 35 cycles using a biotinylated T7 forward primer and T3 reverse primer in buffer with dNTPs and Taq polymerase. FlexMap microsphere beads conjugated with antitag oligonucleotides were then added and allowed to hybridize. Amplicons were captured by streptavidin-phyoerythrin, and 100 events per bead were analyzed for internal bead color and phyoerythrin reporter fluorescence on a Luminex FlexMap 3D analyzer. Data for replicate loadings, expressed in mean fluorescent intensity of at least 100 beads per sample, were scaled to the human transferrin gene and row-normalized for heat map representation using GeneE open software (Broad Institute).
Biochemical Assays
Culture media were collected and either processed immediately or frozen at −20°C until analysis. Albumin content was measured through competitive or sandwich ELISA assays (MP Biomedicals, Fitzgerald, Bethyl Laboratories) using horseradish peroxidase detection and 3,3’,5,5’-tetramethylbenzidine (TMB, Fitzgerald Industries) as a substrate. Urea concentration was determined colorimetrically using diacetylmonoxime with acid and heat (Stanbio Labs)24. To quantify CYP450 activity at 48 hours after small molecule exposure, iHeps cultures were incubated with substrates (coumarin from Sigma for CYP2A6, luciferin-IPA from Promega for CYP3A4) for 4 hours at 37°C; primary human hepatocyte cultures were incubated with 7-benzyloxy-4-trifluoromethylcoumarin (BFC, BDGentest) at 50uM for 1 hr at 37°C in phenol-red free media. Many different CYP450 isoforms process BFC into its fluorescent product of 7-hydroxy-4-trifluoromethylcoumarin (7-HFC). Incubation medium was collected and metabolite concentration quantified via luminescence, or fluorescence after hydrolization of potential metabolite conjugates by β-glucuronidase/arylsulfatase (Roche, IN).
Cell culture
All cells were cultured under standard tissue culture conditions. Details of specific platforms can be found in Supplementary Note 1.
Supplementary Material
Acknowledgements
This research was funded by a Broad Institute SPARC grant (SNB), NIH R01-DK065152 (SNB), NIH R01-DK56966 (SNB), NIH R01 GM089652 (AEC), NSF CAREER award DBI 1148823 (AEC), NIDDK DK085445 (WG), DK55743 (SAD), DK087377 (SAD), HL094857 (SAD), HG006398 (SAD). S.N.B. is a Howard Hughes Medical Institute investigator. We thank H. Fleming for help with manuscript preparation, T. Golub and R. Gould for discussions, and H. Green for providing J2-3T3 fibroblasts. The authors wish to dedicate this paper to the memory of Officer Sean Collier, for his caring service to the MIT community and for his sacrifice.
Footnotes
Author Contributions
J.S. and S.N.B designed all experiments. J.S. performed and analyzed all experiments. D.J.L. helped analyze image-based experiments. N.T.R. helped perform and analyze screening experiments, and analyzed chemical characterization data. R.E.S. helped design, perform and analyze iPS experiments. D.T. developed and implemented the Luminex system. T.E.N., S.A.D., W.G., and A.E.C. contributed reagents and expertise. J.S., S.N.B., and N.T.R. wrote the manuscript. S.N.B. supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Shepard CW, Finelli L, Alter M. Global epidemiology of hepatitis C virus infection. Lancet Infectious Diseases. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Lin HM, et al. Center-specific graft and patient survival rates--1997 United Network for Organ Sharing (UNOS) Report. Jama-Journal of the American Medical Association. 1998;280:1153–1160. doi: 10.1001/jama.280.13.1153. [DOI] [PubMed] [Google Scholar]
- 3.Allen JW, Hassanein T, Bhatia SN. Advances in bioartificial liver devices. Hepatology. 2001;34:447–455. doi: 10.1053/jhep.2001.26753. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DR. Cytochrome P450 and the individuality of species. Archives of Biochemistry and Biophysics. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- 5.Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 6.Mitaka T. The current status of primary hepatocyte culture. International Journal of Experimental Pathology. 1998;79:393–409. doi: 10.1046/j.1365-2613.1998.00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Runge D, Michalopoulos GK, Strom SC, Runge DM. Recent advances in human hepatocyte culture systems. Biochemical and Biophysical Research Communications. 2000;274:1–3. doi: 10.1006/bbrc.2000.2912. [DOI] [PubMed] [Google Scholar]
- 8.Richman RA, Claus TH, Pilkis SJ, Friedman DL. HORMONAL-STIMULATION OF DNA-SYNTHESIS IN PRIMARY CULTURES OF ADULT RAT HEPATOCYTES. Proceedings of the National Academy of Sciences of the United States of America. 1976;73:3589–3593. doi: 10.1073/pnas.73.10.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimaoka S, Nakamura T, Ichihara A. STIMULATION OF GROWTH OF PRIMARY CULTURED ADULT-RAT HEPATOCYTES WITHOUT GROWTH-FACTORS BY COCULTURE WITH NONPARENCHYMAL LIVER-CELLS. Experimental Cell Research. 1987;172:228–242. doi: 10.1016/0014-4827(87)90109-1. [DOI] [PubMed] [Google Scholar]
- 10.Mitaka T, Sattler CA, Sattler GL, Sargent LM, Pitot HC. MULTIPLE CELL-CYCLES OCCUR IN RAT HEPATOCYTES CULTURED IN THE PRESENCE OF NICOTINAMIDE AND EPIDERMAL GROWTH-FACTOR. Hepatology. 1991;13:21–30. [PubMed] [Google Scholar]
- 11.Block GD, et al. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. Journal of Cell Biology. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuguchi T, et al. Enhanced proliferation and differentiation of rat hepatocytes cultured with bone marrow stromal cells. Journal of Cellular Physiology. 2001;189:106–119. doi: 10.1002/jcp.1136. [DOI] [PubMed] [Google Scholar]
- 13.Uyama N, et al. Regulation of cultured rat hepatocyte proliferation by stellate cells. Journal of Hepatology. 2002;36:590–599. doi: 10.1016/s0168-8278(02)00023-5. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi N, et al. Prevention of acute liver failure in rats with reversibly immortalized human hepatocytes. Science. 2000;287:1258–1262. doi: 10.1126/science.287.5456.1258. [DOI] [PubMed] [Google Scholar]
- 15.Castell JV, Jover R, Martinez-Jimenez CP, Gomez-Lechon MJ. Hepatocyte cell lines: their use, scope and limitations in drug metabolism studies. Expert Opinion on Drug Metabolism & Toxicology. 2006;2:183–212. doi: 10.1517/17425255.2.2.183. [DOI] [PubMed] [Google Scholar]
- 16.Nyberg SL, et al. PRIMARY HEPATOCYTES OUTPERFORM HEP G2 CELLS AS THE SOURCE OF BIOTRANSFORMATION FUNCTIONS IN A BIOARTIFICIAL LIVER. Annals of Surgery. 1994;220:59–67. [PMC free article] [PubMed] [Google Scholar]
- 17.Song Z, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Research. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 18.Si-Tayeb K, et al. Highly Efficient Generation of Human Hepatocyte-Like Cells from Induced Pluripotent Stem Cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan GJ, et al. Generation of Functional Human Hepatic Endoderm from Human Induced Pluripotent Stem Cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ploss A, et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nature Biotechnology. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 22.Chen AA, et al. Humanized mice with ectopic artificial liver tissues. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11842–11847. doi: 10.1073/pnas.1101791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. Faseb Journal. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 24.Khetani S, Bhatia SN. Development and characterization of microscale models of rat and human livers. Hepatology. 2007;46:773A–773A. [Google Scholar]
- 25.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology. 2006;7 doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones TR, et al. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. Bmc Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones TR, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1826–1831. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goessling W, et al. Genetic Interaction of PGE2 and Wnt Signaling Regulates Developmental Specification of Stem Cells and Regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.North TE, et al. PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17315–17320. doi: 10.1073/pnas.1008209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 31.Michalopoulos GK. Liver regeneration. Journal of Cellular Physiology. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins GM, Anderson RM. Experimental pathology of liver: restoration of liver in white rat following partial surgical removal. Arch. Pathol. 1931;12:186–202. [Google Scholar]
- 33.Overturf K, et al. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nature Genetics. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 34.Rhim JA, Sandgren EP, Degen JL, Palmiter RD, Brinster RL. Replacement of Diseased Mouse-Liver by Hepatic Cell Transplantation. Science. 1994;263:1149–1152. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 35.Stocker E, WH K, Brau G. Capacity of regeneration in liver epithelia of juvenile, repeated partially hepatectomized rats. Autoradiographic studies after continous infusion of 3H-thymidine. Virchows Arch. B Cell Pathol. 1973;14 [PubMed] [Google Scholar]
- 36.Chen SB, Hilcove S, Ding S. Exploring stem cell biology with small molecules. Molecular Biosystems. 2006;2:18–24. doi: 10.1039/b512000b. [DOI] [PubMed] [Google Scholar]
- 37.North TE, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang WD, et al. Identification of small-molecule inducers of pancreatic beta-cell expansion. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1427–1432. doi: 10.1073/pnas.0811848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michalopoulos G, et al. LIVER-REGENERATION STUDIES WITH RAT HEPATOCYTES IN PRIMARY CULTURE. Cancer Research. 1982;42:4673–4682. [PubMed] [Google Scholar]
- 40.Touboul T, Vallier L, Weber A. Robust differentiation of fetal hepatocytes from human embryonic stem cells and iPS. M S-Medecine Sciences. 2010;26:1061–1066. doi: 10.1051/medsci/201026121061. [DOI] [PubMed] [Google Scholar]
- 41.Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS Cells Can Support Full-Term Development of Tetraploid Blastocyst-Complemented Embryos. Cell Stem Cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Zhao X-y, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:U86–U88. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 43.Azuma H, et al. Robust expansion of human hepatocytes in Fah(-/-)/Rag2(-/-)/Il2rg(-/-) mice. Nature Biotechnology. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Payne CM, et al. Persistence of functional hepatocyte-like cells in immune-compromised mice. Liver International. 2011;31:254–262. doi: 10.1111/j.1478-3231.2010.02414.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, Kim Y, Sharkis S, Marchionni L, Jang Y-Y. In Vivo Liver Regeneration Potential of Human Induced Pluripotent Stem Cells from Diverse Origins. Science Translational Medicine. 2011;3 doi: 10.1126/scitranslmed.3002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jozefczuk J, Prigione A, Chavez L, Adjaye J. Comparative Analysis of Human Embryonic Stem Cell and Induced Pluripotent Stem Cell-Derived Hepatocyte-Like Cells Reveals Current Drawbacks and Possible Strategies for Improved Differentiation. Stem Cells and Development. 2011;20:1259–1275. doi: 10.1089/scd.2010.0361. [DOI] [PubMed] [Google Scholar]
- 47.Behbahan IS, et al. New Approaches in the Differentiation of Human Embryonic Stem Cells and Induced Pluripotent Stem Cells toward Hepatocytes. Stem Cell Reviews and Reports. 2011;7:748–759. doi: 10.1007/s12015-010-9216-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kroon E, et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nature Biotechnology. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.