Abstract
Objective(s)
There is limited information on full-length genome sequences and the early evolution of transmitted HIV-1 subtype C viruses, which constitute the majority of viruses spread in Africa. The purpose of this study was to characterize the earliest changes across the genome of subtype C viruses following transmission, to better understand early control of viremia.
Design
We derived the near full-length genome sequence responsible for clinical infection from five HIV subtype C-infected individuals with different disease progression profiles and tracked adaption to immune responses in the first six months of infection.
Methods
Near full-length genomes were generated by single genome amplification and direct sequencing. Sequences were analyzed for amino acid mutations associated with cytotoxic T-lymphocyte (CTL) or antibody (Ab)-mediated immune pressure, and for reversion.
Results
Fifty-five sequence changes associated with adaptation to the new host were identified with 38% attributed to CTL pressure; 35% to antibody pressure; 16% to reversions and the remainder were unclassified. Mutations in CTL epitopes were most frequent in the first 5 weeks of infection, with the frequency declining over time with the decline in viral load. CTL escape predominantly occurred in nef, followed by pol and env. Shuffling/toggling of mutations was identified in 81% of CTL epitopes with only 7% reaching fixation within the six month period.
Conclusions
There was rapid virus adaptation following transmission, predominantly driven by CTL pressure, with most changes occurring during high viremia. Rapid escape and complex escape pathways provide further challenges for vaccine protection.
Keywords: HIV-1, Africa, genome, acute infection, cytotoxic T-lymphocytes, progression
Introduction
Early host selective pressures drive genetic diversification of the transmitted human immunodeficiency virus (HIV) and potentially influence the course of disease. In heterosexual infection, it is estimated that approximately 80% of individuals are infected with a single virus or virus-infected cell [1,2]. Vaccines that do not block the establishment of this initial infecting virus will need to target the early diversifying virus and thus an understanding of early viral evolution is important. The frequency and speed at which the transmitted virus changes in response to host immune pressures are of key interest to vaccine immunogen design as these provide insights into its strengths and vulnerabilities.
Recent methodologies using single genome amplification (SGA) applied to individuals with acute HIV-1 infection have enabled identification of the sequence of transmitted/founder (t/f) full-length viral genomes [1,3,4]. This approach uses mathematical modeling to derive the sequence of the virus/es responsible for productive clinical infection, and was proven in the SIV (simian immunodeficiency virus) model wherein the derived t/f was identical, or differed by a few nucleotides, to the virus in the inocula [5].
Using this approach, early evolutionary changes following transmission in three subtype B infections from the USA have been mapped [3]. No longitudinal studies elucidating early evolution in full-length subtype C genomes have been reported despite subtype C being the dominant subtype both globally and in Southern Africa where large vaccine and microbicide trials take place [6,7]. Studies in different population settings are essential given that differences in host genetics influence viral evolution [8].
The transmitted virus encounters immune selective pressures almost immediately following infection. Cytotoxic T-lymphocyte (CTL) and neutralizing antibody (Ab) pressure are proven driving forces for viral diversification [9–16]. Evidence of CTL pressure on the viral genome has been identified in the very first weeks of subtype B and C infection [1–3,17–23], and CTL activity has been associated with control of viremia early in infection [24–26]. Most recently, a subtype B full-length genome study using mathematical modeling to determine the killing rate of CTL during acute viremia suggested that these cells have a role in controlling peak viremia [21].
In this study we investigated changes observed across the genomes of t/f subtype C viruses from five heterosexually infected women with differing disease progression profiles. We extrapolated the near full-length genome sequence of the t/f viruses and quantified genetic mutations associated with positive selection from humoral and cellular immune pressures over the first six months following infection.
Methods
Study participants
Samples were obtained from the Centre for the AIDS Program of Research in South Africa (CAPRISA) 002 Acute Infection cohort (Durban, South Africa) [27]. Date of infection was estimated as the midpoint between last negative and first positive HIV antibody test and as 14 days prior for individuals who were RNA positive/antibody negative. Human leukocyte antigen (HLA)-A, B and C types were determined using four digit high resolution HLA typing as described [28]. The study was approved by the Universities of Cape Town, Witwatersrand and KwaZulu Natal.
PCR amplification and sequencing
RNA was extracted from 200–400 µl plasma using the Qiagen Viral RNA Mini Kit (Qiagen, Valencia, CA, USA) and reverse transcribed to cDNA using Superscript III reverse transcriptase and Oligo(dT)20 (Invitrogen, GmbH, Karlsruhe, Germany). Single genome amplification (SGA) [4] and sequencing of near full-length genome amplicons was done using Expand Long Template Taq (Roche Diagnostics, Rotkreuz, Switzerland) as reported [4] with primers described by Rousseau et al. optimized for subtype C [29]. Sequences <6 000 bp were excluded. Salazar-Gonzalez et al. attributed one to five ambiguities within a genome obtained at <20% PCR positivity predominantly to PCR Taq error [3]. We accepted amplicons obtained at <66% positivity with up to six ambiguities. Targeted epitope sequencing was done following gene-specific limiting dilution PCR. Gag and nef clones were generated using limiting dilution PCR and the pGEM-T Easy system as described [30]. Env SGA was described previously [4]. All products were directly sequenced using the ABI 3000 genetic analyser (Applied Biosystems, Foster City, CA, USA) and BigDye terminator reagents.
Sequence analysis
Analyses performed were: sequence alignments, amino acid identity and frequency plots and consensus sequence derivation (BioEdit version 7.0.8.0 [31]); subtyping (REGA HIV Subtyping Tool; http://dbpartners.stanford.edu/RegaSubtyping/); phylogenetic and pair-wise DNA distance analyses (Mega 4 [32]); Highlighter plots (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter); CTL epitope prediction (Epitope Location Finder (ELF) (http://www.hiv.lanl.gov/content/sequence/ELF/epitope_analyzer)and NetMHCpan 2.2 (http://www.cbs.dtu.dk/services/NetMHCpan)[33]); Hypermut detection of APOBEC hypermutation (www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermnut.html Shannon entropies (Entropy One; http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy_one) and mapping of functionally/structurally relevant genome regions (http://www.hiv.lanl.gov/content/sequence/HIV/MAP/landmark). A high average Shannon entropy score was taken as ≥0.25 as described by Bansal et al. for variable HIV proteins [34]. Where an optimal epitope (9 to 11-mer) has not yet been described, the entropy of an 18-mer peptide encompassing the mutating region was used.
Time to most recent common ancestor (MRCA) was determined using BEAST v1.4.7 (Bayesian Evolutionary Analysis Sampling Trees) [35,36] as described previously [1,2]. A relaxed (uncorrelated exponential) molecular clock with general time-reversible substitution model, mean of 2.16 × 10−5 substitutions/site/generation with rates unlinked across codon sites [37] and gamma distribution with four categories and a proportion of invariant sites was used.
Known HLA class I-restricted epitopes or class I HLA-associated polymorphisms were identified using the Los Alamos HIV Molecular Immunology 2008 Compendium (http://www.hiv.lanl.gov/content/immunology/compendium.html) and Matthews et al., 2008 [38]. Subtype C database alignments were obtained from the Los Alamos HIV database (http://www.hiv.lanl.gov/components/sequence/HIV/).
Positive selection and statistical analysis
Non-synonymous to synonymous substitution (dN/dS) rate ratios per codon site were estimated using the MG94xHKY85 codon model [39]. We allowed dS to vary across codon sites and employed the Dual model which takes into account that dS may vary independently of dN [40]. Models were implemented within HyPhy [41] and ensured that correct phylogenetic relationships were used for regions separated by recombination breakpoints [42].
Categorical statistical tests were carried out using the Fisher’s Exact two-tailed test (http://www.graphpad.com/quickcalcs).
Results
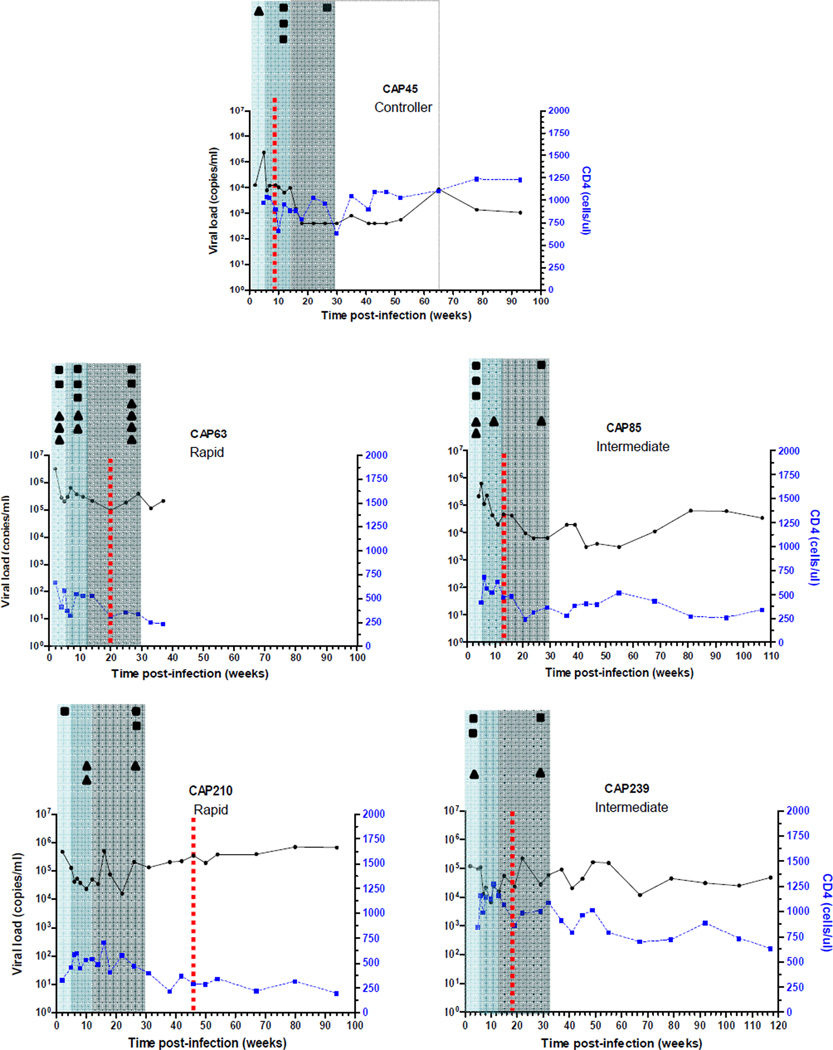
This study characterized virus evolution in five subtype C-infected women recruited two to five weeks following transmission. These women were selected based on infection with a single transmitted/founder virus [2] and clinical disease progression profile (Table 1). One woman was classified as a viremic controller (CAP45; viral loads consistently <2 000 copies/ml and CD4 counts >350 cells/µl in the absence of antiretroviral therapy) [43], two as rapid progressors (CAP63 and CAP210; viral loads >100 000 copies/ml and CD4 counts <350 cells/µl on consecutive visits within the first year of infection) [23,44], and two as intermediate progressors (CAP85 and CAP239; not fitting either controller or rapid status) (Fig.1). A total of 112 near full-length genomes were generated, with an average of nine at screening/enrolment (2–5 weeks post-infection,); six at 3 months (11–13 weeks post-infection); and nine at 6 months (22–29 weeks post-infection). No sequences could be generated at 6 months for viremic controller CAP45 due to low viral loads. Additional sequences (half-genome, SGA and clonal) were generated from various time points ranging from 2 to 117 weeks post-infection (Table 1).
Table 1.
Demographic and sequencing information for five CAPRISA participants
| Participant ID | Gender | Age | Risk factor | Subtype | Disease Progression |
Fiebig Stage1 at first sequenced timepoint |
BEAST mean # days since MRCA (95% CI)2 |
HLA Type | Sample date | Weeks post infection |
Whole
(half) genome sequences |
Sub-genomic clone/SGA sequences |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAP45 | Female | 41 | Heterosexual | C | Slow | I/II | 18 (4–35) | A*23:01, 29:02 B*15:10, 45:01 Cw*06:02, 16:01 |
20-Apr-05 11-May-05 07-June-05 28-June-05 27-Jul-05 04-Apr-06 04-Jul-06 |
2 5 9 12 16 52 65 |
3 6 3 1 3 |
16 1 8 7 |
| CAP63 | Female | 32 | Heterosexual | C | Rapid | III | 30 (10–53) | A*02:01, 23:01 B*45:01 Cw*04:01, 16:01 |
06-Jan-05 09-Mar-05 13-Jul-05 07-Sep-05 |
2 11 29 37 |
11 7 10 5 |
19 26 |
| CAP85 | Female | 24 | Heterosexual | C | Intermediate | V | 53 (20–101) | A*30:02 B*08:01, 45:01 Cw*07:01, 16:01 |
22-Jun-05 18-Aug-13 07-Dec-05 16-Feb-06 10-May-06 07_Jun-06 07-Dec-06 06-Jun-07 |
5 13 29 39 51 55 81 107 |
8 9 7 2 |
21 11 1 9 9 1 7 |
| CAP210 | Female | 43 | Heterosexual | C | Rapid | I/II | 11 (3–25) | A*68:02 B*15:10 Cw*03:04 |
03-May-05 13-Jun-05 21-Sep-05 19-Oct-05 23-Nov-05 |
2 12 22 26 31 |
9 7 11 |
21 8 3 3 |
| CAP239 | Female | 44 | Heterosexual | C | Intermediate | V* | 34 (11–58)* | A*01:23, 29:02 B*42:01, 58:01 Cw*06:02, 17:01 |
19-Jul-05 10-Aug-05 17-Aug-05 21-Sep-05 07-Dec-05 14-Jun-06 11-Jan-07 04-Oct-07 |
2 5 6 11 22 49 79 117 |
2 (3) 8 2 9 0 (4) 3 |
21 75 19 9 9 |
Fiebig stages (Fiebig et al., 2003) are (I/II) HIV RNA positive but antibody negative; (III) ELISA positive but non-reactive Western blot; (V) reactive Western blot without p31 band
Bayesian Evolutionary Analysis Sampling Trees estimated period of infection to observed env diversity from a single infecting strain
Fiebig stage determined for 5 weeks post-infection
Figure 1.
Clinical and immune selection profiles for each of the five women illustrated with viral load and CD4 count graphs. The number of epitopes/genome regions identified as under putative cytotoxic T-lymphocyte pressure (black squares) and antibody pressure (black triangles) are illustrated for approximate time periods post-infection of 0–5 weeks (pale blue shaded region), 5–12 weeks (darker blue shaded region) and 12–29 weeks (grey shaded region). The dashed grey box is used for participant CAP45 as the black square represents an epitope which may have mutated anywhere between 12–65 weeks post-infection. Time of first detection of autologous neutralizing antibodies is indicated by dashed red lines.
Derivation of transmitted/founder (t/f) virus sequences
All sequences were classified as subtype C along the entire length of the genome. Mean intra-participant DNA distances ranged from 0.008 to 0.25% (median 0.03%) at the first timepoint and mean number of days since MRCA ranged from 18–53 days (Table 1) indicating limited sequence diversification since transmission (see Figure S1, Supplemental Digital Content 1, illustrating intraparticipant sequence diversity in a Neighbour-Joining tree). T/f sequences were defined as the consensus of sequences from the earliest timepoint where all genes had an intact open reading frame and no ambiguities [3]. Although CAP63 and CAP85 were classified as infected with a single t/f variant based on env [2], we identified a minor early variant in each (not detected at later timepoints) which, in both cases, differed from the derived t/f at five nucleotide positions, suggesting that these individuals may each have been infected with two very closely related variants.
The majority of early genetic changes are due to CTL pressure or reversion
Using longitudinal near-full length genome and env screening/enrolment SGA sequences, viral evolution from the t/f was analysed for evidence of immune escape or reversion. Substitutions from high frequency/consensus to lower frequency/non-consensus amino acids within or adjacent to known class I HLA-restricted epitopes, or corresponding to known HLA-associated polymorphisms, were classified as CTL pressure [16,19,45,46]. Mutations within the hypervariable loops and potential N-linked glycosylation sites (PNGSs) in env were classified as Ab pressure [9]. Mutations from low/non-consensus to higher frequency/consensus subtype C amino acids within CTL epitopes not restricted by the participants HLA were classified as reversion of transmitted CTL escape mutations [16,45,46]. In addition, clustered mutations within amino acid nine-mers, previously reported to be associated with immune selection [18], or single amino acid mutations persisting to fixation and corresponding to sites under positive selection were identified as putative immune escape.
In viruses from the five women, immune pressure was identified in 55 genome regions (see Figure S2 to S6, Supplemental Digital Content 2, which illustrate genome regions under immune pressure): twenty-one were classified as under CTL pressure (Table 2); nineteen under Ab pressure (Table 3); nine as reversion (see Table S1, Supplemental Digital Content 3, which tabulates genome regions undergoing reversion) and a further six regions contained mutations (clustered within amino acid nine-mers or single persisting to fixation in sites under positive selection) which did not conform to criteria described for CTL or Ab pressure or reversion (Table 3). Identification of regions under immune pressure was supported by selection analysis (for which gag, env and nef supplemental sequences were included), which found a total of 55 sites under positive selection (dN/dS >1) of which 84% (46/55) were situated within genome regions identified as under immune pressure (see Table 2, Table 3, Table S1, Supplemental Digital Content 3 and Figures S2 to S6, Supplemental Digital Content 2, which illustrate positive selection sites corresponding to genome regions under immune pressure). The majority of positively selected sites were identified in env (n=30), followed by gag (n=9), pol (n=7), nef (n=5), rev (n=3) and tat (n=1). APOBEC-mediated G to A hypermutation was identified in 24% (13/55) of regions under immune selection.
Table 2.
Putative cytotoxic T-lymphocyte escape epitopes and polymorphisms
| Participant ID | ORF | Epitope/genome region sequence* | HXB2 psn. | Participant HLA association/s** |
Reference | Time of first
AA change (range)(wks) |
High entropy epitope/peptide (LANL subtype C database) |
Shuffling/Toggling of AA mutations |
|---|---|---|---|---|---|---|---|---|
| CAP45 | Vif | DWHLGHGVSI- | 78–87 | B*15:10 | LANL database# | 12–65 | No | No |
| Rev | IHSISERIL | 52–60 | B*15:10 | LANL database | 5–12 | Yes | Yes | |
| Tat | NCYCKHCSY | 24–32 | A*29:02 | LANL database | 5–12 | Yes | Yes | |
| Nef | EEVGFPVRPQV | 64–74 | B*45:01 | Matthews et al., 2008 | 5–9 | No | Yes | |
| CAP63 | Pol | ALTEICEEM | 188–196 | A*02:01 | LANL database | 5–11 | Yes | Yes |
| Pol | QLTEAVHKI | 522–530 | Predicted A*02:01 | 11–29 | No | Yes | ||
| Vpr | ALIRILQQL | 59–67 | A*02:01 | LANL database | 5–11 | Yes | Yes | |
| Gp41 | SWSNKSEEDIWGNMTWMQ | 102–119 | A*23:01/Cw*04:01 | LANL database | 11–29 | Yes | Yes | |
| Gp41 | LLDSIAITV | 303–311 | A*02:01 | LANL database | 2–4 | Yes | Yes | |
| Nef | ALTSSNTAA | 42–50 | A*02:01 | LANL database | 5–11 | Yes | Yes | |
| Nef | EEVGFPVRPQV | 64–74 | B*45:01 | Matthews et al., 2008 | 0–2 | No | Yes | |
| CAP85 | Pol | KAGYVTDRGRQKVVSLTE | 609–626 | B*08:01 | Matthews et al., 2008 | 0–5 | Yes | Yes |
| Gp41 | RYLGSLVQY | 283–291 | A*30:02 | LANL database | 0–5 | Yes | Yes | |
| Nef | KEVGFPVRPQV | 64–74 | B*45:01 | Matthews et al., 2008 | 0–5 | No | Yes | |
| Nef | YFPDWQNY | 120–127 | A*30:02 | LANL database | 13–29 | No | No | |
| CAP210 | Gag | VHQAISPRTL | 143–152 | B*15:10 | Matthews et al., 2008 | 12–16 | No | No |
| Vif | DWHLGHGVSI | 78–87 | B*15:10 | LANL database | 12–16 | No | Yes | |
| Gp41 | EATDRILEL | 313–321 | Predicted A*68:02 | 2–5 | Yes | Yes | ||
| CAP239 | Gag | TSTLQEQVAW | 240–249 | B*58:01 | Matthews et al., 2008 | 0–2 | No | Yes |
| Pol | IVLPEKESW | 399–407 | B*58:01 | Matthews et al., 2008 | 2–5 | Yes | Yes | |
| Nef | KAAVDLSFF | 82–90 | B*58:01 | Matthews et al., 2008 | 11–22 | Yes | No |
Bold amino acids indicate sites undergoing mutation; underlined amino acids indicate sites evolving under positive selection
Predicted epitopes obtained using Net MHCPan2.0 (www.cbs.dtu.dk/services/NetMHCPan)
Los Alamos National Laboratory (LANL) database (www.hiv.lanl.gov) HIV Molecular Immunology 2008 compendium used
Table 3.
Genome regions under putative antibody-mediated or unclassified immune pressure
| Participant ID | ORF | Genome region sequence* (18 to 20-mer) |
HXB2 position | Putative immune selective pressure |
Time of first mutation in hypervariable loop or PNGS# (range)(wks) |
|---|---|---|---|---|---|
| CAP45 | Gp120 | LTRDGGKTDRNDTEIFRP | 454–470 | Antibody | 0–2 |
| CAP63 | Gp120 | QEIVLENVIENFNMWKND | 82–99 | Antibody | 5–11 |
| Gp120 | LTPLCVTLNCANANITKN | 122–139 | Antibody | 11–29 | |
| Gp120 | MIGEIKNCSFNATTELRD | 147–167 | Antibody | 2–5 | |
| Gp120 | LNNNRSNENSYILINCNS | 184–198 | Antibody | 0–2 | |
| Gp120 | IVHFNQSVKIVCARPHNN | 285–302 | Antibody | 11–29 | |
| Gp120 | IRQAHCNISKTQWNTTLE | 326–343 | Antibody | 11–29 | |
| Gp120 | FNSTYMPNGIHIPNGASEVIT | 396–415 | Antibody | 2–5 | |
| Gp41 | LWSWFNISHWLWYIRIFI | 158–147 | Antibody | 11–29 | |
| Gp41 | IEEEGGEQDNSRSIRLVS | 222–239 | Antibody | 5–11 | |
| CAP85 | Gp120 | DIVPLNNDIGNYSEYRLI | 180–194 | Antibody | 5–13 |
| Gp120 | IVHLNHSVKIVCTRPGNN | 285–302 | Antibody | 0–5 | |
| Gp120 | IRQAHCNISKAEWNNTLE | 326–343 | Antibody | 13–29 | |
| Gp120 | GSSTTTNGSSPITLPCRI | 404–420 | Antibody | 0–5 | |
| Gp120 | RPGGGDMKDNWRSELYKY | 469–486 | Unclassified | 0–5 | |
| Nef | GVGAASQDLGKYGALTSS | 29–46 | Unclassified | 0–5 | |
| CAP210 | Pol | FFRENLAFPEGEARELPS | 1–18 | Unclassified | 12–16 |
| Gp120 | ICSFNATTELRDKKKKEY | 156–173 | Antibody | 5–12 | |
| Gp120 | FNSTHNSTDSTVNSTDST | 391–409 | Antibody | 5–12 | |
| Gp120 | ITCISNITGLLLTRDGGE | 443–460 | Antibody | 22–26 | |
| Nef | SLHGMEDTEREVLQWKFD | 169–186 | Unclassified | 5–12 | |
| CAP239 | Gag | SNPSGPKRPIKCFNCGRE | 382–399 | Unclassified | 2–5 |
| Rev | GRPAEPVPFQLPPIERLH | 65–82 | Unclassified | 2–5 | |
| Gp120 | DIIIRSQNILDNTKTIIV | 269–286 | Antibody | 2–5 | |
| Gp120 | GLLLTWDGGDSKENKTRH | 451–467 | Antibody | 11–22 | |
Bold amino acids indicate sites undergoing mutation; underlined amino acids indicate sites evolving under positive selection; highlighted amino acids indicate sites mutating to result in a change within, or gain/loss of a potential N-linked glycosylation site gain/loss (NXS/Tx, where x is not Proline);
PNGS = potential N-linked glycosylation site
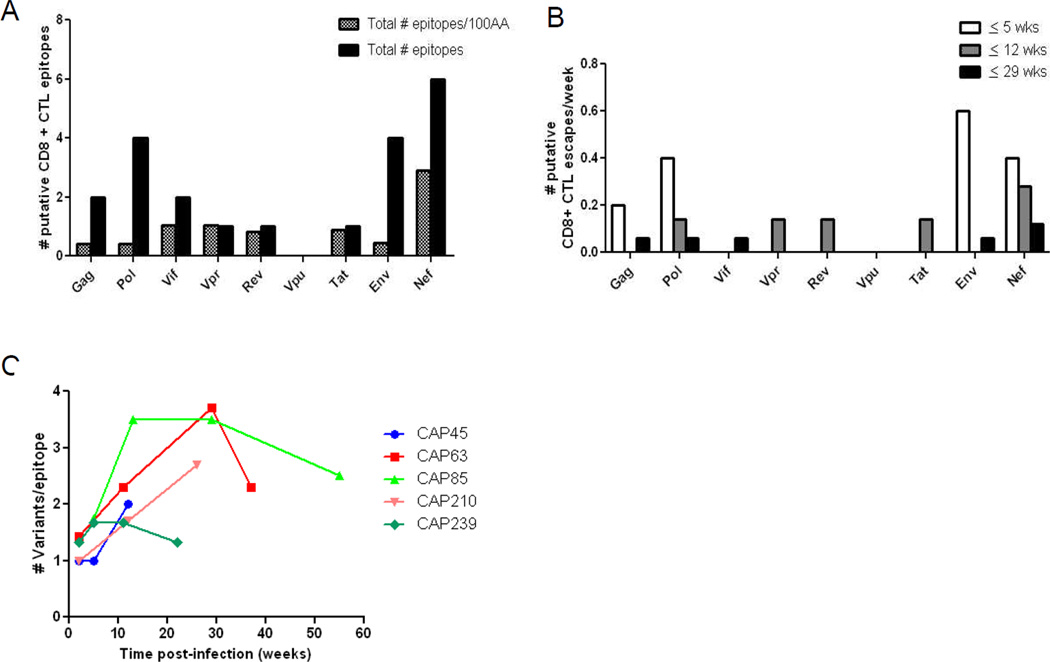
Of the mutating genome regions associated with CTL pressure, nineteen spanned known CTL epitopes, and two spanned predicted epitopes (Table 2). The highest frequency of CTL-driven mutations was located in nef (even when normalizing for amino acid length) (n=6), followed by pol and env (n=4), gag and vif (n=2) and finally rev, tat and vpr (n=1) (Fig. 2A). Reversion was identified most frequently in vpu (n = 3) followed by gag, env and nef (n=2) (see Table S1, Supplemental Digital Content 3).
Figure 2.
Frequency, timing and complexity of putative cytotoxic T-lymphocyte (CTL) escape in the first six months of infection. (A) Total number of putative escapes per gene and per 100 amino acid (AA), (B) total number of putative escapes per week, and (C) total numbers of variants per CTL epitope over time for each of the five women.
CTL escape is most frequent in acute infection
We supplemented near full-length genomes with sub-genomic (gag, env, nef and targeted epitope) sequence data to better elucidate timing of mutations associated with CTL pressure and escape. The majority of mutating epitopes was identified within the first five weeks of infection in structural genes gag, pol, env and nef (Fig. 2B, Fig.1). The earliest of these was identified at two weeks post-infection in the gag HLA B*58:01 restricted TW10 epitope; and the nef HLA B*45:01 restricted EV11 epitope. The frequency of mutation associated with escape slowed over time with an initial 1.6 total escapes/week for the first five weeks of infection, to 0.9 escapes/week between five and twelve weeks post-infection, and 0.4 escapes/week between twelve and twenty-nine weeks post-infection (Fig.2B).
Of the nineteen regions of env with changes in hypervariable loops and PNGSs, mutations in seven (37%) arose in the first five weeks of infection (Fig. 1, Table 3). Since first detection of autologous neutralising antibodies (nAb) for the women in this study ranged from nine to forty-six weeks [44,47] (Fig. 1), these early changes are unlikely to be the result of nAb pressure. However, in one instance, early mutating sites in V5 of participant CAP45 corresponded to sites where neutralization escape mutations (K460D and D462G) were identified later in infection (see Table 3) [44].
Finally, of the nine reversions identified, four occurred in the first five weeks of infection and the remaining between thirteen and twenty-nine weeks post-infection (data not shown).
Shuffling and toggling of amino acid mutations
Shifting of mutations between different positions within an epitope (shuffling) or between different amino acids at the same position (toggling) was observed in 81% (17/21) of mutating CTL epitopes (see Table 2 and Table S2, Supplemental Digital Content 4, illustrating shuffling and toggling in a Nef epitope). For four of the five participants, the number of mutant variants per CTL epitope increased over the six months of infection after which a plateau or decrease was seen owing in part to eventual fixation of escape mutants (Fig. 2C).
Twelve of the 17 CTL epitopes with shuffling/toggling mutations corresponded to subtype C database epitopes with high Shannon entropy (scores ≥ 0.25 [34]). Shuffling/toggling was however not more frequent in epitopes corresponding to high entropy regions than in more conserved regions (p=0.25). We investigated whether mutational shuffling/toggling was due to mutations arising in structurally or functionally essential sites, which when altered may abrogate efficient RNA folding or alter protein structure and function. We therefore compared the proportion of epitopes with shuffling/toggling in functionally or structurally relevant sites with the proportion in sites with no currently known functional or structural significance. We found no significant difference between the two categories (p=0.53), even when including the six genome regions under unclassified immune pressure (p=0.28).
Immune pressure, sequence diversification and disease progression
To determine whether rate of genetic diversification from t/f sequences differed with different disease progression profiles, we compared gene-specific tree lengths using sequences from the first three months of infection. No significant differences between rates of diversification were found (data not shown). Furthermore, no significant association between number of genome regions under immune pressure and viral load over time was identified (data not shown); possibly owing to low participant numbers.
Discussion
Design of a globally relevant HIV-1 vaccine requires an understanding of transmitted viruses and their early immune adaption in different populations. Here we report the first study to comprehensively identify and classify the earliest changes to subtype C transmitted/founder full-length genome viruses. We provide a detailed analysis of the timing, frequency and patterns of these changes in five women with differing clinical disease progression. The predominant and earliest host selective pressure was from cytotoxic T-lymphocytes. Despite increasing breadth of CTL responses over time [23], the frequency of mutations associated with CTL escape declined as viral load declined. We show that complex mutational pathways are used to escape in the majority of epitopes.
We predicted that CTL pressure, or reversion of transmitted escape, accounted for the majority (54%) of changes across the genome, reaffirming the importance of this response in early infection. Supporting our predictions, autologous peptide screening using IFN-γ ELISPOT in three of the five participants confirmed all nine epitopes predicted for these individuals (Liu et al., in press). A further four of the 21 epitopes were likewise confirmed to be responsive (Liu et al., unpublished data). The remaining eight were not confirmed either due to poor cell quality or screening after escape had already occurred. Mutations associated with CTL escape occurred earliest in gag and most frequently in nef, although this was likely influenced by the fact that three of the five individuals were HLA B*45:01 positive and targeted the same nef epitope. A further 35% of changes were associated with Ab pressure. We could not classify 11% of changes which may be associated with novel CTL epitopes [possibly in alternate reading frames [48,49]], compensation of escape, changes in antigen processing, CD4+ T-cell pressure, natural killer (NK)-cell pressure [50], non-neutralizing binding Ab activity, ADCC (antibody-dependant cellular cytotoxicity), viral fitness, or evolutionary drift/hitchhiking. One unclassified mutating region in pol of participant CAP210 was subsequently found to be responsive by IFN-γ ELISPOT and may represent a novel CTL epitope (Liu et al., in press). Another in nef of CAP210 corresponded to a known HLA-DR CD4+ T-cell epitope, however not restricted by the participants HLA-DR (data not shown).
Three findings from this study illustrate the immense pressure the virus is under following infection. Firstly, in these five women escape was rapid and occurred at high frequency in acute infection (< 5 weeks post-infection), with a 4-fold reduction in number of escapes/week over the six month period. Seventy percent of early escape (< 12 weeks post-infection) occurred in high entropy locations compared to only 33% after 12 weeks. These observations, although not statistically significant due to low sample size, consolidate reports that strong CTL pressure and rapid escape prevail early in infection [21,51] and that epitopes with higher entropy escape faster than more conserved epitopes [52]. Thus the observed reduction in mutations associated with CTL escape later in infection seen in these women may be associated with targeting of epitopes that escape slowly.
Secondly, the pathway to CTL escape is complex, with this study showing an unprecedented level (81%) of genome regions under CTL pressure with mutations shifting between different amino acid sites or between different amino acids at the same site resulting in late or no fixation over the period investigated. A recent study proposed that initial mutations are replaced by secondary mutations which are less costly to the virus with respect to its survival [53], however we found no statistical support for more frequent shuffling/toggling in virus regions of structural or functional importance. It may be that this pattern of variation is seen simply because these regions of the genome are able to tolerate multiple changes (supported by the fact that 71% of the epitopes with shuffling/toggling corresponded to high entropy database epitopes). Alternatively, fixation may only occur once mutations are introduced into sites facilitating complete escape as postulated in a detailed study of one participant by Henn et al. [54].
Thirdly, and perhaps surprisingly since neutralizing antibody pressure typically emerges some weeks or months after infection [9,55,56], we identified escape patterns typically associated with Ab pressure as early as two weeks post-infection. Autologous neutralizing Ab (nAb) response data for these five participants indicate the earliest response to have arisen at nine weeks post-infection [44]. These early changes could possibly be attributed to reversion of Ab escape mutations in the donor, early stochastic changes, non-neutralizing binding Ab activity, ADCC, env fitness or as very recently reported in a subtype B study by Bar et al., may in fact be a result of early low level neutralizing antibody responses from which the virus escapes [57]. We saw evidence for this in V5 of CAP45 wherein early changes corresponded to sites where nAb escape later in infection was confirmed in this participant [44].
When examining participant sequence data in the context of disease progression, we found no significant differences in early virus sequence diversification between individuals in our study. However larger studies would be needed to evaluate the role of CTL escape in disease progression. Recent studies revealed that clinical disease profiles are heritable in donor-recipient pairs [58,59] and that viral genotype can be a determinant of viral load set point [60]. Furthermore, transmission of a virus harbouring a mutation with a fitness cost can also contribute to viral control [28,61]. We observed that viremic controller CAP45 was infected with a virus harbouring mutations flanking the B*57/58 Gag TW10 and ISW9 epitopes previously reported to be associated with disease control [28]. It is worth mentioning that the two rapid progressors in this study were HLA-B (CAP63) and HLA-A, B and C (CAP210) homozygous. HLA homozygosity has previously been associated with poor clinical outcome [62].
Numerous CTL studies propose stable CTL epitopes that escape slowly or late in infection due to high fitness costs to be good vaccine targets [21,38,51]. This study suggests that the process of immune escape holds greater complexities which vaccine design strategies will need to take into account. Frequent and persistent shuffling and toggling of mutations within targeted epitopes may indicate high levels of epitope instability early in infection. Early APOBEC-mediated hypermutation, identified in more than a quarter of genome regions under putative immune pressure in this study, represents an efficient mechanism of CTL escape [22]. Not to be discounted is the unclassified 11% of genome changes which demonstrate that much more is at play very early in infection, possibly such as pressure from NK-cells which has recently been emphasized [50]. These additional forces likely play a significant role in shaping the early virus.
Our findings provide novel data on the dynamic interplay between virus and host very early in infection and the complex pathways of escape in response to the earliest immune pressures acting on transmitted/founder subtype C viruses. These processes of immune adaption are likely to pose further challenges that vaccines will need to overcome. Furthermore, this study demonstrates the highly sensitive nature of viral sequencing as a tool for the identification and characterisation of early immune selective pressures that mould early HIV-1 evolution. Examining early host-virus interactions in the context of disease progression will enable us to identify those changes to the virus which are associated with better disease outcome and may therefore be incorporated into vaccine design.
Supplementary Material
Acknowledgements
This work is funded by the National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health (NIH) and the US Department of Health and Human Services (DHHS)(#AI51794, CAPRISA; # DK 49381 (MSC), CHAVI), as well as by the National Research Foundation (# 67385), SA; the South African AIDS Vaccine Initiative; and amfAR grant #106997–43. We thank the clinical staff and participants from the CAPRISA 002 Acute Infection Study. We would like to thank Denis Chopera, Gama Bandawe and Roman Ntale for sub-genomic sequence data and Ziyaad Valley-Omar for assistance with data analysis.
Footnotes
Author contributions are as follows: Melissa-Rose Abrahams: first author, manuscript writing, full-length genome and focused epitope DNA amplification and sequencing, data collation, sequence analyses, statistical analyses; Florette K Treurnicht: study design and coordination, full-length genome DNA amplification and sequencing, sequence analysis; Nobubelo K Ngandu: positive selection analyses and selection methods writing; Sarah A Goodier: full-length genome and focused epitope DNA amplification and sequencing, statistical and sequence analyses; Jinny C Marais: full-length genome sequencing, focused epitope amplification and sequencing, sequence analysis; Helba Bredell: half-genome DNA amplification and sequencing, sequence analysis; Ruwayhida Thebus: full-length genome amplification and sequencing; Debra de Assis Rosa: HLA typing of CAPRISA participants; Koleka Mlisana: clinical site project director; Cathal Seoighe: advising and guidance of clustered mutation and selection analyses; Salim Abdool Karim: CAPRISA study design; Clive M Gray: immunology consultant and assistance with manuscript writing; and Carolyn Williamson: principal investigator, corresponding author, manuscript writing.
References
- 1.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrahams M-R, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping L-H, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 2009;206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pantaleo G. HIV-1 T-cell vaccines: evaluating the next step. Lancet Infect Dis. 2008;8:82–83. doi: 10.1016/S1473-3099(07)70266-9. [DOI] [PubMed] [Google Scholar]
- 7.Abdool Karim Q, Abdool Karim SS, Frohlich Ja, Grobler AC, Baxter C, Mansoor LE, et al. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329:1168–1174. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu XG, Addo MM, Perkins BA, Wej F, Rathod A, Geer SC, et al. Differences in the expressed HLA class I alleles effect the differential clustering of HIV type 1-specific T cell responses in infected Chinese and caucasians. AIDS Res Human Retroviruses. 2004;20:557–564. doi: 10.1089/088922204323087813. [DOI] [PubMed] [Google Scholar]
- 9.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 10.Treurnicht FK, Seoighe C, Martin DP, Wood N, Abrahams M-R, Rosa DDA, et al. Adaptive changes in HIV-1 subtype C proteins during early infection are driven by changes in HLA-associated immune pressure. Virology. 2010;396:213–225. doi: 10.1016/j.virol.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rybarczyk BJ, Montefiori D, Johnson PR, West A, Johnston RE, Swanstrom R. Correlation between env V1 / V2 Region Diversification and Neutralizing Antibodies during Primary Infection by Simian Immunodeficiency Virus sm in Rhesus Macaques. J Virol. 2004;78:3561–3571. doi: 10.1128/JVI.78.7.3561-3571.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 14.Frost SDW, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci USA. 2005;102:18514–9. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlson JM, Brumme ZL. HIV evolution in response to HLA-restricted CTL selection pressures: a population-based perspective. Microbes Infect. 2008;10:455–461. doi: 10.1016/j.micinf.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, Sullivan KMO, et al. Selective Escape from CD8+ T-Cell Responses Represents a Major Driving Force of Human Immunodeficiency Virus Type 1 ( HIV-1 ) Sequence Diversity and Reveals Constraints on HIV-1 Evolution. J Virol. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nature. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 18.Jones NA. Determinants of Human Immunodeficiency Virus Type 1 Escape from the Primary CD8+ Cytotoxic T Lymphocyte Response. J Exp Med. 2004;200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, McNevin J, Cao J, Zhao H, Genowati I, Wong K, et al. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J Virol. 2006;80:9519–9529. doi: 10.1128/JVI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray CM, Mlotshwa M, Riou C, Mathebula T, de Assis Rosa D, Mashishi T, et al. Human immunodeficiency virus-specific gamma interferon enzyme-linked immunospot assay responses targeting specific regions of the proteome during primary subtype C infection are poor predictors of the course of viremia and set point. J Virol. 2009;83:470–478. doi: 10.1128/JVI.01678-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goonetilleke N, Liu MKP, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mlotshwa M, Riou C, Chopera D, de Assis Rosa D, Ntale R, Treunicht F, et al. Fluidity of HIV-1-specific T-cell responses during acute and early subtype C HIV-1 infection and associations with early disease progression. J Virol. 2010;84:12018–12029. doi: 10.1128/JVI.01472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, et al. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 27.van Loggerenberg F, Mlisana K, Williamson C, Auld SC, Morris L, Gray CM, et al. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One. 2008;3:e1954. doi: 10.1371/journal.pone.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chopera DR, Woodman Z, Mlisana K, Mlotshwa M, Martin DP, Seoighe C, et al. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 2008;4:e1000033. doi: 10.1371/journal.ppat.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseau CM, Birditt BA, McKay AR, Stoddard JN, Lee TC, McLaughlin S, et al. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J Virol Methods. 2006;136:118–125. doi: 10.1016/j.jviromet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Chopera DR, Mlotshwa M, Woodman Z, Mlisana K, de Assis Rosa D, Martin DP, et al. Virological and immunological factors associated with HIV-1 differential disease progression in HLA-B 58:01-positive individuals. J Virol. 2011;85:7070–7080. doi: 10.1128/JVI.02543-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 32.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bansal A, Gough E, Sabbaj S, Ritter D, Yusim K, Sfakianos G, et al. CD8 T-cell responses in early HIV-1 infection are skewed towards high entropy peptides. AIDS. 2005;19:241–250. [PubMed] [Google Scholar]
- 35.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthews PC, Prendergast A, Leslie A, Crawford H, Payne R, Rousseau C, et al. Central Role of Reverting Mutations in HLA Associations with Human Immunodeficiency Virus Set Point. J Virol. 2008;82:8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 40.Pond SK, Muse SV. Site-to-site variation of synonymous substitution rates. Mol Biol Evol. 2005;22:2375–2385. doi: 10.1093/molbev/msi232. [DOI] [PubMed] [Google Scholar]
- 41.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 42.Scheffler K, Martin DP, Seoighe C. Robust inference of positive selection from recombining coding sequences. Bioinformatics. 2006;22:2493–2499. doi: 10.1093/bioinformatics/btl427. [DOI] [PubMed] [Google Scholar]
- 43.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. Int J Infect Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 44.Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams M-R, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 2009;5:e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Gladden AD, Altfeld M, Kaldor JM, Cooper Da, Kelleher AD, et al. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J Virol. 2007;81:193–201. doi: 10.1128/JVI.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brumme ZL, Brumme CJ, Carlson J, Streeck H, John M, Eichbaum Q, et al. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J Virol. 2008;82:9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore PL, Gray ES, Choge IA, Ranchobe N, Mlisana K, Abdool Karim SS, et al. The C3-V4 Region Is a Major Target of Autologous Neutralizing Antibodies in Human Immunodeficiency Virus Type 1 Subtype C Infection. J Virol. 2008;82:1860–1869. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bansal A, Carlson J, Yan J, Akinsiku OT, Schaefer M, Sabbaj S, et al. CD8 T cell response and evolutionary pressure to HIV-1 cryptic epitopes derived from antisense transcription. J Exp Med. 2010;207:51–59. doi: 10.1084/jem.20092060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger CT, Carlson JM, Brumme CJ, Hartman KL, Brumme ZL, Henry LM, et al. Viral adaptation to immune selection pressure by HLA class I-restricted CTL responses targeting epitopes in HIV frameshift sequences. J Exp Med. 2010;207:61–75. doi: 10.1084/jem.20091808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:496–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davenport MP, Loh L, Petravic J, Kent SJ. Rates of HIV immune escape and reversion: implications for vaccination. Trends Microbiol. 2008;16:561–566. doi: 10.1016/j.tim.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Ferrari G, Korber B, Goonetilleke N, Liu MKP, Turnbull EL, Salazar-Gonzalez JF, et al. Relationship between functional profile of HIV-1 specific CD8 T cells and epitope variability with the selection of escape mutants in acute HIV-1 infection. PLoS Pathog. 2011;7:e1001273. doi: 10.1371/journal.ppat.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herbeck JT, Rolland M, Liu Y, McLaughlin S, McNevin J, Zhao H, et al. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J Virol. 2011;85:7523–7534. doi: 10.1128/JVI.02697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, et al. Whole Genome Deep Sequencing of HIV-1 Reveals the Impact of Early Minor Variants Upon Immune Recognition During Acute Infection. PLoS Pathog. 2012;8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gray ES, Moore PL, Choge IA, Decker JM, Bibollet-Ruche F, Li H, et al. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007;81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeh WW, Rahman I, Hraber P, Coffey RT, Nevidomskyte D, Giri A, et al. Autologous neutralizing antibodies to the transmitted/founder viruses emerge late after simian immunodeficiency virus SIVmac251 infection of rhesus monkeys. J Virol. 2010;84:6018–6032. doi: 10.1128/JVI.02741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bar KJ, Tsao C-Y, Iyer SS, Decker JM, Yang Y, Bonsignori M, et al. Early Low-Titer Neutralizing Antibodies Impede HIV-1 Replication and Select for Virus Escape. PLoS Pathog. 2012;8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hecht FM, Hartogensis W, Bragg L, Bacchetti P, Grant R, Barbour J, et al. HIV RNA level in early infection is predicted by viral load in the transmission source. AIDS. 2010;24:941–945. doi: 10.1097/QAD.0b013e328337b12e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollingsworth TD, Laeyendecker O, Shirreff G, Donnelly Ca, Serwadda D, Wawer MJ, et al. HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog. 2010;6:e1000876. doi: 10.1371/journal.ppat.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alizon S, von Wyl V, Stadler T, Kouyos RD, Yerly S, Hirschel B, et al. Phylogenetic approach reveals that virus genotype largely determines HIV set-point viral load. PLoS Pathog. 2010;6:e1001123. doi: 10.1371/journal.ppat.1001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miura T, Brumme ZL, Brockman Ma, Rosato P, Sela J, Brumme CJ, et al. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J Virol. 2010;84:7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang J, Costello C, Keet IP, Rivers C, Leblanc S, Karita E, et al. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res Human Retroviruses. 1999;15:317–324. doi: 10.1089/088922299311277. [DOI] [PubMed] [Google Scholar]
- 63.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–189. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.