Abstract
Introduction
Our retrospective cohort study investigated the effect of tumor site and stage on the associations between the allelic variants of glutathione S-transferase (GST) and DNA-repair genes and overall survival (OS) in CRC patients treated with 5-fluorouracil (5-FU)-based adjuvant chemotherapy.
Material and Methods
We genotyped GSTM1, GSTT1, GSTP1 Ile105Val, XRCC1 Arg399Gln, XRCC3 Thr241Met, and XPD Lys751Gln in 491 CRC patients between 1995 and 2001. A Cox proportional-hazards model was used to calculate the hazard ratios (HRs) and 95% confidence intervals (CIs) for the relationships between the allelic variants and OS. Survival analyses were performed for each allelic variant by using the log-rank test and Kaplan-Meier analysis.
Results
The CRC patients with the XPD Gln allelic variants had poorer survival than patients with the Lys/Lys genotype (HR = 1.38, 95% CI = 1.02–1.87), and rectal cancer patients had the poorest survival among them (HR = 1.87, 95% CI = 1.18–2.95). A significantly shorter OS was observed among stage II/III colon cancer patients with the XRCC1 Gln allelic variants (HR = 1.69, 95% CI = 1.06–2.71), compared to those with XRCC1 Arg/Arg genotype. In the combined analysis of the XRCC1 and XPD genes patients with stage II/III tumors, the poorest OS occurred in colon cancer patients with the XRCC1 Gln and XPD Gln allelic variants (HR = 2.60, 95% CI = 1.19–5.71) and rectal cancer patients with the XRCC1 Arg/Arg and XPD Gln allelic variants (HR = 2.77, 95% CI = 1.25–6.17).
Conclusion
The XPD and XRCC1 allelic variants may be prognostic markers for CRC patients receiving 5-FU based chemotherapy. The contributions of the XPD and XRCC1 allelic variants to OS are tumor site- and/or stage-dependent.
Introduction
After surgery, most metastatic colorectal cancer (CRC) patients receive an adjuvant chemotherapy regimen consisting of combination therapy using 5-fluorouracil (FU) and oxaliplatin (FOLFOX) or leucovorin (LV) for 6 to 8 mo to reduce the probability of tumor recurrence and prolong survival [1]–[3]. The functional single-nucleotide polymorphisms (SNPs) in drug-targeted genes, including xenobiotic-metabolizing and DNA-repair genes, correlate with variability in clinical outcome in multiple types of cancer [4]–[8]. The identification of genetic markers may help identify patients who may benefit from chemotherapy and reduce potential toxicity.
Variable chemosensitivity may involve the detoxification pathway, including the glutathione S-transferases (GSTs). The GSTs are multigene families of enzymes that inactivate electrophilic xenobiotics by conjugation with glutathione, preventing DNA damage and adduct formation [9]. Interindividual differences in GST activity may be mediated by genetic polymorphisms [9]–[12]. The structural deletion polymorphisms in GSTM1 and GSTT1 result in the loss of enzyme-catalyzed detoxification activity [10], and are predictors of clinical outcome in gastric cancer patients receiving platinum/5-FU chemotherapy [13]. In addition, the reduced glutathione conjugation resulting from polymorphism in the GSTP1 Ile105Val (rs1695) coding region may be associated with an increased survival in CRC patients treated with oxaliplatin/5-FU [14], [15].
Various DNA-repair enzymes play important roles in preventing treatment resistance and protecting the genome against carcinogenesis [16]–[19]. The expression of the base excision repair (BER) gene family is triggered by internal oxidative stress and DNA damage [20]. The BER pathway involves the X-ray cross-complementing group 1 gene XRCC1 [21]. The XRCC1 protein directly associates with polymerase beta, DNA ligase III, and poly (ADP-ribose) polymerase (PARP) in single-strand break-repair processes that may play a role in tumor cell sensitivity to 5-FU treatment [22], [23]. The XRCC3 protein, a member of the Rad51-related enzyme family, contributes to the maintenance of chromosome stability through DNA double-strand break/recombination repair in homologous recombination [24].
The nucleotide excision repair (NER) gene family functions within a range of structurally unrelated DNA lesions and DNA adducts [16], [17]. The xeroderma pigmentosum complementation group D (XPD) protein, a member of the NER family functions in sensing, binding, and the subsequent recruitment of repair-related factors [5]. Previous studies have shown that the allelic variants XRCC1 Arg399Gln (rs25487), XRCC3 Thr241Met (rs861539), and XPD Lys751Gln (rs13181) are associated with DNA adduct levels and repair capacity [5], [25], [26]. Therefore, these polymorphisms of DNA-repair genes may affect clinical outcomes in cancer patients receiving chemotherapy [7], [13], [20], [27], [28].
Our previous studies have shown that the null genotypes of GSTM1 and GSTT1, GSTP1 Ile105Val, XRCC1 Arg399Gln, XRCC3 Thr241Met, and XPD Lys751Gln allelic variants are associated with significantly increased risks of CRC [29]–[32]. However, other previous reports of associations between GST and DNA-repair allelic variants and clinical outcomes in CRC have been conflicting [6], [11], [33]–[35]. Whether these conflicting findings have been due to tissue-specific differences in gene expression between colonic and rectal tumors is unclear [36] because reports of stratified analyses based on the site of the CRC tumor are scant. Therefore, in our present study, we investigated the relationship between the allelic variants of relevant GST and DNA-repair genes and the chemotherapeutic outcomes in a retrospective CRC cohort who received adjuvant chemotherapy in Taiwan to determine whether differences exist among the associations in the tumor site or pathological stage.
Materials and Methods
Participant selection
Our study was approved by the Institutional Review Board of China Medical University Hospital and Chang Gung Memorial Hospital (CGMH). All patients provided written informed consent before participation in our study. We reviewed 2716 newly diagnosed and histologically confirmed CRC patients who underwent surgery at CGMH between January 1995 and December 2001. We enrolled 499 patients without familial adenomatous polyposis or hereditary nonpolyposis colorectal cancer who underwent 5-FU-based adjuvant chemotherapy as first-line treatment after surgery, and received follow-up examinations every 3 to 6 mo at the outpatient clinic of the Colorectal Section of CGMH.
The postoperative adjuvant chemotherapy regimens included both daily and weekly or monthly treatments. In the daily treatment, tegafur (300–350 mg/m2/d) and levamisole (45 mg/d) were administered orally in 3 doses per day for 28 d, followed by a 7-d rest period, totaling 12 mo. The weekly treatments consisted of one of the following treatment schemes for 6 wk, followed by a 2 wk rest, totaling 12 mo: (1) 5-FU (450 mg/m2/d) and leucovorin (50 mg/d) were administered as an intravenous bolus once weekly; (2) 5-FU (450 mg/m2/d) were administered intravenously once weekly, and levamisole (50 mg) was administered orally 3 times per day for 3 d every 2 wk; or (3) a 24-h intravenous infusion of 5-FU (2600 mg/m2/d) and leucovorin (150 mg/d) was administered once weekly. The monthly treatments consisted of one of the following treatment schemes for 6 mo: (1) a continuous 5 d infusion of 5-FU (800 mg/m2/d) and leucovorin (50 mg/d) was administered once monthly, or (2) patients received a 5 d continuous infusion of 5-FU (800 mg/ m2/d) once monthly combined with levamisole (50 mg) administered orally 3 times per day for 3 d every 2 wk.
Clinical and questionnaire assessment
The clinical data included a medical history, physical examination (including a rectal and perineal examination), and the measurement of carcinoembryonic antigen (CEA). Liver sonograms, chest X-ray, and colonoscopy (or barium enema examination) were performed annually. All patients were followed until May 2008, with a median follow-up time of 48.8 mo (range = 1.5 to 133.3 mo). Overall survival (OS) was measured from the date of surgery until death from any cause. In the calculation of OS, the event date represented the date of death, and the censoring date represented the last known date on which the patient was known to be alive.
DNA extraction and genotyping
The details of the genotyping methods have been described elsewhere [29], [30]. DNA was isolated from peripheral blood leukocytes by using sodium dodecyl sulfate (SDS)-proteinase K-RNase digestion and phenol-chloroform extraction. The GSTM1 and GSTT1 genetic variants were identified using polymerase chain reaction (PCR), and the GSTP1 Ile105Val, XRCC1 Arg399Gln, XRCC3 Thr241Met, and XPD Lys751Gln allelic variants were analyzed using PCR combined with restriction fragment length polymorphism.
The presence or absence of GSTM1, GSTT1, or β-globin (internal control) DNA was detected during multiple PCRs with the following primers: GSTM1 (5′-TGCCCTACTTGATTGATGGG-3′ and 5′-CTGGATTGTAGCAGATCATGC-3′); GSTT1 (5′-TTCCTTACTGGTCCTCACATCTC-3′ and 5′-CACCGGATCATGGCCAGCA-3′); and β-globin (5′-CACAACTGTGTTCACTAGC-3′ and 5′-CAACTTCATCCACGTTCACC-3′). The primer sequences used to detect the genotype corresponding to GSTP1 Ile105Val DNA were 5′-CCTCTCCCTTTCCTCTGTTC-3′ and 5′-CAGGTGAGGGGGACATCT-3. The GSTP1 Ile105Val PCR product was digested using Alw26I (New England Biolabs, Beverly, MA, USA). The primer sequences used to detect the genotype corresponding to XRCC1 Arg399Gln DNA were 5′-TTGTGCTTTCTCTGTGTCCA-3′ and 5′-TCCTCCAGCCTTTTCTGATA-3′. The XRCC1 Arg399Gln PCR product was digested using MspI (New England Biolabs). The primer sequences used to detect the genotype corresponding to XRCC3 Thr241Met DNA were 5′-GCCTGGTGGTCATCGACTC-3′, and 5′-ACAGGGCTCTGGAAGGCACTGCTCAGCTCACGCACC-3′. The XRCC3 Thr241Met PCR product was digested using NcoI (New England Biolabs). The primer sequences used to detect the genotype corresponding to XPD Lys751Gln DNA were 5′-CCTCTCCCTTTCCTCTGTTC-5′, and 5′-CAGGTGAGGGGGACATCT-3′. The XPD Lys751Gln PCR product was digested using PstI (Takara Bio, Shiga, Japan). For quality control, 10% of the PCRs were randomly repeated, and showed 100% concordance for all the allelic variants analyzed. All laboratory personnel were blinded to the survival status of the patient samples.
Statistical analysis
We used χ2-tests to evaluate possible associations between the categorical variables, and continuous variables were analyzed using a Student t test. Allelic frequencies were compared with those expected at the Hardy-Weinberg equilibrium (HWE) by using a χ2 test. Univariate analysis of the Kaplan-Meier estimates and the log-rank test were used to compare the overall survival curves. A Cox proportional-hazards model was used to estimate the hazard ratios (HRs) and the 95% confidence intervals (CIs) for the various allelic variants associated with OS. Age, sex, and the tumor, node, metastasis (TNM) stage were considered potential confounding covariates, and were thus included in the multivariate regression. The Cox proportional-hazards regression model was based on a priori knowledge of factors known to carry prognostic or predictive information to estimate adjusted HRs and 95% CIs. The HRs for differences in OS based on the tumor site and stage were estimated using stratified analysis. All models were examined for adherence to the proportional-hazards assumption by assessing the log-minus-log survival plots and performing the Schoenfeld test. The log-minus-log survival plots and the results of the Schoenfeld tests (P range = 0.22 to 0.90) indicated no violations of the proportionality assumption for the 6 SNPs investigated. All analyses were performed using the SAS statistical software package, version 9.3 for Windows (SAS Institute, Cary, NC, USA). The results of comparisons with a 2 sided P value less than 0.05 were considered to represent statistically significant relationships. To account for multiple comparisons, we calculated the false discovery rate (FDR) by using the PROC MULTTEST in SAS. The statistical power of our analyses was estimated using the PASS statistical software package, version 11 (NCSS, Kaysville, UT, USA).
Results
Demographic and clinical characteristics
Eight CRC patients who failed genotyping were excluded from further analysis. The demographic and clinical characteristics of the 491 patients are summarized in Table 1. The mean age and standard deviation (SD) was 58.5±12.5 y. Our cohort consisted of 260 (52.9%) men, and 168 (35.5%) patients had a family history of cancer. The primary tumor was located in the colon in 283 (57.6%) patients and in the rectum in 208 (42.4%) patients. Multiple primary malignancies occurred in 4.9% of the cohort, and 78.4% had a moderately differentiated tumor. The CEA was present at ≥5 ng/mL in 53.6% of the cohort. In the TNM evaluation, 74 (15.1%), 267 (54.4%), and 150 (30.5%) patients were classified as being stage II, III, and IV, respectively. Compared to patients with rectal cancer, colon cancer patients were younger, were diagnosed more frequently with multiple malignancies, presented more often with moderately differentiated tumors, and were less likely to be TNM stage III (P<0.05). No significant associations were found between sex, family history of cancer, or CEA level and the tumor site.
Table 1. Demographic characteristics, clinical features, and allele distributions of study patients.
| Total | Colon | Rectum | ||
| Variablesa | N (%) | N (%) | N (%) | P |
| Ageb (y) | 58.5±12.5 | 57.1±12.9 | 60.4±11.6 | 0.004c |
| Sex | 0.105d | |||
| Male | 260 (52.9) | 141 (49.8) | 119 (57.2) | |
| Female | 231 (47.1) | 142 (50.2) | 89 (42.8) | |
| Family history of cancer | 0.961d | |||
| No | 305 (64.5) | 175 (64.6) | 130 (64.4) | |
| Yes | 168 (35.5) | 96 (35.4) | 72 (35.6) | |
| Multiplicity | 0.029d | |||
| No | 467 (95.1) | 264 (93.3) | 203 (97.6) | |
| Yes | 24 (4.9) | 19 (6.7) | 5 (2.4) | |
| Histological differentiation | 0.009d | |||
| Well | 59 (12.1) | 38 (13.6) | 21 (10.1) | |
| Moderately | 382 (78.4) | 207 (73.9) | 175 (84.5) | |
| Poorly | 46 (9.5) | 35 (12.5) | 11 (5.4) | |
| Carcinoembryonic antigen | 0.086d | |||
| <5 ng/mL | 219 (46.4) | 117 (43.0) | 102 (51.0) | |
| ≥5 ng/mL | 253 (53.6) | 155 (57.0) | 98 (49.0) | |
| TNM stage | 0.001d | |||
| II | 74 (15.1) | 56 (19.8) | 18 (8.7) | |
| III | 267 (54.4) | 138 (48.8) | 129 (62.0) | |
| IV | 150 (30.5) | 89 (31.4) | 61 (29.3) | |
| GSTM1 | 0.254d | |||
| Null | 286 (58.2) | 171 (60.4) | 115 (55.3) | |
| Present | 205 (41.8) | 112 (39.6) | 93 (44.7) | |
| GSTT1 | 0.317d | |||
| Null | 249 (50.7) | 149 (52.6) | 100 (48.1) | |
| Present | 242 (49.3) | 134 (47.4) | 108 (51.9) | |
| GSTP1 Ile105Val | 0.632d | |||
| Ile/Ile | 336 (68.4) | 191 (67.5) | 145 (69.7) | |
| Ile/Val | 139 (28.3) | 81 (28.6) | 58 (27.9) | |
| Val/Val | 16 (3.3) | 11 (3.9) | 5 (2.4) | |
| XRCC1 Arg399Gln | 0.016d | |||
| Arg/Arg | 249 (50.7) | 159 (56.2) | 90 (43.3) | |
| Arg/Gln | 212 (43.2) | 110 (38.9) | 102 (49.0) | |
| Gln/Gln | 30 (6.1) | 14 (5.0) | 16 (7.7) | |
| XRCC3 Thr241Met | 0.565e | |||
| Thr/Thr | 459 (93.5) | 263 (92.9) | 196 (94.2) | |
| Thr/Met | 32 (6.5) | 20 (7.1) | 12 (5.8) | |
| Met/Met | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| XPD Lys751Gln | 0.327e | |||
| Lys/Lys | 404 (82.3) | 234 (82.7) | 170 (81.7) | |
| Lys/Gln | 84 (17.1) | 46 (16.2) | 38 (18.3) | |
| Gln/Gln | 3 (0.6) | 3 (1.1) | 0 (0.0) | |
Sum may not be equal to the total number (N) because of missing data.
Mean±standard deviation.
Student t-test.
Chi-squared test.
Fisher exact test.
TNM: tumor-node-metastasis.
Genotype frequencies
The genotype distributions of the GST and DNA-repair allelic variants in the CRC patients are also shown in Table 1. The frequencies of the null genotypes of GSTM1 and GSTT1 and the GSTP1 Val, XRCC1 Gln, XRCC3 Met, and XPD Gln allelic variants were 58.2%, 50.7%, 17.4%, 27.7%, 3.3%, and 9.2%, respectively. The allele frequencies of the GSTP1, XRCC1, XRCC3, and XPD allelic variants were at HWE. We found that the distribution of these allelic variants did not vary significantly according to the tumor site, except for the XRCC1 Gln allelic variants (P = 0.016). The XRCC1 Arg/Arg genotype was more prevalent in colon cancer patients (56.2%) than in rectal cancer patients (43.3%).
Association between GSTs and DNA-repair allelic variants and survival
Univariate analysis revealed that the association between the XRCC1 Gln allelic variants and reduced OS was more significant than that of XRCC1 Arg/Arg genotype, with a median survival of 43.0 versus 57.1 mo, respectively (HR = 1.36, 95% CI = 1.07–1.73). However, this association was absent following adjustment for the covariates, which included age, sex, and the stage of disease. The other allelic variants analyzed were not associated with OS in univariate analysis of the Kaplan-Meier estimates (data not shown).
Table 2 shows the adjusted associations between the allelic variants and OS in the multivariate Cox proportional-hazards regression model. Among the 6 allelic variants tracked, reduced OS was associated with heterozygous carriers of XPD Lys/Gln (HR = 1.34, 95% CI = 0.99–1.84, P = 0.062, FDR = 0.372) and homozygous carriers of XPD Gln/Gln (HR = 2.38, 95% CI = 0.75–7.53, P = 0.141). Compared to patients with XPD Lys/Lys genotype, patients with XPD Gln allelic variants were significantly associated with reduced OS, with a median survival of 49.0 versus 47.1 mo, respectively (HR = 1.38, 95% CI = 1.02–1.88, P = 0.039, FDR = 0.372).
Table 2. Cox proportional–hazards analysis of associations between allelic variants of metabolizing and DNA–repair genes and overall survival status among colorectal cancer patients.
| Variables | Patients | Events | MSM | HR (95% CI)a | P | FDR |
| GSTM1 | ||||||
| Null | 286 | 161 | 48.1 | 1 (reference) | ||
| Present | 205 | 107 | 50.1 | 0.81 (0.64–1.04) | 0.100 | 0.400 |
| GSTT1 | ||||||
| Null | 249 | 133 | 49.5 | 1 (reference) | ||
| Present | 242 | 135 | 47.6 | 1.15 (0.90–1.46) | 0.271 | 0.643 |
| GSTP1 Ile105Val | ||||||
| Ile/Ile | 336 | 183 | 48.8 | 1 (reference) | ||
| Ile/Val | 139 | 76 | 49.0 | 1.10 (0.84–1.44) | 0.482 | 0.643 |
| Val/Val | 16 | 9 | 38.9 | 1.12 (0.57–2.20) | 0.749 | 0.850 |
| Ile/Val+ Val/Val | 155 | 85 | 49.0 | 1.10 (0.85–1.43) | 0.459 | 0.643 |
| XRCC1 Arg399Gln | ||||||
| Arg/Arg | 249 | 126 | 57.1 | 1 (reference) | ||
| Arg/Gln | 212 | 126 | 42.3 | 1.12 (0.87–1.44) | 0.389 | 0.643 |
| Gln/Gln | 30 | 16 | 49.0 | 1.06 (0.63–1.79) | 0.827 | 0.850 |
| Arg/Gln + Gln/Gln | 242 | 142 | 43.0 | 1.11 (0.87–1.42) | 0.402 | 0643 |
| XRCC3 Thr241Met | ||||||
| Thr/Thr | 459 | 253 | 48.6 | 1 (reference) | ||
| Thr/Met | 32 | 15 | 49.3 | 1.05 (0.62–1.78) | 0.850 | 0.850 |
| Met/Met | 0 | 0 | 0 | – | – | – |
| XPD Lys751Gln | ||||||
| Lys/Lys | 404 | 216 | 49.0 | 1 (reference) | ||
| Lys/Gln | 84 | 49 | 47.8 | 1.34 (0.99–1.84) | 0.062 | 0.372 |
| Gln/Gln | 3 | 3 | 28.5 | 2.38 (0.75–7.53) | 0.141 | 0.423 |
| Lys/Gln + Gln/Gln | 87 | 52 | 47.1 | 1.38 (1.02–1.88) | 0.039 | 0.372 |
Adjusted for age, sex, and tumor-node-metastasis stage.
MSM: median survival month; FDR: false discovery rate.
Association between the genotype and survival by tumor site and TNM stage
The results of the stratified analyses of OS associated with the GSTs and DNA-repair allelic variants based on the tumor location are shown in Table 3. Rectal cancer patients with the XPD Gln allelic variants had a shorter OS compared with patients with XPD Lys/Lys genotype, with a median survival 44.9 versus 51.6 mo (log-rank test P = 0.230). The multivariate Cox regression model also showed that the XPD Gln allelic variants were associated with an 87% increased risk of shorter OS compared to XPD Lys/Lys genotype (HR = 1.87; 95% CI = 1.18–2.95, P = 0.007, FDR = 0.084). However, no adverse effect of Gln allelic variants on OS was observed in colon cancer patients. No significant association between the other allelic variants and OS was observed based on the tumor site.
Table 3. Cox proportional-hazards analysis of associations between allelic variants and overall survival status among colorectal cancer patients based on tumor location.
| Colon | Rectum | |||||||||||
| Variables | Patients | Events | MSM | HR (95% CI)a | P | FDR | Patients | Events | MSM | HR (95% CI)a | P | FDR |
| GSTM1 | ||||||||||||
| Null | 171 | 95 | 48.1 | 1 (reference) | 115 | 66 | 48.5 | 1 (reference) | ||||
| Present | 112 | 57 | 48.9 | 0.83 (0.59–1.15) | 0.262 | 0.689 | 93 | 50 | 56.7 | 0.82 (0.56–1.19) | 0.287 | 0.689 |
| GSTT1 | ||||||||||||
| Null | 149 | 79 | 49.1 | 1 (reference) | 100 | 54 | 55.8 | 1 (reference) | ||||
| Present | 134 | 73 | 47.1 | 1.21 (0.88–1.68) | 0.240 | 0.689 | 108 | 62 | 48.5 | 1.12 (0.77–1.62) | 0.559 | 0.917 |
| GSTP1 Ile105Val | ||||||||||||
| Ile/Ile | 191 | 102 | 48.1 | 1 (reference) | 145 | 81 | 49.5 | 1 (reference) | ||||
| Ile/Val + Val/Val | 92 | 50 | 48.6 | 1.12 (0.80–1.58) | 0.506 | 0.917 | 63 | 35 | 49.0 | 1.07 (0.72–1.59) | 0.745 | 0.917 |
| XRCC1 Arg399Gln | ||||||||||||
| Arg/Arg | 159 | 75 | 57.1 | 1 (reference) | 90 | 51 | 57.2 | 1 (reference) | ||||
| Arg/Gln + Gln/Gln | 124 | 77 | 36.4 | 1.25 (0.90–1.73) | 0.181 | 0.689 | 118 | 65 | 47.7 | 0.98 (0.68–1.42) | 0.910 | 0.917 |
| XRCC3 Thr241Met | ||||||||||||
| Thr/Thr | 263 | 142 | 48.1 | 1 (reference) | 196 | 111 | 49.0 | 1 (reference) | ||||
| Thr/Met | 20 | 10 | 49.0 | 1.06 (0.56–2.01) | 0.866 | 0.917 | 12 | 5 | 67.4 | 0.95 (0.38–2.39) | 0.917 | 0.917 |
| XPD Lys751Gln | ||||||||||||
| Lys/Lys | 234 | 125 | 48.0 | 1 (reference) | 170 | 91 | 51.6 | 1 (reference) | ||||
| Lys/Gln + Gln/Gln | 49 | 27 | 49.1 | 1.10 (0.72–1.67) | 0.667 | 0.917 | 38 | 25 | 44.9 | 1.87 (1.18–2.95) | 0.007 | 0.084 |
Adjusted for age, sex, and tumor-node-metastasis stage.
MSM: median survival month; HR: hazard ratio; CI: confidence interval; FDR: false discovery rate.
The stratified analyses of the associations between OS and the polymorphisms of the GST and DNA-repair alleles based on the TNM stage showed that both stage II/III and stage IV CRC patients with the XPD Gln allelic variants didn't have a significantly poorer OS, with HRs of 1.43 (P = 0.085) and 1.31 (P = 0.264), respectively, compared to patients with XPD Lys/Lys genotype (Table 4). The association between the XRCC1 Gln allelic variants and reduced OS in stage II/III patients approached significance (HR = 1.41, 95% CI = 1.00–1.99, P = 0.051, FDR = 0.510). The other allelic variants were not associated with OS in stage IV patients.
Table 4. Cox proportional-hazards analysis of associations between allelic variants and overall survival status among colorectal cancer patients based on tumor-node-metastasis stage.
| Stage II/III | Stage IV | |||||||||||
| Variables | Patients | Events | MSM | HR (95% CI)a | P | FDR | Patients | Events | MSM | HR (95% CI)a | P | FDR |
| GSTM1 | ||||||||||||
| Null | 199 | 79 | 59.0 | 1 (reference) | 87 | 82 | 18.7 | 1 (reference) | ||||
| Present | 141 | 49 | 60.4 | 0.80 (0.56–1.15) | 0.224 | 0.713 | 64 | 58 | 20.0 | 0.87 (0.62–1.22) | 0.416 | 0.713 |
| GSTT1 | ||||||||||||
| Null | 176 | 65 | 60.6 | 1 (reference) | 73 | 68 | 20.2 | 1 (reference) | ||||
| Present | 164 | 63 | 59.1 | 1.11 (0.78–1.57) | 0.563 | 0.831 | 78 | 72 | 18.8 | 1.09 (0.78–1.52) | 0.623 | 0.831 |
| GSTP1 Ile105Val | ||||||||||||
| Ile/Ile | 232 | 86 | 60.8 | 1 (reference) | 104 | 97 | 19.2 | 1 (reference) | ||||
| Ile/Val + Val/Val | 108 | 42 | 58.9 | 1.17 (0.81–1.70) | 0.404 | 0.713 | 47 | 43 | 19.9 | 1.00 (0.70–1.43) | 0.992 | 0.992 |
| XRCC1 Arg399Gln | ||||||||||||
| Arg/Arg | 180 | 62 | 60.9 | 1 (reference) | 69 | 64 | 20.0 | 1 (reference) | ||||
| Arg/Gln + Gln/Gln | 160 | 66 | 56.7 | 1.41 (1.00–2.00) | 0.051 | 0.510 | 82 | 76 | 18.8 | 1.02 (0.72–1.43) | 0.930 | 0.992 |
| XRCC3 Thr241Met | ||||||||||||
| Thr/Thr | 314 | 119 | 60.0 | 1 (reference) | 145 | 134 | 19.4 | 1 (reference) | ||||
| Thr/Met | 26 | 9 | 67.4 | 0.91 (0.46–1.79) | 0.780 | 0.936 | 6 | 6 | 17.7 | 1.42 (0.62–3.27) | 0.404 | 0.713 |
| XPD Lys751Gln | ||||||||||||
| Lys/Lys | 275 | 97 | 60.3 | 1 (reference) | 129 | 119 | 19.6 | 1 (reference) | ||||
| Lys/Gln + Gln/Gln | 65 | 31 | 58.6 | 1.43 (0.95–2.14) | 0.085 | 0.510 | 22 | 21 | 17.7 | 1.31 (0.82–2.09) | 0.264 | 0.713 |
Adjusted for age and sex.
MSM: median survival month; HR: hazard ratio; CI: confidence interval; FDR: false discovery rate.
Table 5 shows the associations of the XRCC1 and XPD allelic variants with OS in CRC patients stratified by tumor site and TNM stage. The poorest OS was observed among the stage II/III colon cancer patients with the XRCC1 Gln allelic variants (HR = 1.69, 95% CI = 1.06–2.71, P = 0.028, FDR = 0.252). Although the rectal cancer patients who inherited XPD Gln allelic variants had significantly reduced OS, this negative effect was not more prominent in any specific TNM stage (stage II/III, HR = 1.57, 95% CI = 0.88–2.81; stage IV, HR = 1.98, 95% CI = 0.94–4.17).
Table 5. Tumor site- and tumor-node-metastasis stage-specific hazard ratios for the associations between the XRCC1 Arg399Gln and XPD Lys751Gln allelic variants and overall survival among colorectal cancer patients.
| Colon | Rectum | |||||||||||
| Stage II/III | Stage IV | Stage II/III | Stage IV | |||||||||
| Variables | HR (95% CI)a | P | FDR | HR (95% CI)a | P | FDR | HR (95% CI)a | P | FDR | HR (95% CI)a | P | FDR |
| XRCC1 Arg399Gln | ||||||||||||
| Arg/Gln+Gln/Gln vs Arg/Arg | 1.69 (1.06–2.71) | 0.028 | 0.252 | 1.05 (0.67–1.66) | 0.829 | 0.933 | 1.15 (0.67–1.96) | 0.612 | 0.918 | 0.93 (0.55–1.59) | 0.794 | 0.933 |
| XPD Lys751Gln | ||||||||||||
| Lys/Gln+Gln/Gln vs Lys/Lys | 1.28 (0.72–2.26) | 0.403 | 0.727 | 1.00 (0.54–1.85) | 0.999 | 0.999 | 1.57 (0.88–2.81) | 0.125 | 0.375 | 1.98 (0.94–4.17) | 0.072 | 0.324 |
Adjusted for age and sex.
HR: hazard ratio; CI: confidence interval; FDR: false discovery rate.
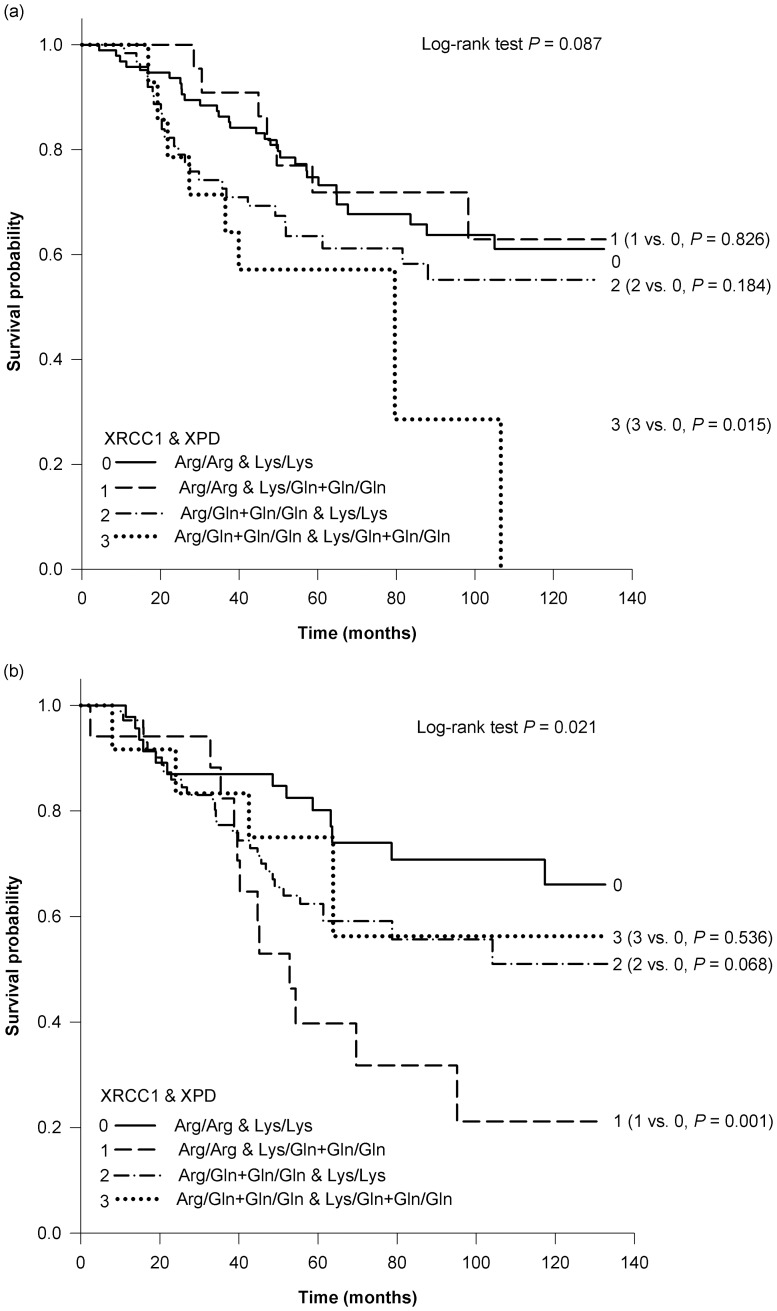
In the combined analysis of the XRCC1 and XPD allelic variants, the Kaplan-Meier survival curves showed differences in OS among the 4 allelic variants analyzed in stage II/III colon cancer patients (log-rank test P = 0.087, Figure 1a). Compared to patients with XRCC1 Arg/Arg and XPD Lys/Lys genotype, those with XRCC1 Gln and XPD Gln allelic variants had poorer OS (log-rank test P = 0.015), with an HR of 2.60 (95% CI = 1.19–5.71, FDR = 0.102, Table 6). Kaplan-Meier analysis showed significant variability in survival (log-rank test P = 0.021) among the subgroups in stage II/III rectal cancer patients (Figure 1b). The XRCC1 Arg/Arg and XPD Gln allelic variants were associated with a significantly poorer OS, compared with that of patients with XRCC1 Arg/Arg and XPD Lys/Lys genotype (log-rank test P = 0.001; HR = 2.77, 95% CI, 1.25–6.17, FDR = 0.102). However, this effect on OS was not observed among stage IV colon or rectal cancer patients with the XRCC1 Arg/Arg and XPD Gln allelic variants. FDR analysis indicated that the combined HRs of the XRCC1 Gln and XPD Gln allelic variants in stage II/III colon cancer patients (HR = 2.60; FDR = 0.102) and the XRCC1 Arg/Arg and XPD Gln allelic variants in stage II/III rectal cancer patients (HR = 2.77; FDR = 0.102) were not significantly influenced by type I error because of multiple comparisons. However, the statistical power of our analysis was determined to be 63% (HR = 2.60) and 49% (HR = 2.77) for these associations.
Figure 1. Kaplan-Meier estimates of the effect of coexisting XRCC1 and XPD allelic variants on overall survival in colorectal cancer patients stratified by tumor site and tumor-node-metastasis stage.
(a) The survival curve of stage II/III colon cancer patients (overall log-rank test P = 0.087). (b) The survival curve of stage II/III rectal cancer patients (overall log-rank test P = 0.021).
Table 6. Tumor site- and tumor-node-metastasis stage-specific hazard ratios for the associations between the coexisting XRCC1 Arg399Gln and XPD Lys751Gln allelic variants and overall survival among colorectal cancer patients.
| Colon | Rectum | ||||||||||||
| Stage II/III | Stage IV | Stage II/III | Stage IV | ||||||||||
| XRCC1 | XPD | HR (95% CI)a | P | FDR | HR (95% CI)a | P | FDR | HR (95% CI)a | P | FDR | HR (95% CI)a | P | FDR |
| Arg/Arg | Lys/Lys | 1 (reference) | 1 (reference) | 1 (reference) | 1 (reference) | ||||||||
| Arg/Arg | Lys/Gln + Gln/Gln | 1.03 (0.45–2.35) | 0.945 | 0.967 | 0.98 (0.41–2.38) | 0.967 | 0.967 | 2.77 (1.25–6.17) | 0.012 | 0.102 | 1.63 (0.59–4.46) | 0.345 | 0.690 |
| Arg/Gln + Gln/Gln | Lys/Lys | 1.54 (0.90–2.62) | 0.115 | 0.324 | 1.05 (0.64–1.72) | 0.863 | 0.967 | 1.73 (0.88–3.38) | 0.110 | 0.324 | 0.91 (0.50–1.64) | 0.744 | 0.967 |
| Arg/Gln + Gln/Gln | Lys/Gln + Gln/Gln | 2.60 (1.19–5.71) | 0.017 | 0.102 | 1.07 (0.44–2.56) | 0.885 | 0.967 | 1.43 (0.46–4.44) | 0.535 | 0.917 | 2.40 (0.76–7.59) | 0.135 | 0.324 |
Adjusted for age and sex.
HR: hazard ratio; CI: confidence interval; FDR: false discovery rate.
Discussion
We found a significant relationship between the XPD Lys751Gln allelic variant and OS for CRC patients, particularly for rectal cancer patients, whereas XRCC1 Arg399Gln Gln allele correlated with reduced OS in stage II/III colon cancer patients. In addition, the poorest OS was present among the stage II/III colon cancer patients with both XRCC1 Gln and XPD Gln allelic variants and among the stage II/III rectal cancer patients with both the XRCC1 Arg/Arg and XPD Gln allelic variants. However, significant associations were not observed between OS and the other GST and DNA-repair gene allelic variants in CRC patients receiving 5-FU-based chemotherapy.
The XPD gene is an important DNA-repair gene that codes for enzymes that recruit NER in the repair of a wide range of DNA lesions [16], [17]. Mutations that alter the amino acid sequence impact the interactions of XPD enzyme with other members, such as XPA, ERCC1, and replication protein A of the NER complex, resulting in different DNA-repair activities [18]. The XPD Lys751Gln polymorphism converts the basic amino acid, Lys, to the polar amino acid, Gln, at approximately 50 bp upstream from the poly(A) signal, which may affect the function of the XPD protein [4]. Patients with the XPD Lys/Lys genotype have sub-optimal DNA-repair activity, and are more sensitive to chemotherapy [5], [6].
Previous studies have shown that the XPD Lys751Gln allelic variant may be associated with various clinical outcomes in CRC patients receiving chemotherapy [6], [24], [34], [37]. Park et al [6] found that advanced-stage CRC patients with XPD Gln/Gln genotype treated with chemotherapy tended to have progressive disease and significantly reduced survival, compared to patients with XPD Lys allelic variants (P = 0.002). A previous study of XPD Lys751Gln in 166 advanced CRC patients receiving FOLFOX therapy found a negative relationship between XPD Gln/Gln genotype and adverse progression-free survival (HR = 2.21, 95% CI = 1.17–4.17, P = 0.01) [34]. Our results are consistent with previous studies that have shown a negative association between the XPD Gln allelic variants and OS. However, 2 studies have reported no significant relationship between XPD Lys751Gln and clinical outcomes in CRC patients [7], [33].
We demonstrated that the reduction in OS associated with XPD Lys751Gln Gln allelic variants were improved in rectal cancer patients. In the contrast, Duldulao et al. [38] observed that XPD Lys/Lys genotype was significantly associated with increased toxicity to neoadjuvant chemoradiotherapy in 132 stage II/III rectal cancer patients in the United States. However, Cecchin et al [39] reported no association between the XPD Lys751Gln allelic variant and the tumor regression grade in 238 rectal cancer patients treated with neoadjuvant chemoradiotherapy in Italy. Different ethnicities, treatment regiments, and outcome measurements may have contributed to these inconsistencies.
The association between the XRCC1 Arg399Gln allelic variant and OS has been previously reported [35], [37]. Artac et al [37] recruited 43 metastatic CRC patients who received irinotecan-based therapy, and found that OS is associated with XRCC1 Gln/Gln genotype (HR = 2.85, P = 0.04). In a study of stage II/III rectal cancer patients in Spain, patients with XRCC1 Arg/Arg had a greater probability of a positive response to chemoradiotherapy than those with XRCC1 Arg/Gln (OR = 4.18, 95% CI = 1.62–10.7) [35]. In our study, we observed that only the XRCC1 Gln allelic variants were significantly associated with reduced OS in stage II/III colon cancer patients.
In contrast, Huang et al [22] showed that XRCC1 Gln/Gln genotype was significantly associated with favorable OS (HR = 0.15, 95% CI = 0.04–0.57) and progression-free survival (HR = 0.31, 95% CI = 0.10–0.91) in metastatic CRC patients receiving FOLFOX-4 chemotherapy. Previous reports have suggested that the XRCC1 Gln allelic variants are deficient in DNA-repair activity, leading to increased chromosomal damage [27], [40], [41]. Thus, suboptimal repair activity in tissues favors carcinogenesis, but may ensure tumor sensitivity to drug or ionizing treatment [42]. However, other studies have shown that the XRCC1 Arg399Gln allelic variant was not significantly associated with the outcome [7], [22] or response to chemotherapy [33], [34], [43].
In our analysis of the associations between the allelic variants of GST and DNA-repair genes and the OS stratified by tumor site and stage, we found that the XPD 751Gln and the XRCC1 399Gln allelic variants were significantly associated with reduced OS for stage II/III rectal cancer and colon cancer, respectively. Although previous studies have not shown that specific DNA repair activity may vary between the colon and the rectum, a high proportion of rectal tumors has been shown to have reduced levels of thymidylate synthase, based on enzyme activity assays [44], [45]. Despite the relatively small number of cases and the lack of statistical significance for the results, our findings support an association between polymorphisms in DNA-repair genes and OS based on specific tumor locations among CRC patients treated with 5-FU chemotherapy. The prognostic value of variability in DNA-repair activity based on a CRC tumor site warrants further study.
Similar to the findings of previous studies, we found no association between OS and the XRCC3 Thr241Met, GSTM1, GSTT1, and GSTP1 Ile105Val allelic variants [14], [35], [39], [46]. A meta-analysis of 13 independent studies that included 1234 advanced or metastatic CRC patients found no significant association between the GSTP1 Ile105Val allelic variant and tumor response [46]. However, Stoehlmacher et al [11] observed that GSTP1 Val/Val genotype was associated with increased survival in CRC patients following combination therapy with oxaliplatin and 5-FU. The association between the GSTP1 Ile105Val allelic variant and the chemotherapeutic outcome in CRC also warrants further study.
There are potential limitations to our findings. First, all of our participants received 5-FU-based chemotherapy. Thus, we did not investigate the genotypes associated with clinical outcomes in an untreated control group. Second, we did not examine mRNA expression or perform immunohistochemistry analysis of tumor tissues. Therefore, our analysis was not free of potential biases, and does not account for the loss of heterozygosity in the tumor. However, immunohistochemistry is a subjectively semiquantitative method, and is limited by the sensitivity of the antibodies and tissue-handling techniques used. Third, our study design lacked statistical power to support the significance of the association between the XPD Ly751Gln and XRCC1 Arg399Gln allelic variants and the OS in CRC patients treated with 5-FU-based chemotherapy (49% and 63%, respectively). Nevertheless, our study included newly diagnosed and histologically confirmed CRC patients, a relatively large sample, and a long-term follow-up. Thus, our results may shed light on the value of the XPD Ly751Gln and XRCC1 Arg399Gln allelic variants as prognostic markers for CRC of various tumor sites and stages.
In conclusion, we evaluated multiple xenobiotic-metabolizing and DNA-repair genetic polymorphisms as prognosticators of OS in CRC patients receiving 5-FU-based chemotherapy. Our results showed that the XPD and XRCC1 allelic variants were tumor site- and/or stage-dependently associated with OS for CRC patients receiving 5-FU-based chemotherapy. Further studies are warranted to identify the underlying biochemical mechanisms affected by the mutations in the allelic variants, and to validate the roles of XPD and XRCC1 genetic polymorphisms as predictors of chemotherapeutic outcome in CRC patients.
Acknowledgments
We express our sincere appreciation to the study participants. We also wish to thank the staffs from Chang Gung Memorial Hospital.
Funding Statement
The authors' study was supported by grants from Taipei Medical University (TMU98-AE1-B05), the National Science Council, Taiwan (NSC97-2314-B-039-020-MY3 and NSC97-2314-B-038-050-MY3), China Medical University (CMU97-259 and CMU97-156), Taiwan Department of Health Clinical Trial and Research Center of Excellence (DOH102-TD-B-111-004), and the Center of Excellence for Cancer Research of the Taiwan Department of Health at Taipei Medical University (DOH101-TD-C-111-008). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Moore HC, Haller DG (1999) Adjuvant therapy of colon cancer. Semin Oncol 26: 545–555. [PubMed] [Google Scholar]
- 2. Soong R, Shah N, Salto-Tellez M, Tai BC, Soo RA, et al. (2008) Prognostic significance of thymidylate synthase, dihydropyrimidine dehydrogenase and thymidine phosphorylase protein expression in colorectal cancer patients treated with or without 5-fluorouracil-based chemotherapy. Ann Oncol 19: 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stewart JM, Zalcberg JR (1998) Update on adjuvant treatment of colorectal cancer. Curr Opin Oncol 10: 367–374. [DOI] [PubMed] [Google Scholar]
- 4. Dybdahl M, Vogel U, Frentz G, Wallin H, Nexo BA (1999) Polymorphisms in the DNA repair gene XPD: correlations with risk and age at onset of basal cell carcinoma. Cancer Epidemiol Biomarkers Prev 8: 77–81. [PubMed] [Google Scholar]
- 5. Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, et al. (2000) XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis 21: 551–555. [DOI] [PubMed] [Google Scholar]
- 6. Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, et al. (2001) A Xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res 61: 8654–8658. [PubMed] [Google Scholar]
- 7. Ott K, Rachakonda PS, Panzram B, Keller G, Lordick F, et al. (2011) DNA repair gene and MTHFR gene polymorphisms as prognostic markers in locally advanced adenocarcinoma of the esophagus or stomach treated with cisplatin and 5-fluorouracil-based neoadjuvant chemotherapy. Ann Surg Oncol 18: 2688–2698. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Wu J, Zhong R, Wu C, Zou L, et al.. (2012) Multi-loci analysis reveals the importance of genetic variations in sensitivity of platinum-based chemotherapy in non-small-cell lung cancer. Mol Carcinog. [DOI] [PubMed]
- 9. Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA (1998) Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 19: 275–280. [DOI] [PubMed] [Google Scholar]
- 10. Meyer DJ, Coles B, Pemble SE, Gilmore KS, Fraser GM, et al. (1991) Theta, a new class of glutathione transferases purified from rat and man. Biochem J 274 (Pt 2): 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stoehlmacher J, Park DJ, Zhang W, Groshen S, Tsao-Wei DD, et al. (2002) Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst 94: 936–942. [DOI] [PubMed] [Google Scholar]
- 12. Ruzzo A, Canestrari E, Maltese P, Pizzagalli F, Graziano F, et al. (2007) Polymorphisms in genes involved in DNA repair and metabolism of xenobiotics in individual susceptibility to sporadic diffuse gastric cancer. Clin Chem Lab Med 45: 822–828. [DOI] [PubMed] [Google Scholar]
- 13. Wang Z, Chen JQ, Liu JL, Qin XG, Huang Y (2012) Polymorphisms in ERCC1, GSTs, TS and MTHFR predict clinical outcomes of gastric cancer patients treated with platinum/5-Fu-based chemotherapy: a systematic review. BMC Gastroenterol 12: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen YC, Tzeng CH, Chen PM, Lin JK, Lin TC, et al. (2010) Influence of GSTP1 I105V polymorphism on cumulative neuropathy and outcome of FOLFOX-4 treatment in Asian patients with colorectal carcinoma. Cancer Sci 101: 530–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romero A, Martin M, Oliva B, de la Torre J, Furio V, et al. (2012) Glutathione S-transferase P1 c.313A > G polymorphism could be useful in the prediction of doxorubicin response in breast cancer patients. Ann Oncol 23: 1750–1756. [DOI] [PubMed] [Google Scholar]
- 16. Shuck SC, Short EA, Turchi JJ (2008) Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res 18: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rouillon C, White MF (2011) The evolution and mechanisms of nucleotide excision repair proteins. Res Microbiol 162: 19–26. [DOI] [PubMed] [Google Scholar]
- 18. Melis JP, Luijten M, Mullenders LH, van Steeg H (2011) The role of XPC: implications in cancer and oxidative DNA damage. Mutat Res 728: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S, Tang D, Xue K, Xu L, Ma G, et al. (2002) DNA repair gene XRCC1 and XPD polymorphisms and risk of lung cancer in a Chinese population. Carcinogenesis 23: 1321–1325. [DOI] [PubMed] [Google Scholar]
- 20. Mahimkar MB, Samant TA, Kannan S, Tulsulkar J, Pai PS, et al. (2012) Polymorphisms in GSTM1 and XPD genes predict clinical outcome in advanced oral cancer patients treated with postoperative radiotherapy. Mol Carcinog 51 Suppl 1E94–103. [DOI] [PubMed] [Google Scholar]
- 21. Hitomi K, Iwai S, Tainer JA (2007) The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 6: 410–428. [DOI] [PubMed] [Google Scholar]
- 22. Huang MY, Huang ML, Chen MJ, Lu CY, Chen CF, et al. (2011) Multiple genetic polymorphisms in the prediction of clinical outcome of metastatic colorectal cancer patients treated with first-line FOLFOX-4 chemotherapy. Pharmacogenet Genomics 21: 18–25. [DOI] [PubMed] [Google Scholar]
- 23. Geng L, Huehls AM, Wagner JM, Huntoon CJ, Karnitz LM (2011) Checkpoint signaling, base excision repair, and PARP promote survival of colon cancer cells treated with 5-fluorodeoxyuridine but not 5-fluorouracil. PLoS One 6: e28862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruzzo A, Graziano F, Loupakis F, Santini D, Catalano V, et al. (2008) Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFIRI chemotherapy. Pharmacogenomics J 8: 278–288. [DOI] [PubMed] [Google Scholar]
- 25. Spitz MR, Wu X, Wang Y, Wang LE, Shete S, et al. (2001) Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res 61: 1354–1357. [PubMed] [Google Scholar]
- 26. Matullo G, Peluso M, Polidoro S, Guarrera S, Munnia A, et al. (2003) Combination of DNA repair gene single nucleotide polymorphisms and increased levels of DNA adducts in a population-based study. Cancer Epidemiol Biomarkers Prev 12: 674–677. [PubMed] [Google Scholar]
- 27. Huang Y, Li L, Yu L (2009) XRCC1 Arg399Gln, Arg194Trp and Arg280His polymorphisms in breast cancer risk: a meta-analysis. Mutagenesis 24: 331–339. [DOI] [PubMed] [Google Scholar]
- 28. Liu L, Yuan P, Wu C, Zhang X, Wang F, et al. (2011) Assessment of XPD Lys751Gln and XRCC1 T-77C polymorphisms in advanced non-small-cell lung cancer patients treated with platinum-based chemotherapy. Lung Cancer 73: 110–115. [DOI] [PubMed] [Google Scholar]
- 29. Yeh CC, Hsieh LL, Tang R, Chang-Chieh CR, Sung FC (2005) Vegetable/fruit, smoking, glutathione S-transferase polymorphisms and risk for colorectal cancer in Taiwan. World J Gastroenterol 11: 1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yeh CC, Sung FC, Tang R, Chang-Chieh CR, Hsieh LL (2005) Polymorphisms of the XRCC1, XRCC3, & XPD genes, and colorectal cancer risk: a case-control study in Taiwan. BMC Cancer 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yeh CC, Sung FC, Tang R, Chang-Chieh CR, Hsieh LL (2007) Association between polymorphisms of biotransformation and DNA-repair genes and risk of colorectal cancer in Taiwan. J Biomed Sci 14: 183–193. [DOI] [PubMed] [Google Scholar]
- 32. Yeh CC, Lai CY, Hsieh LL, Tang R, Wu FY, et al. (2010) Protein carbonyl levels, glutathione S-transferase polymorphisms and risk of colorectal cancer. Carcinogenesis 31: 228–233. [DOI] [PubMed] [Google Scholar]
- 33. Martinez-Balibrea E, Abad A, Aranda E, Sastre J, Manzano JL, et al. (2008) Pharmacogenetic approach for capecitabine or 5-fluorouracil selection to be combined with oxaliplatin as first-line chemotherapy in advanced colorectal cancer. Eur J Cancer 44: 1229–1237. [DOI] [PubMed] [Google Scholar]
- 34. Ruzzo A, Graziano F, Loupakis F, Rulli E, Canestrari E, et al. (2007) Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol 25: 1247–1254. [DOI] [PubMed] [Google Scholar]
- 35. Lamas MJ, Duran G, Gomez A, Balboa E, Anido U, et al. (2012) X-ray cross-complementing group 1 and thymidylate synthase polymorphisms might predict response to chemoradiotherapy in rectal cancer patients. Int J Radiat Oncol Biol Phys 82: 138–144. [DOI] [PubMed] [Google Scholar]
- 36. Wolpin BM, Meyerhardt JA, Mamon HJ, Mayer RJ (2007) Adjuvant treatment of colorectal cancer. CA Cancer J Clin 57: 168–185. [DOI] [PubMed] [Google Scholar]
- 37. Artac M, Bozcuk H, Pehlivan S, Akcan S, Pehlivan M, et al. (2010) The value of XPD and XRCC1 genotype polymorphisms to predict clinical outcome in metastatic colorectal carcinoma patients with irinotecan-based regimens. J Cancer Res Clin Oncol 136: 803–809. [DOI] [PubMed] [Google Scholar]
- 38. Duldulao MP, Lee W, Nelson RA, Ho J, Le M, et al. (2013) Gene polymorphisms predict toxicity to neoadjuvant therapy in patients with rectal cancer. Cancer 119: 1106–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cecchin E, Agostini M, Pucciarelli S, De Paoli A, Canzonieri V, et al. (2011) Tumor response is predicted by patient genetic profile in rectal cancer patients treated with neo-adjuvant chemo-radiotherapy. Pharmacogenomics J 11: 214–226. [DOI] [PubMed] [Google Scholar]
- 40. Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, et al. (2006) Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res 12: 2101–2108. [DOI] [PubMed] [Google Scholar]
- 41. Matullo G, Guarrera S, Carturan S, Peluso M, Malaveille C, et al. (2001) DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int J Cancer 92: 562–567. [DOI] [PubMed] [Google Scholar]
- 42. Hu JJ, Smith TR, Miller MS, Mohrenweiser HW, Golden A, et al. (2001) Amino acid substitution variants of APE1 and XRCC1 genes associated with ionizing radiation sensitivity. Carcinogenesis 22: 917–922. [DOI] [PubMed] [Google Scholar]
- 43. Chua W, Goldstein D, Lee CK, Dhillon H, Michael M, et al. (2009) Molecular markers of response and toxicity to FOLFOX chemotherapy in metastatic colorectal cancer. Br J Cancer 101: 998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamada T, Tanaka N, Yokoi K, Seya T, Kanazawa Y, et al. (2008) Correlation between clinical pathologic factors and activity of 5-FU-metabolizing enzymes in colorectal cancer. J Nippon Med Sch 75: 23–27. [DOI] [PubMed] [Google Scholar]
- 45. Ohrling K, Edler D, Hallstrom M, Ragnhammar P (2008) Expression of thymidylate synthase in liver and lung metastases of colorectal cancer and their matched primary tumours. Anticancer Res 28: 1741–1747. [PubMed] [Google Scholar]
- 46. Ye F, Liu Z, Tan A, Liao M, Mo Z, et al. (2013) XRCC1 and GSTP1 polymorphisms and prognosis of oxaliplatin-based chemotherapy in colorectal cancer: a meta-analysis. Cancer Chemother Pharmacol 71: 733–740. [DOI] [PubMed] [Google Scholar]