Abstract
In the Drosophila oocyte, gurken (grk) mRNA encodes a secreted TGF-α signal that specifies the future embryonic dorso-ventral axes by altering the fate of the surrounding epithelial follicle cells. We previously identified a number of RNA binding proteins that associate specifically with the 64 nucleotide grk localization signal, including the Drosophila orthologue of polypyrimidine tract-binding protein (PTB), Hephaestus (Heph). To test whether Heph is required for correct grk mRNA or protein function, we used immunoprecipitation to validate the association of Heph with grk mRNA and characterized the heph mutant phenotype. We found that Heph is a component of grk mRNP complexes but heph germline clones show that Heph is not required for grk mRNA localization. Instead, we identify a novel function for Heph in the germline and show that it is required for proper Grk protein localization. Furthermore, we show that Heph is required in the oocyte for the correct organization of the actin cytoskeleton and dorsal appendage morphogenesis. Our results highlight a requirement for an mRNA binding protein in the localization of Grk protein, which is independent of mRNA localization, and we propose that Heph is required in the germline for efficient Grk signalling to the somatic follicle cells during dorso-ventral patterning.
Introduction
The intracellular transport and subsequent localized translation of messenger RNA is a powerful and general strategy used to temporally and spatially restrict specific proteins to their site of function [1], [2], [3], [4], [5], [6]. Asymmetric protein distribution arising from the translation of localized mRNAs govern the regulation of cell polarity [7], [8], [9] and the segregation of cell fate and embryonic polarity determinants in oocytes and embryos [10], [11], [12]. In the developing Drosophila oocyte, the localization of gurken (grk), oskar (osk), bicoid (bcd) and nanos (nos) mRNAs plays a fundamental role in the determination of the axes of the oocyte and future embryo. grk encodes a TGF-α family signal [13] that acts as a secreted oocyte ligand for the Drosophila epidermal growth factor receptor (EGFR) [14], [15], [16]. Localization of grk mRNA and local translation of Grk at the dorso-anterior corner of the oocyte in mid-oogenesis [13], [17], [18] results in the localized activation of EGFR in the overlying follicle cells, inducing them to adopt a dorsal fate, thereby defining the dorsal-ventral axis of the egg and future embryo [19], [20]. Concomitant with grk localization to the future dorsal anterior region of the oocyte, osk mRNA accumulates at the posterior pole. Locally translated Osk protein nucleates the assembly of the germ plasm, which determines germ cell fate in the embryo [21], [22]. Osk-dependent assembly of germ plasm is also required for the posterior localization and translation of nos mRNA, which is in turn required for abdomen formation in the embryo [23], [24], [25].
The transport of mRNA cargoes to their cellular destinations occurs in large ribonucleoprotein (RNP) complexes termed transport particles or granules [17], [18], [26], [27], [28]. These complexes coordinate and control multiple steps including transport of the mRNA along the cytoskeleton by molecular motors, translational repression of the localizing mRNA, in addition to its anchoring and translational activation upon reaching the final destination [4]. The composition, organization and dynamics of such complexes therefore dictate the behaviour and fate of the target RNA in the cell. However, many questions remain unanswered about these transport RNPs and their association with localized mRNAs. Although many of the known proteins that function in grk mRNA localization and translational regulation have been identified through genetic studies [13], [29], [30], [31], [32], [33], we recently identified, using complementary biochemical approaches, new proteins that could potentially function in grk regulation. We used GRNA chromatography to identify proteins that can associate specifically with the grk localization signal (GLS) [34], a 64-nucleotide stem loop structure in the coding region of the grk transcript that is necessary and sufficient for dorso-anterior localization of the RNA [35]. We identified a number of known factors previously shown to be required for grk mRNA localization and translational regulation [13], [29], [30], [31], [33] as well as a number of other RNA binding proteins that were not previously implicated in the function of grk. These included Syncrip, which we showed was required for grk mRNA localization and translational regulation, and the Drosophila orthologue of polypyrimidine tract-binding protein (PTB), Hephaestus (Heph), which was previously only known to play a role in osk mRNA localization.
Polypyrimidine Tract Binding (PTB) protein is an hnRNP family member that is thought to act as an RNA chaperone with the ability to bind RNA in a sequence specific manner and to restructure the RNA substrate [36], [37], [38], [39], [40], [41]. It acts to regulate a number of forms of RNA metabolism, including negative and positive regulation of alternative splicing [39], [40], [42], [43], [44], [45], [46], 3′-end processing [47], stabilization [48], miRNA-mediated gene regulation [49], translation [50], [51], [52], [53], and, of particular relevance to this work, mRNA localization and localized translation [54], [55], [56], [57], [58]. Drosophila PTB is encoded by the hephaestus (heph) locus, and shares over 50% amino acid identity and 60% similarity with vertebrate forms of PTB (mammalian PTB/hnRNPI and Xenopus VgRBP60). Genetic screens and developmental studies indicate that Drosophila Heph/PTB regulates spermatogenesis in adult males [59], [60], [61], [62], embryonic development [63] and sensory bristle and wing margin development during larval and pupal stages [64], [65], [66]. Work by Besse et al. [55] identified Heph as a key structural component of osk RNP complexes in the Drosophila oocyte, required for efficient osk mRNA localization and also translational repression of osk mRNA during localization. Heph is required indirectly for osk localization through the properly timed organization of the oocyte microtubule (MT) cytoskeleton, but directly controls Osk translation through the multimerization of individual osk mRNA molecules and the formation of high order translationally repressed osk RNP complexes.
As Heph associates with grk and GLS RNA in vitro we set out to test the hypothesis that Heph is a component of a grk RNP at one or a number of stages of oogenesis and plays a role in grk localization, anchoring, or localized Grk translation. We show using RNA immunoprecipitation (RNA IP) and analysis of heph mutant germline clones that Heph is a component of grk mRNP complexes but is not required for grk mRNA localization. Instead, Heph is required in the germline for proper Grk protein dorso-anterior localization and for efficient Grk signalling from the germline to the somatic follicle cells.
Results
Heph is a Component of grk RNP Complexes
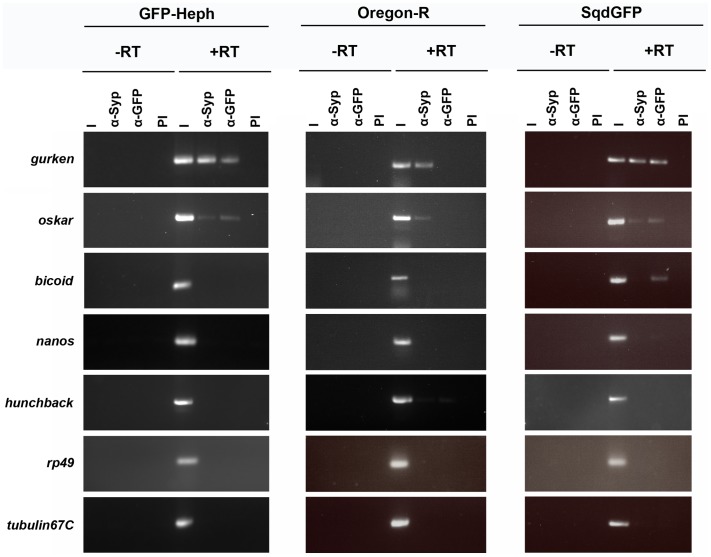
GRNA chromatography studies have shown that Heph associates in vitro with grk, and more specifically, the GLS (Table S1) [34]. It has also been previously reported that Heph/PTB represses the translation of osk by promoting the formation of large, high-order osk containing messenger ribonucleoprotein (mRNP) complexes [55]. To ask whether Heph is a component of grk RNPs in the germline, we carried out RNA IP experiments using anti-GFP antibodies and previously characterized GFP-PTB/Heph protein-trap ovaries [55]. RT-PCR showed that grk and osk mRNAs are present in the fractions precipitated from GFP-PTB/Heph protein-trap ovaries and from SqdGFP control ovaries using anti-GFP and control anti-Syncrip antibodies, but not in immunoprecipitates using control rabbit serum, or in anti-GFP immunoprecipitates from wild-type control females (Figure 1). No specific enrichment of highly expressed rp49 and tubulin67C mRNAs, of the localized bcd and nos mRNAs, or of the non-localized hunchback mRNA control from the initial GRNA chromatography experiments [34] was observed in the GFP-PTB/Heph precipitated fractions. These results, combined with the earlier GRNA results [34] indicate that Heph is a component of grk mRNP complexes in vivo.
Figure 1. Heph associates with grk and osk mRNAs.
RT-PCR amplification of mRNAs recovered in fractions immunoprecipitated from GFP-Heph protein trap, SqdGFP control or Oregon-R control ovarian extracts using anti-GFP and control anti-Syp or pre-immune rabbit serum. I = Input RNA (10%); PI = control rabbit serum; −/+RT = minus/plus reverse transcriptase.
To test where Heph protein is expressed in oogenesis we stained wild type egg chambers with anti-PTB/Heph antibodies. We found that the protein is localized in the cytoplasm of the follicular epithelial cells throughout oogenesis (Figure S1A–E). In the germline, Heph is present throughout the oocyte and nurse cell cytoplasm with an accumulation at the posterior pole of the oocyte from around stage 9 of oogenesis onward (Figure S1C–E). These signals are reduced in hephe1 hypomorphic mutant germline clones (Figure S1H and I), confirming that the protein detected in wild-type egg chambers is indeed specifically Heph. The posterior localization of Heph is particularly strong in late stage 10 egg chambers (Figure S1D–E), and reflects Heph’s role as a key structural component of osk mRNP complexes [55]. Despite a general signal throughout the oocyte cytoplasm, Heph does not accumulate at the dorso-anterior cap and so does not colocalize with localized grk mRNA (Figure S1C–G). This indicates that the interaction of Heph with grk mRNA may be transient and occur at earlier stages of grk localization before the RNA reaches the dorso-anterior cap.
The pattern of localization observed here is consistent with that of both endogenous Heph and GFP-PTB/Heph previously shown by Besse et al. [55]. In these previous studies endogenous Heph and GFP-PTB/Heph were also additionally present in the nuclei of both the nurse cells and the follicular epithelial cells throughout oogenesis, and may not have been seen here due to the use of different antibodies or conditions for immunostaining. However, in both studies it is clear that there is no enrichment of Heph at the dorso-anterior cap.
heph Mutant Eggs have Patterning and Morphological Defects
To investigate the role of Heph in grk mRNA regulation, three different alleles [55],[64] belonging to the same lethal complementation group were used (Table 1). These alleles also fail to complement the male sterility of the original P-element-induced allele heph2 [59]. heph03429 is a P-element insertion that maps to a large intron. hephe2 is an EMS-induced mis-sense mutation that behaves as a null allele and corresponds to a deletion covering RRM 1, RRM 2 and part of RRM 3, whilst hephe1 substitutes a conserved glycine (G) residue to a glutamine (Q) residue in RRM 1 [64]. heph03429 and hephe1 are hypomorphic alleles that strongly affect Heph expression [55]. Due to the lethality of these alleles, it was necessary to make germline clones using the FLP-dominant female sterile (DFS) technique [67],[68]. As previously observed [55], germ cells homozygous for hephe2 fail to produce differentiated egg chambers, suggesting that Heph function is required for early germ cell maintenance, whilst germ cells homozygous for heph03429 and hephe1 do support development until later stages of oogenesis.
Table 1. Alleles associated with the heph locus.
| allele | mutagen | mutation | viable | complementation deficiency DfG45 | complementation heph03429 | origin |
| hephe2 | EMS | deletion of RRMs 1, 2 and part of 3 | no | no | no | Dansereau et al. [64] |
| hephe1 | EMS | point mutation (G to Q in RRM1) | no | no | no | Dansereau et al. [64] |
| heph03429 | P-element | insertion at position 27756259 (intron) | no | no | – | Besse et al. [55] |
A number of alleles map to the heph locus. Two EMS-induced alleles, hephe1 and hephe2, alter RRM domains. hephe2 is a deletion of several coding exons including the coding region for RRM1, RRM2 and part of RRM3. hephe1 is a mis-sense mutation that changes a glycine (G) to a glutamine (Q) in RRM1. The P-element allele heph03429 [64] maps to a heph intron.
doi:10.1371/journal.pone.0069978.t001
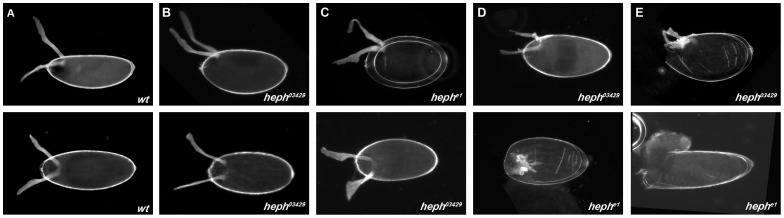
Local translation of Grk at the dorso-anterior of the oocyte is required for dorso-ventral axis determination in both the eggshell and the embryo. Grk signalling causes the overlying follicle cells to adopt a dorsal cell fate, allowing at a later stage for the correct specification and positioning of anterior and dorsal chorion structures, such as the dorsal appendages. Analysis of patterning and dorsal appendage morphogenesis in the eggshell is therefore a sensitive way in which to investigate grk localization, Grk translation and Grk signalling within the egg chamber. Analysis of heph mutant eggs revealed a number of eggshell phenotypes. None of the eggs laid were fully wild-type (class A) (Figure 2A–E and Table 2), and most were shorter and rounder than wild-type eggs (class B–D) (Figure 2B–D, and Table 2). There were a number of phenotypes seen in the dorsal appendages; wild-type length and morphology (class B), broader than wild-type with ragged edges (class C), shorter with ragged edges, resembling ‘antlers’, joined at the centre by additional dorsal appendage material (class D). Within class B and C, a number of eggs were observed where inter-dorsal appendage spacing was greater than that observed in wild-type eggs. A small number of eggs were also fully open at the anterior (class E), resembling eggs laid by cup mutants. The distribution of the numbers of eggs in the different classes varies for hephe1 and heph03429 eggs. The majority of hephe1 eggs were class D, whilst heph03429 eggs were class B or C, suggesting that hephe1 mutant eggs have a slightly more severe phenotype, and is probably a reflection of the difference in the molecular nature of the two alleles.
Figure 2. heph mutant eggs have patterning defects.
(A) Wild-type egg with two dorsal appendages that mark the dorsal anterior surface. (B–D) Eggs are shorter and rounder in the heph mutant alleles and have a range of dorsal appendage defects from (B) spacing defects (C) broad with ragged edges to (D) shorter with ‘antlers’, joined by additional dorsal appendage material. (E) A small number of eggs are fully open at the anterior. See Table 2 for quantitation of eggshell phenotypes.
Table 2. Quantitation of eggshell phenotypes observed for hephe1 and heph03429 germline clones.
| Genotype | A (wild-type) | B | C | D | E |
| OrR (n = 153) | 100 | 0 | 0 | 0 | 0 |
| hephe1 (n = 111) | 0 | 24.3 | 10.8 | 56.8 | 8.1 |
| heph03429 (n = 135) | 0 | 42.2 | 42.2 | 8.9 | 6.7 |
Numbers are given as percentages of the total number of eggs counted. Oregon R (OrR) served as a wild-type control strain.
doi:10.1371/journal.pone.0069978.t002
The eggshells shown in Figure 2B–D have characteristics of those with dorso-ventral patterning defects. The eggs are short and round, the dorsal appendages are broader than wild-type, there is an increase in the inter-appendage space between the two dorsal appendages, and there is ectopic dorsal appendage material at the dorsal midline. The dorsal appendages of these eggs also resemble the ‘moose antlers’ seen in eggs that develop from bullwinkle (bwk) mutant germline clone egg chambers [69],[70], where the follicle cell migrations that form the dorsal appendages are abnormal. The lack of wild-type heph function in the germline also results in a number of phenotypes that may reflect defects in the germline actin cytoskeleton. The eggs resemble the short eggs seen for mutants such as chickadee [71] and quail [72], where actin dynamics or regulation is perturbed. Staining of heph mutant germline clones with phalloidin shows that the dense bundles of actin filaments that ordinarily form in the nurse cell cytoplasm at stage 10, prior to dumping at stage 11, do not form in a small number of heph mutant germline clones (Figure S2), and aberrant actin patches are also observed in the oocyte cytoplasm (Figure S2B). It is unclear whether these actin patches are free-floating, detached ring canals, or the accumulation of actin spheres. Such actin spheres are observed in kinesin heavy chain (khc) [73] and also Bic-C mutant egg chambers [74]. A partial dumpless phenotype (due to incomplete transfer of nurse cell contents into the oocyte at stage 11) is also observed. The eggshells shown in Figure 2E are fully open at the anterior, and resemble the eggs laid by cup mutant females. Such defects may result from insufficient oocyte growth and the subsequent misalignment of the follicle cell migration that would cover the anterior of the oocyte (for review of eggshell patterning see [75]). In summary, examination of the eggshells of heph germline clones reveals multiple roles for heph in the female germline, ranging from regulation of dorso-ventral patterning and correct migration of the somatic follicle cells, to organization of the actin cytoskeleton in the germline.
grk mRNA Localization is not Perturbed in heph Mutant Oocytes
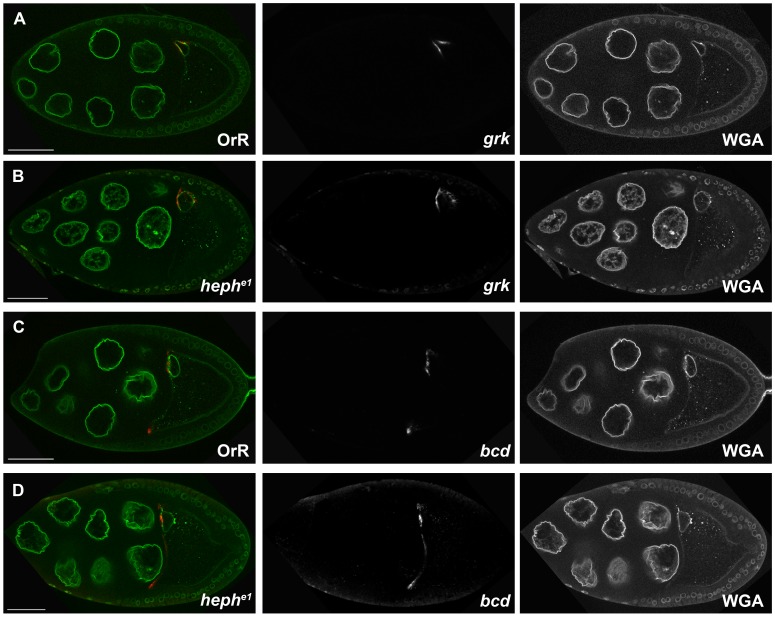
In an effort to understand the nature of the dorso-ventral patterning defects seen in heph mutant eggs, and the presence of Heph in grk mRNP complexes, we investigated the distribution of grk mRNA in heph mutant egg chambers. In wild-type egg chambers, the pattern of grk mRNA localization changes during the progression of oogenesis. In early oogenesis (stages 1–7), grk is localized in a posterior crescent between the nucleus and the overlying follicle cells. During stages 7 and 8, grk begins to accumulate around the new dorso-anterior position of the nucleus as well as forming a transient ring at the anterior of the oocyte. From stages 9 to 10B, grk is tightly localized in a dorso-anterior cap near the oocyte nucleus (Figure 3A and Figure S3). grk mRNA localization in hephe1 and heph03429 oocytes was analysed by in situ hybridization and found to be identical to the wild-type pattern of localization at all stages of oogenesis (shown for hephe1 oocytes Figure 3B and Figure S3). Fluorescently labelled full-length grk RNA also localized to the dorso-anterior cap when injected into hephe1 and heph03429 stage 9 oocytes in an in vivo injection assay [18], indicating that Heph is not required in the oocyte cytoplasm for grk localization (data not shown). bcd localization was also unaffected in heph mutant germline clones (Figure 3C and D). This was to be expected as RNA IP experiments (Figure 1) indicate that Heph is not a component of bcd mRNP complexes.
Figure 3. grk mRNA localization is not perturbed in heph mutant oocytes.
In situ hybridization using a grk or bcd probe was carried out on Oregon-R (OrR) (A) and (C), or hephe1 germline clone egg chambers (B) and (D). Egg chambers were also stained with fluorescent Wheat Germ Agglutinin (WGA) conjugate in order to label nuclear membranes. Scale bar 50 µm.
Grk Protein is Mislocalized in heph Mutant Oocytes
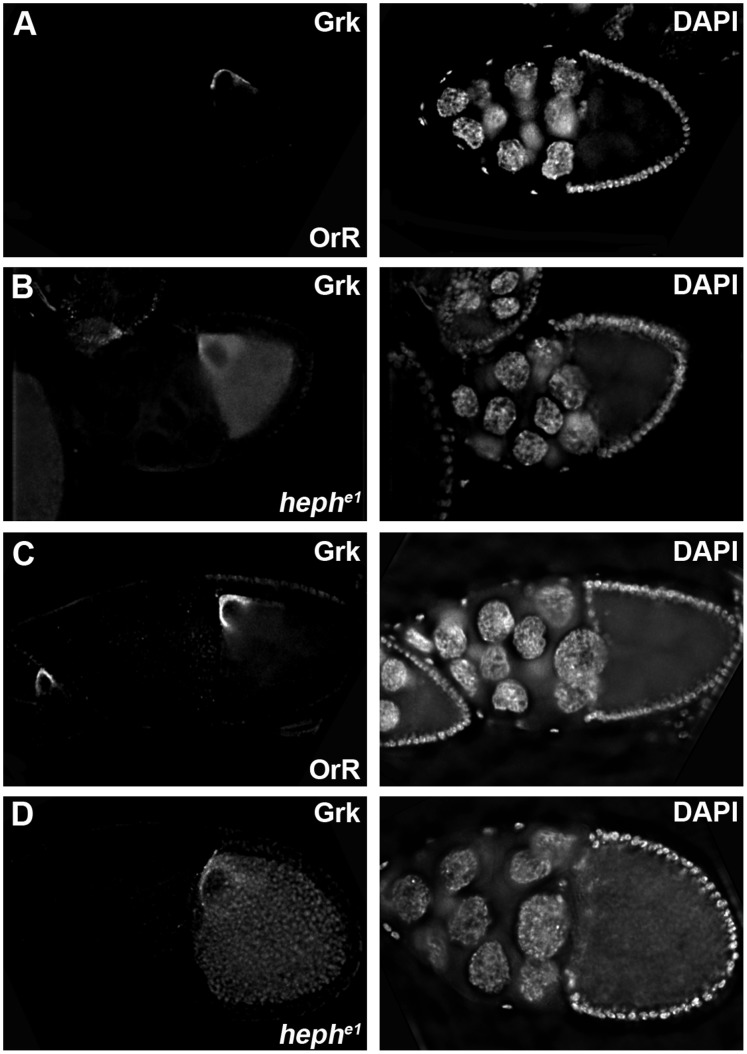
Next we investigated the distribution of Grk protein. Interestingly, in contrast to the normal localization of grk mRNA, we found an abnormal distribution of Grk protein in heph germline clones. During early and mid-oogenesis the distribution of Grk protein mirrors that of grk mRNA, reflecting the two signalling events in which Grk acts to polarize both the follicular epithelium and the future embryo. At stage 6, Grk is enriched at the posterior of the oocyte (Figure S4A), whilst by stage 9 Grk is at the dorso-anterior cap (Figure S4C). By the later stages of oogenesis (stages 10–12) the distribution of Grk protein extends over approximately half the length of the dorsal midline of the oocyte to form an elongated anterior to posterior stripe [76] (Figure 4A and C) where Grk is present in the oocyte cytoplasm close to the nucleus, in neighbouring follicle cells, and in the extracellular space between the cortices of the oocyte and the follicular epithelium [77].
Figure 4. Grk protein is mislocalized in heph mutant stage 10 oocytes.
Stage 10A (A) and stage 10B (C) Oregon-R (OrR) egg chambers, and stage 10A (B), and stage 10B (D) hephe1 germline clone egg chambers were stained with anti-Gurken and DAPI. In hephe1 germline mutant egg chambers at stages 10A and 10B Grk protein is evenly distributed throughout the oocyte.
In heph mutants, posterior Grk localization and signalling at stage 7 of oogenesis is normal as the nucleus migrates correctly in all heph mutant oocytes. Grk protein localization is also generally normal up to stage 9 of oogenesis (Figure S4), although in a small number of mutant oocytes we see mild defects where Grk protein is present throughout the oocyte cytoplasm in addition to being at the dorso-anterior (Figure S4E and F). In stage 10A and 10B heph mutant oocytes this mislocalization of Grk in the oocyte cytoplasm is more pronounced and evident in large proportions of heph mutant oocytes (Figure 4B and D). In 42.9% heph03429 stage 10A (n = 35) oocytes and 66.7% of 10B (n = 36) oocytes Grk was dispersed throughout the oocyte cytoplasm. The percentage of oocytes with this defect was higher in the hephe1 mutant where 96.9% stage 10A (n = 32) and 97.3% stage 10B (n = 37) oocytes showed Grk throughout the oocyte cytoplasm (Figure 4B and D). The fact that the total levels of Grk protein are unaffected in heph mutant ovaries (Figure S5), leads us to conclude that the phenotype is due to mislocalization of Grk protein, and not to changes in the expression level of Grk. This mislocalization is not a result of its mRNA mislocalization, indicating that Heph is having a specific effect on Grk protein localization.
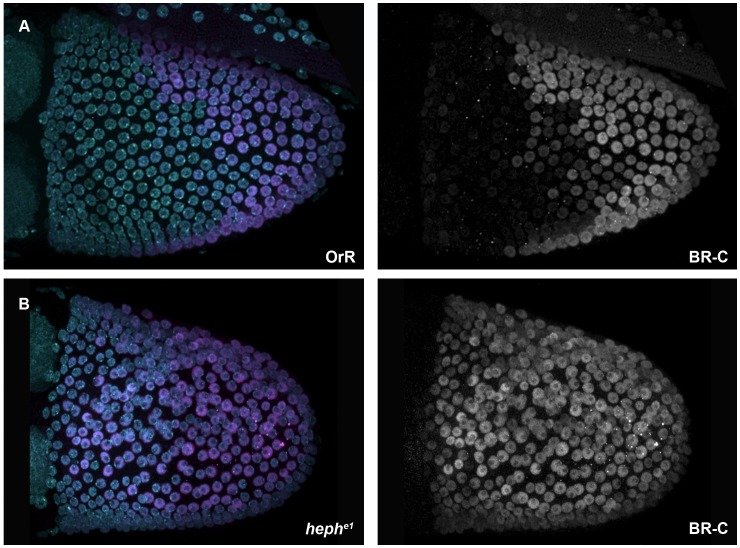
BR-C Protein is Expressed in all Follicle Cells in heph Mutant Stage 10 Egg Chambers
The mislocalization of Grk protein could potentially be due to defects in Grk cleavage or secretion from the oocyte. To visualize and assess whether Grk can be secreted and therefore signal to the neighbouring follicle cells in heph mutant germline clones, we examined the expression of the Broad-Complex (BR-C) locus. BR-C encodes a group of four alternatively spliced zinc finger transcription factor isoforms whose elevated expression from stage 10 of oogenesis marks the follicle cells that will later form the dorsal appendages [78]. Analysis of BR-C expression in multiple mutants supports a mechanism whereby BR-C is induced above a critical level of EGFR activation, but is repressed at a higher level [79],[80]. This repression results in a lack of BR-C staining at the dorsal midline (the ‘dorsal gap’) that receive the highest level of Grk signal (Figure 5A) and ultimately in the formation of two separate dorsal appendages. In approximately 63% (n = 11) hephe1 germline mutant egg chambers BR-C is detected in all dorsal follicle cells, and there is no dorsal gap (Figure 5B). This is consistent with a number of the observed eggshell phenotypes including changes in the inter-appendage space between the two dorsal appendages, the presence of ectopic dorsal appendage material at the dorsal midline and defects in dorsal appendage morphogenesis. It is also consistent with the defects in Grk signalling in these egg chambers, and could indicate that follicle cells in the dorsal midline are not receiving the high levels of Grk signalling as they would in wild-type egg chambers, but are receiving an intermediate level of signalling normally seen by cells either side of the midline.
Figure 5. BR-C protein is expressed in all follicle cells in heph mutant stage 10 egg chambers.
Stage 10 Oregon-R (OrR) egg chambers (A) and hephe1 germline clone egg chambers (B) were stained with anti-Broad-core and DAPI. In stage 10 OrR egg chambers, BR-C is not detected in the follicle cells at the dorsal anterior region (the ‘dorsal gap’) (A), whilst in hephe1 germline mutant egg chambers BR-C is detected in all dorsal follicle cells (B).
Discussion
We identified Heph as a factor able to associate in vitro with grk and GLS-containing RNA [34] leading us to hypothesize that it is a trans-acting factor required for the localization, anchoring or translational control of grk mRNA in the Drosophila oocyte. This hypothesis was supported indirectly by the fact that a function for the Heph/PTB in the formation of high-order osk mRNP complexes and the subsequent translational repression of osk in the oocyte was also previously characterized [55]. Using RNA IP experiments we have shown here that Heph binds specifically to grk and osk mRNAs. These results are consistent with the previous in vitro RNA-pulldown experiments [34] and therefore suggest that Heph is a component of the grk mRNP complexes, in addition to complexes containing osk [55]. Heph does not colocalize with grk mRNA at the dorso-anterior corner of the oocyte, suggesting that this interaction occurs at earlier stages of grk localization before the RNA reaches its final destination, and may also be transient. This is in contrast to the case of osk, where Heph/PTB can be visualized at the posterior pole where it colocalizes with osk mRNA [55].
Our examination of heph mutant germline clones revealed multiple functions for heph during oogenesis in addition to the previously characterized roles of Heph in early germ cell maintenance and osk translational repression [55]. Follicle cell phenotypes observed in mutant germline clones indicate that Heph may be involved in signalling between the germline and the somatic follicle cells, and in the subsequent patterning and morphogenesis of the dorsal appendages. We have shown that Heph is not required for grk mRNA localization, despite the interaction of Heph with grk mRNP complexes. Nor is Heph required for Grk protein localization and signalling in the earlier stages of oogenesis. However, Heph is required for correct Grk protein localization and Grk signalling in stage 10 oocytes as assessed by Grk and BR-C expression.
It remains unclear why loss of heph function would result in the mislocalization of Grk protein during stage 10 of oogenesis when grk mRNA localization is unaffected. The dispersal of Grk throughout the oocyte in heph mutants suggests that Heph could function in the trafficking of Grk protein after translation [74],[77],[81],[82],[83],[84],[85]. There are a number of previously studied mutants, such as rab6 [83],[86], and syntaxin 1A (Syx1A) [84] in which grk mRNA localization is unaffected, but Grk protein is mislocalized. Furthermore, in these mutants, as is the case with heph, only the dorso-anterior localization and signalling of Grk protein is affected, and not the posterior localization or signalling during earlier stages of oogenesis. This is thought to be a consequence of a lower threshold for EGFR activation by Grk signalling in the posterior follicle cells compared with the dorso-anterior follicle cells [74]. In null germline clones of the small GTPase Rab6, Grk mislocalizes to droplet-like structures throughout the ooplasm [83],[84] that colocalize with post-Golgi vesicle markers [83]. In hypomorphic germline clones of the Syx1A t-SNARE protein, Grk protein is also diffused throughout the ooplasm and partially colocalizes with Rab6 [84]. Localization of Grk protein at the dorso-anterior of the oocyte therefore depends upon membrane trafficking, and it is possible that Heph also functions to regulate and restrict Grk protein trafficking only to the dorso-anterior corner of the oocyte. With reduced Heph activity, follicle cells other than those at the dorso-anterior corner would then receive ectopic low-level Grk signalling which could explain the BR-C expression pattern and eggshell phenotypes that we observed.
The underlying molecular cause for a Grk mislocalization or trafficking defect in heph mutants is also uncertain. It is possible that Heph directly regulates the expression of mRNAs encoding proteins that are required to act in the secretory pathway, such as Rab6 and Syx1A. Vesicular trafficking is also coupled with actin cytoskeletal dynamics [87],[88], and it is possible that Grk mislocalization or trafficking defects could be due to defects in the actin cytoskeleton. The lack of wild-type heph function in the female germline results in egg chamber and eggshell phenotypes consistent with defects in actin regulation. The short eggs have a partial dumpless phenotype that is presumably caused by the lack of the dense bundles of actin filaments that ordinarily form in the nurse cell cytoplasm prior to dumping. This phenotype is seen in a small number of mutant germline clones, but it is possible that a higher number of clones have more subtle defects in actin filament organization that were missed in our analysis. Actin regulation by Heph has also been observed in Drosophila spermatogenesis [61], where loss of a male specific Heph isoform in the testes results in disruption of the actin cones of the spermatid individualization complex and prevents proper individualization of spermatids. Phenotypes seen in heph mutants during embryonic development can also be attributed to defects in actin regulation [63]. heph mutant embryos are defective in dorsal closure, and actin accumulates to high levels in a disorganized fashion in dorso-lateral regions. Whether Heph directly acts on mRNAs that encode cytoskeletal components is unknown. Indeed, mammalian PTB regulates members of the actin cytoskeleton, including α- and β-tropomyosin and α-actinin [89],[90].
There is a precedent for an RNA-binding protein such as Heph to regulate actin organization and vesicle trafficking within the oocyte. Bicaudal-C (Bic-C) is a KH-type RNA-binding protein that is also required for translational repression of unlocalized osk [91]. In Bic-C mutant egg chambers, Grk is mislocalized during mid-oogenesis and sequestered within actin-coated structures that contain distinct sets of secretory proteins in addition to microfilament interacting proteins [74],[92]. Bic-C is thought to affect the expression of a number of proteins during oogenesis and several Bic-C RNA targets encode proteins that are predicted to be involved in vesicle trafficking and/or organization of the actin-cytoskeleton [93]. Interestingly, like Heph, Bic-C was also found to associate in vitro with grk in GRNA chromatography studies (Supplementary Table 1) [34], although analysis of hatching frequency and viability of progeny from trans-heterozygotes of Bic-C and heph mutant alleles do not reveal any genetic interaction between Bic-C and heph (data not shown).
Our results highlight several perhaps unexpected issues. First, it is likely that many more factors are required for various aspects of grk mRNA localization, translational regulation and Grk signalling, including previously identified factors as well as unknown ones. Second, it is clear from our work that additional genetics studies are required to characterize the factors identified biochemically as binding to localized transcripts in the oocyte. Third, there are several aspects of Heph function in oogenesis that are not fully understood. In order to elucidate the primary functions of Heph in the germline, and the observed heph mutant phenotypes, future experiments will have to uncover the RNA targets for Heph. This will provide insight into the pathways that are perturbed in heph mutants and may well discover some unexpected mechanisms that could explain the complexity of the heph phenotype. For example, Heph is also required indirectly for osk localization through the properly timed organization of the oocyte microtubule (MT) cytoskeleton, and in heph mutants there is a delay in focusing the microtubule plus-ends at the posterior pole of the oocyte [55]. It is possible that this defect in the efficient establishment of microtubule polarity in the oocyte is a consequence of defects in actin organization [94],[95] and/or vesicular trafficking [86], or could indicate a more direct role in regulating the expression of proteins that organize the microtubule cytoskeleton. Finally, one particularly intriguing question relates to why Heph is present in grk mRNP complexes if it is a regulator of trafficking and/or of the actin cytoskeleton as we hypothesize. As a component of the grk RNP, it is possible that Heph could act at the interface of grk localization and the secretory pathway in order to promote efficient and restricted local secretion of Grk protein, providing an example of how interaction between secretory and mRNA processing pathways can promote efficient protein trafficking [74],[85].
Materials And Methods
Genetics
Stocks were raised on standard cornmeal-agar medium at 25°C. The wild-type control strain was Oregon R (OrR). The P(PZ}heph[03429] (Bloomington Stock Center) had been recombined onto the FRT82B chromosome by Besse et al. [55], and was a gift from A. Ephrussi. The FRT82B-recombined EMS mutant alleles hephe1 and hephe2 were characterized by Dansereau et al. [64] and were a gift from W. Brook. Germline clones of heph were generated using the FLP/FRT dominant female sterile technique [67],[68]. Females heterozygous for the FRT82B-recombined hephe1, hephe2 and heph03429 alleles were mated to P{ry+, FRT}82B P{w+ovoD1}/TM3 sb males and the progeny third instar larvae were heat shocked at 37°C for 2 h each day for 3 days. hephe1 and heph03429/ovoD1 females were collected and mated to OrR males. GFP-PTB/Heph was a GFP protein trap line [55] and was a gift from A. Ephrussi. SquidGFP was also a GFP protein trap line from A. Debec.
Eggshell Preparation
Axis specification defects were studied in eggs from heph mutant germline clones. For eggshell preparation, freshly laid eggs were mounted in a 1∶1 mixture of lactic acid:Hoyer’s medium and incubated overnight at 65°C.
Immunoprecipitation
Rabbit anti-GFP (Invitrogen), rabbit anti-Syp (S. McDermott, unpublished), and control rabbit pre-immune sera were cross-linked to Protein A Sepharose using standard protocols, and the beads equilibrated in IP buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% NP-40, 10% glycerol and Complete EDTA-free protease inhibitor). For RNA immunoprecipitation all steps were carried out in the presence of RNasin (Promega). 20 µl of a 50% slurry was added to 500 µg precleared ovarian lysate, and the mixture incubated overnight at 4°C with mixing. The beads were then rinsed once with cold IP buffer and washed four times for 5 min each at 4°C. After the final wash, immunocomplexes were eluted from the beads by the addition of 100 µl extraction buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA and 1.3% SDS) plus 100 U RNasin and incubation for 30 min at 65°C. Following centrifugation, the elution step was repeated and the two supernatants combined. RNA was extracted from input samples and immunoprecipitates using Trizol LS reagent (Invitrogen). Following DNase (Ambion) treatment, immunoprecipitated RNA and 10% total RNA were then used as a template for cDNA synthesis in combination with SuperScript III (Invitrogen) RT and random hexamer primers (Invitrogen). cDNA was then used directly as a template for PCR with primers as described in Supporting Information.
Western Blotting
30 to 50 µg of cleared ovarian lysate was loaded per well and ran on NuPAGE Novex 4–12% Bis–Tris Gels (Invitrogen). Following electrophoresis, proteins were transferred to Polyvinylidene Fluoride (PVDF) (Bio-Rad) membranes using the XCell II Blot Module (Invitrogen) according to the manufacturer’s instructions. Western blotting was performed using standard protocols. The mouse anti-Grk mAb1D12 concentrate (Developmental Studies Hybridoma Bank) was used at 1∶5000 and the rabbit anti-tubulin (Sigma) at 1∶2000. Rat anti-PTB/Heph was a gift from A. Ephrussi and was used at 1∶2000. Immunoreactive bands were visualized with HRP-conjugated sheep anti-mouse, and rat and donkey anti-rabbit secondary antibodies (GE Healthcare Life Sciences) at 1∶5000 and with enhanced chemiluminescence (GE Healthcare Life Sciences).
Immunohistochemistry and in situ Hybridization
Ovaries were prepared for immunostaining as previously described (MacDougall et al., 2003). The following primary antibodies were used: mouse anti-Broad-core supernatant mAb25E9.D7 (1∶100) (Developmental Studies Hybridoma Bank), mouse anti-Grk mAb1D12 concentrate (1∶300) (Developmental Studies Hybridoma Bank) and rabbit anti-PTB/Heph (1∶500) (gift from A. Ephrussi). Alexa-conjugated secondary goat antibodies (1∶500) (Invitrogen) were used. Ovaries were also stained with phalloidin to label F-actin (Invitrogen) and with 10 µg/mL DAPI to label nuclei. In situ hybridization was performed as previously described (Wilkie et al., 1999). Digoxigenin-labeled antisense probes corresponding to either gurken (T. Schüpbach) or bicoid (C. Nüsslein-Volhard) full-length coding sequences were detected using sheep HRP-conjugated anti-DIG antibody (1∶500; Roche) followed by fluorescent tyramide signal amplification (Perkin Elmer). Ovaries were also stained with a fluorescent Wheat Germ Agglutinin conjugate (1 µg/mL) (Invitrogen) to label nuclear membranes.
Synthesis of Fluorescently Labeled Capped RNA and Injection
RNA was transcribed in vitro using UTP-Alexa 546 or UTP-Cy3 (Wilkie and Davis, 2001) and a plasmid containing the full-length grk cDNA (T. Schupbach). Ovaries were dissected in Series 95 halocarbon oil (KMZ Chemicals) and egg chambers adhered to coverslips using tungsten needles. Fluorescently labeled RNA (250–500 ng/ml) was injected into oocytes using Femtotip needles (Eppendorf). At least two batches of RNA were injected for each experiment on separate occasions.
Imaging and Deconvolution
A Leica SP5 confocal microscope and a widefield DeltaVision microscope (Applied Precision, Olympus IX70, and Roper Coolsnap HQ) were used to image fixed material. DeltaVision images were acquired with Olympus 20x/0.75, 40x/0.95, 60x/0.9 or 100x/1.4 objective lenses and then deconvolved.
Supporting Information
Distribution of Heph during Drosophila oogenesis. Stages 4–6 (A), stage 8 (B), stage 9 (C) and stage 10B (D and E) egg chambers stained with anti-Heph antibodies and DAPI. (E) is an enlargement of the oocyte shown in the egg chamber in (D). (F) is an enlargement of the dorso-anterior corner (white box) of the stage 9 oocyte in (C) whilst (G) is an enlargement of the dorso-anterior corner (white box) of the stage 10B oocyte in (D). Heph is detected in the cytoplasm of the follicular epithelial cells throughout oogenesis. In the germline, Heph is present throughout the oocyte cytoplasm with an accumulation at the posterior pole of the oocyte from around stage 9 of oogenesis onward (C–E). However, Heph does not accumulate at the dorso-anterior cap coincident with grk mRNA (C–G). Scale bars in A, B, and E represent 60 µm. Scale bars in C and D represent 90 µm. (H) is the same wild-type egg chamber shown in (D) for comparison with (I), which is a hephe1 germline clone egg chamber also stained with anti-Heph antibodies. Oocyte and nurse cell signals are lost in hephe1 mutant egg chambers, indicating that the signal is specific.
(TIF)
Actin structures are disrupted in heph03429 and hephe1 germline clone egg chambers. Wild-type Oregon-R (OrR) (A), heph03429 germline clone (B), and hephe1 germline clone egg chambers (C) were stained with Phalloidin to label F-actin.
(TIF)
grk mRNA localization is not perturbed in stage 10 heph mutant oocytes. In situ hybridization using a grk was carried out on Oregon-R (OrR) (A), or hephe1 germline clone egg chambers (B). Egg chambers were also stained with DAPI to label DNA.
(TIF)
Grk protein localization in heph mutant stage 6 to stage 9 oocytes. Stage 6 (A), stage 8 (B), and stage 9 (C) wild-type Oregon-R (OrR) egg chambers, and stage 6 (D), stage 8 (E), and stage 9 (F) hephe1 germline clone egg chambers were stained with anti-Gurken and DAPI.
(TIF)
Levels of Grk protein are unaffected in heph mutant ovaries. Western blot showing relative levels of Grk and Heph in wild-type Oregon-R (OrR) ovarian extracts and in ovarian extracts prepared from heph germline clones. Tubulin was used as a loading control.
(TIF)
16 trans-acting factors identified by GRNA affinity chromatography to associate with 5′ORF RNA. Heph/PTB is highlighted in bold. Adapted from McDermott et al. [34].
(DOC)
Acknowledgments
We are grateful to Flybase, the Bloomington Drosophila Stock Center and the Developmental Studies Hybridoma Bank for reagents. We are also very grateful to all those who provided fly strains and antibodies, and extend thanks to David Finnegan (University of Edinburgh, UK) in whose laboratory this work was finished.
Funding Statement
This work was supported by a Wellcome Trust Senior Research Fellowship to ID (081858) and a Wellcome Trust PhD studentship to SMM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Blower MD, Feric E, Weis K, Heald R (2007) Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol 179: 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holt CE, Bullock SL (2009) Subcellular mRNA localization in animal cells and why it matters. Science 326: 1212–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, et al. (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187. [DOI] [PubMed] [Google Scholar]
- 4. Martin KC, Ephrussi A (2009) mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meignin C, Davis I (2010) Transmitting the message: intracellular mRNA localization. Curr Opin Cell Biol 22: 112–119. [DOI] [PubMed] [Google Scholar]
- 6. Mili S, Moissoglu K, Macara IG (2008) Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453: 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Condeelis J, Singer RH (2005) How and why does beta-actin mRNA target? Biol Cell 97: 97–110. [DOI] [PubMed] [Google Scholar]
- 8. Horne-Badovinac S, Bilder D (2008) Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genet 4: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Z, Wang L, Hays TS, Cai Y (2008) Dynein-mediated apical localization of crumbs transcripts is required for Crumbs activity in epithelial polarity. J Cell Biol 180: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Broadus J, Fuerstenberg S, Doe CQ (1998) Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature 391: 792–795. [DOI] [PubMed] [Google Scholar]
- 11. King ML, Messitt TJ, Mowry KL (2005) Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell 97: 19–33. [DOI] [PubMed] [Google Scholar]
- 12. Kugler JM, Lasko P (2009) Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly (Austin) 3: 15–28. [DOI] [PubMed] [Google Scholar]
- 13. Neuman-Silberberg FS, Schupbach T (1993) The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 75: 165–174. [PubMed] [Google Scholar]
- 14. Gonzalez-Reyes A, Elliott H, St Johnston D (1995) Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature 375: 654–658. [DOI] [PubMed] [Google Scholar]
- 15. Roth S, Neuman-Silberberg FS, Barcelo G, Schupbach T (1995) cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell 81: 967–978. [DOI] [PubMed] [Google Scholar]
- 16. Schupbach T (1987) Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell 49: 699–707. [DOI] [PubMed] [Google Scholar]
- 17. Jaramillo AM, Weil TT, Goodhouse J, Gavis ER, Schupbach T (2008) The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J Cell Sci 121: 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. MacDougall N, Clark A, MacDougall E, Davis I (2003) Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev Cell 4: 307–319. [DOI] [PubMed] [Google Scholar]
- 19. Nilson LA, Schupbach T (1999) EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol 44: 203–243. [DOI] [PubMed] [Google Scholar]
- 20. van Eeden F, St Johnston D (1999) The polarisation of the anterior-posterior and dorsal-ventral axes during Drosophila oogenesis. Curr Opin Genet Dev 9: 396–404. [DOI] [PubMed] [Google Scholar]
- 21. Ephrussi A, Lehmann R (1992) Induction of germ cell formation by oskar. Nature 358: 387–392. [DOI] [PubMed] [Google Scholar]
- 22. Kim-Ha J, Smith JL, Macdonald PM (1991) oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66: 23–35. [DOI] [PubMed] [Google Scholar]
- 23. Ephrussi A, Dickinson LK, Lehmann R (1991) Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50. [DOI] [PubMed] [Google Scholar]
- 24. Wang C, Dickinson LK, Lehmann R (1994) Genetics of nanos localization in Drosophila. Dev Dyn 199: 103–115. [DOI] [PubMed] [Google Scholar]
- 25. Gavis ER, Lehmann R (1994) Translational regulation of nanos by RNA localization. Nature 369: 315–318. [DOI] [PubMed] [Google Scholar]
- 26. Weil TT, Forrest KM, Gavis ER (2006) Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev Cell 11: 251–262. [DOI] [PubMed] [Google Scholar]
- 27. Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C (2007) Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell 13: 523–538. [DOI] [PubMed] [Google Scholar]
- 28. Krichevsky AM, Kosik KS (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32: 683–696. [DOI] [PubMed] [Google Scholar]
- 29. Kelley RL (1993) Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes Dev 7: 948–960. [DOI] [PubMed] [Google Scholar]
- 30. Norvell A, Kelley RL, Wehr K, Schupbach T (1999) Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev 13: 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caceres L, Nilson LA (2009) Translational repression of gurken mRNA in the Drosophila oocyte requires the hnRNP Squid in the nurse cells. Dev Biol 326: 327–334. [DOI] [PubMed] [Google Scholar]
- 32. Goodrich JS, Clouse KN, Schupbach T (2004) Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development 131: 1949–1958. [DOI] [PubMed] [Google Scholar]
- 33. Geng C, Macdonald PM (2006) Imp associates with squid and Hrp48 and contributes to localized expression of gurken in the oocyte. Mol Cell Biol 26: 9508–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McDermott SM, Meignin C, Rappsilber J, Davis I (2012) Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification. Biology Open 1: 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I (2005) gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev Cell 9: 51–62. [DOI] [PubMed] [Google Scholar]
- 36. Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE (2003) The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell 11: 757–771. [DOI] [PubMed] [Google Scholar]
- 37. Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, et al. (2005) Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309: 2054–2057. [DOI] [PubMed] [Google Scholar]
- 38. Auweter SD, Oberstrass FC, Allain FH (2007) Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein-RNA complexes of weak affinities. J Mol Biol 367: 174–186. [DOI] [PubMed] [Google Scholar]
- 39. Garcia-Blanco MA, Jamison SF, Sharp PA (1989) Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev 3: 1874–1886. [DOI] [PubMed] [Google Scholar]
- 40. Perez I, Lin CH, McAfee JG, Patton JG (1997) Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3: 764–778. [PMC free article] [PubMed] [Google Scholar]
- 41. Singh R, Valcarcel J, Green MR (1995) Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176. [DOI] [PubMed] [Google Scholar]
- 42. Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, et al. (2010) The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA 107: 1894–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lou H, Helfman DM, Gagel RF, Berget SM (1999) Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol 19: 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Southby J, Gooding C, Smith CW (1999) Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of alpha-actinin mutally exclusive exons. Mol Cell Biol 19: 2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV (2012) Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev 26: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng S, Gray EE, Chawla G, Porse BT, O'Dell TJ, et al.. (2012) PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci 15: 381–388, S381. [DOI] [PMC free article] [PubMed]
- 47. Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, et al. (2004) Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol 24: 4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fred RG, Tillmar L, Welsh N (2006) The role of PTB in insulin mRNA stability control. Curr Diabetes Rev 2: 363–366. [DOI] [PubMed] [Google Scholar]
- 49. Engels B, Jannot G, Remenyi J, Simard MJ, Hutvagner G (2012) Polypyrimidine tract binding protein (hnRNP I) is possibly a conserved modulator of miRNA-mediated gene regulation. PLoS ONE 7: e33144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim JH, Paek KY, Ha SH, Cho S, Choi K, et al. (2004) A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol Cell Biol 24: 7878–7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim DY, Woo KC, Lee KH, Kim TD, Kim KT (2010) hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb alpha via IRES-mediated translation. Nucleic Acids Res 38: 7068–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ohno S, Shibayama M, Sato M, Tokunaga A, Yoshida N (2011) Polypyrimidine tract-binding protein regulates the cell cycle through IRES-dependent translation of CDK11(p58) in mouse embryonic stem cells. Cell Cycle 10: 3706–3713. [DOI] [PubMed] [Google Scholar]
- 53. Kim YK, Hahm B, Jang SK (2000) Polypyrimidine tract-binding protein inhibits translation of bip mRNA. J Mol Biol 304: 119–133. [DOI] [PubMed] [Google Scholar]
- 54. Babic I, Sharma S, Black DL (2009) A role for polypyrimidine tract binding protein in the establishment of focal adhesions. Mol Cell Biol 29: 5564–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Besse F, Lopez de Quinto S, Marchand V, Trucco A, Ephrussi A (2009) Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev 23: 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, et al. (1999) A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell 4: 431–437. [DOI] [PubMed] [Google Scholar]
- 57. Lewis RA, Gagnon JA, Mowry KL (2008) PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol Cell Biol 28: 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ma S, Liu G, Sun Y, Xie J (2007) Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim Biophys Acta 1773: 912–923. [DOI] [PubMed] [Google Scholar]
- 59. Castrillon DH, Gonczy P, Alexander S, Rawson R, Eberhart CG, et al. (1993) Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135: 489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Robida MD, Singh R (2003) Drosophila polypyrimidine-tract binding protein (PTB) functions specifically in the male germline. EMBO J 22: 2924–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Robida M, Sridharan V, Morgan S, Rao T, Singh R (2010) Drosophila polypyrimidine tract-binding protein is necessary for spermatid individualization. Proc Natl Acad Sci USA 107: 12570–12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schulz C, Kiger AA, Tazuke SI, Yamashita YM, Pantalena-Filho LC, et al. (2004) A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics 167: 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wesley CS, Guo H, Chaudhry KA, Thali MJ, Yin JC, et al. (2011) Loss of PTB or negative regulation of Notch mRNA reveals distinct zones of Notch and actin protein accumulation in Drosophila embryo. PLoS ONE 6: e21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dansereau DA, Lunke MD, Finkielsztein A, Russell MA, Brook WJ (2002) Hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster. Development 129: 5553–5566. [DOI] [PubMed] [Google Scholar]
- 65. Davis MB, Sun W, Standiford DM (2002) Lineage-specific expression of polypyrimidine tract binding protein (PTB) in Drosophila embryos. Mech Dev 111: 143–147. [DOI] [PubMed] [Google Scholar]
- 66. Norga KK, Gurganus MC, Dilda CL, Yamamoto A, Lyman RF, et al. (2003) Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr Biol 13: 1388–1396. [DOI] [PubMed] [Google Scholar]
- 67. Chou TB, Noll E, Perrimon N (1993) Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119: 1359–1369. [DOI] [PubMed] [Google Scholar]
- 68. Chou TB, Perrimon N (1996) The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144: 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rittenhouse KR, Berg CA (1995) Mutations in the Drosophila gene bullwinkle cause the formation of abnormal eggshell structures and bicaudal embryos. Development 121: 3023–3033. [DOI] [PubMed] [Google Scholar]
- 70. Tran DH, Berg CA (2003) bullwinkle and shark regulate dorsal-appendage morphogenesis in Drosophila oogenesis. Development 130: 6273–6282. [DOI] [PubMed] [Google Scholar]
- 71. Cooley L, Verheyen E, Ayers K (1992) chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell 69: 173–184. [DOI] [PubMed] [Google Scholar]
- 72. Robinson DN, Cooley L (1997) Genetic analysis of the actin cytoskeleton in the Drosophila ovary. Annu Rev Cell Dev Biol 13: 147–170. [DOI] [PubMed] [Google Scholar]
- 73. Januschke J, Gervais L, Dass S, Kaltschmidt JA, Lopez-Schier H, et al. (2002) Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr Biol 12: 1971–1981. [DOI] [PubMed] [Google Scholar]
- 74. Kugler JM, Chicoine J, Lasko P (2009) Bicaudal-C associates with a Trailer Hitch/Me31B complex and is required for efficient Gurken secretion. Dev Biol 328: 160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Berg CA (2005) The Drosophila shell game: patterning genes and morphological change. Trends Genet 21: 346–355. [DOI] [PubMed] [Google Scholar]
- 76. Neuman-Silberberg FS, Schupbach T (1996) The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech Dev 59: 105–113. [DOI] [PubMed] [Google Scholar]
- 77. Pizette S, Rabouille C, Cohen SM, Therond P (2009) Glycosphingolipids control the extracellular gradient of the Drosophila EGFR ligand Gurken. Development 136: 551–561. [DOI] [PubMed] [Google Scholar]
- 78. Deng WM, Bownes M (1997) Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development 124: 4639–4647. [DOI] [PubMed] [Google Scholar]
- 79. Yakoby N, Lembong J, Schupbach T, Shvartsman SY (2008) Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development 135: 343–351. [DOI] [PubMed] [Google Scholar]
- 80. Cheung LS, Schupbach T, Shvartsman SY (2011) Pattern formation by receptor tyrosine kinases: analysis of the Gurken gradient in Drosophila oogenesis. Curr Opin Genet Dev 21: 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Herpers B, Rabouille C (2004) mRNA localization and ER-based protein sorting mechanisms dictate the use of transitional endoplasmic reticulum-golgi units involved in gurken transport in Drosophila oocytes. Mol Biol Cell 15: 5306–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bokel C, Dass S, Wilsch-Brauninger M, Roth S (2006) Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development 133: 459–470. [DOI] [PubMed] [Google Scholar]
- 83. Januschke J, Nicolas E, Compagnon J, Formstecher E, Goud B, et al. (2007) Rab6 and the secretory pathway affect oocyte polarity in Drosophila. Development 134: 3419–3425. [DOI] [PubMed] [Google Scholar]
- 84. Tian AG, Tamori Y, Huang YC, Melendez NT, Deng WM (2013) Efficient EGFR signaling and dorsal-ventral axis patterning requires syntaxin dependent Gurken trafficking. Dev Biol 373: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wilhelm JE, Buszczak M, Sayles S (2005) Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell 9: 675–685. [DOI] [PubMed] [Google Scholar]
- 86. Coutelis JB, Ephrussi A (2007) Rab6 mediates membrane organization and determinant localization during Drosophila oogenesis. Development 134: 1419–1430. [DOI] [PubMed] [Google Scholar]
- 87. Stamnes M (2002) Regulating the actin cytoskeleton during vesicular transport. Curr Opin Cell Biol 14: 428–433. [DOI] [PubMed] [Google Scholar]
- 88. Fricke R, Gohl C, Dharmalingam E, Grevelhorster A, Zahedi B, et al. (2009) Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr Biol 19: 1429–1437. [DOI] [PubMed] [Google Scholar]
- 89. Matlin AJ, Southby J, Gooding C, Smith CW (2007) Repression of alpha-actinin SM exon splicing by assisted binding of PTB to the polypyrimidine tract. RNA 13: 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, et al. (2005) Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans 33: 457–460. [DOI] [PubMed] [Google Scholar]
- 91. Saffman EE, Styhler S, Rother K, Li W, Richard S, et al. (1998) Premature translation of oskar in oocytes lacking the RNA-binding protein bicaudal-C. Mol Cell Biol 18: 4855–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Snee MJ, Macdonald PM (2009) Bicaudal C and trailer hitch have similar roles in gurken mRNA localization and cytoskeletal organization. Dev Biol 328: 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, et al. (2007) Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell 13: 691–704. [DOI] [PubMed] [Google Scholar]
- 94. Dahlgaard K, Raposo AA, Niccoli T, St Johnston D (2007) Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell 13: 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang Y, Riechmann V (2008) Microtubule anchoring by cortical actin bundles prevents streaming of the oocyte cytoplasm. Mech Dev 125: 142–152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of Heph during Drosophila oogenesis. Stages 4–6 (A), stage 8 (B), stage 9 (C) and stage 10B (D and E) egg chambers stained with anti-Heph antibodies and DAPI. (E) is an enlargement of the oocyte shown in the egg chamber in (D). (F) is an enlargement of the dorso-anterior corner (white box) of the stage 9 oocyte in (C) whilst (G) is an enlargement of the dorso-anterior corner (white box) of the stage 10B oocyte in (D). Heph is detected in the cytoplasm of the follicular epithelial cells throughout oogenesis. In the germline, Heph is present throughout the oocyte cytoplasm with an accumulation at the posterior pole of the oocyte from around stage 9 of oogenesis onward (C–E). However, Heph does not accumulate at the dorso-anterior cap coincident with grk mRNA (C–G). Scale bars in A, B, and E represent 60 µm. Scale bars in C and D represent 90 µm. (H) is the same wild-type egg chamber shown in (D) for comparison with (I), which is a hephe1 germline clone egg chamber also stained with anti-Heph antibodies. Oocyte and nurse cell signals are lost in hephe1 mutant egg chambers, indicating that the signal is specific.
(TIF)
Actin structures are disrupted in heph03429 and hephe1 germline clone egg chambers. Wild-type Oregon-R (OrR) (A), heph03429 germline clone (B), and hephe1 germline clone egg chambers (C) were stained with Phalloidin to label F-actin.
(TIF)
grk mRNA localization is not perturbed in stage 10 heph mutant oocytes. In situ hybridization using a grk was carried out on Oregon-R (OrR) (A), or hephe1 germline clone egg chambers (B). Egg chambers were also stained with DAPI to label DNA.
(TIF)
Grk protein localization in heph mutant stage 6 to stage 9 oocytes. Stage 6 (A), stage 8 (B), and stage 9 (C) wild-type Oregon-R (OrR) egg chambers, and stage 6 (D), stage 8 (E), and stage 9 (F) hephe1 germline clone egg chambers were stained with anti-Gurken and DAPI.
(TIF)
Levels of Grk protein are unaffected in heph mutant ovaries. Western blot showing relative levels of Grk and Heph in wild-type Oregon-R (OrR) ovarian extracts and in ovarian extracts prepared from heph germline clones. Tubulin was used as a loading control.
(TIF)
16 trans-acting factors identified by GRNA affinity chromatography to associate with 5′ORF RNA. Heph/PTB is highlighted in bold. Adapted from McDermott et al. [34].
(DOC)