Abstract
Cryptococcus is an emerging global health threat that is annually responsible for over 1,000,000 infections and one third of all AIDS patient deaths. There is an ongoing outbreak of cryptococcosis in the western United States and Canada. Cryptococcosis is a disease resulting from the inhalation of the infectious propagules from the environment. The current and most frequently used animal infection models initiate infection via liquid suspension through intranasal instillation or intravenous injection. These models do not replicate the typically dry nature of aerosol exposure and may hinder our ability to decipher the initial events that lead to clearance or the establishment of infection. We have established a standardized aerosol model of murine infection for the human fungal pathogen Cryptococcus. Aerosolized cells were generated utilizing a Collison nebulizer in a whole-body Madison Chamber at different humidity conditions. The aerosols inside the chamber were sampled using a BioSampler to determine viable aerosol concentration and spray factor (ratio of viable aerosol concentration to total inoculum concentration). We have effectively delivered yeast and yeast-spore mixtures to the lungs of mice and observed the establishment of disease. We observed that growth conditions prior to exposure and humidity within the Madison Chamber during exposure can alter Cryptococcus survival and dose retained in mice.
Introduction
Cryptococcosis is an opportunistic fungal infection caused by Cryptococcus and which is a predominant cause of morbidity and mortality in immunocompromised and HIV-positive hosts [1]. Cryptococcus is responsible for approximately one-third of all AIDS related deaths and a more prevalent cause of HIV-related mortality in sub-Saharan Africa than tuberculosis [1]. In other developing countries infections as a result of Cryptococcus are second only to tuberculosis and frequently co-occur together [1]. The development of new antifungals, highly active antiviral treatment (HAART), and combination therapy regimens has increased long-term survival rates, but infections remain difficult to treat and frequently reoccur. Cryptococcus is now increasingly associated with a rising number of infections in seemingly healthy humans and animals worldwide [2]–[9].
Three closely related species/varieties, Cryptococcus neoformans var. neoformans, C. neoformans var. grubii and C. gattii are predominantly responsible for most human and animal infections. C. neoformans var. grubii is the most prevalent clinical isolate worldwide. However, C. neoformans var. neoformans is more prevalent in some regions, such as Europe. C. gattii is the most prevalent cause of cryptococcosis in immunocompetent hosts. However, in some regions of the world, C. gattii is now increasingly identified as the cause of infections in immunocompromised hosts [10]–[17]. Most cryptococcal infections result from the inhalation of infectious particles (yeast cells or spores) from the environment. Development of overt disease can be the result of acute, latent, or chronic infections. In nature C. neoformans var. neoformans is most frequently isolated from avian habitats and guano, and C. gattii is most frequently isolated from trees and soil. C. neoformans var. grubii, the most prevalent worldwide, has been frequently associated with avian habitats, and also trees and soil. Frequent isolation of Cryptococcus from plants and mating in association with plants, trees, soil, and avian guano suggest the importance of the environmental niche for both the development of infectious propagules (i.e. spores) and the maintenance of infectious reservoirs [18]–[24]. Cryptococcus is a prominent environmental organism, and humans are frequently exposed to its airborne infectious propagules through inhalation [25], [26]. How the environmental growth conditions and airborne infectious route influences risk and development of disease is presently unknown.
In laboratory conditions, both yeast cells and spores are effective at initiating disease but differ in size and mass [22], [23], [27]–[29]. Due to the small size of spores, it is presumed that spores are more effectively aerosolized in nature and thus more efficiently enter the bronchial tubes and alveolar spaces within the lungs [27], [29]. Additionally, spores are implicated in promoting infection because, in contrast to yeast cells, they do not require opsonization for phagocytosis by macrophages [29]. Macrophages play pivotal roles in the early host defense against Cryptococcus [30]. Ongoing outbreaks of C. gattii in the United States and Canada have renewed interest to better understand the acquisition and development of cryptococcosis.
Infections resulting from the inhalation of infectious yeast cells or spores are predominantly modeled utilizing tail-vein injection or intranasal instillation in the murine model host [31]–[33]. Although both methodologies cause morbidity and mortality, neither truly mimics the inhalation of infectious particles because these methodologies require yeast and/or spores to be administered within a suspension. The inoculation route, high dosages of cells utilized, and initial fluid solution could alter initial immune response and downstream sequela [34]–[38]. Appropriate control of cryptococcal infections depends on an intact immune system including macrophages, neutrophils, T cells, and the appropriate production and activity of cytokines [39]–[45]. Mounting evidence suggests that there are significant differences in risk to infection, development of disease, progression and host survival between the different Cryptococcus species [30], [37], [38], [46]–[48]. Current murine models are most frequently used but fall short of replicating the actual aerosol route of infection and may hinder our ability to decipher initial events that lead to the clearance or establishment of latent or progressive infection. Therefore these models may not accurately reflect the virulence properties of spores versus yeast, mating type a versus α strains, or the differences between molecular types or species [33], [49]–[53].
Aerosol infection models for Cryptococcus or other fungi have rarely been utilized [50], [53]–[56]. Historic applications of this approach include crude exposure from soil inoculated with Cryptococcus, utilization of a jet nebulizer, and infection in a Henderson Chamber [50], [54], [57]. Information extracted from the soil seeding infection was limited because infection rates and development of disease were variable, mice were exposed for multiple days, and the dosages of cells the mice were exposed to versus the dose initially retained in the lungs were not delineated. Infection initiated through the Henderson Chamber resulted in more consistent infection rates and development of disease in comparison to crude soil exposure [50]. Only 20% of mice exposed to aerosolized Cryptococcus basidiospores via a jet nebulizer succumbed to infection [53]. Variable infection rates and disease development were likely due to differences in the inoculum used, method of aerosolization, duration of exposure, host genotype, and strain of Cryptococcus utilized. These early studies were successful at initiating infection, but computer-controlled facilities were not available, Cryptococcus species were not yet taxonomically differentiated, and mating and the production of spores versus yeast had not been fully appreciated [50], [58].
The Madison Aerosol Chamber (College of Engineering Shop, University of Wisconsin) is now the standard apparatus for aerosol exposure of primary pathogens designated by the National Institute of Allergy and Infectious Diseases. Significant technological advancements now allow computer-controlled devices within the Madison Chamber to obtain samples pre- and post-nebulization from the exposure chamber (BioSampler), to adjust and monitor relative humidity within the chamber, and to collect pertinent data on mass and size of aerosolized cells exposed to mice [59]. These data can determine the effects of temperature, relative humidity, and the nebulization process on cells. In addition, we can calculate the dose that the mice were exposed to during a given time period, which facilitates the establishment of defined aerobiology exposure parameters for a given pathogen [59]. This study is the first contemporary application of the Madison Chamber to establish a well validated aerosol model of murine infection of Cryptococcus. We define the preparation of inoculum, concentration of inoculum, particle size distributions consistent with efficient aerosolization, and defined aerobiology exposure parameters for the effective and consistent delivery of aerosolized Cryptococcus cells to mice. A well validated inhalation model has considerable potential to facilitate novel insights into the primary exposure route of Cryptococcus in humans, time course of disease development, environmental conditions enhancing risk to acquisition, the role of spores and/or yeast in the infection process, and virulence of divergent mating types and genotypes which could lead to novel preventative strategies or therapeutic modalities.
Materials and Methods
Cryptococcus Strains, Growth Media, and Inoculum Preparation
Experiments were performed with C. neoformans var. neoformans JEC21; C. neoformans var. grubii H99α, KN99a, YSB119α, and KN99a NEO1; and C. gattii EJB18 (Table S1). For preliminary aerosolization tests we varied growth condition(s), exposure time, and humidity to increase the viable number of cells that mice would be exposed to within the whole-body exposure apparatus (dose presented) as determined from the viable BioSampler densities. We aimed to obtain a calculated dose presented of greater than 100 cells/mouse to maximize dose delivered and retention within the lungs. In various trials, cells were (1) grown in yeast peptone dextrose broth (YPD; 1% yeast extract, 2% peptone, 2% glucose) and prepared directly from the liquid preparation; (2) grown in YPD and subjected to natural evaporation and drying to mimic desiccation in nature, before being resuspended in water for nebulization; or (3) subcultured in YPD broth and then inoculated on agar for growth, mating, and the production of dry yeast and/or spores. Although cells were initially grown by different methods all Cryptococcus strains were collected and 20 mL of 1×108 cell/mL suspension were used as standard inoculum density for each aerobiology exposure. To confirm the density of each cell preparation prior to nebulization (Pre-nebulization, CFU/mL) in the Madison Chamber 500 µL aliquots from every final preparation were taken and diluted on YPD agar in triplicate and colony-forming units (CFUs) were determined and compared to the expected 1×108 cell/mL. In total, eleven trial experiments were performed, eight without mice, and three utilizing mice (Table S2). Broth-grown Cryptococcus cells were passaged twice in 250 mL flasks containing 25 mL of YPD broth with shaking at 30°C overnight (8–12 hrs). Cells were then collected by centrifugation, washed, and resuspended in 5 mL autoclaved millipore water. Cell density was determined by hemacytometer and 20 mL of a 1×108 cell/mL suspension were prepared for each strain. Cells subjected to drying prior to use in the Madison Chamber were grown in identical conditions to broth grown cells, but after the initial collection by centrifugation and resuspension in 5 mL water, cells were poured into a sterile petri plate, with the lid placed slightly ajar and allowed to air dry in a BSLII hood overnight. Cells were then collected, counted with a hemacytometer, and diluted to the standard density.
In other trials Cryptococcus cells were grown on V8 agar (pH 5) or Arabidopsis leaf agar (20 g chopped Arabidopsis thaliana leaves/L, 0.1% glucose, 2% agar), which mimics plant substrates in the environment where Cryptococcus is known to grow and can mate. Cryptococcus strains were grown and subcultured twice in 10 mL YPD broth and collected by centrifugation, washed twice with 5 mL autoclaved millipore water, and resuspended in 5 mL autoclaved water. Then 100 µL of each strain was spread individually onto V8 agar or Arabidopsis agar plates and incubated at room temperature in the dark for two to three weeks. The colonies were scraped from the plates, washed, suspended in 5 mL water, counted with a hemacytometer, and diluted to the standard cell density.
To induce mating for the collection of yeast-spore mixtures H99α/KN99a or YSB119α/KN99a NEO1 strains were grown in combination on V8 agar (pH 5). Cells were subcultured and prepared for plating as above, but after suspension in 5 mL water, equal proportions (1.5 mL) of a cells were mixed with α cells and 30 to 40 individual 15 µL volumes were spotted on four to six V8 agar plates. Plates were incubated in a dark room at room temperature for three to four weeks until prolific sporulation was confirmed by light microscopy. Mating colonies (including yeast and spores) were scraped and washed with autoclaved water from each plate and resuspended in autoclaved water. The suspension was centrifuged, washed, re-suspended in 5 mL water, counted with a hemacytometer, and diluted to the standardized inoculum cell density.
Aerosol Exposure
Cryptococcus aerosols were generated using a 6-jet Collison nebulizer (CN25, BGI Inc.), operated at an air-flow-rate of 13±1 lpm (19±1 PSI) and connected to a whole-body Madison Exposure Chamber (Figure S1). In trials utilizing humidified air, the nebulizer air was mixed with humidified air (37±1 lpm) as it entered the Madison Chamber. Humidified air was generated by two humidifiers connected in series (ZAB2-DT-S1-400-7, Lab Commerce Inc.). Humidity in the chamber was controlled from 30%–100% relative humidity (RH) and was measured in real-time during the aerosol exposure. Nebulized and humidified air flow rates were controlled with AeroMP (Aerosol Management Platform, Biaera Technologies, LLC) [57]. The Madison Chamber was operated under negative pressure at −13±1 inches of water column using a vacuum pump. Cryptococcus aerosols inside the Madison Chamber were sampled using an SKC BioSampler (SKC Inc, Model 225–9595) and aerodynamic particle sizer (APS, TSI Inc.). The start and stop operation of APS and BioSampler was controlled by AeroMP [57]. Particle size distribution by count and mass were measured by the APS at two-minute intervals for 30 seconds for the duration of the aerosol exposure. AeroMP was used to calculate the count mean aerodynamic diameter (CMAD) and mass mean aerodynamic diameter (MMAD). MMAD is used to describe a polydisperse aerosol particle size above and below which 50% of the mass of the particles is contained. The geometric standard deviation (GSD) was calculated using the particle size distributions by mass [60]. Aliquots obtained post-nebulization and from the BioSampler were dilution plated in triplicate on YPD agar, incubated at room temperature for two to four days and the average CFUs/mL was calculated. This data was utilized to determine the effect of nebulization on the viability of Cryptococcus cells by comparing the CFUs prior to nebulization and post-nebulization. Post-nebulization to Pre-nebulization ratio equal to or greater than one indicates 100% survival post nebulization, whereas a ratio value less than one indicates decreased viability post-nebulization. Viable aerosol density (Ca) was used to calculate spray factor (Fs), the ratio of viable aerosol concentration in the chamber to the starting inoculum density in the nebulizer [57], [59].
Viable aerosol concentration (Ca) was calculated as  where Csam is concentration in BioSampler, VBS is the volume of liquid in the BioSampler after aerosol exposure, td is aerosol exposure time duration, and QBS is BioSampler air flow rate.
where Csam is concentration in BioSampler, VBS is the volume of liquid in the BioSampler after aerosol exposure, td is aerosol exposure time duration, and QBS is BioSampler air flow rate.
Spray factor was calculated as 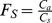 where Ca is viable aerosol concentration and CS is nebulizer sample concentration.
where Ca is viable aerosol concentration and CS is nebulizer sample concentration.
The dose presented (Dp) was calculated using Guyton’s formula.
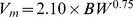 where BW is weight of mice in grams and Vm is minute volume.
where BW is weight of mice in grams and Vm is minute volume.
 where VE is the exposure volume and td is exposure time.
where VE is the exposure volume and td is exposure time.
 where DP is dose presented and Ca is the viable aerosol concentration.
where DP is dose presented and Ca is the viable aerosol concentration.
A total of 15 mL sterile filtered water was used in the BioSampler to capture aerosolized samples used to determine the cell density and viability of aerosolized Cryptococcus inside the Madison Chamber. This density was used to calculate the dose presented (Dp) to mice using Guyton’s formula [61]. GraphPad Prism version 6.00 for Windows (GraphPad Software, San Diego California USA) was used for data analysis to construct graphs. The P-values for linear regression analysis were calculated using the F-test. Two-way ANOVA was used to compare CFUs.
Animals
All animal studies were conducted in the Division of Laboratory Animal Resources (DLAR) facilities at Duke University Medical Center (DUMC) and were handled according to the guidelines defined by the United States Animal Welfare Act and in full compliance with the DUMC Institutional Animal Care and Use Committee (IACUC). The aerosol infection models were reviewed and approved by the DUMC IACUC under IACUC protocol #A185-11-07. Six week old A/Jcr mice (Cat. No. 01A24, NCI-Frederick) or 10-week-old C57BL/6 mice (Cat. No. 00064, Jackson Labs) were used. Mice were acclimatized in the facility for one week prior to aerosol challenge and were housed in hermetically sealed cages at 21°C and 50% humidity with a 12 hr light/12 hr dark.
To simulate aerosol generation in nature, and with the aim of increasing the dose presented to the mice, we air dried cells grown in broth or grew cells on agar. After observing an increased dose presented, we extended the aerosol trials to include mice. For proof of principle, eight C57BL/6 mice were used to confirm that we were able to deliver and retain viable Cryptococcus cells within mouse lungs (dose retained). Dose retained was determined from viable CFUs from the lung homogenates. Four mice each were exposed to aerosolized C. gattii strain EJB18 grown in YPD broth and air-dried or cells grown on Arabidopsis agar. Mice were exposed to aerosolized Cryptococcus in the whole-body exposure chamber for 20 min. at standardized aerobiology conditions (13 lpm air-flow-rate, 19 PSI, and 70% RH). At one hour post-exposure, mice were humanely euthanized, and lung, spleen, and brain tissues were sterilely collected in a BLSII hood and homogenized in 2 mL of 1× phosphate buffered saline (PBS). Then 200 µL directly from stock and from a 1∶10 dilution was plated on YPD agar in triplicate, incubated two to four days, and CFUs were calculated to determine the doses retained in the mouse lungs. Dose retained within the lungs is a function of aerodynamic particle size, dose exposed to, and respiratory rate and volume in the animal model.
We extended our validated protocol to assess the delivery of aerosolized yeast and spores. Eight A/Jcr mice per group were exposed to aerosolized H99 (α), KN99 (a), or a yeast-spore mixture obtained from mated mixtures in the whole-body exposure chamber for one hour at standardized aerobiology conditions (13 lpm air flow-rate, 19 PSI, and 70% RH). Four mice per group were humanly euthanized and lung, spleen, and brain tissues were sterilely collected at one hour and three weeks post exposure and CFUs were determined as previously described.
In an additional experiment, we utilized previously constructed C. neoformans var. grubii strains YSB119 (α) and KN99a NEO1, which contain selectable markers for nourseothricin (NAT) or neomycin (NEO) resistance to determine if we could track the contribution of spores versus yeast throughout the aerosolization process and to the infection process (Table S2). We aimed to determine if spores survived nebulization, aerosolized, or were retained within the lungs better than yeast cells. Strains were grown individually in YPD broth under drug selection and spot plated on V8 agar (without drug addition) alone or in combination as described to induce mating. After independent culture, YSB119 and KN99a NEO1 cells were harvested from V8 plates and diluted to 1×108 cell/mL. Then 10 mL of each cell preparation were mixed together to constitute the mixed inoculum without spores. Pre-nebulization, post-nebulization, BioSampler, and mouse tissue CFUs were determined by plating on YPD agar, YPD containing NAT (100 mg/mL), YPD containing NEO (200 mg/mL), and YPD containing both NAT and NEO. All statistical analyses were carried out using Prism (Version 5.04). Linear regression was used to assess correlations, and the Student’s T-test was used for pairwise comparisons.
Results
Effects of Growth Media and Humidity on Spray Factor and Viable Aerosol Concentration
Spray factor, the ratio of viable aerosol output divided by the original suspension concentration, throughout the 11 trial experiments varied from to 4×10−11 to 1.4×10−7 (Table S2). Higher spray factor was correlated with increased dispersal and exposure to infectious particles during aerosol exposure. Increased Spray factor was observed for cells of C. neoformans var. grubii (H99) and C. gattii (EJB18) grown on Arabidopsis leaf (p = 0.003) and V8 agar (p = 0.008) in comparison to YPD broth-grown cells at 70% and 95% relative humidity (RH) (Figure 1a and Table S2). A significantly higher log spray factor for broth-grown cells was observed at 95% RH in comparison to aerosolization at 70% RH (Figure 1a, p<0.01). Growth on agar and increased humidity increased the viable aerosol concentration (BioSampler and dose presented) of C. neoformans var. grubii (H99) and C. gattii (EJB18) inside the Madison Chamber. At 70% (p = 0.03) and 95% RH (p = 0.005), growth on agar resulted in significantly higher values for viable aerosols than the broth-grown cells (Figure 1b). Viable BioSampler concentrations in preliminary experiments at 70% RH ranged from 3.0×101 cell/mL (broth-grown) to 8.5×105 cells/mL (Arabidopsis agar-grown). A significant positive correlation between RH and spray factor was observed for C. neoformans var. grubii (Figure 2a, p = 0.005, R2 = 0.802). A similar but non-significant trend was also observed in C. gattii (Figure 2c, p = 0.06, R2 = 0.532) and within a single trial with C. neoformans var. neoformans (JEC21) cells (Table S2, Trial #9 data).
Figure 1. Growth on agar increases spray factor and viable aerosols for Cryptococcus during aerosolization in the Madison Chamber.
(A) Log spray factor of C. neoformans var. grubii (H99) and C. gattii (EJB18) plotted in respect to growth condition and relative humidity. High humidity increases spray factor for broth-grown cells. (B) Log aerosol concentration of C. neoformans var. grubii (H99) and C. gattii (EJB18) plotted in respect to growth condition and relative humidity. Growth on agar increased log aerosol concentrations. Triangle, square, or circles represent growth conditions denoted in the x-axis for each individual exposure Mean values (n = 3–7, +/− SEM) are plotted. * indicates p<0.01.
Figure 2. Aerosolization at high humidity increases spray factor and viable aerosols for Cryptococcus during aerosolization in the Madison Chamber.
(A and C ) Log spray factor plotted against relative humidity for broth-grown (A) C. neoformans var. grubii H99 (p = 0.0005, R2 = 0.802) and (C) C. gattii EJB18 (p = 0.06, R2 = 0.534). Spray factor increases with relative humidity. (B and D) Log aerosol concentration plotted against relative humidity for broth-grown (B) C. neoformans var. grubii H99 (p = 0.011, R2 = 0.57) and (D) C. gattii EJB18 (p = 0.12, R2 = 0.402). Log aerosol concentration increases with relative humidity. Filled circles and squares represent C. neoformans var. grubii (H99) and open circles and squares represent C. gattii EJB18.
Increased BioSampler concentrations were obtained with increasing humidity and increasing duration of exposure (Table S2). A direct and significant correlation between increased humidity and increased aerosol concentration was observed for H99 (Figure 2b, p = 0.011, R2 = 0.57). A non-statistical trend was observed for C. gattii EJB18 (Figure 2d, p = 0.12, R2 = 0.402) and C. neoformans var. neoformans (JEC21, Table S2, trial #9). Aerosol concentration of H99 and EJB18 showed greater stability at high humidity and greater variability was observed at lower humidity (Figure 2b and 2d). The highest and most consistent aerosol concentrations were obtained from growth on V8 agar media and aerosolization at 95% humidity for one hour (Figure 1 and Table S2).
Aerosol Delivery to Mice
Mice were exposed to aerosols in three independent trial experiments (Table S2, Trial #5, #8, and #11). Delivery of aerosolized Cryptococcus cells to the lungs of mice was successful in all three trials. In the first preliminary mouse exposure, only 50% (EJB18, YPD-dried) to 75% (EJB18, Arabidopsis leaf agar) of mice exposed to aerosolized C. gattii at 70% RH for 20 minutes retained viable C. gattii one hour post exposure (Table S2). We observed that 100% of mice retain viable C. neoformans var. grubii cells within the lungs when mice were exposed for one hour at 95% RH. C. neoformans var. grubii H99α, KN99a, or the mated mixture with spores were aerosolized in individual exposures and the average doses retained were 2.07×102 cells/lung, 3.52×102 cells/lung, and 4.88×102 cells/lung, respectively (Figure 3). Although the dose retained for the mated mixtures with spores was 1.4× (KN99a) and 2.4× (H99α) higher than yeast alone, this difference was not statistically significant. Spleen and brain tissues were not colonized by Cryptococcus one hour post-exposure (Figure 3a). Over the 24 day duration of the experimental exposure, 100% of the mice displayed patterns of weight loss and significant increase of tissue burdens (lung, brain, and spleen), and one mouse expired prior to the 24 day time-point, consistent with disease development (Figure 3b). Additionally, we used C. neoformans var. grubii strains YSB119α and KN99a NEO1 with drug resistance markers nourseothricin (NAT, YSB119α) or neomycin (NEO, KN99a NEO1) (Table S1). To consolidate exposures, we mixed YSB119α and KN99a NEO1 in equal portions to use as the yeast only control for the mated mixtures containing yeast and spores. Similarly, we observed a lower dosed retained in mice exposed to yeast alone (8.8×101 cells/mouse) versus the yeast and spore mixtures (1.5×102 cells/mouse) (Figure 4). Although dose retained for the mated mixtures with spores was 1.7× higher than yeast alone (YSB119α and KN99a NEO1), this difference was not statistically significant. We were able to track the presence of NAT-, NEO-, and NAT- and NEO- resistant colonies from aerosolization to post-nebulization. Doubly-drug resistant colonies represented less than 0.001% of the cells obtained pre- and post-nebulization. We also noticed a large disparity between total CFUs on YPD agar and total CFUs of NEO-resistant cells, indicating a generalized loss of the NEO resistance marker (data not shown). After 3–4 weeks incubation we observed a loss of NEOR cells in the KN99a NEO1 strain incubated alone or in combination with YSB119α. After growth on V8 only 35% of the non-mated NEO marked parental strains retained NEO resistance. In contrast, 83% of NEOR cells isolated from mating colonies retained the NEO selective marker. After observing the loss of NEO resistance in the KN99a NEO1 strain we confirmed that the NEO marker was episomal and not integrated by PCR, which prevented the extension of this experiment to explore the role of spores in the infection process. In summary, we obtained NAT- and NEO-resistant colonies but did not observe double-drug resistant colonies in the BioSampler or from the lung tissues of mice.
Figure 3. Aerosol delivery of C. neoformans var. grubii yeast and yeast-spore mixtures.
(A) C. n.var. grubii, H99 (MATα), KN99a, and a mated mixture containing spores was effectively delivered to mice as assayed by dose retained in lungs (95% RH and 1 hour exposure). (A) Increased tissue burden, dissemination to the brain, and (B) decreased weight was observed at 24 days post-exposure and demonstrates the developmental sequelae of cryptococcosis. No Cryptococcus colonization was obtained from brain or spleen tissues 1 hour post exposure. No significant differences in CFUs were observed between H99 (MATα), KN99a, or mated mixtures (with spores) at any 1 or 25 days post exposure. Mean value (n = 3–4, +/− SEM) are plotted.
Figure 4. Aerosol delivery of C. neoformans var. grubii (YSB119α and KN99a NEO1) yeast and yeast-spore mixtures.
Animal exposure was performed utilizing Cryptococcus strains containing different drug resistance markers. NATR (Red), NEOR (Green), and NATR+NEOR (Purple) resistant colonies were obtained pre- and post- nebulization in similar proportions indicating both yeast and spores equally survive nebulization. NATR+NEOR resistant colonies were not observed from the BioSampler or lung tissues and we observed a net loss of total cell density (Blue) from post-nebuliztion to BioSampler, to dose presented, and to dose retained.
Aerodynamic Particle Size, Concentration, and Stability during Aerosol Exposure
Aerodynamic particle characteristics mass mean aerodynamic diameter (MMAD), count mean aerodynamic diameter (CMAD), and geometric standard deviation (GSD) were determined in 8 out of the 11 independent trial exposures involving C. neoformans var. neoformans, C. neoformans var. grubii, and C. gattii (Figure S2). The MMAD distributions were similar but variable among trials and ranged from 1.0 µm–4.0 µm (Figure S2 and Table S2). However, the MMAD indicates that the particle size was consistently in the respirable range of less than 5 µm except for trial 6 in which the particle sizes ranged from 9.7 µm to 17.2 µm. The observed MMAD, CMAD, and GSD for C. neoformans and C. gattii were similar. MMAD of C. gattii ranged from 1.0 µm to 4 µm, CMAD 0.67 to 0.90, and GSD ranged from 1.2 to 2.95. The MMAD of C. neoformans var grubii ranged from 1.6 µm to 2.1 µm, CMAD 0.78 to 0.83, and GSD ranged from 1.8 to 2.74. In general, the observed MMAD was most consistent at 95% humidity. Particle size distribution by count and mass was variable and ranged up to 10 µm, but a higher concentration and mass of particles less than 5 µm were observed (Figure S3 to S6). The particle density and mass profiles for C. gattii (EJB18) were consistent over increased aerosolization time from 20 to 60 minutes (Figure S3). Agar grown C. gattii EJB18 exhibited one major particle diameter peak around 3 µm (Figure S4a, b, c) in contrast to the two peaks (∼1.5 µm and 3 µm) observed for broth grown cells (Figure S3). Agar grown Cryptococcus cells (Figure S4a, b, c) exhibited more consistent aerodynamic particle diameter, mass, and density profiles versus YPD-broth (Figure S4d, e) or YPD-dried cells (Figure S4f). During aerosolization of C. neoformans var. grubii (H99) particle density, mass, and size profiles remained consistent between 45% to 95% humidity (Figure S5). Broth-grown H99 cells (Figure S5) display a broad aerodynamic particle range in contrast to C. gattii EJB18 broth grown (Figure S3) or agar grown EJB18 cells (Figure S4). Particle mass, size, and density distributions were similar between H99 yeast cells and the mated mixture containing yeast and spores (Figure S6). The size of aerosolized C. neoformans var. grubii particles was stable during 20 minutes of exposure at 45%, 70% and 95% humidity (Figure S7a). The coefficient of variance of MMAD was 3.12% (45% RH), 3.54% (70% RH), and 3.83% (95% RH). The concentration of particles generated was more consistent at 70% and 95% RH than with 45% RH (Figure S7b). The coefficient of variance related to cell density was higher for 45% RH than 70% RH and 95% RH.
Discussion
This is the first published report demonstrating the use of the Madison Chamber, coupled sampling port, and software for the aerosol delivery of Cryptococcus to mice. We demonstrated the ability to aerosolize viable C. neoformans and C. gattii cells in the respirable range and produced adequate delivery to initiate disease in the murine host. The multiplex sampling port and coupled software interface allowed measurement of viable aerosol concentration, spray factor, dose presented and particle size characteristics. These data were essential to define the aerobiology parameters for optimal and consistent delivery of Cryptococcus to murine lungs and to understand the characteristics of aerosolized Cryptococcus from generation to delivery and retention.
Preliminary trials were performed to confirm the viability of Cryptococcus cells post-nebulization and the quantity of viable cells in the respirable range (<10 µm) for deposition in the lungs. Initial inoculum density was maximized to 1×108 cells/mL in a 20 mL volume because the viability of Cryptococcus post-nebulization had not been previously determined, and yeast cells were larger (up to 10 µm) than typical aerosolized pathogens [55], [59]. Our inoculum density is similar to previous reports used to initiate C. neoformans, Aspergillus, and bacterial aerosol infections, but our experiments utilized greater volume than previously reported [50], [53], [54], [56], [59]. The density of viable Cryptococcus yeast cells post-nebulization was equal to or greater than the inoculum density pre-nebulization. High viability post-nebulization was correlated with high spray factor. We observed a higher spray factor for Cryptococcus than previously reported for the primary bacterial pathogens Yersinia pestis, Bacillus anthracis, and Mycobacterium tuberculosis [59], [62]–[65]. Cryptococcus is an environmental organism that has been isolated from plants, soil, and guano. Cryptococcus may survive nebulization better than other organisms because in the environment, it can survive harsh conditions for long periods of time, withstanding desiccation, natural irradiation, passage through hosts, and temperature fluctuations, possibly due to its polysaccharide capsule and ability to melanize [66]. The small increases in cell density observed post-nebulization are most likely an artifact from loss of water volume during the nebulization process and not a result of increasing viability or replication. In all except one trial, MMAD was less than 5 µm and within the respirable range. In the outlier, Trial #6, MMAD ranged from 9 µm to 17 µm. We suspect that the unusual MMAD observed may be the result of inefficient desiccation of yeast cells prior to re-suspension, non-standard growth conditions, or unexplained measurement malfunction during aerosolization.
The initial two trials utilizing broth grown cells resulted in very low and inconsistent dose presented (<70 cells/mouse). Sukroongreung et al. (1989) estimated the spore load inhaled by a mouse in 15 minutes of exposure at 103 to 106 cells, which exceeded what we observed [53]. Effective aerosol delivery of bacteria to murine lungs has been reported with doses presented in the range of 100 to 600 cells/mouse [59]. Aerosol delivery of Aspergillus fumigatus conidia resulted in a dose retained of 103 cells/mouse [55], [56]. We aimed to obtain a calculated dose presented greater than 100 cells/mouse to maximize dose delivered and retention within the mouse lungs because Cryptococcus yeast cells are larger than Aspergillus conidia and most bacterial cells delivered through aerosol challenge, 1 µm to 10 µm versus 0.9 µm to 2 µm, respectively [23], [67], [68]. This principle was confirmed since the observed range of MMAD, CMAD, and GSD for of aerosolized Cryptococcus cells was larger and more variable than previously reported for various bacteria and fungi [59], [67]–[69]. We observed a broadened aerodynamic size range for C. gattii comprised of two distinct peak concentrations in comparison to C. neoformans, which displayed a smaller size range with one single broad peak size range. These observations appear consistent with previous morphological descriptions detailing the larger and more elongated yeast and spore morphology of C. gattii in contrast to the more consistent and round cell morphology of C. neoformans [70].
In an attempt to maximize the dose presented, we grew Cryptococcus cells under different laboratory conditions that would more closely mimic conditions in nature, resulting in substrate-grown and/or desiccated cells. Cells were grown in YPD broth and subjected to natural desiccation overnight or grown on Arabidopsis agar. During the third trial, we were able to monitor relative humidity by an additional integrated package that was previously unavailable. We observed increased viable BioSampler concentrations, which resulted in a calculated dose presented that attained our predicted cut off value of >100 cells/mouse for both YPD-dried and Arabidopsis agar-grown cells. The increase in the calculated dose presented could be attributed to both 70% relative humidity and/or changes in the cell preparation methodology, and additional experimentation will be necessary to establish the most important factor. Both relative humidity and cell growth conditions are known to impact survival and infectivity for bacteria and viruses [71]–[78]. Additional trial exposures (without mice) were conducted to expand the aerosol model to C. neoformans and attempt to determine whether humidity was an important factor for a successful Cryptococcus aerosol model. We observed a linear increasing trend for C. neoformans var. grubii in BioSampler cell densities and dose presented that were positively correlated with increased humidity. These experiments in combination with additional experiments confirmed that increased humidity during aerosoliation increased viable aerosol and dose presented for C. neoformans var. neoformans (JEC21), C. neoformans var. grubii (H99), and C. gattii (EJB18). Following this finding, all aerosolization experiments were conducted at optimal humidity (RH = 95%). Humidity has been implicated to be a key factor in the viability and transmission of influenza (virus) and Francisella tularensis (bacteria) [77]–[79]. Low humidity is associated with increased transmission and high humidity is associated with lower transmission of influenza [77], [79]. Increased environmental isolation of Cryptococcus gattii has been associated with the flowering of Eucalyptus in Australia and during dry, hot summers in Vancouver, BC and the pacific northwestern USA [24]. The finding that humidity can influence viability and transmission of infectious propagules for fungal, viral, and bacterial pathogens suggests factors that may contribute to seasonal patterns of disease incidence.
Following the preliminary experiments, we immediately moved to conducting whole-body aerosol exposures in mice (Trial #5) upon observing a dose presented >100 cells/mouse in order to test the developmental criteria of the Cryptococcus aerosol exposure model. Mice were exposed to C. gattii cells from YPD-dried or Arabidopsis agar preparations. The dose presented from Arabidopsis agar preparations were slightly higher than those observed from the mice exposed to broth-grown and dried cells. The ability to precisely determine viable aerosols presented in the BioSampler, dose presented, and dose retained in this experiment utilizing the Madison Chamber sets these studies apart from all previously published reports on the aerosolization of Cryptococcus. Soil seeding and four-day exposure limited determining dose exposed and dose retained in mice that were exposed to Cryptococcus via soil exposure methodology. Exposure via the jet nebulizer or Henderson Chamber is much more similar to exposure via the Madison Chamber and hence is more readily comparable. Smith et al (1964) reported that mice received ∼104 viable cells, but this value far exceeds CFUs obtained post mouse expiration and there is no methodology on the specifics of how this value was calculated. It is likely that what Smith et al. (1964) describes as “viable cells received” is comparable to either our BioSampler densities or viable dose presented to mice, and the former is most likely [50]. Our BioSampler densities were ∼103 cells/mL to 105 cells/mL which is similar to the observed “viable cells received” reported.
In our study, 50% of the mice exposed to the YPD-dried preparation had quantifiable dose retained in their lungs versus 75% of the mice exposed to the Arabidopsis agar preparation. These rates of initial colonization are similar to the 67% to 100% terminal infection reported by Smith et al. in 1964 and much higher than the 20% mortality rate obtained by Sukroongreung et al. (1998) [50], [53]. In contrast, aerosol delivery of Aspergillus to immuno-compromised mice resulted in much higher lung burden (dose retained) but resulted in only 60 to 70% mortality [55]. CFUs observed in the lungs were variable between 0–50 cells/mouse, and only a fraction of the dose presented was retained in the lungs. Some cells may be lodged in the upper respiratory system (nose, mouth, or throat), and may not immediately contribute to the dose retained. Cells deposited in the upper and lower respiratory tracts could be cleared by exhalation, killed by host defense mechanisms, or contribute to later development of infection. Actual dose presented and dose retained for each mouse was not addressed in Fu et al. (2012) but average CFUs in lung, blood, and brain tissues were determined at three and seven days post-exposure [54]. Dose retained at one-hour post-exposure in our experiments were much lower than CFUs obtained in other reports at three days, seven days, or post-expiration. In previous reports, the time-point chosen to assay dose retained varies from immediately post-exposure to 24 hrs depending on the organism. Our aim was to obtain the viable number of Cryptococcus cells that reached the lungs, minimizing the potential effects of killing by the host’s immunological system, replication, or dissemination of Cryptococcus. We defined the dose retained at the one-hour time-point because previous studies have indicated that macrophages can quickly initiate killing within two hours and significant differences in killing of Cryptococcus can be observed within one to four hours of in vitro incubation with macrophages and neutrophils [19], [40], [44]. Furthermore, previous reports have established that development of disseminated Cryptococcus does not often occur within hours of intranasal exposure because macrophages are implicated in the exit of Cryptococcus from the lungs, dissemination through the blood, passage through the blood brain barrier, and colonization of the brain [80]–[85]. We expect the initial dose retained to increase over time when Cryptococcus is able to survive and replicate within the host.
Additional animal exposures were performed with mated and non-mated C. neoformans var. grubii H99 (α) and KN99 (a) in order to determine if the low dose retained was sufficient to initiate disease development and whether the presence of spores enhanced Cryptococcus viability during aerosoliation, increased dose retained, or enhanced development of disease. H99α and KN99a yeast were aerosolized separately or from a mated mixture containing hyphal fragments, yeast, and spores. No significant differences were observed in pre-nebulization, post-nebulization, BioSampler, spray factor, or dose presented between α or a yeast cells alone or in combination with spores. We observed higher but non-significant dose retained in mice exposed to the mated mixtures (yeast+spores). Over time, it is possible that these small differences could be important for disease progression and the development of systemic cryptococcosis. As expected, spleen and brain tissues were not colonized by Cryptococcus at one hour post exposure. Over the 24-day duration of the experiment, 100% of mice displayed patterns of weight loss and significant increase of tissue burdens (lung, brain, and spleen), and one mouse expired prior to the 25-day time-point, consistent with disease development. The observed increase in CFUs, dissemination to the brain, weight loss, and death indicate progressive disease development consistent with murine infection models. We did not observe any significant differences in viable densities post-nebulization or particle size, mass, or density distribution of yeast cells or mated mixtures containing spores. Our data suggests that both yeast cells and spores are similar in their ability to survive nebulization and can share overlapping particle size, mass, and density characteristics during aerosolization. Increased relative humidity may contribute to increased viability of desiccated yeast cells post-nebulization. Our data supports the idea that in nature, both yeast cells and spore can serve as infectious particles and that risk to Cryptococcus infection may differ seasonally with varying environmental factors such as humidity. Additional research will be needed to determine if differences exist in the ability of spores versus yeast cells to traverse into the lower respiratory tract, or to enter, survive, replicate, or exit macrophages contributing to enhanced infectivity or dissemination. We demonstrated that at 95% relative humidity, we obtained 100% infection of mice, which exceeds infection efficiency reported for aerosol delivery via crude soil seeding, jet nebulizer, or Henderson apparatus [50], [53], [54].
Conclusions
We developed a well-validated aerosol model for Cryptococcus utilizing the Madison Chamber and coupled instrumentation. Our data suggest a novel role of relative humidity in the viability and infectivity of Cryptococcus. We define inoculum growth, cell density, and aerobiology delivery parameters for consistent delivery of low numbers of Cryptococcosis to murine lungs.
Supporting Information
Madison Chamber and whole body exposure system. (A and C) Overview of the Aerosol exposure system contained within the Class III BioSafety Cabinet (2). (B) The Madison Chamber (1) is attached to a Class III BioSafety Cabinet (2). AeroMP (3) collects data from the chamber pressure sensor (3), Relative humidity and temperature probe (4), and (5) Aerodynamic particle sizer (APS). AeroMP (3) also controls the nebulizer flow, dilution airflow (6), humidifier, BioSampler, and APS. Mice were exposed in a whole body exposure apparatus (7) housed within the Madison Chamber. Directional flow is delineated with red arrows, Collison nebulizer (8), and Impinger (9).
(PNG)
Mass Median Aerodynamic Diameter (MMAD) of Cryptococcus during aerosolization in the Madison Chamber. MMAD of all trials appeared relatively consistent between 1.5 µm and 4.0 µm except for trial #6. MMAD for trial #11 was not plotted because APS was not functional. No significant differences were observed in the MMAD of C. neoformans var. grubii (H99) and C. gattii (EJB18).
(TIF)
Aerodynamic particle characteristics of C. gattii remain consistent during aerosolization over different lengths of time. Trial #2 (A) EJB18 30 min.; (B) EJB18 45 min.; (C) EJB18 60 min. YPD-broth grown and RH undetermined. Particle mass (open blue triangle) and particle density (open red circle) plotted respective to particle diameter.
(TIF)
Aerodynamic particle characteristics of C. gattii during aerosolization. Agar grown cells display more consistent particle size, mass, and density in comparison to cells grown in YPD-broth. The aerodynamic properties of cells grown in broth with drying are more consistent then cells prepared for aerosolization without drying. (A) EJB18 Arabidopsis agar, Trial #3; (B) EJB18 Arabidopsis agar, Trial #3; (C) EJB18 Arabidopsis agar, Trial #5; (D) EJB18 YPD-dried; Trial # 5 (E) EJB18 YPD-dried; Trial #10 (F) EJB18 YPD-broth 45% RH. Particle mass (open blue triangle) and particle density (open red circle) plotted respective to particle diameter.
(TIF)
Aerodynamic particle characteristics of C. neoformans var. grubii remain stable during aerosolization at different relative humidity. Trial #7 YPD-broth grown (A) H99α 45% RH; (B) H99α 70% RH; (C) H99α 95% RH. Particle mass (open blue triangle) and particle density (open red circle) plotted respective to particle diameter.
(TIF)
Aerodynamic particle characteristics of C. neoformans var. grubii yeast cells and mated mixtures containing spores are similar. (E) H99α, Trial #8; (F) Mated mixture H99α+KN99a+Spores, Trial #8. Particle mass (open blue triangle) and particle density (open red circle) plotted respective to particle diameter. Cells were V8 agar grown and aerosolized at 95% relative humidity.
(TIF)
Cryptococcus particle size stability and density during aerosol exposure. Aerodynamic mass distribution was measured using the Aerodynamic Particle Sizer (APS) connected to the Madison Chamber. Sampling was done for 30 seconds, every 2 minutes for 20 minutes during Trial #10a. During aerosolization MMAD (A) and aerosol concentration (B) of C. neoformans var. grubii (H99) were measured. MMAD remains consistent when aerosolized at different relative humidity. Particle density changes over the 20 min exposure when aerosolized at 45% humidity but remains constant when aerosolized at 70% and 95% relative humidity. Circle 45% RH, square 70% RH, triangle 95% RH.
(TIF)
Strains utilized in this study.
(DOC)
Aerosol experiments. Eleven independent aerosol trial experiments were performed to optimize viable aerosol, dose presented and dose retained in the mouse model host. Three independent trials included mice (Trials #5, #8, and #11).
(DOC)
Acknowledgments
We are indebted to Sarah Seay and Ching-Ju Chen from the Animal Core at Duke RBL, and Wenjun Li and Anna Floyd-Averette from the Heitman laboratory for assistance with animal handling and necropsies. We also thank Scott Alderman for his assistance in developing the standard operating procedures for handling Cryptococcus and for decontamination of the aerosol exposure chamber.
Funding Statement
The work was support by the 2010–11 Dean Voucher Program, UC6 AI058607 (Regional Biocontainment Laboratory at Duke), and NIH/NAID R37 AIO39115-15 to JH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, et al. (2009) Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23: 525–530. [DOI] [PubMed] [Google Scholar]
- 2. Hagen F, Colom MF, Swinne D, Tintelnot K, Iatta R, et al. (2012) Autochthonous and dormant Cryptococcus gattii infections in Europe. Emerging Infectious Diseases 18: 1618–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, et al. (2011) Clonality and alpha-a recombination in the Australian Cryptococcus gattii VGII population–an emerging outbreak in Australia. PLOS ONE 6: e16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sellers B, Hall P, Cine-Gowdie S, Hays AL, Patel K, et al. (2012) Cryptococcus gattii: An emerging fungal pathogen in the Southeastern United States. The American Journal of the Medical Sciences 343: 510–511. [DOI] [PubMed] [Google Scholar]
- 5. Loperena-Alvarez Y, Ren P, Li X, Schoonmaker-Bopp DJ, Ruiz A, et al. (2010) Genotypic characterization of environmental isolates of Cryptococcus gattii from Puerto Rico. Mycopathologia 170: 279–285. [DOI] [PubMed] [Google Scholar]
- 6. Cogliati M, Chandrashekar N, Esposto MC, Chandramuki A, Petrini B, et al. (2012) Cryptococcus gattii serotype-C strains isolated in Bangalore, Karnataka, India. Mycoses 55: 262–268. [DOI] [PubMed] [Google Scholar]
- 7. McCulloh RJ, Phillips R, Perfect JR, Byrnes EJ, 3rd, Heitman J, et al (2011) Cryptococcus gattii genotype VGI infection in New England. The Pediatric Infectious Disease Journal 30: 1111–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baro T, Torres-Rodriguez JM, Morera Y, Alia C, Lopez O, et al. (1999) Serotyping of Cryptococcus neoformans isolates from clinical and environmental sources in Spain. Journal of Clinical Microbiology 37: 1170–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iatta R, Hagen F, Fico C, Lopatriello N, Boekhout T, et al. (2011) Cryptococcus gattii Infection in an immunocompetent patient from Southern Italy. Mycopathologia 174: 87–92. [DOI] [PubMed] [Google Scholar]
- 10. Chaturvedi S, Dyavaiah M, Larsen RA, Chaturvedi V (2005) Cryptococcus gattii in AIDS patients, southern California. Emerging Infectious Diseases 11: 1686–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Albuquerque PC, Rodrigues ML (2012) Research trends on pathogenic Cryptococcus species in the last 20 years: a global analysis with focus on Brazil. Future Microbiology 7: 319–329. [DOI] [PubMed] [Google Scholar]
- 12. Tay ST, Rohani MY, Soo Hoo TS, Hamimah H (2009) Epidemiology of cryptococcosis in Malaysia. Mycoses 53: 509–514. [DOI] [PubMed] [Google Scholar]
- 13. Litvintseva AP, Thakur R, Reller LB, Mitchell TG (2005) Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. Journal of Infectious Diseases 192: 888–892. [DOI] [PubMed] [Google Scholar]
- 14. Kang Y, Tanaka H, Moretti ML, Mikami Y (2009) New ITS genotype of Cryptococcus gattii isolated from an AIDS patient in Brazil. Microbiology and Immunology 53: 112–116. [DOI] [PubMed] [Google Scholar]
- 15. Aminnejad M, Diaz M, Arabatzis M, Castaneda E, Lazera M, et al. (2012) Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia 173: 337–346. [DOI] [PubMed] [Google Scholar]
- 16. Capoor MR, Mandal P, Deb M, Aggarwal P, Banerjee U (2008) Current scenario of cryptococcosis and antifungal susceptibility pattern in India: a cause for reappraisal. Mycoses 51: 258–265. [DOI] [PubMed] [Google Scholar]
- 17. Chen SC, Slavin MA, Heath CH, Playford EG, Byth K, et al. (2012) Clinical manifestations of Cryptococcus gattii infection: determinants of neurological sequelae and death. Clinical Infectious Diseases 55: 789–798. [DOI] [PubMed] [Google Scholar]
- 18. Xue C, Tada Y, Dong X, Heitman J (2007) The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host and Microbe 1: 263–273. [DOI] [PubMed] [Google Scholar]
- 19. Springer DJ, Ren P, Raina R, Dong Y, Behr MJ, et al. (2010) Extracellular fibrils of pathogenic yeast Cryptococcus gattii are important for ecological niche, murine virulence and human neutrophil interactions. PLOS ONE 5: e10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Botes A, Boekhout T, Hagen F, Vismer H, Swart J, et al. (2009) Growth and mating of Cryptococcus neoformans var. grubii on woody debris. Microbial Ecology 57: 757–765. [DOI] [PubMed] [Google Scholar]
- 21. Nielsen K, De Obaldia AL, Heitman J (2007) Cryptococcus neoformans mates on pigeon guano: Implications for the realized ecological niche and globalization. Eukaryotic Cell 6: 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruiz A, Bulmer GS (1981) Particle size of airborn Cryptococcus neoformans in a tower. Applied Environmental Microbiology 41: 1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neilson JB, Fromtling RA, Bulmer GS (1977) Cryptococcus neoformans: size range of infectious particles from aerosolized soil. Infection and Immunity 17: 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, et al. (2007) Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Applied Environmental Microbiology 73: 1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen LC, Goldman DL, Doering TL, Pirofski L, Casadevall A (1999) Antibody response to Cryptococcus neoformans proteins in rodents and humans. Infection and Immunity 67: 2218–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramsay S (2001) C. neoformans common in urban children. Lancet 357: 1507. [DOI] [PubMed] [Google Scholar]
- 27. Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J (2009) Spores as infectious propagules of Cryptococcus neoformans . Infection and Immunity 77: 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM (2009) Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryotic Cell 8: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giles SS, Dagenais TR, Botts MR, Keller NP, Hull CM (2009) Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans . Infection and Immunity 77: 3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, et al. (2009) The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proceedings of the National Academy of Sciences 106: 12980–12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staib F, Mishra SK (1975) Selective involvement of the brain in experimental murine cryptococcosis. I. Microbiological observations. Zentralbl Bakteriol [Orig A] 232: 355–364. [PubMed] [Google Scholar]
- 32. Lim TS, Murphy JW, Cauley LK (1980) Host-etiological agent interactions in intranasally and intraperitoneally induced cryptococcosis in mice. Infection and Immunity 29: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carroll SF, Guillot L, Qureshi ST (2007) Mammalian model hosts of cryptococcal infection. Comparative Medicine 57: 9–17. [PubMed] [Google Scholar]
- 34. Geunes-Boyer S, Oliver TN, Janbon G, Lodge JK, Heitman J, et al. (2009) Surfactant protein D increases phagocytosis of hypocapsular Cryptococcus neoformans by murine macrophages and enhances fungal survival. Infection and Immunity 77: 2783–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ganendren R, Carter E, Sorrell T, Widmer F, Wright L (2006) Phospholipase B activity enhances adhesion of Cryptococcus neoformans to a human lung epithelial cell line. Microbes and Infection 8: 1006–1015. [DOI] [PubMed] [Google Scholar]
- 36. Gross NT, Camner P, Chinchilla M, Jarstrand C (1998) In vitro effect of lung surfactant on alveolar macrophage defence mechanisms against Cryptococcus neoformans . Mycopathologia 144: 21–27. [DOI] [PubMed] [Google Scholar]
- 37. Mershon KL, Vasuthasawat A, Lawson GW, Morrison SL, Beenhouwer DO (2009) Role of complement in protection against Cryptococcus gattii infection. Infection and Immunity 77: 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ngamskulrungroj P, Chang Y, Sionov E, Kwon-Chung KJ (2012) The primary target organ of Cryptococcus gattii is different from that of Cryptococcus neoformans in a murine model. mBio 3: e00103–00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mednick AJ, Feldmesser M, Rivera J, Casadevall A (2003) Neutropenia alters lung cytokine production in mice and reduces their susceptibility to pulmonary cryptococcosis. European Journal of Immunology 33: 1744–1753. [DOI] [PubMed] [Google Scholar]
- 40. Wozniak KL, Vyas JM, Levitz SM (2006) In vivo role of dendritic cells in a murine model of pulmonary cryptococcosis. Infection and Immunity 74: 3817–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marroni M, Pericolini E, Cenci E, Bistoni F, Vecchiarelli A (2007) Functional defect of natural immune system in an apparent immunocompetent patient with pulmonary cryptococcosis. Journal of Infection 54: e5–e8. [DOI] [PubMed] [Google Scholar]
- 42. Voelz K, Lammas DA, May RC (2009) Cytokine signaling regulates the outcome of intracellular macrophage parasitism by Cryptococcus neoformans . Infection and Immunity 77: 3450–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weinberg PB, Becker S, Granger DL, Koren HS (1987) Growth inhibition of Cryptococcus neoformans by human alveolar macrophages. American Review of Respiratory Disease 136: 1242–1247. [DOI] [PubMed] [Google Scholar]
- 44. Srinivas U, Kar R, Saxena R, Prasad PH (2008) Cryptococcal neoformans profiles in peripheral blood neutrophils: an unusual presentation. Indian Journal of Pathology Microbiology 51: 296–297. [DOI] [PubMed] [Google Scholar]
- 45. Johnston SA, May RC (2013) Cryptococcus interactions with macrophages: evasion and manipulation of the phagosome by a fungal pathogen. Cellular Microbiology 15: 403–411. [DOI] [PubMed] [Google Scholar]
- 46. Schoffelen T, Illnait-Zaragozi MT, Joosten LA, Netea MG, Boekhout T, et al. (2013) Cryptococcus gattii induces a cytokine pattern that is distinct from other cryptococcal species. PLOS ONE 8: e55579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wright L, Bubb W, Davidson J, Santangelo R, Krockenberger M, et al. (2002) Metabolites released by Cryptococcus neoformans var. neoformans and var. gattii differentially affect human neutrophil function. Microbes and Infection 4: 1427–1438. [DOI] [PubMed] [Google Scholar]
- 48. Cheng PY, Sham A, Kronstad JW (2009) Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans . Infection and Immunity 77: 4284–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guillot L, Carroll SF, Badawy M, Qureshi ST (2008) Cryptococcus neoformans induces IL-8 secretion and CXCL1 expression by human bronchial epithelial cells. Respiratory Research 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith CD, Ritter R, Larsh HW, Furcolow ML (1964) Infection of white swiss mice with airborne Cryptococcus neoformans . Journal of Bacteriology 87: 1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kwon-Chung KJ, Edman JC, Wickes BL (1992) Genetic association of mating types and virulence in Cryptococcus neoformans . Infection and Immunity 60: 602–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barchiesi F, Cogliati M, Esposto MC, Spreghini E, Schimizzi AM, et al. (2005) Comparative analysis of pathogenicity of Cryptococcus neoformans serotypes A, D and AD in murine cryptococcosis. Journal of Infection 51: 10–16. [DOI] [PubMed] [Google Scholar]
- 53. Sukroongreung S, Kitiniyom K, Nilakul C, Tantimavanich S (1998) Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans . Medical Mycology 36: 419–424. [PubMed] [Google Scholar]
- 54. Fu M, Wang Y, Lin Y, Zhang L, Luo W, et al. (2012) Fungal infection induced by Cryptococcus neoformans aerosol inhalation in immunosuppressed Balb/c mice. Journal of Southern Medical University 32: 169–172. [PubMed] [Google Scholar]
- 55. Sheppard DC, Rieg G, Chiang LY, Filler SG, Edwards JE Jr, et al. (2004) Novel inhalational murine model of invasive pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy 48: 1908–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheppard DC, Graybill JR, Najvar LK, Chiang LY, Doedt T, et al. (2006) Standardization of an experimental murine model of invasive pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy 50: 3501–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hartings JM, Roy CJ (2004) The automated bioaerosol exposure system: preclinical platform development and a respiratory dosimetry application with nonhuman primates. Journal of Pharmacological and Toxicological Methods 49: 39–55. [DOI] [PubMed] [Google Scholar]
- 58. Henderson DW (1952) An apparatus for the study of airborne infection. The Journal of Hygiene 50: 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saini D, Hopkins GW, Chen CJ, Seay SA, Click EM, et al. (2011) Sampling port for real-time analysis of bioaerosol in whole body exposure system for animal aerosol model development. Journal of Pharmacological and Toxicological Methods 63: 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinds WC (1999) Aerosol Technology: Properties, Behavior, and Measurement of Airborne Particles. Canada: John Wiley & Sons. 504 p.
- 61. Guyton AC (1947) Analysis of respiratory patterns in laboratory animals. The American Journal of Physiology 150: 78–83. [DOI] [PubMed] [Google Scholar]
- 62. Heine HS, Bassett J, Miller L, Hartings JM, Ivins BE, et al. (2007) Determination of antibiotic efficacy against Bacillus anthracis in a mouse aerosol challenge model. Antimicrobial Agents and Chemotherepy 51: 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Agar SL, Sha J, Foltz SM, Erova TE, Walberg KG, et al. (2008) Characterization of a mouse model of plague after aerosolization of Yersinia pestis CO92. Microbiology 154: 1939–1948. [DOI] [PubMed] [Google Scholar]
- 64. Agar SL, Sha J, Foltz SM, Erova TE, Walberg KG, et al. (2009) Characterization of the rat pneumonic plague model: infection kinetics following aerosolization of Yersinia pestis CO92. Microbes and Infection 11: 205–214. [DOI] [PubMed] [Google Scholar]
- 65. Nuermberger EL, Yoshimatsu T, Tyagi S, Bishai WR, Grosset JH (2004) Paucibacillary tuberculosis in mice after prior aerosol immunization with Mycobacterium bovis BCG. Infection and Immunity 72: 1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McFadden DC, Casadevall A (2001) Capsule and melanin synthesis in Cryptococcus neoformans . Medical Mycology 39 Suppl 119–30. [PubMed] [Google Scholar]
- 67. Deacon LJ, Pankhurst LJ, Drew GH, Hayes ET, Jackson S, et al. (2009) Particle size distribution of airborne Aspergillus fumigatus spores emitted from compost using membrane filtration. Atmospheric Environment 43: 5698–5701. [Google Scholar]
- 68. Meklin T, Reponen T, Toivola M, Koponen V, Husman T, et al. (2002) Size distributions of airborne microbes in moisture-damaged and reference school buildings of two construction types. Atmospheric Environment 36: 6031–6039. [Google Scholar]
- 69.Nedeljkovic-Davidovic A (2008) Fluence response curves for Bacillus subtilis spores applied to UVC lamps testing: The Pennsylvania State University. 85 p.
- 70. Kwon-Chung KJ (1976) A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 68: 943–946. [PubMed] [Google Scholar]
- 71.Tang JW (2009) The effect of environmental parameters on the survival of airborne infectious agents. Journal of the Royal Society Interface Suppl 6: S737–746. [DOI] [PMC free article] [PubMed]
- 72. Stone RC, Johnson DL (2002) A note on the effect of nebulization time and pressure on the culturability of Bacillus subtilis and Pseudomonas fluorescens . Aerosol Science and Technology 36: 536–539. [Google Scholar]
- 73. Kim S, Ramakrishnan MA, Raynor P, Goyal S (2007) Effects of humidity and other factors on the generation and sampling of a coronavirus aerosol. Aerobiologia 23: 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Marthi B, Fieland VP, Walter M, Seidler RJ (1990) Survival of bacteria during aerosolization. Applied Environmental Microbiology 56: 3463–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McDevitt JJ, Rudnick SN, Radonovich LJ (2012) Aerosol susceptibility of influenza virus to UV-C light. Applied Environmental Microbiology 78: 1666–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bresee J, Hayden FG (2013) Epidemic influenza–responding to the expected but unpredictable. N Engl J Med 368: 589–592. [DOI] [PubMed] [Google Scholar]
- 77. Yang W, Elankumaran S, Marr LC (2012) Relationship between humidity and influenza A viability in droplets and implications for influenza’s seasonality. PLOS ONE 7: e46789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Faith SA, Smith LP, Swatland AS, Reed DS (2012) Growth conditions and environmental factors impact aerosolization but not virulence of Francisella tularensis infection in mice. Frontiers in Cellular and Infection Microbiology 2: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lowen AC, Mubareka S, Steel J, Palese P (2007) Influenza virus transmission is dependent on relative humidity and temperature. PLOS Pathog 3: 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kechichian TB, Shea J, Del Poeta M (2007) Depletion of alveolar macrophages decreases the dissemination of a glucosylceramide-deficient mutant of Cryptococcus neoformans in immunodeficient mice. Infection and Immunity 75: 4792–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Alvarez M, Casadevall A (2006) Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Current Biology 16: 2161–2165. [DOI] [PubMed] [Google Scholar]
- 82. Feldmesser M, Casadevall A (1997) Effect of serum IgG1 to Cryptococcus neoformans glucuronoxylomannan on murine pulmonary infection. Journal of Immunology 158: 790–799. [PubMed] [Google Scholar]
- 83. Gross NT, Nessa K, Camner P, Chinchilla M, Jarstrand C (1997) Interaction between Cryptococcus neoformans and alveolar macrophages. Journal of Medical and Veterinary Mycology 35: 263–269. [PubMed] [Google Scholar]
- 84. Osterholzer JJ, Milam JE, Chen GH, Toews GB, Huffnagle GB, et al. (2009) The role of dendritic cells and alveolar macrophages in regulating early host defense against pulmonary infection with Cryptococcus neoformans . Infection and Immunity 77: 3749–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Charlier C, Chretien F, Baudrimont M, Mordelet E, Lortholary O, et al. (2005) Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. American Journal of Patholology 166: 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Madison Chamber and whole body exposure system. (A and C) Overview of the Aerosol exposure system contained within the Class III BioSafety Cabinet (2). (B) The Madison Chamber (1) is attached to a Class III BioSafety Cabinet (2). AeroMP (3) collects data from the chamber pressure sensor (3), Relative humidity and temperature probe (4), and (5) Aerodynamic particle sizer (APS). AeroMP (3) also controls the nebulizer flow, dilution airflow (6), humidifier, BioSampler, and APS. Mice were exposed in a whole body exposure apparatus (7) housed within the Madison Chamber. Directional flow is delineated with red arrows, Collison nebulizer (8), and Impinger (9).
(PNG)
Mass Median Aerodynamic Diameter (MMAD) of Cryptococcus during aerosolization in the Madison Chamber. MMAD of all trials appeared relatively consistent between 1.5 µm and 4.0 µm except for trial #6. MMAD for trial #11 was not plotted because APS was not functional. No significant differences were observed in the MMAD of C. neoformans var. grubii (H99) and C. gattii (EJB18).
(TIF)
Aerodynamic particle characteristics of C. gattii remain consistent during aerosolization over different lengths of time. Trial #2 (A) EJB18 30 min.; (B) EJB18 45 min.; (C) EJB18 60 min. YPD-broth grown and RH undetermined. Particle mass (open blue triangle) and particle density (open red circle) plotted respective to particle diameter.
(TIF)
Aerodynamic particle characteristics of C. gattii during aerosolization. Agar grown cells display more consistent particle size, mass, and density in comparison to cells grown in YPD-broth. The aerodynamic properties of cells grown in broth with drying are more consistent then cells prepared for aerosolization without drying. (A) EJB18 Arabidopsis agar, Trial #3; (B) EJB18 Arabidopsis agar, Trial #3; (C) EJB18 Arabidopsis agar, Trial #5; (D) EJB18 YPD-dried; Trial # 5 (E) EJB18 YPD-dried; Trial #10 (F) EJB18 YPD-broth 45% RH. Particle mass (open blue triangle) and particle density (open red circle) plotted respective to particle diameter.
(TIF)
Aerodynamic particle characteristics of C. neoformans var. grubii remain stable during aerosolization at different relative humidity. Trial #7 YPD-broth grown (A) H99α 45% RH; (B) H99α 70% RH; (C) H99α 95% RH. Particle mass (open blue triangle) and particle density (open red circle) plotted respective to particle diameter.
(TIF)
Aerodynamic particle characteristics of C. neoformans var. grubii yeast cells and mated mixtures containing spores are similar. (E) H99α, Trial #8; (F) Mated mixture H99α+KN99a+Spores, Trial #8. Particle mass (open blue triangle) and particle density (open red circle) plotted respective to particle diameter. Cells were V8 agar grown and aerosolized at 95% relative humidity.
(TIF)
Cryptococcus particle size stability and density during aerosol exposure. Aerodynamic mass distribution was measured using the Aerodynamic Particle Sizer (APS) connected to the Madison Chamber. Sampling was done for 30 seconds, every 2 minutes for 20 minutes during Trial #10a. During aerosolization MMAD (A) and aerosol concentration (B) of C. neoformans var. grubii (H99) were measured. MMAD remains consistent when aerosolized at different relative humidity. Particle density changes over the 20 min exposure when aerosolized at 45% humidity but remains constant when aerosolized at 70% and 95% relative humidity. Circle 45% RH, square 70% RH, triangle 95% RH.
(TIF)
Strains utilized in this study.
(DOC)
Aerosol experiments. Eleven independent aerosol trial experiments were performed to optimize viable aerosol, dose presented and dose retained in the mouse model host. Three independent trials included mice (Trials #5, #8, and #11).
(DOC)