Summary
Lipid metabolism is tightly controlled by the nutritional state of the organism. Nutrient-rich conditions increase lipogenesis whereas nutrient deprivation promotes fat oxidation. In this study, we identify the mitochondrial sirtuin, SIRT4, as a novel regulator of lipid homeostasis. SIRT4 is active in nutrient-replete conditions to repress fatty acid oxidation while promoting lipid anabolism. SIRT4 deacetylates and inhibits malonyl CoA decarboxylase (MCD), an enzyme that produces acetyl CoA from malonyl CoA. Malonyl CoA provides the carbon skeleton for lipogenesis and also inhibits fat oxidation. Mice lacking SIRT4 display elevated MCD activity and decreased malonyl CoA in skeletal muscle and white adipose tissue. Consequently, SIRT4 KO mice display deregulated lipid metabolism leading to increased exercise tolerance and protection against diet-induced obesity. In sum, this work elucidates SIRT4 as an important regulator of lipid homeostasis, identifies MCD as a novel SIRT4 target, and deepens our understanding of the malonyl CoA regulatory axis.
Introduction
According to the bioenergetic demands of the organism, tissues must appropriately adjust their metabolism to either store lipids or catabolize fatty acids to generate more energy (Duncan et al., 2007; Long and Zierath, 2006). This balance between lipid anabolic and catabolic processes is coordinately and precisely regulated, in part by the cellular levels of the metabolite malonyl CoA (Saggerson, 2008; Saha and Ruderman, 2003). To undergo β-oxidation, fatty acids must cross both the inner and outer mitochondrial membranes, and this rate-limiting step is catalyzed by carnitine palmitoyltransferase 1 (CPT1), which is allosterically inhibited by malonyl CoA. In addition, malonyl CoA serves as the chain-elongating unit for fatty acid synthesis. Thus, regulation of malonyl CoA levels provides a means to control the balance between fat synthesis and fat oxidation. Two enzymes regulate cellular malonyl CoA levels: acetyl CoA carboxylase (ACC) converts acetyl CoA to malonyl CoA and malonyl CoA decarboxylase (MCD) converts it back to acetyl CoA. The regulation of ACC activity through phosphorylation by AMPK is well characterized (Hardie, 2011), whereas the regulation of MCD activity is much less studied.
Sirtuins are NAD+-dependent deacylases and ADP-ribosyltransferases involved in many biological processes, mediating adaptive responses to the cellular environment (Houtkooper et al., 2012; Lombard et al., 2011). Proteomic surveys revealed that a majority of mitochondrial metabolic enzymes are differentially acetylated according to the nutritional state of the cell (Wang et al., 2010; Yang et al., 2011; Zhao et al., 2010) suggesting that acetylation may regulate global cellular metabolism and coordinates fuel switching. Three of the mammalian sirtuins (SIRT3, SIRT4 and SIRT5) are located in the mitochondria and may play roles as sensors of energy status in this organelle (Houtkooper et al., 2012; Lombard et al., 2011). SIRT4 is one of the least characterized mitochondrial sirtuins (Haigis et al., 2006; Ahuja et al., 2007). Previously, it was shown that SIRT4 represses fat catabolism (Nasrin et al., 2010), but the direct substrates involved and the physiological significance remain unknown. Importantly, the role of SIRT4 in lipid synthesis and storage has never been investigated.
In this study, we demonstrate a novel function for SIRT4 in the regulation of lipid metabolism. We find that SIRT4 represses fatty acid oxidation in skeletal muscle and stimulates lipogenesis in white adipose tissue (WAT), indicating that SIRT4 can regulate the balance between fat oxidation and fat synthesis. To achieve this regulation, SIRT4 directly binds, deacetylates and represses malonyl CoA deacarboxylase (MCD). As a consequence, SIRT4 KO mice display deregulation of crucial physiologic aspects of lipid metabolism leading to increased tolerance to exercise challenge and protection against diet-induced obesity.
Results
SIRT4 promotes lipid synthesis and represses fatty acid oxidation
To assess the role of SIRT4 on lipid homeostasis, we initially examined its role in de novo lipid synthesis. As WAT is the major organ responsible for lipid synthesis and storage, we tested the role of SIRT4 on lipogenesis in the mouse adipocyte cell line, F442A. We found that overexpression of SIRT4 leads to an increase in lipogenesis, as measured by [14C]-acetate incorporation into the lipid fraction (Figure 1A), and an increase in accumulation of trigycerides (TG) and stored lipids (Figure 1B and 1C). This increase was not observed in cells overexpressing the catalytic mutant of SIRT4, SIRT4H162Y (Ahuja et al., 2007). To confirm these results using a primary model, we examined lipogenesis using adipocytes freshly isolated from WAT from SIRT4 WT and KO mice. As observed in F442A cells, lipid synthesis was decreased in SIRT4 KO primary adipocytes (Figure 1D). Together, these results suggest that SIRT4 activity promotes fat anabolism.
Figure 1. SIRT4 regulates lipid metabolism.
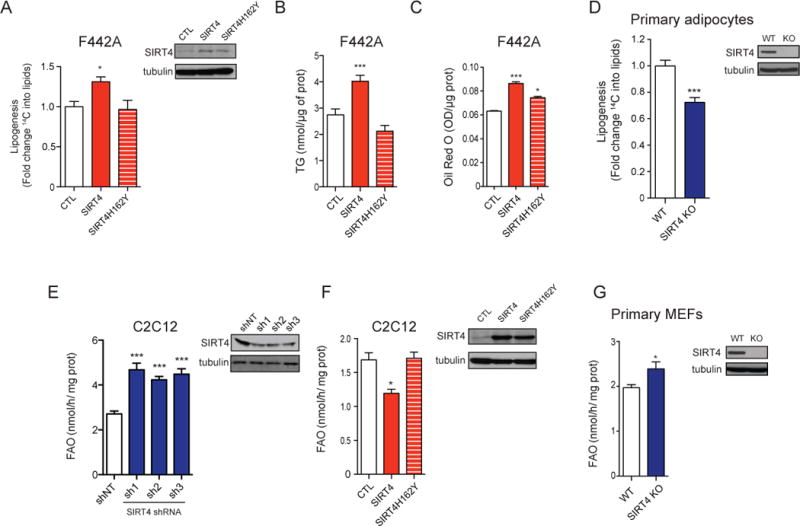
(A) Lipogenesis was measured using 14C-acetate (n=3), (B) Triglyceride levels (n=6), and (C) Oil Red O staining (n=6) were determined using F442A adipocytes stably expressing empty vector control (CTL, open bar), SIRT4 (red bar) or the catalytic inactive mutant of SIRT4, SIRT4H162Y (red striped bar). (D) Lipogenesis was measured using 14C-acetate in WT (open bar) and SIRT4 KO (blue bar) primary adipocyte lines (n=3). (E) Fatty acid oxidation (FAO) was measured in C2C12 cells expressing control shRNA (shNT, open bar) or shRNAs targeted against SIRT4 (blue bars) (n=3). (F) FAO was measured in C2C12 cells overexpressing empty vector control, SIRT4 or SIRT4H162Y (n=3). (G) FAO was determined using WT and SIRT4 KO primary MEF lines (n=3). Levels of SIRT4 protein were determined by Western blotting using antibodies to SIRT4 and tubulin as a loading control. In each panel, data represent mean ± SEM. (*) p < 0.05; (**) p < 0.01, (***) p < 0.001.
To assess the role of SIRT4 in lipid catabolism, we measured fatty acid oxidation in C2C12 cells, a mouse myocyte cell line. As muscles rely primarily on fatty acids for energy, myocytes provide a good model to study fatty acid oxidation. We found that the oxidation of palmitate, a saturated long chain fatty acid, was significantly higher in C2C12 cells in which SIRT4 expression was stably reduced by lentiviral expression of three independent shRNAs against SIRT4 compared to control cells (Figure 1E). Conversely, palmitate oxidation was diminished in C2C12 cells stably overexpressing SIRT4, but not in cells overexpressing SIRT4H162Y (Figure 1F). Similarly, fatty acid oxidation was elevated in primary SIRT4 KO mouse embryonic fibroblasts (MEFs) (Figure 1G). These data demonstrate for the first time that SIRT4 promotes lipogenesis and represses fatty acid oxidation, indicating that SIRT4 may coordinate the balance between lipid catabolic and anabolic pathways.
SIRT4 represses Malonyl CoA Decarboxylase (MCD)
As SIRT4 represses palmitate oxidation and stimulates lipogenesis, we reasoned that SIRT4 regulates an enzyme positioned at the interface between oxidative and synthetic pathways. One major branch point in lipid homeostasis is the interconversion of acetyl CoA to malonyl CoA, a metabolite that inhibits fat oxidation, while promoting fat synthesis (Figure 2A). Two enzymes regulate cellular malonyl CoA levels: acetyl CoA carboxylase (ACC) converts acetyl CoA to malonyl CoA, and malonyl CoA decarboxylase (MCD) converts it back to acetyl CoA. Together these enzymes constitute a highly responsive control system modulating the levels of malonyl CoA and thereby the rate of fatty acid catabolism or synthesis (Figure 2A). To test this idea, we determined whether SIRT4 physically binds with ACC or MCD in co-immunoprecipitation studies. We did not detect an interaction between SIRT4 and ACC (Figure S1A). While MCD localizes both to the cytosol and the mitochondrial matrix, in skeletal muscle the majority of MCD activity resides within mitochondria (Kerner and Hoppel, 2002). Thus, we investigated and observed a physical interaction between SIRT4 and SIRT4H162Y with mitochondrial MCD (Figure 2B and 2C). We confirmed that SIRT4 and MCD colocalize in mitochondria using confocal microscopy (Figure 2D). In contrast to SIRT4, the other mitochondrial sirtuins, SIRT3 and SIRT5, showed no detectable physical association with MCD in control immunoprecipitations under these conditions, further indicating a specific interaction between SIRT4 and MCD.
Figure 2. SIRT4 interacts and represses MCD activity.
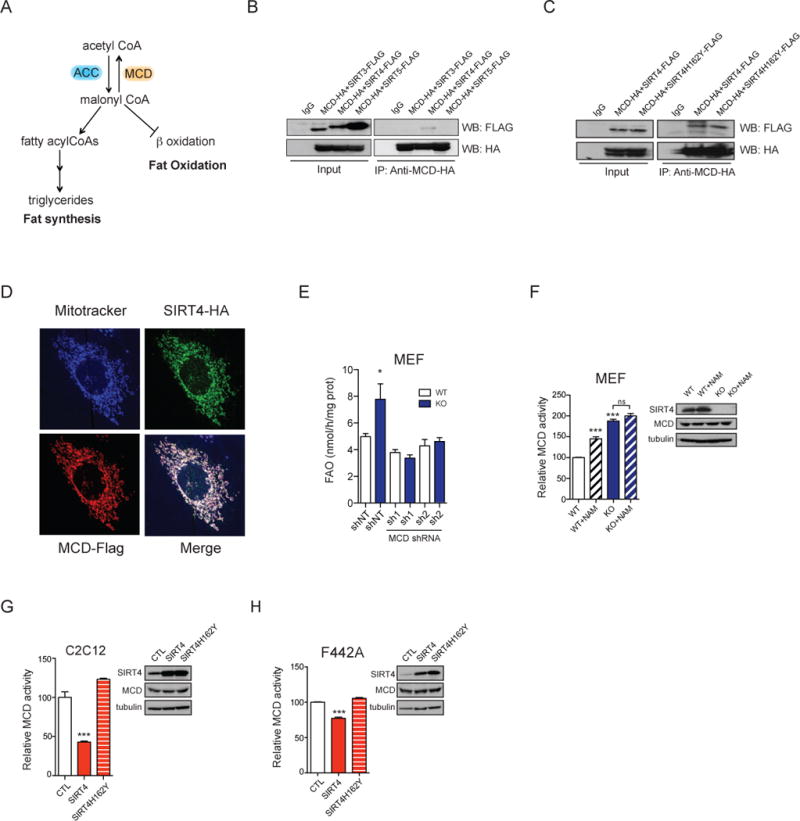
(A) Schematic of the regulation of lipid homeostasis and malonyl CoA by acetyl-CoA carboxylase (ACC) and malonyl CoA decarboxylase (MCD). (B) Sirtuin-MCD interactions were assessed by cotransfecting expression vectors for SIRT3, SIRT4 or SIRT5 (FLAG-tagged at the C-terminus) with an expression vector for C-terminal HA-tagged MCD in HEK293T cells. HA-tagged MCD was immunoprecipitated and interactions were detected by immunoblotting with antibodies against FLAG. (C) Expression vectors containing SIRT4 or SIRT4H162Y (FLAG-tagged) were co-transfected with C-terminal HA-tagged MCD in HEK293T cells, and SIRT4-MCD binding was assessed by immunoprecipitation of MCD-HA and Western blotting with FLAG antibodies. (D) The subcellular localization of SIRT4-HA (green) and MCD-FLAG (red) stably overexpressed in immortalized MEFs was examined by immunofluorescence using HA and FLAG antibodies and the mitochondrial marker Mitotracker (pseudo-colored in blue). (E) FAO rates were assessed in WT and SIRT4 KO primary MEFs treated with control shRNA (shNT) or two shRNAs against MCD (sh1 and sh2) as indicated (n=3). (F) Relative MCD activity in SIRT4 WT and SIRT4 KO immortalized MEFs treated or not with nicotinamide (NAM; striped bars) (n=3). (G–H) Relative MCD activity in C2C12 (n=3) (G) or in F442A cells (n=3) (H) overexpressing empty vector control, SIRT4 or SIRT4H162Y. Levels of SIRT4 and MCD proteins were determined by Western blotting using antibodies for SIRT4 and MCD and tubulin as a loading control. In each panel, data represent mean ± SEM. (*) p < 0.05; (**) p < 0.01, (***) p < 0.001. See also Figure S1.
We next investigated the importance of MCD activity in SIRT4-mediated regulation of lipid homeostasis. We observed that overexpression of MCD in C2C12 and F442A cells increases fatty acid oxidation rates and repressed lipogenesis, respectively (Figure S1B–1D). Thus, MCD overexpression phenocopied SIRT4 deletion. To assess directly whether SIRT4 regulates lipid homeostasis through MCD, we analyzed fatty acid oxidation in SIRT4 WT and KO MEFs in which MCD expression was stably reduced by lentiviral expression of shRNA against MCD (Figure S1E). Reduction of MCD abrogated the increased fatty acid oxidation found in SIRT4 KO cells to levels comparable to WT cells (Figure 2E), demonstrating that the increased fatty acid oxidation in SIRT4 KO cells required MCD activity.
Based on these findings, we hypothesized that MCD activity would be elevated in SIRT4 KO cells, decreasing malonyl CoA levels, subsequently increasing fat oxidation. In agreement with this model, MCD activity was elevated 2-fold in SIRT4 KO MEFs compared to WT MEFs (Figure 2F). Next, to confirm that acute modulation of SIRT4 activity can regulate MCD, we treated cells with a pan-sirtuin inhibitor nicotinamide (NAM). We found that MCD activity was significantly increased in the WT cells treated with NAM compared to the untreated cells. This is due to inhibition of SIRT4, as NAM had no further effect in SIRT4 KO cells (Figure 2F). To confirm these findings, we further examined the effect of SIRT4 on MCD activity in muscle and adipocyte cells, using C2C12 and F442A cell lines stably expressing WT or H162Y SIRT4. In both cell types, overexpression of SIRT4 resulted in reduced MCD activity, whereas overexpression of the catalytic mutant had no effect (Figure 2G and 2H). Taken together, these data suggest that SIRT4 represses MCD activity in muscle and adipocyte cell lines.
SIRT4 deacetylates MCD
Our results demonstrate that in cells SIRT4 binds MCD and represses its enzymatic activity. We sought to assess whether this repression is mediated by post-translational modification. Although they belong to the same protein family, sirtuins exhibit several enzymatic activities, including deacetylation, deacylation, and ADP-ribosylation. Previous reports found that SIRT4 is a weak ADP-ribosyltransferase but not a BSA or histone deacetylase (Ahuja et al., 2007; Haigis et al., 2006; Schwer et al., 2002). To identify posttranslational modifications of MCD, we immunoprecipitated MCD from C2C12 cells and analyzed posttranslational modifications by mass spectrometry. MCD was previously shown to be acetylated on 6 different lysines (K58, K167, K210, K316, K388, and K444) (Nam et al., 2006). Our analysis identified additional acetylation on lysine residue 471 (Figure S2A), but did not detect ADP-ribosylation, malonylation or succinylation. Notably, K471 is one of the most conserved lysines of MCD and is invariant from D.Rerio to humans (Figure S2B). Hence, we speculated that SIRT4 may have a substrate-specific deacetylase activity, as has been demonstrated for SIRT6 and SIRT7 (Barber et al., 2012; Zhong et al., 2010).
To test whether MCD acetylation level is regulated by SIRT4, we stably expressed a FLAG-tagged murine MCD in MEFs and treated cells with or without NAM. MCD acetylation was measured after immunoprecipitation of MCD by western blotting with anti-acetyl lysine antibody. We found that NAM treatment increased MCD acetylation levels (Figure 3A). We also detected an increase in the acetylation level of MCD in the SIRT4 KO cells compared to the WT cells (Figure 3B). Furthermore, SIRT4 overexpression reduced MCD acetylation (Figure 3C). These data demonstrate that SIRT4 regulates the acetylation levels of MCD in cells.
Figure 3. SIRT4 deacetylates MCD.
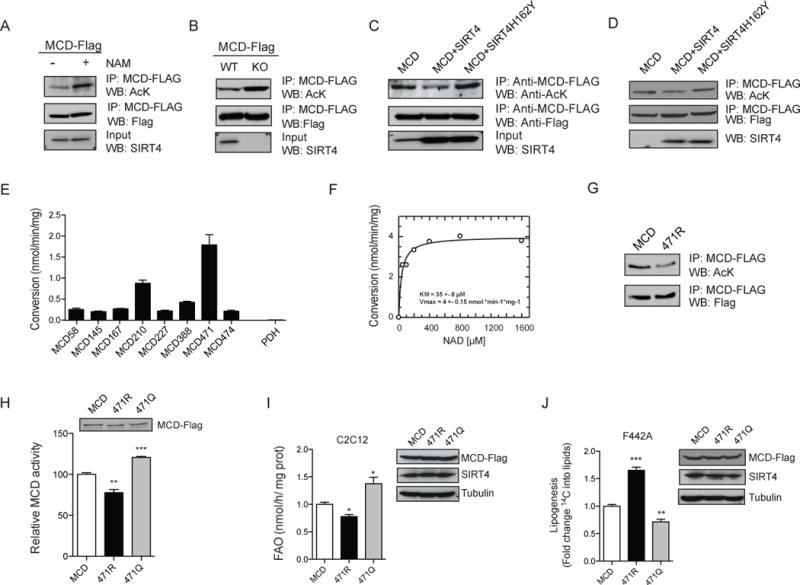
(A) MCD acetylation was measured in WT immortalized MEFs before or after treatment with NAM. FLAG-tagged MCD was stably overexpressed in WT MEFs treated with (+) or without (−) NAM and immunoprecipitated using antibodies against FLAG. MCD acetylation levels were assessed with antibodies against acetyl-lysine (AcK). (B) MCD acetylation was assessed using WT and SIRT4 KO MEFs as described for panel A. (C) MCD acetylation was measured in C2C12 cells stably overexpressing FLAG-tagged MCD and SIRT4 or SIRT4H162Y. After immunoprecipitation of MCD with anti-FLAG antibodies, acetylation was measured as for panel A. (D) In vitro deacetylation assay was performed using immunopurified MCD and SIRT4. FLAG-MCD was immunoprecipitated from MEFs and incubated with FLAG-SIRT4 and FLAG-SIRT4H162Y immunoprecipitated from HEK293 cells and MCD acetylation status assessed by Western blot. (E) Recombinant SIRT4 was incubated with synthesized acetylated peptides of MCD and peptide deacetylation was assessed using mass spectrometry. Acetylated peptide from pyruvate dehydrogenase (PDH) was included as a negative control (n=3). (F) Acetylated peptide was incubated with SIRT4 and NAD+ concentrations were varied as indicated. Peptide deacetylation levels were analyzed by LC-MS. (G) Constructs encoding MCD, MCD K471R or MCD K471Q were expressed in HEK 293T cells and MCD activity was measured (n=4). (H–I) Retrovirus used to generate stable C2C12 (H) and F442A (I) cell lines overexpressing MCD, MCD K471R or MCD K471Q where FAO rates and lipogenesis were assessed (n=3). In each panel, data represent mean ± SEM. (*) p < 0.05; (**) p < 0.01, (***) p < 0.001. See also Figure S2 and S6.
To test whether SIRT4 can directly deacetylate MCD, we assessed the ability of SIRT4 to deacetylate MCD protein in vitro. We incubated MCD-FLAG with SIRT4-FLAG and SIRT4H162Y immunoprecipitated from HEK293T cells in the presence of NAD+. We observed that SIRT4 directly deacetylates MCD in vitro, whereas SIRT4H162Y does not (Figure 3D). Since SIRT4 interacts with SIRT3 (Ahuja et al., 2007), we wanted to exclude the possibility that the observed deacetylation was due to a contamination by SIRT3. We performed in vitro deacetylation assays using SIRT4 immunoprecipitated from HEK293T cells where SIRT3 was stably reduced by shRNA (Figure S2C) and immunoprecipitated MCD from SIRT3 KO cells. We then observed that SIRT4 isolated from SIRT3-deficient cells could deacetylate MCD (Figure S2D). Next, to determine whether the deacetylation of MCD by SIRT4 regulates its activity, we measured MCD activity after in vitro deacetylation by SIRT4. MCD activity was reduced after incubation with SIRT4, but not with SIRT4H162Y (Figure S2E). Together, these results demonstrate for the first time that SIRT4 has a substrate-specific deacetylase activity, and that MCD is a novel target of SIRT4.
To elucidate which residues on MCD are deacetylated by SIRT4, we performed a series of mass spectrometry-based deacetylation assays using chemically synthesized acetylated peptides of MCD. We found that SIRT4 deacetylated lysine 471 of MCD with the highest efficiency but presented little activity against other acetylated MCD peptides (Figure 3E, Figure S2F and Table S1). Providing an additional negative control, SIRT4 did not deacetylate pyruvate dehydrogenase (PDH), which is readily deacetylated by SIRT3 (data not shown), suggesting that SIRT4 deacetylase activity is substrate-specific (Figure 3E), and also not due to contaminant deacetylase activity. Moreover, this activity was NAD+-dependent and the SIRT4 used in this assay showed a Km for NAD+ comparable to that of other sirtuins (Figure 3F). Next we investigated the contribution of K471 to the total acetylation level and enzymatic activity of MCD. We tested the acetylation level of K471R MCD mutant, which cannot be acetylated on that residue, and found that K471R MCD was notably less acetylated than wild-type MCD (Figure 3G). To confirm the significance of K471 deacetylation to MCD enzyme function, we tested the enzymatic activity of K471R and K471Q MCD mutants, which mimic constitutive deacetylation and acetylation, respectively. K471R MCD had a reduced enzymatic activity (Figure 3H), whereas the K471Q variant had elevated activity (Figure 3H). These data indicate that SIRT4 possesses direct deacetylase activity on K471 of MCD in vitro and acetylation of this residue accounts for a significant contribution to the total level of MCD acetylation and activity.
Finally, we examined the consequences of K471 acetylation status on palmitate oxidation and lipogenesis. In accordance with the effect observed on its enzymatic activity, the K471R MCD variant diminished fat oxidation whereas K471Q MCD enhanced fat oxidation (Figure 3I). Similarly, lipogenesis was promoted by K471R MCD and repressed by K471Q MCD (Figure 3J). Of note, mutation of K210 in MCD, which was a weaker peptide SIRT4 substrate, did not alter fat oxidation or lipogenesis (Figure S2D and S2E). These results show that the acetylation status of the K471 residue of MCD regulates both lipogenesis and FAO.
SIRT4 deacetylates MCD in vivo during the fed state
We next sought to understand the physiological relevance of the biochemical regulation of MCD by SIRT4. During periods of nutritional abundance, when surplus metabolic intermediates funnel into fatty acid synthesis and energy storage, the steady-state levels of malonyl CoA rise (Saggerson, 2008). This in turn prevents entry of fatty acids into mitochondria and dampens fat oxidation, while promoting fat synthesis (Figure 4A, left panel). Conversely, in the fasted state, when malonyl CoA is low, fatty acids are transported into the mitochondria and undergo β-oxidation (Figure 4A, right panel). Thus, malonyl CoA level is tightly linked to the nutritional state of the organism. As malonyl CoA is the substrate for MCD, we hypothesized that the regulation of MCD by SIRT4 might be linked to nutritional status of mice. To test this idea, we measured SIRT4 protein levels during the fed and fasting state in WT mice. SIRT4 levels decreased with fasting in muscle (Figure 4B) and WAT (Figure 4C), supporting the idea that SIRT4 may be important for the regulation of lipid metabolism during nutrient rich conditions.
Figure 4. SIRT4 deacetylates MCD in vivo during the fed state controlling malonyl CoA levels.
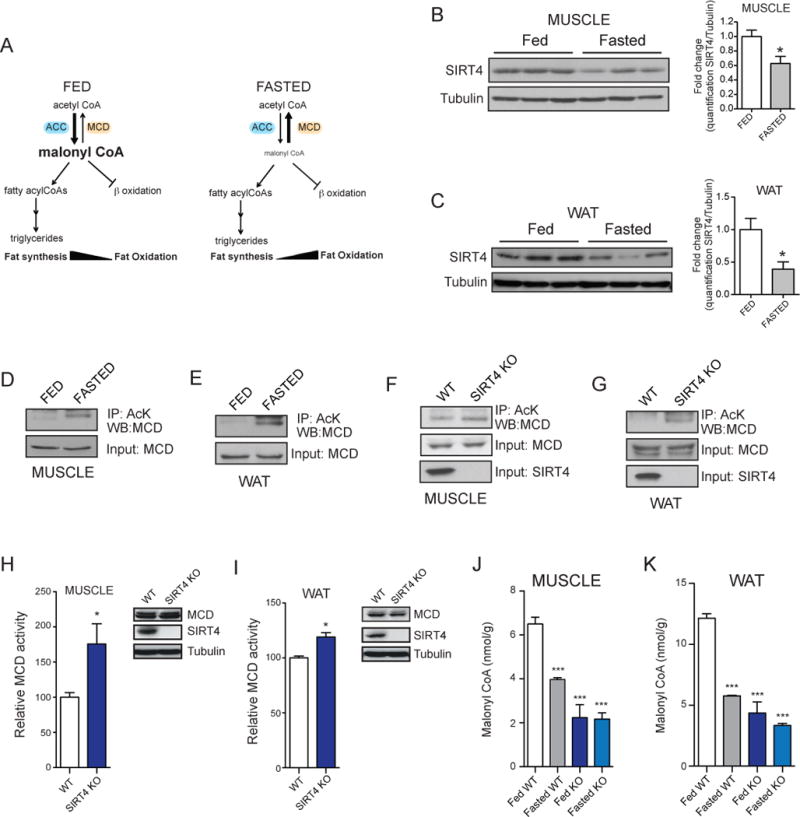
(A) Schematic of the regulation of lipid homeostasis during fed and fasted state. (B–C) Muscle (soleus) and WAT were harvested from mice under fed or fasted conditions and analyzed for SIRT4 expression by Western blot and normalized to tubulin. Images were quantified (right panels) for the SIRT4/tubulin ratio using Image J. (D–E) Muscle (soleus) and WAT extracts from WT mice fed or fasted were immunoprecipitated with a monoclonal AcK antibody and blotted for MCD. (F–G) Muscle (soleus) and WAT extracts from WT and SIRT4 KO fed mice were immunoprecipitated with a monoclonal AcK antibody and blotted for MCD. (H–I) Relative MCD activity in muscle (quadriceps) and WAT from WT and SIRT4 KO fed mice (n=4 per genotype). (K–L) Malonyl CoA levels were measured in muscle (quadriceps) and WAT from WT (open and grey bars) and SIRT4 KO (dark and light blue bars) mice under fed and fasted conditions (n=4 per genotype, per condition). In each panel, data represent mean ± SEM. (*) p < 0.05; (**) p < 0.01, (***) p < 0.001.
As SIRT4 represses MCD activity via deacetylation, we reasoned that MCD acetylation would be regulated by nutrient status. We assessed the acetylation level of MCD in muscle and WAT in WT mice under fed and fasted conditions. Proteins were immunoprecipitated with monoclonal anti-acetyl-lysine antibody and analyzed by western blot with MCD antibody. We found that MCD was deacetylated in muscle and WAT of fed mice (Figure 4D and 4E). Importantly, MCD is hyperacetylated in muscle and WAT of SIRT4 KO mice compared to WT (Figure 4F and 4G), demonstrating that SIRT4 is necessary for MCD deacetylation in vivo under fed conditions.
To determine whether SIRT4 represses MCD activity in vivo, we measured MCD activity in muscle and WAT of WT and SIRT4 KO mice in the fed state and found that, as in cells, MCD activity was significantly increased in SIRT4 KO compared to WT tissues (Figure 4H and 4I). Since MCD regulates lipid metabolism by lowering malonyl CoA levels, we hypothesized that SIRT4 may control malonyl CoA levels in response to nutrient availability. As expected, in both muscle and WAT of WT mice, malonyl CoA levels diminished with fasting (Figure 4J and 4K). Strikingly, SIRT4 deletion reduced malonyl CoA levels in both muscle and WAT during the fed state and abolished the switch between high and low malonyl CoA levels in the fed versus fasted state (Figure 4J and 4K). Thus, these studies identify SIRT4 as a physiological regulator of malonyl CoA levels in vivo.
Alteration of lipid metabolism in SIRT4 KO mice
As MCD activity and malonyl CoA levels were altered in SIRT4 KO mouse, we examined potential physiological indicators of dysregulation in lipid metabolism. We measured the lipid composition of WAT and skeletal muscle isolated from WT and SIRT4 KO mice under fed and fasted conditions and observed that the levels of triglycerides and phospholipids in SIRT4 KO mice under fed conditions trended to the levels observed in WT and SIRT4 KO mice under fasting conditions (Figure 5A–C). Moreover, the differences between fed and fasted levels of triglycerides and phospholipids were blunted in SIRT4 KO tissues. These observations indicated that SIRT4 loss in WAT and muscle may in part mimic a fasted state of lipid metabolism during fed conditions, and dampens the physiological switch to low nutrient conditions.
Figure 5. SIRT4 KO mice display an altered lipid metabolism in vivo.
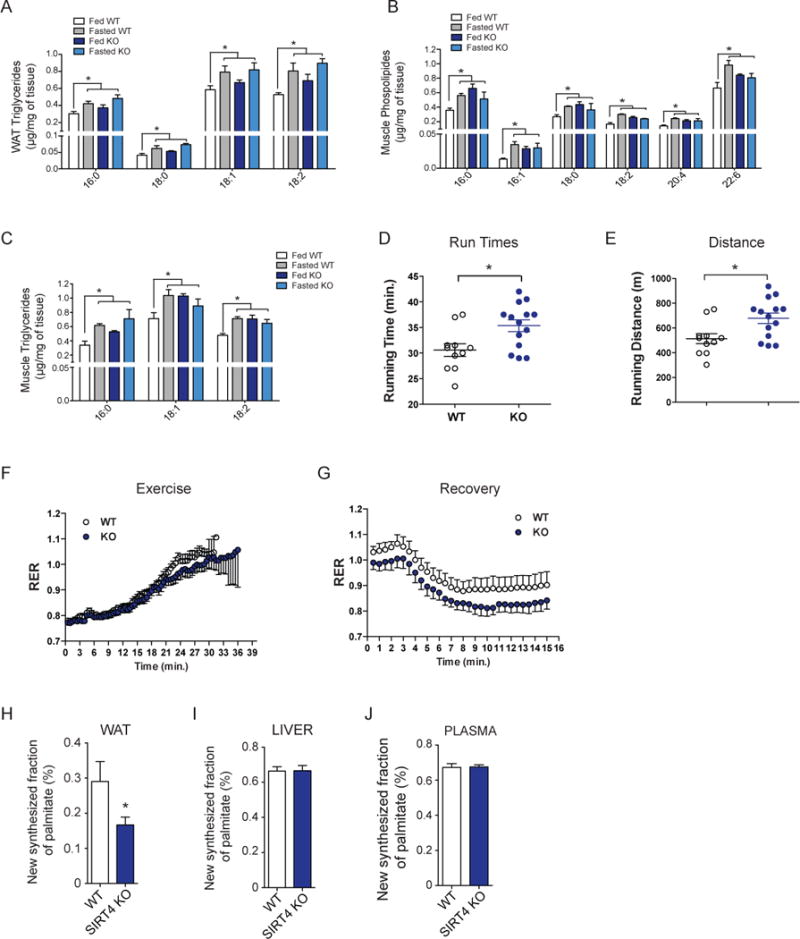
(A) Triglyceride composition from WAT from WT and SIRT4 KO fed and fasted mice (n=3 mice per genotype, per condition). (B–C) Skeletal Muscle (quadriceps) phospholipid and triglyceride composition from WT and SIRT4 KO fed and fasted mice (n=3 mice per genotype, per condition). (D–E) Exercise tolerance assays were performed on WT and SIRT4 KO mice (n=11–14 per genotype). (F–G) RER in WT and SIRT4 KO mice during exercise (F) and recovery (G) (n=11–14). (H–J) De novo lipogenesis in vivo was measured by determining incorporation of deuterated water into palmitate in WAT (H), liver (I), and plasma (J) (n=6 per genotype). In each panel, data represent mean ± SEM. (*) p < 0.05; (**) p < 0.01, (***) p < 0.001. See also Figure S3.
We next examined the physiological relevance of low malonyl CoA levels in fat oxidation and lipid synthesis. Exercise capacity is one well-established physiological readout of fatty acid oxidation. For example, AMPK activation increases skeletal muscle oxidative capacity and exercise endurance in mice (Narkar et al., 2008; Thomson et al., 2007). As SIRT4 loss increased MCD activity resulting in low malonyl CoA level, we speculated that a metabolic shift toward lipid utilization in SIRT4 KO mice might augment exercise performance. Using untrained SIRT4 KO and WT littermate controls, we found that SIRT4 KO mice ran 20% further distance and longer running times during a graded, maximal treadmill challenge (Figure 5D and 5E). Whole body indirect calorimetry showed that the respiratory exchange ratio (RER) of SIRT4 KO mice trended lower at high workloads and throughout 15 min of recovery (Figure 5F and G), suggesting increased lipid oxidation during these periods. We did not detect any major changes in the fiber types of SIRT4 WT and KO mice (Figure S3A). As well, blood glucose and lactate levels, measured 15 min after exercise, were similar between genotypes (Figure S3B). To test the possibility that these phenotypes stem from a general increase in mitochondrial content or function, we examined mitochondrial DNA content, but did not observe differences in muscle or WAT of SIRT4 WT and KO mice (Figure S3C). In addition, we did not detect gross abnormalities in mitochondrial ultrastructure analyzed by electron microscopy (Figure S3D). Finally, we do not see a significant difference in mitochondrial and fat oxidation gene expression in the muscle of SIRT4 KO mice compared to WT muscle (Figure S3E). Thus, these results are consistent with an increase in fat oxidation in SIRT4 KO mice, leading to higher exercise capacity.
To examine whether SIRT4 loss affected lipid synthesis in vivo, we monitored de novo lipogenesis by measuring incorporation of deuterated water into palmitate, as previously described (Edmond et al., 1998; Lee et al., 1994). Deuterium atoms exchange with hydrogen atoms on different carbon positions of glucose metabolites as they go through the glycolytic/gluconeogenic pathways and the TCA cycle (Radziuk and Pye, 2002), and the accumulation of these deuterated metabolites can contribute to deuterium labeling of fatty acids during de novo lipogenesis. The extent of labeled palmitate, representing de novo lipid synthesis, can be measured by GC-MS. We observed a 50% reduction in the percentage of newly synthesized lipids in SIRT4 KO WAT compared to WT tissue, highlighting the importance of SIRT4 to de novo lipogenesis in vivo (Figure 5H). This effect is not observed in liver and plasma where we found that palmitate synthesis was identical in WT and SIRT4 KO animals (Figure 5I and 5J). Malonyl CoA levels were also not different in livers of SIRT4 KO mice (Figure S3F), consistent with previous studies identifying a major role for MCD in organs other than the liver (Ruderman et al., 2003). Overall, these results suggest that SIRT4 plays a major role in lipid homeostasis by repressing fat oxidation and promoting lipid anabolism in vivo.
SIRT4 deletion protects against dietary induced obesity
Increased lipogenesis can contribute to obesity. We therefore investigated whether the decrease in lipogenesis observed in SIRT4 KO mice would correspond with a change in weight gain. On a standard low fat diet (LFD), SIRT4 KO mice have normal growth curves (Figure 6A), and do not display differences in adipose fat mass (Figure S4A–4D) or serum lipid profiles (Figure S4E–4H). Strikingly, when SIRT4 KO mice were placed on a high fat diet (HFD), their weight gain remained similar to the mice under LFD, and was significantly less than the weight gain of WT mice under HFD (Figure 6A). Thus, we show for the first time that loss of SIRT4 protects mice from HFD-induced obesity.
Figure 6. SIRT4 KO mice are protected from diet-induced weight gain.
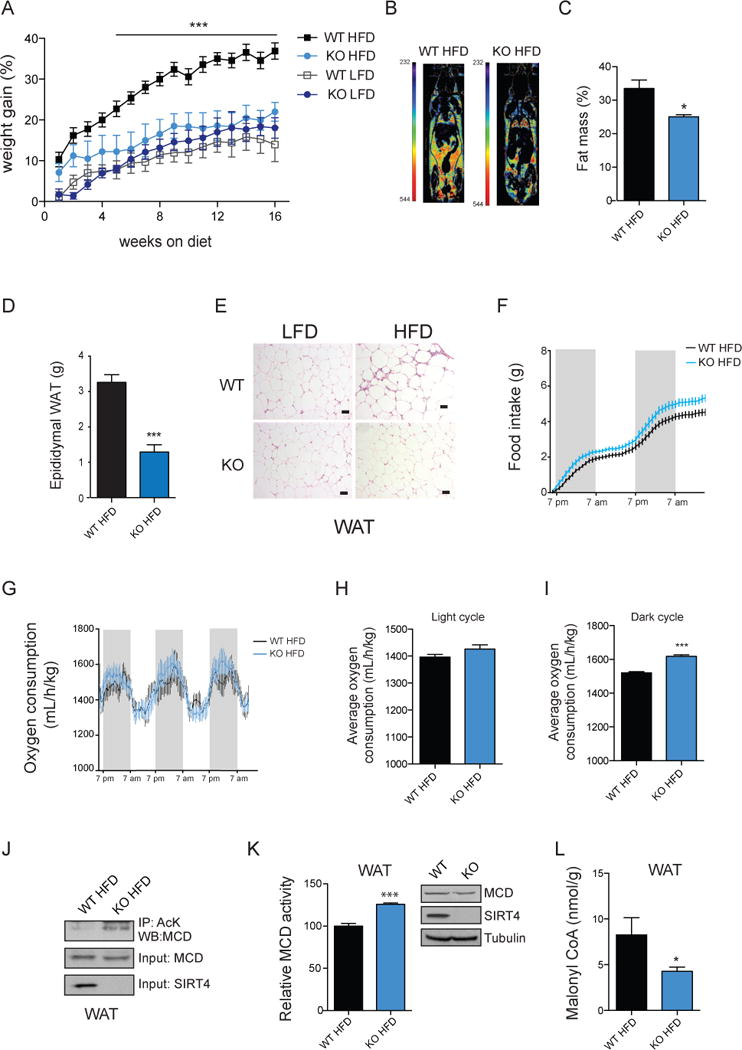
(A) Body weight of WT and SIRT4 KO mice on a low fat diet (LFD) and high fat diet (HFD) (n=10–12 per genotype). (B) Representative images of CT-scan of WT and SIRT4 KO mice on a HFD with fat mass highlighted. Red represents the highest value in the range of WAT density and in blue the lowest. (C) Percentage of fat mass analyzed by CT-scan of WT and SIRT4 KO mice on a HFD (n=6 per genotype). (D) Epididymal WAT weights from WT and SIRT4 KO mice on a HFD (n=6 per genotype). (E) Representative hematoxylin and eosin staining slides of WAT of WT and SIRT4 KO mice under LFD and HFD. Scale bar is 50 μM. (F) Food intake in WT and SIRT4 KO mice on a HFD (n=6 per genotype). (G–I) Energy expenditure in WT and SIRT4 KO mice on a HFD (n=6 per genotype). (J) WAT extracts from WT and SIRT4 KO mice under HFD were immunoprecipitated with a monoclonal AcK antibody and Western blotted for MCD. (K) MCD activity in WAT of WT and SIRT4 KO mice under HFD. (L) Malonyl CoA levels in WAT of WT and SIRT4 KO mice under HFD. In each panel, data represent mean ± SEM. (*) p < 0.05; (**) p < 0.01, (***) p < 0.001. See also Figures S4 and S5.
To better understand the mechanism of protection against obesity in SIRT4 KO mice, we assessed the body composition of animals fed a HFD using computed tomography (CT) scans. The percentage of fat mass was significantly lower in SIRT4 KO mice compared to control animals (Figure 6B and 6C). By contrast, absence of SIRT4 did not appear to affect the percentage of brown adipose tissue (BAT) (Figure S4I). In addition, while liver weight was not affected by SIRT4 loss (Figure S4J), weight of epididymal WAT was significantly lower in the SIRT4 KO mice compared to WT animals (Figure 6D). We found that WAT in SIRT4 KO mice appears normal and we did not detect increased signs of apoptosis, structural alterations, or inflammation (Figure 6E and S4K–L).
To test whether protection from adiposity was due to difference in food intake, we analyzed food consumption. SIRT4 KO mice ate equivalent amounts (or slightly more) as WT controls (Figure 6F). Likewise, we did not detect differences in their respiratory exchange ratio or physical activity (Figure S4M and S4N). By contrast, analysis of energy expenditure revealed that SIRT4 KO mice have a significant increase in energy expenditure during the dark cycle (Figure 6G–6I); this increased energy burning phenotype is consistent with our cellular data and provides an explanation for the protection against a HFD. As the dark cycle corresponds to the fed state in mice, this result also supports the role of SIRT4 function during nutrient abundance. Interestingly, despite protection against diet-induced obesity, the SIRT4 KO mice were equally susceptible to glucose and insulin intolerance when compared to their WT counterparts (Figure S5A–E).
Finally, we assessed MCD acetylation levels in SIRT4 WT and KO mice under HFD in WAT. As expected, we found that MCD was hyperacetylated in SIRT4 KO mice compared to WT mice (Figure 6J). Measurement of MCD activity demonstrated an elevated activity (Figure 6K and S5F) and consistently malonyl CoA levels (Figure 6L and S5G) were lower in SIRT4 KO compared to WT mice. These data demonstrate that SIRT4 loss increases MCD acetylation and activity during a high fat dietary challenge.
Discussion
In this study, we identify a novel role for the mitochondrial sirtuin, SIRT4, in the metabolic reprogramming towards anabolic lipid processes to promote lipogenesis while inhibiting fatty acid oxidation (Figure 1). Mechanistically, SIRT4 mediates this switch in lipid homeostasis by binding, deacetylating K471, and inhibiting MCD activity (Figures 2 and 3). Interestingly, only a few cases of protein deacetylation have been described to result in decreased enzymatic activity (Zhao et al., 2010; Kim et al., 2012). K471 is at close distance to the malonyl CoA entry point (PDB: 2YGW) and its acetylation might help gate the ligand entry (Figure S6). In addition, we discovered that SIRT4 regulates MCD activity and malonyl CoA levels in vivo (Figure 4). SIRT4 represses MCD in the fed state (Figure 4), promoting lipid synthesis and storage while inhibiting lipid catabolism. SIRT4 KO mice are deficient for this switch between fed and fasted states (Figure 5), and demonstrate decreased lipogenesis in vivo (Figure 5). As a consequence, when placed under HFD, SIRT4 KO mice are resistant to diet induced obesity (Figure 6).
Sirtuins possess multiple enzymatic activities, and this is the first study to demonstrate that SIRT4 possesses deacetylase activity. SIRT5 was reported to have deacetylase activity (Nakagawa et al., 2009), but recent work demonstrated that SIRT5 is also a demalonylase and desuccinylase (Du et al., 2011; Peng et al., 2011). Similarly, SIRT6 was initially thought to function solely as an ADP-ribosyltransferase, but later studies identified a substrate-specific deacetylase activity (Zhong et al., 2010). No deacetylase activity was reported for SIRT4 using histone or BSA substrates (Haigis et al., 2006; Ahuja et al., 2007; Schwer et al., 2002). However, our data suggests that the absence of deacetylase activity for SIRT4 may have been a reflection of the lack of an appropriate substrate. Here, we show for the first time that SIRT4 has a susbstrate-specific deacetylase activity, as SIRT4 deacetylates MCD directly and regulates the levels of its acetylation in cells and organs. In the future, it will be interesting to assess whether SIRT4 possesses other deacylase activities.
Maintenance of metabolic homeostasis requires a coordinated regulation of energy intake, storage, and expenditure. Metabolic pathways are designed to sense incoming nutritional and environmental cues and to respond appropriately. Due to their dependency on NAD+, sirtuins are critical modulators of metabolism, sensing changes in metabolic cues in order to exert adaptive responses (Houtkooper et al., 2012). Our work reveals that SIRT4 is a novel regulator of the switch between anabolic and catabolic pathways through the regulation of MCD. It was proposed that AMPK may regulate MCD activity, but this regulation remains unclear (Kuhl et al., 2006; Habinowski et al., 2001; Sambandam et al., 2004; Saha et al., 2000). We uncovered a distinct regulation of MCD via acetylation. Thus, it will be interesting for future studies to examine how MCD acetylation may synergize with differences in AMPK activity.
Our data suggest that the mitochondrial form of MCD is important for control of lipid metabolism, shedding new light on the significance of mitochondrial malonyl CoA. Our findings indicate that influencing mitochondrial malonyl CoA can affect both fat oxidation and lipogenesis. Indeed, we show that changes in mitochondrial MCD activity clearly affect total malonyl CoA levels (2–3 fold in SIRT4 KO muscle and WAT). However, little is known about how the mitochondrial pool of malonyl CoA regulates these pathways. Of note, a recent study similarly showed that changes in mitochondrial lipogenesis protected against diet-induced obesity (Smith et al., 2012). It will be important for future studies to examine the mechanisms that control mitochondrial versus cytoplasmic malonyl CoA, such as involvement of transporters or shuttle systems, and dissect their role in lipid homeostasis.
Strikingly, loss of SIRT4 increases exercise capacity. Exercise training activates a number of pathways that contribute to metabolic reprogramming of lipid handling (Bassel-Duby and Olson, 2006). Of relevance, decreased malonyl CoA has been observed in muscle after exercise, increasing fat oxidation (Dean et al., 2000; Hutber et al., 1997). A study showed that these changes were associated with increased MCD activity (Kuhl et al., 2006). In line with these observations, we show that deletion of SIRT4 leads to decreased malonyl CoA levels and increased exercise capacity, suggesting that SIRT4 might play a role in metabolic reprogramming during exercise training.
The adaptations that improve exercise performance are typically expected to protect against obesity and related metabolic disorders, making it important to identify proteins involved in this reprogramming to treat metabolic diseases. However, the glucose homeostasis of SIRT4 KO mice on a HFD resemble that of WT mice on a HFD (Figure S6), demonstrating that while SIRT4 loss protects against diet-induced adiposity, it does not protect the animals against metabolic dysfunction that arises with high-fat challenge. Thus, SIRT4 loss appears to uncouple metabolic fitness from obesity. Interestingly, and in line with our observation, deletion of MCD protects against insulin resistance (Koves et al., 2008). Likewise, increased mitochondrial flux and fatty acid oxidation are associated with some models of insulin resistance (Sunny et al., 2011; Koves et al., 2008). To understand this uncoupling, it will be important for future studies to probe the role of SIRT4 and MCD in glucose and insulin homeostasis and to assess further the potential therapeutic consequences of modulating SIRT4 or MCD function. In sum, our work uncovers a novel feature of lipid metabolic reprogramming mediated by SIRT4 deacetylation of MCD.
Experimental procedures
Cell culture
Mouse embryonic fibroblasts (MEFs) were isolated from SIRT4 WT and KO littermate embryos as described (Xu, 2005) and immortalized using the SV40 large T antigen. F442A cells were differentiated as previously described (Djian et al., 1985). Isolation of primary adipocytes was performed as previously described (Eguchi et al., 2011). All primary cell cultures were performed using a minimum of 2 independently generated cell lines per genotype. For NAM treatments, cells were incubated overnight with a final concentration of 20 mM NAM. Lentiviral shRNA against SIRT4 clones were purchased from Openbiosystems and lentiviral shRNA against MCD were obtained from The RNAi Consortium (TRC) at the Broad Institute/Harvard. Stable knock-down cell lines were generated according to TRC instructions. MCD cDNA was purchased from Openbiosystems and cloned into pBabe vector for stable expression and pcDNA (HA tag) for transient transfection. Cells overexpressing SIRT4 and SIRT4H162Y were generated by retroviral infection by pBabe vector. Commercial antibodies were used to analyze acetyl-lysine (ImmuneChem for westen blotting and PTM biolabs for immunoprecipitation), and antibodies raised against murine SIRT4 were described previously (Haigis et al., 2006).
Measurement of de novo lipogenesis in cells, triglyceride content and Oil Red O staining
For the measurement of lipogenesis, F442A cells and primary adipocytes were placed overnight in low glucose low serum media, then labeled with 1-14C acetic acid (Perkin Elmer) while stimulated with insulin and high glucose for 1 hour. Cells were washed twice with PBS before lysis in 0.5% Triton X-100. The lipid fraction was extracted by the addition of chloroform and methanol (2:1 v/v), followed by the addition of water. Samples were centrifuged and 14C incorporation was measured in the bottom, lipid-containing phase using a scintillation counter. Each condition was normalized to protein concentrations. Triglycerides were measured using the adipogenesis assay kit from Biovison (K610-100) and Oil Red O staining were performed using the adipogenesis assay kit from Millipore (ECM950).
Fatty acid oxidation
C2C12 cells were differentiated in 2% Horse Serum media and incubated overnight in culture medium containing 100 mM palmitate (C16:0) and 1 mM carnitine. In the final 2 hours of incubation, cells were pulsed with 1.7 μCi [9, 10(n)-3H]palmitic acid (GE Healthcare), and the medium was collected and eluted on ion exchange columns packed with DOWEX 1X2-400 resin (Sigma) to analyze the released 3H2O, formed during cellular oxidation of [3H]palmitate. For FAO assays in MEFs, primary MEFs were used below passage 5.
MCD activity assay and determination of Malonyl CoA levels
MCD activity was tested using a radiochemical assay (Kerner and Hoppel, 2002). Briefly, protein lysates are incubated with [2-14C]malonyl CoA which is decarboxylated by MCD into [2-14C]acetyl-CoA, which is converted to [2-14C]acetylcarnitine in the presence of excess L-carnitine (Sigma) and carnitine acetyltransferase (Roche). The positively charged radiolabeled product, acetylcarnitine, is separated from negatively charged excess radiolabeled substrate through an exclusion column and the radioactivity is measured by scintillation counting. MCD activity assays were performed on 50 μg of protein from cell or tissue lysates. Malonyl CoA was determined by the method previously described (McGarry et al., 1978; Saha et al., 1995) to measure malonyl CoA-dependent incorporation of [3H]acetyl CoA into fatty acids, and results were normalized by protein content (Linn, 1981).
Animal Studies
Studies were performed according to protocols approved by the Institutional Animal Care and Use Committee, the Standing Committee on Animals at Harvard. 3 to 4 month SIRT4 WT and KO male littermates (n=6) fed a normal chow diet (PicoLab Diet 5053) were used for all LFD studies. For fasting experiments, food was removed just before the dark cycle (7 pm) and mice were subjected to fasting conditions overnight before sacrifice. For lipid composition, organs were analyzed by the Vanderbilt Mouse Metabolic Phenotyping Center (MMPC) using GCMS. For HFD experiments, SIRT4 WT and KO male littermates (n=6) mice were fed D12492 from Research diets for 16 weeks and studies were performed with 2–3 separate cohorts of mice. Body weight was measured weekly. CT scan studies were performed at the Longwood SAIF (Boston). SIRT4 WT and KO male mice fed a HFD (n=6) were used. Results were analyzed using InVivoScope software.
Exercise tolerance assays
Male SIRT4 KO and littermate controls (n=11–14) were habituated to the metabolic treadmills (Columbus Instruments) for 3 days prior to exercise testing. Each 15 min habituation session consisted of a 5 min exploration session at 0 m/min, 5 min at 6 m/min and 5 min at 15 m/min at a constant 10 degree incline. A mild electrical stimulus was applied to trained mice to remain on the moving treadmill belt. O2 and CO2 gas sensors (Columbus Instruments) were calibrated before every test and testing started at 8AM each morning following removal of food for 1hr. After collection of resting gasses, the treadmill was started at a speed of 6 m/min. and was increased by 3 m/min every 3 min until mice could no longer maintain the set workload. Animals were allowed to recover for 15 min in the metabolic treadmills after which blood glucose (Accu-Check Aviva) and lactate (Nova Biomedicals) were obtained from tail blood. All procedures were approved by Duke University Institutional Animal Care and Use Committee.
Lipogenesis in vivo
De novo lipogenesis studies were done by measuring palmitate (C16:0) enrichments using gas chromatography (GC)-electron impact ionization (EI) mass spectrometry (MS) as previously described (Lee et al., 1994) (Edmond et al., 1998). Briefly, mice were injected with 6% body water of deuterated water and follow with 4% D20 in the drinking water for 14 days. After harvesting the organs, petroleum ether extractions of the fatty acids were performed after acidification of the saponified tissue. Fatty acid methyl esters (FAME) were prepared by adding 200 mL methanolic HCl (Supelco) to the dried fatty acid fraction, and then heated at 100 °C for at least 1 hour. FAME was then dried with nitrogen, re-dissolved in hexane as solvent and analyzed by GC/MS.
Statistical Analysis
Analyses were performed using an unpaired Student’s t-test and ANOVA test for lipid composition. Significant differences are indicated by a single asterisk when p<0.05, double asterisks when p<0.01 and triple asterisk when p<0.001. All experiments were performed at least two to three times.
Supplementary Material
Highlights.
– SIRT4 represses fatty acid oxidation and stimulates lipogenesis.
– SIRT4 deacetylates and represses MCD regulating malonyl coA levels in vivo.
– SIRT4 loss increases exercise capacity and protects against diet-induced obesity.
Acknowledgments
We thank Hong Lu for technical assistance and Carla Harris (Vanderbilt MMPC Lipid Lab, DK59637) for lipid composition analysis. We thank the Nikon Imaging Center at Harvard Medical School. We thank Lydia Finley and the members of the Haigis lab for helpful discussions and constructive comments on the paper. G.L. was supported by Human Frontier Science Program, N.J.G. by National Science Fondation graduate research fellowship, V.C.J.d.B. by grants from the Netherlands Organization for Scientific Research (grants 916.10.065 and 825.07.005) and MD by NIDDK grant F32DK093256. I.J.K. was supported by grants DK58132-01A2, U19AI091175-01 and P60DK020541 (Einstein DRTC). N.B.R. and A.K.S. were supported by USPHS grants RO1DK19514 and RO1DK67509. TK was supported by Ellison Foundation grant AG-NS-0548-09. DM was supported by NIH grants 1RO1HL101189 and 2PO1DK05398. M.C.H. was supported by funding from NIH grant AG032375, Ellison foundation AG-NS-0573-09, Glenn Foundation for Medical Research and funding from SIRTRIS-GSK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Dean D, Daugaard JR, Young ME, Saha A, Vavvas D, Asp S, Kiens B, Kim KH, Witters L, Richter EA, et al. Exercise diminishes the activity of acetyl-CoA carboxylase in human muscle. Diabetes. 2000;49:1295–1300. doi: 10.2337/diabetes.49.8.1295. [DOI] [PubMed] [Google Scholar]
- Djian P, Phillips M, Green H. The activation of specific gene transcription in the adipose conversion of 3T3 cells. J Cell Physiol. 1985;124:554–556. doi: 10.1002/jcp.1041240327. [DOI] [PubMed] [Google Scholar]
- Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E, Sul HS. Regulation of lipolysis in adipocytes. Annu Rev Nutr. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmond J, Higa TA, Korsak RA, Bergner EA, Lee WN. Fatty acid transport and utilization for the developing brain. J Neurochem. 1998;70:1227–1234. doi: 10.1046/j.1471-4159.1998.70031227.x. [DOI] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habinowski SA, Hirshman M, Sakamoto K, Kemp BE, Gould SJ, Goodyear LJ, Witters LA. Malonyl-CoA decarboxylase is not a substrate of AMP-activated protein kinase in rat fast-twitch skeletal muscle or an islet cell line. Arch Biochem Biophys. 2001;396:71–79. doi: 10.1006/abbi.2001.2589. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Sensing of energy and nutrients by AMP-activated protein kinase. Am J Clin Nutr. 2011;93:891S–896. doi: 10.3945/ajcn.110.001925. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012 doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutber CA, Rasmussen BB, Winder WW. Endurance training attenuates the decrease in skeletal muscle malonyl-CoA with exercise. J Appl Physiol. 1997;83:1917–1922. doi: 10.1152/jappl.1997.83.6.1917. [DOI] [PubMed] [Google Scholar]
- Kerner J, Hoppel CL. Radiochemical malonyl-CoA decarboxylase assay: activity and subcellular distribution in heart and skeletal muscle. Anal Biochem. 2002;306:283–289. doi: 10.1006/abio.2002.5696. [DOI] [PubMed] [Google Scholar]
- Kim EY, Kim WK, Kang HJ, Kim JH, Chung SJ, Seo YS, Park SG, Lee SC, Bae KH. Acetylation of malate dehydrogenase 1 promotes adipogenic differentiation via activating its enzymatic activity. J Lipid Res. 2012;53:1864–1876. doi: 10.1194/jlr.M026567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Kuhl JE, Ruderman NB, Musi N, Goodyear LJ, Patti ME, Crunkhorn S, Dronamraju D, Thorell A, Nygren J, Ljungkvist O, et al. Exercise training decreases the concentration of malonyl-CoA and increases the expression and activity of malonyl-CoA decarboxylase in human muscle. Am J Physiol Endocrinol Metab. 2006;290:E1296–1303. doi: 10.1152/ajpendo.00341.2005. [DOI] [PubMed] [Google Scholar]
- Lee WN, Bassilian S, Guo Z, Schoeller D, Edmond J, Bergner EA, Byerley LO. Measurement of fractional lipid synthesis using deuterated water (2H2O) and mass isotopomer analysis. Am J Physiol. 1994;266:E372–383. doi: 10.1152/ajpendo.1994.266.3.E372. [DOI] [PubMed] [Google Scholar]
- Linn TC. Purification and crystallization of rat liver fatty acid synthetase. Arch Biochem Biophys. 1981;209:613–619. doi: 10.1016/0003-9861(81)90320-9. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Tishkoff DX, Bao J. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb Exp Pharmacol. 2011;206:163–188. doi: 10.1007/978-3-642-21631-2_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long YC, Zierath JR. AMP-activated protein kinase signaling in metabolic regulation. J Clin Invest. 2006;116:1776–1783. doi: 10.1172/JCI29044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry JD, Stark MJ, Foster DW. Hepatic malonyl-CoA levels of fed, fasted and diabetic rats as measured using a simple radioisotopic assay. The Journal of biological chemistry. 1978;253:8291–8293. [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HW, Lee GY, Kim YS. Mass spectrometric identification of K210 essential for rat malonyl-CoA decarboxylase catalysis. J Proteome Res. 2006;5:1398–1406. doi: 10.1021/pr050487g. [DOI] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010 doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.012658. M111 012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziuk J, Pye S. Quantitation of basal endogenous glucose production in Type II diabetes: importance of the volume of distribution. Diabetologia. 2002;45:1053–1084. doi: 10.1007/s00125-002-0841-6. [DOI] [PubMed] [Google Scholar]
- Ruderman NB, Park H, Kaushik VK, Dean D, Constant S, Prentki M, Saha AK. AMPK as a metabolic switch in rat muscle, liver and adipose tissue after exercise. Acta Physiol Scand. 2003;178:435–442. doi: 10.1046/j.1365-201X.2003.01164.x. [DOI] [PubMed] [Google Scholar]
- Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- Saha AK, Kurowski TG, Ruderman NB. A malonyl-CoA fuel-sensing mechanism in muscle: effects of insulin, glucose, and denervation. Am J Physiol. 1995;269:E283–289. doi: 10.1152/ajpendo.1995.269.2.E283. [DOI] [PubMed] [Google Scholar]
- Saha AK, Ruderman NB. Malonyl-CoA and AMP-activated protein kinase: an expanding partnership. Mol Cell Biochem. 2003;253:65–70. doi: 10.1023/a:1026053302036. [DOI] [PubMed] [Google Scholar]
- Saha AK, Schwarsin AJ, Roduit R, Masse F, Kaushik V, Tornheim K, Prentki M, Ruderman NB. Activation of malonyl-CoA decarboxylase in rat skeletal muscle by contraction and the AMP-activated protein kinase activator 5-aminoimidazole-4-carboxamide-1-beta -D-ribofuranoside. The Journal of biological chemistry. 2000;275:24279–24283. doi: 10.1074/jbc.C000291200. [DOI] [PubMed] [Google Scholar]
- Sambandam N, Steinmetz M, Chu A, Altarejos JY, Dyck JR, Lopaschuk GD. Malonyl-CoA decarboxylase (MCD) is differentially regulated in subcellular compartments by 5′AMP-activated protein kinase (AMPK). Studies using H9c2 cells overexpressing MCD and AMPK by adenoviral gene transfer technique. Eur J Biochem. 2004;271:2831–2840. doi: 10.1111/j.1432-1033.2004.04218.x. [DOI] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Witkowski A, Moghul A, Yoshinaga Y, Nefedov M, de Jong P, Feng D, Fong L, Tu Y, Hu Y, et al. Compromised mitochondrial Fatty Acid synthesis in transgenic mice results in defective protein lipoylation and energy disequilibrium. PLoS One. 2012;7:e47196. doi: 10.1371/journal.pone.0047196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunny NE, Parks EJ, Browning JD, Burgess SC. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab. 2011;14:804–810. doi: 10.1016/j.cmet.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DM, Porter BB, Tall JH, Kim HJ, Barrow JR, Winder WW. Skeletal muscle and heart LKB1 deficiency causes decreased voluntary running and reduced muscle mitochondrial marker enzyme expression in mice. Am J Physiol Endocrinol Metab. 2007;292:E196–202. doi: 10.1152/ajpendo.00366.2006. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005;Chapter 28 doi: 10.1002/0471142727.mb2801s70. Unit 28 21. [DOI] [PubMed] [Google Scholar]
- Yang L, Vaitheesvaran B, Hartil K, Robinson AJ, Hoopmann MR, Eng JK, Kurland IJ, Bruce JE. The fasted/fed mouse metabolic acetylome: N6-acetylation differences suggest acetylation coordinates organ-specific fuel switching. J Proteome Res. 2011;10:4134–4149. doi: 10.1021/pr200313x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


