Abstract
While glaciers become increasingly recognised as a habitat for diverse and active microbial communities, effects of their climate change-induced retreat on the microbial ecology of glacier-fed streams remain elusive. Understanding the effect of climate change on microorganisms in these ecosystems is crucial given that microbial biofilms control numerous stream ecosystem processes with potential implications for downstream biodiversity and biogeochemistry. Here, using a space-for-time substitution approach across 26 Alpine glaciers, we show how microbial community composition and diversity, based on 454-pyrosequencing of the 16S rRNA gene, in biofilms of glacier-fed streams may change as glaciers recede. Variations in streamwater geochemistry correlated with biofilm community composition, even at the phylum level. The most dominant phyla detected in glacial habitats were Proteobacteria, Bacteroidetes, Actinobacteria and Cyanobacteria/chloroplasts. Microorganisms from ice had the lowest α diversity and contributed marginally to biofilm and streamwater community composition. Rather, streamwater apparently collected microorganisms from various glacial and non-glacial sources forming the upstream metacommunity, thereby achieving the highest α diversity. Biofilms in the glacier-fed streams had intermediate α diversity and species sorting by local environmental conditions likely shaped their community composition. α diversity of streamwater and biofilm communities decreased with elevation, possibly reflecting less diverse sources of microorganisms upstream in the catchment. In contrast, β diversity of biofilms decreased with increasing streamwater temperature, suggesting that glacier retreat may contribute to the homogenisation of microbial communities among glacier-fed streams.
Keywords: Biofilm, glacier-fed streams, microbial diversity, climate change, metacommunity ecology
Introduction
Glacial ecosystems, at the interface between the cryosphere, hydrosphere, pedosphere and the biosphere, are particularly prone to impacts of climate change (Brown et al., 2007a; Milner et al., 2009; Finn et al., 2010; Woodward et al., 2010; Jacobsen et al., 2012). Glacier-fed streams constitute a prominent geomorphological and ecological component of the glacier foreland, and integrate upstream catchment processes. Changes in runoff following glacier retreat shift the relative contributions of icemelt, snowmelt and groundwater to stream discharge (Brown et al., 2007a; Milner et al., 2009), thereby influencing streamwater temperature, electrical conductivity and channel stability in glacier-fed streams (Hannah et al., 2007; Brown et al., 2007a; Milner et al. 2009). The close hydroecological coupling between glaciers and glacier-fed streams makes biodiversity in these streams particularly susceptible to glacier retreat (Brown et al., 2007a; Finn et al., 2010; Jacobsen et al., 2012). For instance, reduced glacial runoff accompanying glacier retreat facilitates the displacement of cold-adapted invertebrate species by range expansion of less cryophilic species towards higher elevations (Brown et al., 2007a; Milner et al., 2009; Finn et al., 2010). Ultimately, such shifts in species distribution may threaten the biodiversity in isolated high-elevation ecosystems with yet unknown implications for downstream biodiversity (Hughes et al., 2009; Finn et al., 2011).
Ice ecosystems harbour complex microbial communities (Simon et al., 2009; Anesio and Laybourn-Parry, 2012), which have been proposed as sentinels of climate change and the general attrition of the cryosphere (Vincent, 2010). The most abundant bacterial phyla detected in glacier ice are Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes as well as photosynthetic Cyanobacteria (Simon et al., 2009; Xiang et al., 2009; Anesio and Laybourn-Parry, 2012; Edwards et al., 2011). While the microbial diversity and activity in glacier ice was the focus of several recent studies (Hodson et al., 2008; Simon et al., 2009; Xiang et al., 2009), the implications of glacier retreat on the microbial communities in glacier-fed streams—most of them contained in benthic biofilms—remain elusive. Filling this knowledge gap is important, given that biofilms, even in glacier-fed streams (Battin et al., 2004), orchestrate numerous ecosystem processes and contribute to large-scale carbon fluxes (Battin et al., 2008). Biofilms are remarkably diverse, partly because they assemble from different sources of biodiversity (for example, soil and groundwater) within the catchment (Hullar et al., 2006; Besemer et al., 2012). Species sorting from these sources can occur through local environmental conditions and is increasingly understood as a key mechanism underlying biofilm community assembly (Besemer et al., 2012).
As glaciers recede, they change the hierarchical habitat template of the riverine landscape where environmental processes operating at local and regional scales differentially affect life in the glacier-fed streams (Ward et al., 2002; Brown et al., 2007a; Milner et al., 2009). Melting glaciers mobilise ice-locked organic matter with implications for downstream carbon cycling (Singer et al., 2012) and heterotrophic activity (Milner et al., 2009). To explore how biofilm biodiversity and community composition in glacier-fed streams might respond to environmental changes as induced by glacier retreat, we surveyed 26 glaciers and their streams in the Austrian Alps. Substituting space for time, we investigated the relationship between microbial community composition and environmental variables apparently linked to glacial retreat. Alluding to studies on invertebrate communities (Brown et al., 2007a; Milner et al., 2009; Finn et al., 2010), we hypothesise an increase of α diversity—the local richness in a given glacier-fed stream—with increasing deviation of the stream environment from a typical glacier-fed stream. Along the same line, we also postulate a decrease of β diversity—the variation of communities among glacier-fed streams.
Material and methods
Study sites and sample collection
The study sites covered 26 glaciers located along the main chain of the Austrian Alps (Silvretta, Ötztaler Alps, Stubaier Alps, Zillertaler Alps, Venediger Group, Granatspitz Group, Glockner Group, Hochalmspitze and Ankogel; geographically ranging from 10.162–13.278°E, 46.778–47.135°N and 2100–2880 m above sea level; Supplementary Figure 1, Supplementary Table 1). Sampling was performed within 45 days in July and August 2010, a period with almost completely snow-free glaciers. We sampled glacier-fed streams within 10 m downstream of the glacier terminus to capture the physicochemical signatures from glacial and non-glacial sources upstream in the catchment (Ward et al., 2002; Brown et al., 2007a). Benthic biofilms were obtained by rigorously shaking ∼80 stones (diameter ∼2 cm) successively in sterile 50 ml tubes. This field procedure removed most of the biofilm from the stones. The supernatant was then filtered (0.2μm filters, GSWP Millipore, Billerica, MA, USA) using sterile equipment. Two to three litres of streamwater were filtered through sterile 0.2-μm filters (GSWP) and all filters were immediately frozen in liquid nitrogen in the field. Streamwater pH, electrical conductivity and water temperature were measured in situ using WTW probes (pH320, Cond340i).
As a putatively prominent source of microbial diversity to the stream, we also sampled subsurface ice (0.3–0.5 m depth) from 14 randomly chosen sites across the ablation zone of each of the 26 glaciers. We first removed the upper ice layer (∼20–30 cm) using sterile (ethanol-flamed) ice picks and then collected ice from beneath into sterile Whirl-Paks (Nasco, Salida, CA, USA). These samples were transported frozen to the next base camp. There, ice was melted within 1 hour and ∼10 l of icemelt were filtered through sterile 0.2-μm filters (GSWP), which were immediately frozen in liquid nitrogen.
Streamwater dissolved organic carbon and nutrients
Sreamwater was analysed for dissolved organic carbon (DOC) concentration using a Sievers 900 TOC Analyser (GE Analytical Instruments, Boulder, CO, USA) operated with an inorganic carbon removal unit. Prior to injection, DOC samples (GFF-filtered, pre-combusted) were automatically acidified in the analyser as recommended by the manufacturer. We determined a method detection limit of the Sievers 900 according to US EPA guidelines and found it typically <6 μg C l−1. The detection limit was calculated as three (approximate critical z-value for 99.9% confidence)-times the standard deviation of nine lab replicates of low DOC water (average DOC concentration: 29.71±1.43 μg C l−1). Concentrations of N-NH4, N-NO2 and N-NO3 in the streamwater were determined using Continuous Flow Analysis (Alliance instruments, Salzburg, Austria). For further analyses, we used ‘total nitrogen', which represents the summation of N-NH4, N-NO2 and N-NO3.
DNA extraction and PCR amplification
Genomic DNA was extracted using the PowerSoil DNA extraction kit (MoBio, Carlsbad, CA, USA) according to the manufacturer's recommendations. The V4–V5 regions of the 16S rRNA gene was amplified in 25-μl PCR reactions containing 0.5 μmol l−1 of each primer (Thermo Scientific, Waltham, MA, USA), 0.2 mmol l−1dNTPs (Thermo Scientific), 40 μg bovine serum albumin (Thermo Scientific), 4 mmol l−1 MgCl2 (Thermo Scientific), 1 U Taq-DNA Polymerase with the recommended PCR buffer (Thermo Scientific) and 4 μl DNA extract (2–4 ng DNA). Primers used for amplification of the 16S rRNA gene were the universal 515F 5′-GTGNCAGCMGCCGCGGTAA-3′ and 926R 5′-CCGYCAATTYMTTTRAGTTT-3′ (Quince et al., 2011), containing the 454 Titanium A and B adaptors, respectively. For each sample, two different sample-specific barcodes contained in the forward primer were employed to reduce barcode-specific bias (Berry et al., 2011).
Samples were amplified using an initial denaturing step of 2 min at 94 °C, followed by 30 cycles of 30 s denaturation at 94 °C, 30 s annealing at 56 °C, 1 min elongation at 72 °C and a final elongation for 10 min at 72 °C. Each PCR reaction included a negative control. PCR products were run on a 1% agarose gel and purified using the Gel Extraction Kit (Qiagen, Hilden, Germany). The purified PCR products were quantified by gel using the Gel Doc XR+System (BioRad, Hercules, CA, USA) and pooled equimolar for pyrosequencing.
454-pyrosequencing
Amplicons were sequenced on a GS FLX Titanium Sequencer in Liverpool (Centre for Genomic Research, University of Liverpool, UK). Raw output files were filtered and de-noised using the software package AmpliconNoiseV1.0 (Quince et al., 2011). After pre-clustering the sequences with PyroNoise (AmpliconNoiseV1.0), PCR single-base errors were corrected via SeqNoise (AmpliconNoiseV1.0), a sequence-based clustering method that performs alignment of the sequences. Chimeras were finally identified and removed with the Perseus algorithm at an intercept of α=−7.5 and a coefficient of β=0.5 (Quince et al., 2011). This filtering and de-noising steps reduced the number of 658 592 flowgrams to 384 320 reads (read length 400 bp). A complete linkage algorithm on a 97% identity level clustered these reads to operational taxonomic units (OTUs) (AmpliconNoiseV1.0) and taxonomic assignments were determined by using a naive Bayesian rDNA classifier (Ribosomal Database Project; Wang et al., 2007) at a 70% confidence threshold.
Data analysis
Dissimilarity matrices of the community composition were calculated by applying the relative abundance-based Bray–Curtis dissimilarity index. Based on these matrices, non-metric multidimensional scaling analysis was used to visualise differences in community composition, and a non-parametric permutational analysis of variance Anderson (2001) to test differences in community composition among habitats (using the R-packages ‘vegan' and ‘ellipse', Oksanen et al., 2011; Murdoch and Chow, 2007). A concomitant test for dispersion homogeneity (the multivariate analogue of variance homogeneity) is provided by the analysis of β diversity (see below).
A principal component analysis was performed to illustrate the relationship and importance of z-transformed environmental variables. Further, after Hellinger-transformation of the microbial community composition data, we performed a redundancy analysis to determine a potential effect of environmental variables on community composition (Legendre et al., 2011; using the R-package ‘vegan': Oksanen et al., 2011). The explanatory values of environmental variables were further explored by running a forward selection test included in the ‘packfor' package (Blanchet et al., 2008). Additionally, these variables were tested for correlations with the relative abundances of the most abundant bacterial families. These Pearson correlation coefficients were visualised in a heat map.
We used Diversity Estimation software (Quince et al., 2008), to estimate the ‘true richness' of the communities by fitting the 454-pyrosequencing-obtained OTU abundances to a Sichel distribution (Sichel, 1974). For comparison, all samples were rarefied to the lowest number of reads obtained from an individual sample (1007 reads; vegan: Oksanen et al., 2011). Using multiple linear regression, we tested for correlations between environmental variables and microbial richness.
To estimate the probability that a biofilm community represented a random sample of the respective streamwater community, we performed a random resampling procedure on the samples from each glacier, using functions of the R-packages vegan, ecodist and gdata (Goslee and Urban, 2007; Oksanen et al., 2011; Warnes et al., 2011; Besemer et al., 2012). Each tested sample pair consisted of the biofilm and the suspended community from one glacier-fed stream. Individual reads were sampled from the suspended community (the probability of each OTU to be sampled being its relative abundance) with replacement until the number of OTUs in this randomly assembled community equalled the richness of the respective biofilm community. This procedure was repeated to yield 1000 random assemblages. The probability of the biofilm community to fall within the distribution of these random assemblages was calculated as the percentage of the distances of the random assemblages to their centroid, which were as high as or higher than the distance of the biofilm community to the centroid. As a conservative approach, the biofilm community data set was reduced to those OTUs, which also occurred in the respective streamwater community.
Dispersion is an expression of average community dissimilarity, equivalent to average Bray–Curtis dissimilarity, and serves as a measure of spatial community variation or β diversity (Anderson et al., 2006a). Dispersion was computed as the average distance of communities to the respective group (that is, the habitat) centroid in the ordination space resulting from a principal coordinate analysis (aka metric scaling) of the Bray–Curtis dissimilarity matrix (Anderson, 2006b). Differences in dispersion among habitats were tested using a permutation procedure (Anderson, 2006b) with functions of vegan in R (Oksanen et al., 2011).
The relationship between selected environmental variables (for example, streamwater temperature) and β diversity of streamwater and biofilm communities was tested by multiple linear regression on distance matrices (Lichstein, 2006; Ptacnik et al., 2010). For this, the Bray–Curtis dissimilarity matrix was unfolded to a vector of pairwise dissimilarities between any two communities and analysed as dependent on pairwise average temperature and pairwise temperature difference in a multiple linear regression model. In this analysis, the hypothesis of interest concerns the effect of average temperature on dissimilarity, but temperature differences must be included as a covariate to account for environmental differentiation simultaneously affecting β diversity; significances are computed by permutation (see Supplementary Information for details). Additionally, as a graphical analysis, we adapted a local polynomial regression fitting procedure (LOESS; Cleveland et al., 1992) to estimate a ‘local' β diversity at a given temperature of interest. Based on a neighbourhood-approach, we identified subsets of sites with similar temperatures, whose communities were then included in the computation of β diversity as an average dissimilarity in a weighted manner (see Supplementary Information for details). In addition to temperature, these analyses were performed on elevation, electrical conductivity and pH, which were previously found to be significantly correlated to streamwater and biofilm community compositions.
Data analysis and visualisation was performed in SPSS (IBM, Armonk, NY, USA), SigmaPlot and R 2.13.0 (R Development Core Team, 2011).
Results
Microbial community composition and taxonomy
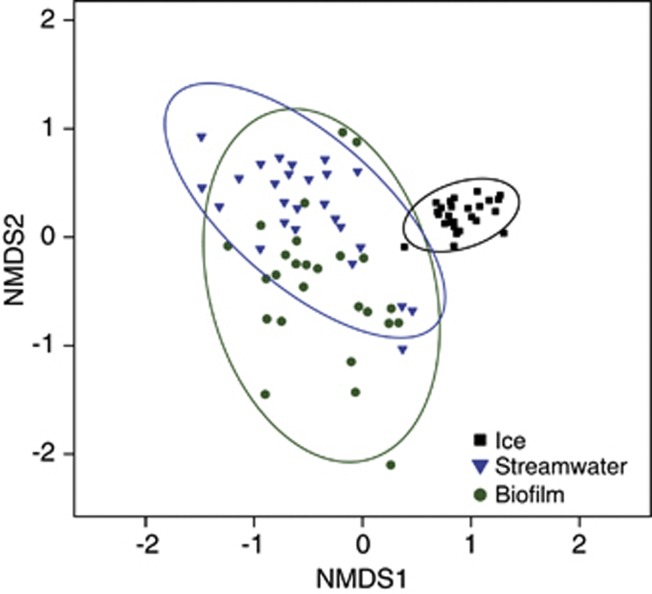
The 454-pyrosequencing data set consisted of 3489±1493 (average±s.d.) reads per sample, which clustered into 11 032 different OTUs. The original pyrosequencing output files are available at the NCBI Sequence Read Archive under the accession number SRX196420. Of all found OTUs, 2.9, 53.6 and 13.2% were unique to glacial ice, streamwater and biofilms, respectively (Supplementary Figure 2). In total, 7.4% of all OTUs were found in all three habitats, 19.8% were shared by biofilm and streamwater communities and 0.6% occurred exclusively in biofilms and glacial ice. Non-metric multidimensional scaling revealed differences in community composition among all three habitats (Figure 1), which were further identified as significant by permutational analysis of variance (pseudo-F=11.326, d.f.1=2, d.f.2=75, P<0.001). However, these differences in community compositions were confounded with differences in dispersion among habitats (β diversity, see below). While glacial ice communities were relatively constrained with little variation among glaciers, communities in the streamwater and biofilms varied broadly and overlapped (Figure 1).
Figure 1.
Community composition in glacial ice, streamwater and biofilms. Non-metric multidimensional scaling analysis of microbial communities based on Bray–Curtis dissimilarities in 26 glacial ecosystems (stress=0.16). Ellipses represent the 95% confidence interval.
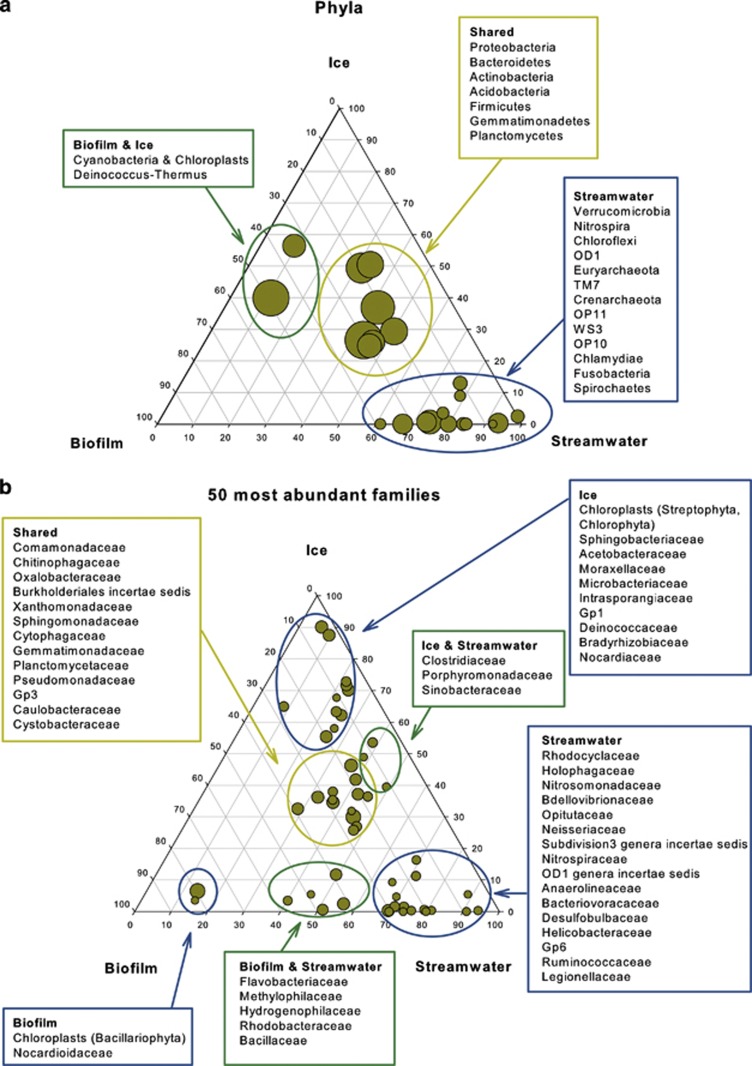
Taxonomic analyses of our sequence data revealed that Proteobacteria, Bacteroidetes and Actinobacteria were the dominant phyla in ice, streamwater and biofilms, whereas Cyanobacteria/chloroplasts were abundant only in glacial ice and biofilm communities (Figure 2a). Several less abundant phyla as Verrucomicrobia or Nitrospira were primarily found in the streamwater, while they were rarely or not detected in biofilms and ice, respectively. Comamonadaceae, Chitinophagaceae and Oxalobacteraceae were the most abundant families detected in all three habitats, while Sphingobacteriaceae and Acetobacteraceae were primarily identified in the ice (Figure 2b). Streamwater communities were characterised by numerous families, many of which occurred neither in ice nor in biofilms (Figure 2b, Supplementary Figure 2).
Figure 2.
Distribution of taxonomic groups in glacial ice, streamwater and biofilm. The percentage of (a) phyla and (b) the 50 most abundant families associated with each habitat is visualised in ternary plots. The position in the triangle indicates the relative abundance of each taxon among the three habitats; the size of the circle represents the relative abundance of taxa.
Relationship between environmental variables and microbial community composition
To identify the potential effect of streamwater geochemistry and glacial characteristics on microbial community composition and diversity in glacier-fed streams, we first tested the relationship of the variables among study sites. Principal component analysis revealed streamwater electrical conductivity and pH to form a major environmental gradient across the glacier-fed streams. This gradient was also associated with differences in elevation and glacial coverage (% glaciation of the catchment) of the respective glaciers, and all four variables together accounted for most of the environmental variation among glacier-fed streams (Supplementary Figure 3; Supplementary Table 2).
Using redundancy analysis, we found that the physicochemical environment significantly explained variation in community composition in streamwater (F=1.399, P<0.01, n=26) and in biofilms (F=1.419, P<0.01, n=26). In the next step, forward variable selection found that biofilm community composition was significantly explained by streamwater electrical conductivity (P<0.05), pH (P<0.05) and streamwater temperature (P<0.05). Likewise, electrical conductivity (P<0.01) and pH (P<0.01) were related to streamwater community composition, whereas streamwater temperature was not. Elevation explained variation in community composition of streamwater (P<0.01), but not of biofilms. Glacial coverage was neither related to biofilm, nor to streamwater communities (data not shown).
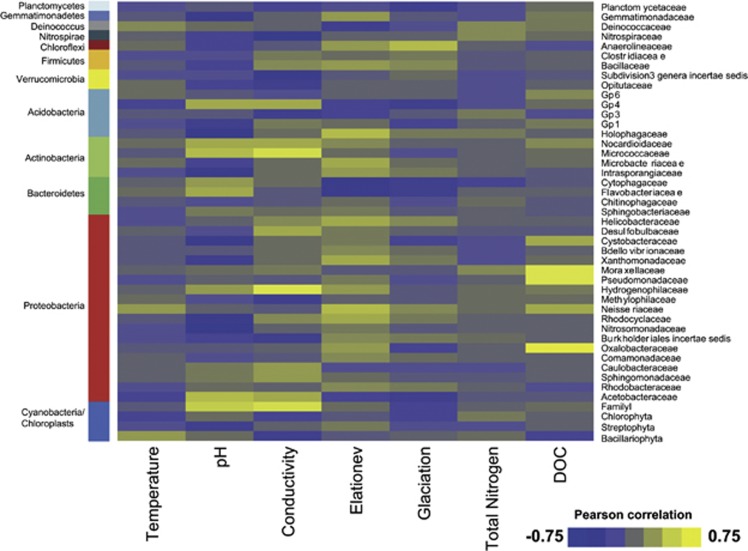
Variation in electrical conductivity and pH correlated with the relative abundance of microbial families and to some extent even of phyla in the biofilms (Figure 3). For instance, Actinobacteria, Nitrospira and Verrucomicrobia varied with electrical conductivity, whereas Acidobacteria, Gemmatimonadetes and Proteobacteria were related to changes in pH (Supplementary Table 3).
Figure 3.
Heat map of Pearson correlations between environmental variables and families detected in biofilms. Colours represent the r-values of Pearson correlations between relative abundances of the 45 most abundant bacterial families and environmental parameters. Families were ordered according to taxonomic affiliations. We found several families that were strongly correlated to pH and electrical conductivity. Positive correlations with DOC were only found with Proteobacteria. Streamwater temperature and total nitrogen were only weakly related to the abundance of taxonomic families.
To better understand these differing community composition patterns, we looked for indications of species sorting as a possible mechanism of community assembly. We tested whether biofilms were a product of purely stochastic immigration from the source community suspended in the streamwater, by comparing biofilm communities with random samples of the streamwater communities. Resampling revealed that in all 26 glacial ecosystems, the biofilm communities significantly differed from the centroid of the random assemblages (probability of the biofilm community to fall within the distribution of the random assemblages P<0.001; Supplementary Figure 4).
Relationship between environmental variables and microbial biodiversity
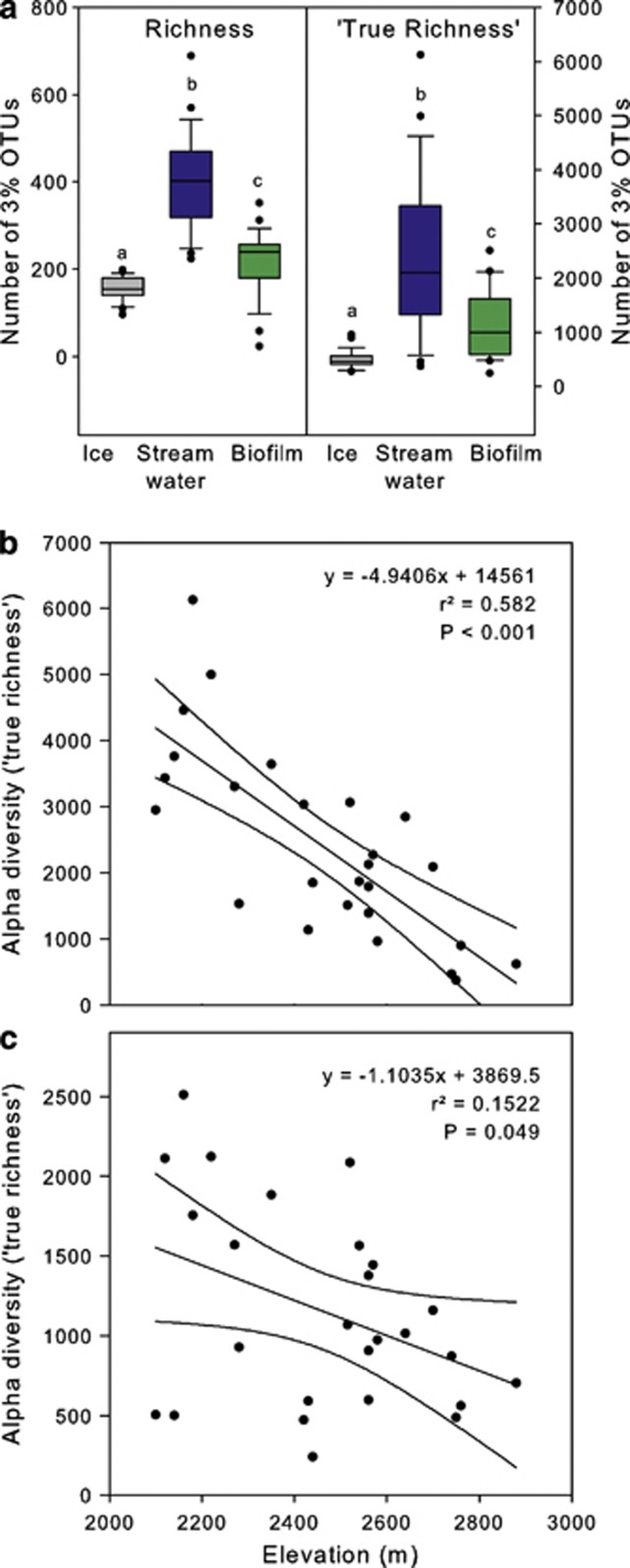
α diversity was highest in streamwater, intermediate in biofilms and lowest in glacial ice, as revealed by 'true richness' (Quince et al., 2008) and rarefied richness (Figure 4a). Based on our results from community composition, we investigated the relationship between elevation, electrical conductivity, pH, streamwater temperature and α diversity. Multiple linear regressions revealed elevation and electrical conductivity as main variables explaining the ‘true richness' in streamwater (adjusted r2=0.67, F=10.508, P<0.001, n=26) and biofilms (adjusted r2=0.43, F=3.883, P<0.05, n=26). Using simple linear regression, it was found that richness of both streamwater and biofilm communities decreased with higher elevation, though for biofilms this relationship was weak (Figures 4b and c); electrical conductivity was not significantly related to the richness of streamwater communities, while biofilm richness decreased with electrical conductivity (r2=0.18, P<0.05, n=26; data not shown). The simultaneous decrease of biofilm biodiversity with elevation and electrical conductivity seems counterintuitive, as a higher contribution of glacial meltwater to streamflow results in lower electrical conductivity. This pattern is likely driven by an imprint of geology, particularly the prevalence of carbonate minerals, among our study streams, as indicated by a significant correlation between electrical conductivity and pH (r=0.56, P<0.01, n=26).
Figure 4.
Microbial α diversity. (a) Boxplot representation of rarefied richness (all samples rarefied to 1007 reads) and ‘true richness' estimated by fitting Sichel distribution curves to the abundance distributions obtained from the 454-pyrosequencing data. Median (line), 1st and 3rd quartile (box margins), 5 and 95% percentiles (whiskers), outliers (points). Different letters on each box represent significant differences between habitats as determined by Wilcoxon tests followed by Bonferroni correction (P<0.01). (b,c) Microbial richness decreased with higher elevation. Linear regression include 95% confidence intervals in the (b) streamwater and (c) biofilms.
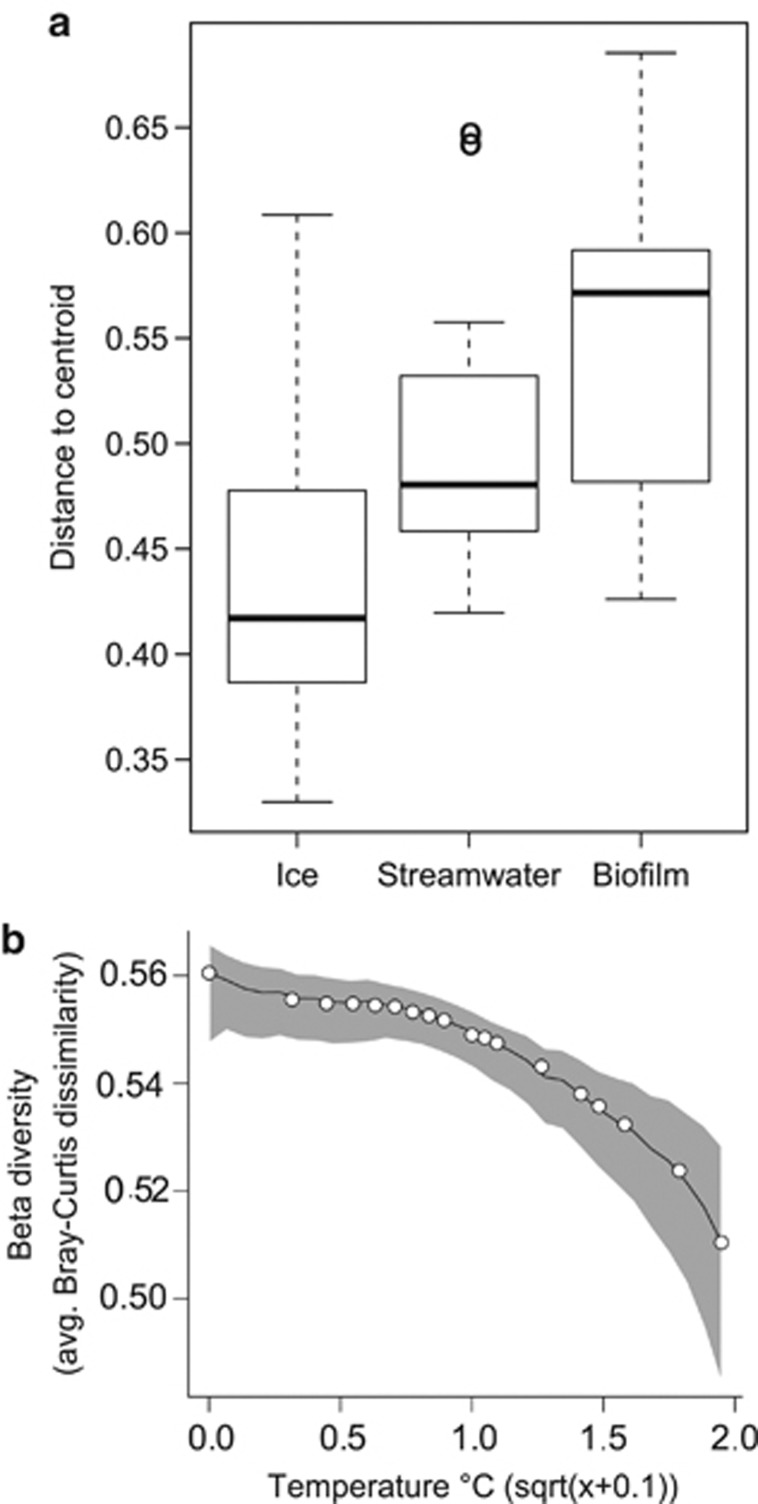
In agreement with patterns of community composition, β diversity among glacial ecosystems was highest in biofilms, intermediate in streamwater and lowest in ice (Figure 5a). We found a significant decrease of biofilm β diversity with streamwater temperature (Figure 5b); electrical conductivity, pH and elevation were not significantly correlated with biofilm β diversity (Supplementary Figure 5). Microbial β diversity in streamwater did not correlate with any of the tested variables (data not shown).
Figure 5.
Spatial variation of microbial communities. (a) β diversities of the three habitats as the average distance of communities to the group (that is, habitat) centroid after principal coordinate analysis of the Bray–Curtis dissimilarity matrix. β diversity differed significantly among habitats (pseudo-F=17.082, d.f.1=2, d.f.2=75, P<0.001 for all pairwise comparisons). (b) Graphical analysis of biofilm β diversity as a function of streamwater temperature by local polynomial regression fitting. Multiple linear regression on distance matrices revealed β diversity to decline significantly with increasing streamwater temperature (pseudo-t=−4.77, P<0.05). The shaded area gives a bootstrap confidence interval for the generated trendline. See Materials and methods and Supplementary Information for computational details.
Discussion
Glaciers retreat worldwide, making it necessary to better understand the impacts of retreating glaciers on glacial ecosystems. Our study streams encompassed a gradient of elevation, glacial coverage and streamwater electrical conductivity, suggesting distinct hydrological settings of the glacier-fed streams at the catchment scale as a result of variable contributions of icemelt, snowmelt and groundwater to streamflow (Brown et al., 2003; Brown et al., 2007a; Milner et al., 2009). Additionally, catchment geology and hydrochemical processes may imprint the observed environmental gradient, as indicated by the positive correlation of streamwater electrical conductivity and pH. At local scale, depending on geomorphology (for example, slope and slope breaks), the ablation zone of some of our study glaciers was fragmented following thinning, and meltwater therefore flowed over bare rock exposed to solar radiation (Supplementary Figure 6). This caused elevated and outlying streamwater temperatures in these systems and weakened the relationship between streamwater temperature and glacial coverage across all sites. The physicochemical signatures in glacier-fed streams thus reflected catchment scale and local processes operating along the various flow paths (that is, supraglacial, englacial, subglacial and non-glacial).
The most abundant phyla identified in our study streams are typical for freshwater ecosystems (Tamames et al., 2010). Their regular occurrence in other ice ecosystems (Simon et al., 2009; Xiang et al., 2009; Anesio and Laybourn-Parry, 2012) suggests that they comprise cryophilic taxa, as also supported by cultivation-dependent approaches (Cheng and Foght, 2007; Loveland-Curtze et al., 2009). Streamwater electrical conductivity and pH were associated with community composition in biofilms at the level of OTUs, family and even of phyla (Figure 3, Supplementary Table 3). These results are in line with soil studies (Fierer and Jackson, 2006) suggesting that pH—a prime physiological control on single-celled organisms—structures microbial communities even at the phylum level (for example, Acidobacteria, Proteobacteria), and with studies on invertebrate communities in glacier-fed streams (Brown et al., 2007b). Certain biofilm taxa, some of them (for example, Nitrospira) having critical roles in biogeochemistry, seem particularly prone to physicochemical shifts. Streamwater temperature, known to influence benthic microbial community composition in streams (Hullar et al., 2006), appeared to further affect biofilm community composition, probably at a more local scale given the pattern of streamwater temperature among our study streams. Expected long-term changes of physiochemical conditions following glacial retreat could potentially have widespread implications for glacial stream ecosystem functions. Owing to their sessile mode of life, biofilm microorganisms are likely even more susceptible to such changes compared with transient cells in the streamwater.
Despite the cold and mainly oligotrophic conditions in glacial ecosystems, our ‘true richness' estimates for streamwater and biofilm communities were comparable to those from non-glacial streams (Besemer et al., 2012). In line with the present study, Besemer et al. (2012) reported higher microbial richness in streamwater than in biofilm communities. We attribute the differences in α diversity between ice, streamwater and biofilms to various degrees of environmental harshness (sensu Jacobsen and Dangles, 2011) and metacommunity (that is, a set of local communities linked by dispersal) size (Leibold et al., 2004), which interact to determine biodiversity. In fact, glacial ice is a harsh but comparatively constant environment (Hodson et al., 2008) harbouring a constrained microbial community, which may be particularly well adapted to its environment; cell immigration is assumedly dominated by atmospheric deposition (Hervas and Casamayor, 2009). Glacier-fed streams, however, are dynamic with pronounced temporal fluctuations (Brown et al., 2007a; Milner et al., 2009), and collect microorganisms from various glacial (for example, subglacial, englacial and supraglacial runoff; Anesio and Laybourn-Parry, 2012) and non-glacial sources (for example, groundwater; Brown et al., 2007a; Milner et al., 2009). In fact, microorganisms from cryoconite holes (that is, holes with high microbial activity at the glacier surface, which form around solar-heated debris; Edwards et al., 2011), the englacial environment (Anesio and Laybourn-Parry, 2012), and also from groundwater, atmospheric deposition (Hervas and Casamayor, 2009), residual snow (Hervas and Casamayor, 2009) and adjacent soils and rocks (Bardgett et al., 2007; Schütte et al., 2010) may form a metacommunity that potentially contributes to the community in the glacier-fed streams. We suggest that decreasing α diversity in the streamwater with elevation may be due, at least in part, to less diverse sources of microorganisms upstream in the catchment. Apparently glacial ice contributed only marginally to the microorganisms in the streamwater, whose α diversity may therefore increase as other compartments of the metacommunity (for example, groundwater, soils) gain importance. Similarly, the low commonality of taxa both at the OTU (Supplementary Figure 2) and family level (Figure 2b) between ice and biofilm communities suggests minor contributions of the ice communities to biofilm assembly in the glacier-fed streams.
Differing patterns of community composition and diversity suggest deviating assembly mechanisms in biofilm and streamwater communities. In the frame of metacommunity theory (Leibold et al., 2004), species sorting, where the local environment and biotic interactions select from the metacommunity, would be a candidate mechanism for biofilm community assembly (Besemer et al., 2012). In contrast, streamwater communities could likely be explained by mass effects, determined by large cell influx rates and short residence times limiting the influence of environmental factors. The local environment in the glacier-fed streams would thus select microorganisms from the streamwater for biofilm formation, the diversity of which is influenced by the size of the metacommunity upstream. The latter may be affected, for instance, by the diversity of hydrological flow paths upstream and the various habitats they connect, and also by a possible catchment scale imprint on geochemistry (for example, electrical conductivity, pH). We found that stochastic immigration of OTUs from the streamwater into the biofilms was unlikely to explain the observed community composition of biofilms, thus supporting the assumption that species sorting has a role in biofilm community assembly. Collectively, these findings suggest that the various components of the hierarchical habitat template differentially influence the composition of microbial communities in glacier-fed streams, which complies with general stream ecology (Ward et al., 2002).
Ice, streamwater and biofilm communities exhibited different levels of spatial variation among all study sites, likely reflecting community responses to various degrees of environmental variation in these habitats, and different assembly mechanisms. The relatively small range of streamwater temperature was apparently sufficient to influence the spatial variation of biofilm communities among glacier-fed streams. We suggest therefore that sensitive warming of glacier-fed streams, driven by streamflow shifts from glacial to non-glacial sources or local flow over exposed bedrock in a fragmenting glacial landscape, could have an impact on the distribution of microbial diversity in these systems (Vincent, 2010).
Substituting space for time, our findings suggest biofilms in glacier-fed streams as sentinels of glacier retreat and expand the current knowledge on the threatened biodiversity in the European Alps (Brown et al., 2007a; Finn et al., 2010; Jacobsen et al., 2012). Invertebrate α diversity is thought to increase as glaciers recede, because of species migrating upstream (Brown et al., 2007a; Milner et al., 2009; Finn et al., 2010). Microorganisms, however, primarily disperse downstream with water flow. Therefore, we suggest that both shifts in the metacommunity upstream of the glacier-fed stream and changes of the local physicochemical environment, as induced by glacial retreat, alter biodiversity and composition of the streamwater communities and, via species sorting, also of biofilm communities. Glacier retreat may increase microbial α diversity in glacier-fed streams, while concomitantly reducing β diversity, and thereby contributing to the homogenisation of biofilm communities among glacier-fed streams—similar to the patterns observed for invertebrates (Jacobsen and Dangles, 2011; Jacobsen et al., 2012). Spatially isolated and sporadically occurring invertebrate populations face an elevated risk of local extinction as glaciers recede, which ultimately reduces their spatial variation (Finn et al., 2012; Jacobsen and Dangles, 2011; Jacobsen et al., 2012). The responsiveness of biofilms to the local environment in glacier-fed streams invokes a similarly patchy distribution susceptible to homogenisation following glacier retreat. These findings call for more research exploring the implications of microbial biodiversity shifts in glacier-fed streams for ecosystem processes and downstream biogeochemistry.
Acknowledgments
We are grateful to K Wagner and T Urich for support in the laboratory and to C Preiler, L Hartmann, B Eichelberger, B Preiler, I Hödl, and L Nicklas for assistance in the field. C Kroisleitner provided glacier coverage data. C Quince and M Bengtsson helped with data analysis. The manuscript benefited from comments by H Peter, R Sommaruga and three anonymous reviewers. Financial support came from the Austrian Science Fund (START Y420-B17) to TJB.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson MJ, Ellingsen KE, Mcardle BH. Multivariate dispersion as a measure of beta diversity. Ecol Lett. 2006a;9:683–693. doi: 10.1111/j.1461-0248.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Anderson MJ. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006b;62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- Anesio AM, Laybourn-Parry J. Glaciers and ice sheets as a biome. Trends Ecol Evol. 2012;27:219–225. doi: 10.1016/j.tree.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Bardgett RD, Richter A, Bol R, Garnett MH, Bäumler R, Xu X, et al. Heterotrophic microbial communities use ancient carbon following glacial retreat. Biol Lett. 2007;3:487–490. doi: 10.1098/rsbl.2007.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battin JT, Wille A, Psenner R, Richter A. Large-scale environmental controls on microbial biofilms in high-alpine streams. Biogeoscience. 2004;1:159–171. [Google Scholar]
- Battin TJ, Kaplan LA, Findlay S, Hopkinson CS, Marti E, Packman AI, et al. Biophysical controls on organic carbon fluxes in fluvial networks. Nat Geosci. 2008;1:95–100. [Google Scholar]
- Berry D, Mahfoudh KB, Wagner M, Loy A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol. 2011;77:7846–7849. doi: 10.1128/AEM.05220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer K, Peter H, Logue JB, Langenheder S, Lindström ES, Tranvik LJ, et al. Unraveling assembly of stream biofilm communities. ISME J. 2012;6:1459–1468. doi: 10.1038/ismej.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet FG, Legendre P, Borcard D. Forward selection of explanatory variables. Ecology. 2008;89:2623–2632. doi: 10.1890/07-0986.1. [DOI] [PubMed] [Google Scholar]
- Brown LE, Hannah DM, Milner AM. Alpine stream habitat classification: An alternative approach incorporating the role of dynamic water source contributions. Arct Antarct Alp Res. 2003;35:313–322. [Google Scholar]
- Brown LE, Hannah DM, Milner AM. Vulnerability of alpine stream biodiversity to shrinking glaciers and snowpacks. Glob Change Biol. 2007a;13:958–966. [Google Scholar]
- Brown LE, Milner AM, Hannah DM. Groundwater influence on alpine stream ecosystems. Freshwater Biol. 2007b;52:878–890. [Google Scholar]
- Cheng SM, Foght JM. Cultivation-independent and –dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol Ecol. 2007;59:318–330. doi: 10.1111/j.1574-6941.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Cleveland WS, Grosse E, Shyu WM.1992Local regression modelsChambers JM, Hastie TJ, (eds)Statistical Models in S Wadsworth & Brooks/Cole, Pacific Grove, CA, USA [Google Scholar]
- Edwards A, Anesio AM, Rassner SM, Sattler B, Hubbard B, Perkins WT, et al. Possible interactions between bacterial diversity, microbial activity and supraglacial hydrology of cryoconite holes in Svalbard. ISME J. 2011;5:150–160. doi: 10.1038/ismej.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn DS, Bonada N, Múrria C, Hughes JM. Small but mighty: headwaters are vital to stream network biodiversity at two levels of organization. J N Am Benthol Soc. 2011;30:963–980. [Google Scholar]
- Finn DS, Khamis K, Milner AM. Loss of small glaciers will diminish beta diversity in Pyrenean streams at two levels of biological organization. Global Ecol Biogeogr. 2012;22:40–51. [Google Scholar]
- Finn DS, Räsänen K, Robinson CT. Physical and biological changes to a lengthening stream gradient following a decade of rapid glacial recession. Glob Change Biol. 2010;16:3314–3326. [Google Scholar]
- Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw. 2007;22:1–19. [Google Scholar]
- Hannah DM, Brown LE, Milner AM, Gurnell AM, McGregor GR, Petts GE, et al. Integrating climate–hydrology–ecology for alpine river systems. Aquatic Conserv: Mar Freshw Ecosyst. 2007;17:636–656. [Google Scholar]
- Hervas A, Casamayor EO. High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol Ecol. 2009;67:219–228. doi: 10.1111/j.1574-6941.2008.00617.x. [DOI] [PubMed] [Google Scholar]
- Hodson A, Anesio AM, Tranter M, Fountain A, Osborn M, Priscu J, et al. Glacial ecosystems. Ecol Monogr. 2008;78:41–67. [Google Scholar]
- Hughes JM, Schmidt DJ, Finn DS. Genes in streams: using DNA to understand the movement of freshwater fauna and their riverine habitat. Bioscience. 2009;59:573–583. [Google Scholar]
- Hullar MAJ, Kaplan LA, Stahl DA. Recurring seasonal dynamics of microbial communities in stream habitats. Appl Environ Microbiol. 2006;72:713–722. doi: 10.1128/AEM.72.1.713-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen D, Dangles O. Environmental harshness and global richness patterns in glacier-fed streams. Global Ecol Biogeogr. 2011;21:647–656. [Google Scholar]
- Jacobsen D, Milner AM, Brown LE , Dangles O. Biodiversity under threat in glacier-fed river systems. Nature Clim Change. 2012;2:361–364. [Google Scholar]
- Legendre P, Oksanen J, Braak CFJ. Testing the significance of canonical axes in redundancy analysis. Methods Ecol Evol. 2011;2:269–277. [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, et al. The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett. 2004;7:601–613. [Google Scholar]
- Lichstein JW. Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant Ecol. 2006;188:117–131. [Google Scholar]
- Loveland-Curtze J, Miteva VI, Brenchley JE. Herminiimonas glaciei sp. nov., a novel ultramicrobacterium from 3042 m deep Greenland glacial ice. Int J Syst Evol Microbiol. 2009;59:1272–1277. doi: 10.1099/ijs.0.001685-0. [DOI] [PubMed] [Google Scholar]
- Milner AM, Brown LE, Hannah DM. Hydroecological response of river systems to shrinking glaciers. Hydrol Process. 2009;23:62–77. [Google Scholar]
- Murdoch D, Chow ED.2007. Ellipse: functions for drawing ellipses and ellipse-like confidence regions. R Package Version 0.3 – 5.
- Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RB, Simpson GL, et al. 2011. Vegan: community Ecology Package. R Package Version 1.17-11.
- Ptacnik R, Andersen T, Brettum P, Lepistö L, Willén E. Regional species pools control community saturation in lake phytoplankton. Proc R Soc B. 2010;277:3755–3764. doi: 10.1098/rspb.2010.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. ISMEJ. 2008;2:997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, AT; 2011. [Google Scholar]
- Schütte UME, Abdo Z, Foster J, Ravel J, Bunge J, Solheim B, et al. Bacterial diversity in a glacier foreland of the high Arctic. Mol Ecol. 2010;19:54–66. doi: 10.1111/j.1365-294X.2009.04479.x. [DOI] [PubMed] [Google Scholar]
- Sichel H. On a distribution representing sentence-length in written prose. J R Stat Soc A. 1974;137:25–34. [Google Scholar]
- Simon C, Wiezer A, Strittmatter AW, Daniel R. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl Environ Microbiol. 2009;75:7519–7526. doi: 10.1128/AEM.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer GA, Fasching C, Wilhelm L, Niggemann J, Steier P, Dittmar T, et al. Biogeochemically diverse organic matter in Alpine glaciers and its downstream fate. Nature Geosci. 2012;5:710–714. [Google Scholar]
- Tamames J, Abellán JJ, Pignatelli M, Camacho A, Moya A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010;10:85. doi: 10.1186/1471-2180-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent WF. Microbial ecosystem responses to rapid climate change in the Arctic. ISME J. 2010;4:1089–1091. doi: 10.1038/ismej.2010.108. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JV, Tockner K, Arscott DB, Claret C. Riverine landscape diversity. Freshwater Biol. 2002;47:517–539. [Google Scholar]
- Warnes GR, Bolker B, Gorjanc G, Grothendieck G, Korosec A, Lumley T, et al. 2011. gdata: Various R programming tools for data manipulation. R Package Version 2.8.2..
- Woodward G, Perkins DM, Brown LE. Climate change and freshwater ecosystems: impacts across multiple levels of organization. Phil Trans R Soc B. 2010;365:2093–2106. doi: 10.1098/rstb.2010.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Shang T, Chen Y, Jing Z, Yao T. Dominant bacteria and biomass in the Kuytun 51 Glacier. Appl Environ Microbiol. 2009;75:7287–7290. doi: 10.1128/AEM.00915-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.