Abstract
Despite the ubiquity of chemoautotrophic symbioses at hydrothermal vents, our understanding of the influence of environmental chemistry on symbiont metabolism is limited. Transcriptomic analyses are useful for linking physiological poise to environmental conditions, but recovering samples from the deep sea is challenging, as the long recovery times can change expression profiles before preservation. Here, we present a novel, in situ RNA sampling and preservation device, which we used to compare the symbiont metatranscriptomes associated with Alviniconcha, a genus of vent snail, in which specific host–symbiont combinations are predictably distributed across a regional geochemical gradient. Metatranscriptomes of these symbionts reveal key differences in energy and nitrogen metabolism relating to both environmental chemistry (that is, the relative expression of genes) and symbiont phylogeny (that is, the specific pathways employed). Unexpectedly, dramatic differences in expression of transposases and flagellar genes suggest that different symbiont types may also have distinct life histories. These data further our understanding of these symbionts' metabolic capabilities and their expression in situ, and suggest an important role for symbionts in mediating their hosts' interaction with regional-scale differences in geochemistry.
Keywords: symbiosis, hydrothermal vents, metatranscriptomics, chemoautotrophy, Alviniconcha
Introduction
Endosymbioses among marine invertebrates and chemoautotrophic bacteria have a key role in the ecology and biogeochemistry of deep-sea hydrothermal vents and similar environments. Symbionts derive energy by oxidizing reduced compounds (for example, sulfide, methane, hydrogen) and fix inorganic carbon, providing nutrition to their hosts (Felbeck, 1981; Fisher and Childress, 1984; Childress et al., 1986, 1991; Girguis and Childress, 2006; Petersen et al., 2011). Much is known about host biochemical and morphological adaptations to both their symbionts and the environment (reviewed in Stewart et al., 2005; Childress and Girguis, 2011).
Surprisingly less, however, is known about the relationship between symbiont physiology and the environment, in particular how variations in environmental geochemistry influence symbiont metabolic activity and, in turn, how this affects the ecology of the animal–microbe association. The symbioses between the deep-sea snail Alviniconcha and its chemoautotrophic symbionts afford a unique opportunity to examine these relationships. Alviniconcha are provannid gastropods that are indigenous to vents in the Western Pacific and Indian Ocean and harbor chemoautotrophic symbionts within host cells located in the gill (Suzuki et al., 2006). At the Eastern Lau Spreading Center in the southwestern Pacific (ELSC, Supplementary Figure S1), genetically distinct Alviniconcha ‘types' (likely cryptic species) form associations with three lineages of chemoautotrophic Proteobacteria: two γ-proteobacteria (termed γ-1 and γ-Lau) and an ɛ-proteobacterium (Beinart et al., 2012). A recent study of these host-symbiont associations (hereafter referred to as holobionts), found striking patterns of distribution along the ∼300 km length of the ELSC, wherein snails hosting ɛ-proteobacteria dominated the northern vent fields and those hosting γ-proteobacteria dominated the southern fields (Beinart et al., 2012). Vent fluids also showed marked changes in geochemistry along this range, with substantially elevated hydrogen and hydrogen sulfide concentrations in the northernmost fields (Mottl et al., 2011; Beinart et al., 2012). This unprecedented pattern of holobiont distribution across this 300 km spreading center suggests a link between symbiont physiology and the environment; namely that differences in the availability of reduced compounds may influence the realized niche of the holobionts as a function of their symbionts' metabolic capacity.
Our understanding of these observed patterns would be facilitated by analyses that reveal the symbionts' physiological poise in situ. Transcriptomic studies have been used to relate changes in gene expression to environmental conditions (Gracey, 2007; Gracey et al., 2008). Previous studies have also used transcriptomics to study gene expression in both host and chemoautotrophic symbionts (Harada et al., 2009; Stewart et al., 2011; Wendeberg et al., 2012). However, using transcriptomics to study patterns of gene expression in the deep sea (that is, in situ) is especially challenging. Organisms are typically held in ambient seawater during sampling and recovery, so their transcriptional profiles likely change in the hours between collection and preservation (Wendeberg et al., 2012).
To better examine the relationship between Alviniconcha symbiont physiology and the observed regional-scale differences in geochemistry, we developed a novel in situ sampling and preservation system that allowed us to quickly homogenize and preserve holobionts at the seafloor, simultaneously stopping transcription and stabilizing nucleic acids for downstream analysis. Using this device, we collected individual Alviniconcha from four vent fields spanning the previously observed geochemical gradient along the ELSC. Gene expression analyses revealed key differences among symbiont types in the expression of genes relating to hydrogen and sulfur oxidation. In contrast, similarities in patterns of gene expression relating to nitrogen metabolism—which deviate from canonical models of nitrate reduction—suggest a potentially unique strategy of nitrogen utilization that is shared among these symbionts. We also observed differences in gene expression that may be relevant to the maintenance and transmission of these associations. Our results clearly illustrate differences in physiological poise among these symbiont types, underscore the likely role of symbiont physiology in structuring holobiont distribution, and provide insights into how these phylogenetically distinct symbionts have evolved to exploit resources and niches in these highly dynamic environments.
Materials and methods
Instrument design
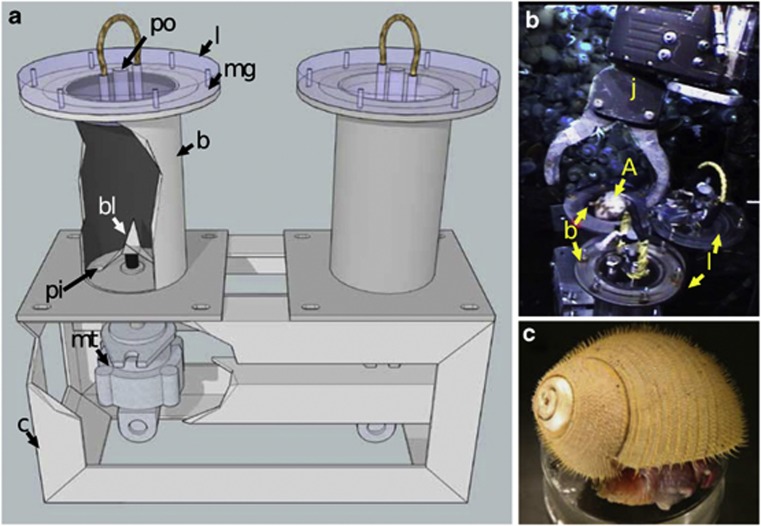
To preserve holobiont RNA in situ, we designed a sample container/homogenizer termed the In Situ Mussel And Snail Homogenizer (ISMASH; Figures 1a and b). The ISMASH consists of a 1 l stainless steel blending cylinder, the bottom of which contains a rotating blade assembly and preservative inlet. The top is open to permit sample insertion but is sealed post-collection via a magnetically-latched lid equipped with a one-way check valve. The ISMASH can be operated at any depth attainable by the submersible. During operations, the ISMASH is deployed open and empty on the submersible's working platform. When individual specimens are deposited in the cylinder, the operator places the magnetic lid on the cylinder, and RNALater (Ambion Inc., Grand Island, NY, USA) is pumped in from the bottom, displacing the less-dense seawater through the lid's check valve. After pumping sufficient RNALater to ensure thorough flushing, a hydraulic motor actuates the blade assembly and the sample is homogenized. For a complete instrument description and operational procedures, see Supplementary Information.
Figure 1.
The In Situ Mussel and Snail Homogenizer (ISMASH). Cutaway schematic of the ISMASH design (a) and a photograph of the ROV JASON II manipulator arm depositing an Alviniconcha into the ISMASH body (b), with parts labeled: po, preservative outlet; l, magnetic sealing acrylic lid; mg, magnets; b, blender body; bl, blade assembly; pi, preservative inlet; mt, motor; c, chassis; j, ROV JASON II manipulator arm; A, Alviniconcha. An Alviniconcha snail, photograph taken shipboard (c).
Sample collection
Alviniconcha holobionts (Figure 1c) were collected along the ELSC using the ROV Jason II aboard the R/V Thomas G Thompson during expedition TM-235 in 2009. One snail was collected randomly from among large aggregations at each of the four vent fields (Kilo Moana and Tow Cam in the north, ABE and Tu'i Malila in the south; Table 1, Supplementary Figure S1), then homogenized in situ. Time limitations prohibited additional sampling. Homogenization was completed within 4–10 min of collection. Upon recovery, the ∼1 l homogenates were carefully transferred to sterile glass jars, incubated overnight at 4 °C, and then frozen and maintained at −20° for ∼5 months before extraction.
Table 1. Sample information for each metatranscriptome: vent field, collection details, and host and symbiont type.
| Vent field | Dive | Date, time (GMT) | Latitude, longitude | Depth (m) | Host type (accession) | Symbiont type |
|---|---|---|---|---|---|---|
| Kilo Moana | J2-433 | 6/6/2009, 05:49 | 20 03.227 S, 176 8.008 W | 2615 | HT-II (JX134579) | ɛ |
| Tow Cam | J2-432 | 6/5/2009, 11:58 | 20 18.973 S, 176 8.195 W | 2722 | HT-II (JX134578) | ɛ |
| ABE | J2-431 | 6/3/2009, 14:29 | 20 45.794 S, 176 11.478 W | 2146 | HT-I (JX134580) | γ-1 |
| Tu'i Malila | J2-430 | 6/2/2009, 09:50 | 21 59.363 S, 176 34.105 W | 1869 | HT-III (JX134581) | γ-1/ γ-Lau |
Nucleic acid extraction, library preparation, and sequencing
Before extraction, homogenates were thawed and rehomogenized in a clean, sterile blender (Waring Inc., Torrington, CT, USA) to maximize uniformity. One 2-ml aliquot was taken from each sample and centrifuged at 14 000 × g for 10 min in a refrigerated centrifuge. RNA was extracted from the pellet using TRIzol (Invitrogen Inc., Grand Island, NY, USA) per the manufacturer's protocol. After each extraction, RNA was assessed with an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA) to determine concentration and integrity.
To maximize mRNA representation in our metatranscriptomic libraries, we preferentially removed eukaryotic and bacterial ribosomal RNA (rRNA) using sample-specific rRNA probes as in Stewart et al. (2010). Each of the four libraries was sequenced in a separate, gasketed quadrant using Titanium chemistry, yielding a full plate on a Roche Genome Sequencer FLX (Roche Inc., Basel, Switzerland).
DNA was extracted from an additional 2 ml aliquot per sample using a DNeasy Blood & Tissue kit (Qiagen Inc., Venlo, Netherlands) per the manufacturer's protocol. DNA samples were used to assess host and symbiont identities, as well as microbial diversity (while the gill endosymbionts are typically monocultures, our technique would also include any epibionts or other microbes associated with the snail). We confirmed host genotypes by sequencing 500 bp of the mitochondrial cytochrome oxidase I gene (as in Beinart et al., 2012; Table 1). The identity and abundances of the major symbiont types were assessed using SYBR Green qPCR assays on a Mx3005P real-time thermal cycler (Stratagene Inc., Santa Clara, CA, USA), using previously described primers and protocols (Beinart et al., 2012). Microbial diversity was assessed via pyrosequencing of the V1–V3 region of the bacterial 16S rRNA gene (Dowd et al., 2008).
Metatranscriptomic analyses
Sequences were filtered of rRNA sequences using BLASTN (cutoff bit score=50) against a custom database of rRNA sequences derived from the SILVA LSU and SSU databases and microbial genomes (Pruesse et al., 2007). Non-rRNA reads were further filtered for potentially artifactual duplicate sequences (reads of equal length sharing 100% sequence identity) using custom scripts as in the study of Gomez-Alvarez et al. (2009). The remaining reads were annotated using BLASTX against the NCBI nr database (as of 28 April 2011; bit score cutoff=50). BLASTX results were examined for taxonomic representation, gene content, and functional pathways in MEGAN4 (Huson et al., 2011).
Given the representation of both host and symbiont sequences in the libraries, we analyzed reads annotated as eukaryotic or bacterial separately. Each sample's eukaryotic and bacterial transcriptomes were normalized to the total number of non-rRNA reads assigned to their respective taxonomic division in MEGAN. Reads not assigned a division-level taxonomic identification were excluded from further analysis.
To validate the results from MEGAN, we submitted our entire raw transcriptomic data set and, separately, the fraction of non-rRNA reads identified as bacterial in origin to MG-RAST (Meyer et al., 2008). There was broad agreement between results derived from MEGAN and those from MG-RAST. Unless otherwise specified, all described results are derived from manual searches of the bacterial data sets with SEED/Subsystems annotations in MG-RAST (e-value=10−4). All eight datasets are now publicly available on MG-RAST (Table 2).
Table 2. Metatranscriptome sequence characteristics.
| Kilo Moana | Tow Cam | ABE | Tu'i Malila | |
|---|---|---|---|---|
| Total reads | 199 679 | 165 105 | 182 311 | 171 587 |
| rRNA reads | 102 675 | 83 436 | 67 669 | 135 141 |
| Non-rRNA reads | 97 004 | 81 669 | 114 642 | 36 446 |
| Taxon-assigned proteins | 35 497 | 35 763 | 48 666 | 17 408 |
| % Eukaryotic | 71% | 60% | 58% | 37% |
| (MG-RAST accn.) | (4492532.3) | (4492531.3) | (4492530.3) | (4492529.3) |
| % Bacterial | 27% | 38% | 39% | 59% |
| (MG-RAST accn.) | (4491348.3) | (4491346.3) | (4491347.3) | (4491344.3) |
| % γ-1a (16S) | 0/0% | 0/0% | 87/98% | 33/48% |
| % γ-Laua (16S) | 0/0% | 0/0% | 0/0% | 59/51% |
| % ɛa (16S) | 84/100% | 98/100% | 0/2% | 0/1% |
Proportion of symbiont 16S rRNA gene copies in ISMASH DNA as determined via 454 pyrosequencing/quantitative PCR.
Bacterial 16S rRNA gene diversity from amplicon pyrosequencing was assessed in QIIME v1.4.0 (Caporaso et al., 2010). Raw pyrosequencing flowgrams were denoised using the QIIME Denoiser (Reeder and Knight, 2010), then sequences were filtered for chimeras using the de novo implementation of the UCHIME chimera checker (Edgar et al., 2011). Sequences were then clustered at 97% identity using UCLUST (Edgar, 2010), and the resulting operational taxonomic units were analyzed using the default QIIME pipeline.
Results and discussion
Transcriptome characteristics and taxonomic composition
The ISMASH was highly effective at preserving RNA in situ. Assessments of extracted RNA showed good preservation, with clearly defined eukaryotic and prokaryotic rRNA peaks (Supplementary Figure S2). Rapid in situ homogenization likely facilitated the penetration of preservative throughout the tissues of these large-shelled organisms. More importantly, in situ homogenization arrests metabolism and alleviates concerns about transcriptional changes that might arise if specimens are simply submerged in a preservative in situ, or are recovered in seawater and preserved on board ship.
Between 160 000 and 200 000 metatranscriptomic reads were recovered from each sample (Table 2). Of these, 37–79% matched the rRNA database. Between 10–27% of the total reads (17 000 and 49 000) matched to protein-coding genes in the NCBI nr database. In total, we recovered between 9450 and 18 915 putatively bacterial and between 6434 and 28 412 putatively eukaryotic protein-coding transcripts per sample. Approximately 50–60% of transcripts identified as bacterial in origin were successfully assigned a functional annotation, compared with just 12–13% of eukaryotic transcripts. Consistent with our objective of examining symbiont gene expression, subsequent analyses focused solely on genes of bacterial origin. Expression data presented hereafter are normalized to the total number of protein-coding transcripts assigned to bacteria in each sample, and scaled to represent a 10 000 read library.
Host genotyping and assessment of the symbiont compositions from DNA extracts demonstrated that the distribution of sampled holobionts along the spreading center—as well as the observed specificity among host and symbiont types—was concordant with previous results (Beinart et al., 2012). Partial Alviniconcha mitochondrial cytochrome oxidase I gene sequences showed that all four sampled snails had ⩾99% identity to genotypes previously described from the ELSC (Table 1). Taxonomic assignment of bacterial 16S rRNA genes amplified from DNA extracts revealed that 84–98% of bacteria in each sample matched Alviniconcha symbiont reference sequences, with the most abundant operational taxonomic units in each sample matching reference sequences at 99–100% identity (Table 2). ɛ-proteobacterial symbionts dominated the host type II samples from the two northern fields (Kilo Moana and Tow Cam), and γ-proteobacterial symbionts dominated the host type I and type III samples from the two southern fields (ABE and Tu'i Malila, respectively). Though three of the four samples were dominated by a single symbiont phylotype, the Tu'i Malila sample appeared to simultaneously host both γ-1 and γ-Lau phylotypes (Table 2).
To help confirm that the overall taxonomic composition of the data set was reflected in key metabolic pathways, taxonomic assignments for genes involved in sulfur metabolism, hydrogen oxidation, and nitrogen metabolism were manually reviewed in MEGAN (Supplementary Table S1). Despite the uncertainties associated with taxonomy assignment to individual short reads, there was very little evidence for expression of ɛ-proteobacterial transcripts in γ-dominated samples, or vice versa.
Sulfur metabolism
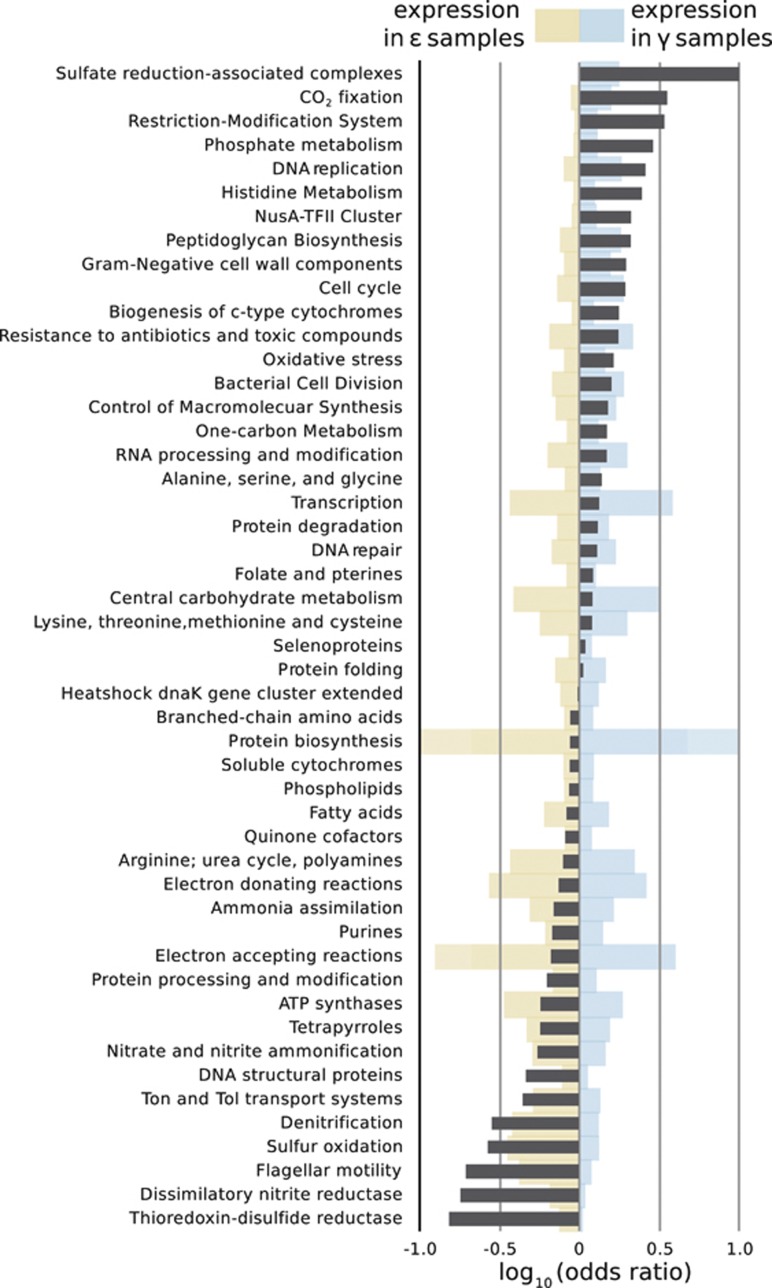
Sulfur oxidation genes were well represented in all metatranscriptomes, constituting 2–3% of the total bacterial transcripts in each sample (Figure 2, Supplementary Table S1). The γ-dominated samples showed expression of sulfur oxidation genes and pathways typical of chemoautotrophic γ-proteobacteria (Figure 3, Supplementary Table S1). From both γ-dominated samples, we recovered transcripts from the core periplasmic Sox genes, soxXYZ, though not the soxCD genes. The Sox complex without SoxCD is utilized for the incomplete oxidation of sulfide or thiosulfate, resulting in the deposition of periplasmic elemental sulfur granules (Grimm et al., 2008; Ghosh and Dam, 2009). The γ-dominated metatranscriptomes also contained transcripts for sulfide:quinone (oxido)reductases (Sqr) and sulfide dehydrogenases (Fcc), which oxidize sulfide to elemental sulfur in the periplasm (Frigaard and Dahl, 2008; Ghosh and Dam, 2009). Consistent with the expression of these genes and pathways, we frequently observe elemental sulfur granules in the gills of Alviniconcha hosting γ-proteobacteria. The further oxidation of this elemental sulfur to sulfate is thought to involve additional pathways (Ghosh and Dam, 2009), such as reverse dissimilatory sulfate reduction (rDSR). Indeed, the DSR genes dsrABCEFJKMLOPS were highly expressed in both γ-dominated samples, with dsrFHR genes present in at least one γ-dominated sample (dsrEFHR abundances determined via MG-RAST IMG annotation). The presence of dsrEFH transcripts indicates use of this pathway for the oxidation of reduced sulfur species. Here, the reverse DSR pathway is likely coupled to sulfite oxidation via an indirect pathway involving APS reductase and sulfate adenylyltransferase, both of which were identified in the γ-dominated metatranscriptomes.
Figure 2.
Summarized differences in expression of the most abundant categories of symbiont genes. Only gene categories with membership comprising >0.5% of the total data set are represented. Blue- and yellow-shaded bars indicate relative levels of expression in γ- or ɛ-dominated metatranscriptomes, respectively, on a linear scale. Dark gray bars indicate the base-10 logarithm of the odds ratio ((Gγ/Tγ)/(Gɛ/Tɛ), where G=no. reads in that category and T=total no. of reads). Positive log (odds ratios) indicate genes more likely to be expressed in γ-dominated metatranscriptomes (‘Protein biosynthesis' abundance and ‘Sulfate reduction' log ratio bars exceed the axis limits in the figure at the scale presented). Genes are summarized by the Level 2 of the SEED Subsystems ontology as annotated in MEGAN4.
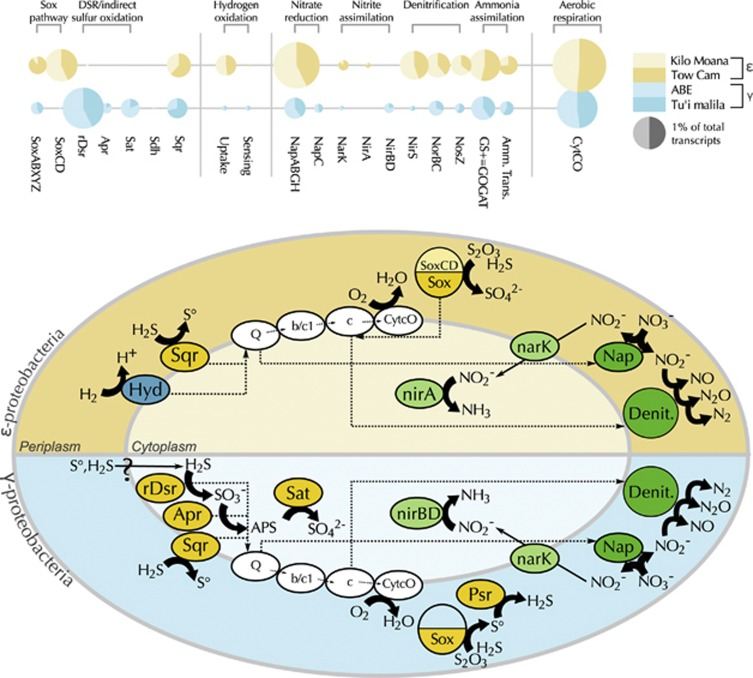
Figure 3.
Energy metabolism pathways and levels of expression in γ- and ɛ-dominated metatranscriptomes. (a) Relative abundance of genes involved in sulfur oxidation, hydrogen oxidation, nitrogen reduction and assimilation, and aerobic respiration. Circle area reflects total normalized expression for each gene category for γ- or ɛ-dominated metatranscriptomes, respectively. Circles are divided according to relative contribution of each individual sample. ɛ-dominated metatranscriptomes are shaded yellow, while γ-dominated metatranscriptomes are shaded blue. (b) Energy metabolism models for γ- and ɛ-proteobacterial symbionts represented in a stylized cell. The upper, yellow half shows the model for ɛ-symbionts, while the blue lower half shows the model for γ-symbionts. Proteins and complexes are colored by metabolic category: yellow=sulfur metabolism; white=aerobic respiration; green=nitrogen metabolism; blue=hydrogen oxidation. Arrows show general direction of electron flux. Sox, Sox multienzyme complex; rDsr, reverse dissimilatory sulfur reduction pathway; Apr, adenylylsulfate reductase; Sat, sulfur adenylyltransferase; Sdh, sulfite dehydrogenase; Sqr, sulfide quinone (oxido)reductase; Psr, polysulfide reductase; Hyd, hydrogenase; Nap, periplasmic nitrate reductase; NarK, nitrate/nitrite transporter; NirA, ferredoxin-dependent nitrite reductase; NirBD, NADH-dependent siroheme nitrite reductase; NirS, membrane-bound respiratory nitrite reductase; Nor, nitric oxide reductase; Nos, nitrous oxide reductase; GS+GOGAT, glutamine synthetase+glutamate synthase; Amm. Trans., ammonium transporter; Q, quinone; b/c1, cytochrome bc1; c, cytochrome c; CytCO, cytochrome c oxidase.
Notably, the expression of an incomplete Sox complex along with reverse DSR and indirect sulfite oxidation pathways is common among γ-proteobacterial chemosynthetic endosymbionts (Harada et al., 2009; Markert et al., 2011; Stewart et al., 2011). We detected transcripts from both pathways in the metatranscriptome derived from a snail dominated by the γ-1 phylotype, suggesting that both pathways are present and being expressed in this symbiont. However, as the other metatranscriptome was derived from a mixed symbiont community of two γ-proteobacterial phylotypes, we cannot say whether these pathways are expressed by one or both of the γ-proteobacterial symbionts.
In contrast to the γ-dominated metatranscriptomes, the ɛ-dominated metatranscriptomes from the northern two vent fields lacked reverse DSR transcripts, but were replete with complete Sox–multienzyme complex and Sqr transcripts (Figure 3, Supplementary Table S1). The full Sox–multienzyme complex is employed by ɛ-proteobacteria for the complete oxidation of sulfide, thiosulfate, and/or elemental sulfur (Yamamoto and Takai, 2011). Additionally, we detected high expression of sqr genes, despite the fact that the ɛ-proteobacterial symbiont, like its free-living relatives from the genus Sulfurimonas, has not been observed to form visible sulfur granules (Sievert et al., 2008). Many ɛ-proteobacterial genomes encode genes for Sqr in addition to the Sox complex, though the role of Sqr in sulfur oxidation and its relationship to the Sox pathway is still uncertain in these symbionts and other ɛ-proteobacteria.
Hydrogen metabolism
Previous work has shown that Alviniconcha hosting ɛ-proteobacterial symbionts dominate at the northern vent fields, where hydrogen concentrations are highest, suggesting that hydrogen might serve as an electron donor for these symbionts (Beinart et al., 2012). The metatranscriptomes from the two ɛ-dominated, northernmost samples revealed the potential for respiratory hydrogen oxidation, with both samples expressing genes allied to Group 1 NiFe-hydrogenases (∼0.5% of all transcripts, Figure 3 and Supplementary Table S1). This group of enzymes, often called uptake hydrogenases, are membrane-bound, respiratory enzymes that oxidize hydrogen and donate electrons to the quinone pool (Vignais and Billoud, 2007). Hydrogen oxidation transcripts for Group 1 NiFe-hydrogenases were also recovered—though with much lower representation—from the γ-dominated metatranscriptomes (Figure 3).
The presence of hydrogenase transcripts in all four of the Alviniconcha metatranscriptomes indicates that hydrogen could be an electron donor in both the γ- and ɛ-proteobacterial symbionts of Alviniconcha. However, the difference in expression of hydrogenases between the ɛ- and the γ-dominated individuals suggests that hydrogen oxidation potentially has a larger role in the energy metabolism of the holobionts with ɛ-proteobacteria (Figure 3, Supplementary Table S1). Phylogenetic analysis of the Alviniconcha ɛ-proteobacterial endosymbionts from the ELSC shows that they are closely allied to members of the genus Sulfurimonas (Beinart et al., 2012), many of which are able to utilize both hydrogen and reduced sulfur compounds as electron donors (Nakagawa et al., 2005). A recent study employed a suite of molecular, physiological, and geochemical approaches to show that vent mussels with γ-proteobacterial symbionts can oxidize hydrogen to support carbon fixation (Petersen et al., 2011). Future studies will use similar approaches to elucidate the degree to which these Alviniconcha ɛ-proteobacterial endosymbionts rely on hydrogen for energy production.
Carbon fixation
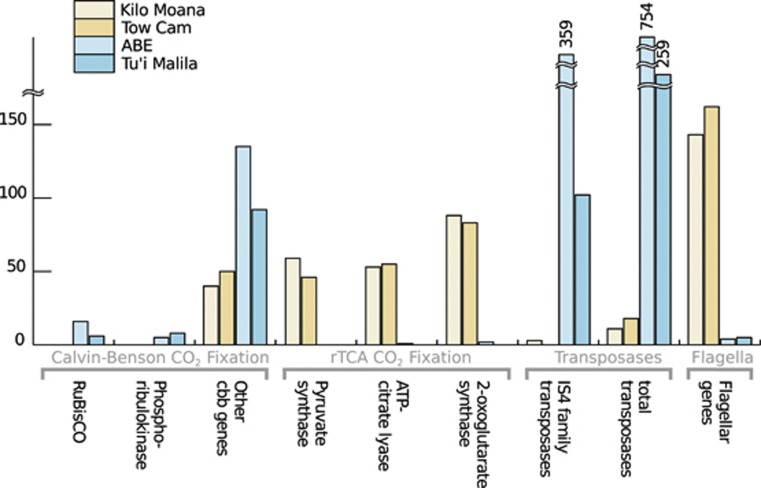
Transcript representation and abundances revealed clear differences in carbon fixation pathways between γ- and ɛ-dominated individuals (Figure 4, Supplementary Table S1). Key genes of the Calvin-Benson-Bassham cycle, including those encoding Form II Ribulose-1,5-bisphosphate carboxylase/oxygenase and phosphoribulokinase, were enriched in γ-dominated metatranscriptomes. In contrast, the three key genes associated with the reductive tricarboxylic acid (rTCA) cycle (ATP citrate lyase, 2-oxoglutarate oxidoreductase, and fumarate reductase) were found only in snails hosting ɛ-proteobacteria. These patterns are consistent with previous studies of carbon fixation in both free-living and symbiotic chemoautotrophs of these two major bacterial groups (Hugler et al., 2005; Takai et al., 2005; Woyke et al., 2006; Nakagawa and Takai, 2008; Sievert et al., 2008; Hügler et al., 2011). Both the Calvin-Benson-Bassham cycle with Form II Ribulose-1,5-bisphosphate carboxylase/oxygenase and the reductive tricarboxylic acid cycle are typically associated with autotrophs from low-oxygen environments (Berg, 2011), suggesting that the symbionts of Alviniconcha are experiencing such conditions. This may be due to low environmental oxygen concentrations around Alviniconcha or limited provisioning to the symbionts by the snails' oxygen-binding proteins hemocyanin and hemoglobin (Wittenberg and Stein, 1995).
Figure 4.
Summarized differences in expression of genes involved in carbon fixation, transposase and flagellar genes. Transcript abundance is normalized to 10 000 per sample.
Nitrogen metabolism
All organisms need nitrogen for growth and biosynthesis, requiring the assimilation of an exogenous source of nitrogen. At vents, dissolved organic nitrogen (for example, free amino acids) is quite low (Johnson et al., 1986), while inorganic nitrogen compounds are typically abundant. Some vent fluids contain nℳ to μℳ concentrations of dissolved ammonium, which is easily assimilated by many organisms. However, there are no data on ammonium concentrations at the ELSC (Tivey, 2007). Nitrate, however, is typically very abundant in seawater surrounding vents (occurring at ∼40 μℳ; Johnson et al., 1986).
Nitrate can be used both as a primary nitrogen source for biosynthesis and growth and as a respiratory terminal electron acceptor. Assimilatory nitrate reduction canonically utilizes the cytoplasmic nitrate reductase Nas. Dissimilatory nitrate reduction (DNR) frequently utilizes the membrane-bound respiratory nitrate reductase Nar, but in some bacteria may also be catalyzed via a periplasmic–enzyme complex (Nap) (Potter et al., 2001). The nitrite generated by DNR may be further reduced to ammonia via dissimilatory nitrate reduction to ammonia (DNRA), which may then be utilized for biosynthesis (though Nap has not typically been associated with nitrate assimilation, Berks, 1995). In both assimilatory nitrate reduction and DNRA, the resulting ammonium is typically assimilated by the glutamine synthetase-glutamate synthase or glutamate dehydrogenase pathways (Reitzer, 2003).
In all of the metatranscriptomes, we found the expression of some genes typically associated with both assimilatory and dissimilatory nitrate reduction, as well as for ammonium assimilation. Curiously, though, we did not find evidence for complete expression of any of the canonical pathways. Instead, periplasmic nitrate reductase (Nap) appeared to catalyze nitrate reduction as the first step for both assimilation and respiration.
Genes involved in ammonium assimilation comprised a substantial portion of all metatranscriptomes, indicating that all Alviniconcha symbionts were poised to assimilate ammonium either from the reduction of nitrate or from the environment (Figure 3, Supplementary Table S1). Both ɛ- and γ-proteobacterial symbionts showed substantial expression of glutamine synthetase-glutamate synthase and ammonium transporters. As mentioned above, all symbionts were poised for nitrate reduction in the periplasm via the Nap complex. We did not detect the periplasmic nitrite reductase Nrf, which is typically the next step in Nap-catalyzed DNRA (Potter et al., 1999). However, both γ- and ɛ-dominated transcriptomes showed expression of the narK nitrite/nitrate transporter and cytoplasmic ammonifying nitrite reductases, representing a potential mechanism for assimilation of periplasmically-reduced nitrate via the shuttling of nitrite into the cytoplasm and subsequent reduction to ammonium and assimilation by glutamine synthetase-glutamate synthase (Figure 3, Supplementary Table S1). This model represents an alternative to the typical pathway for DNRA.
The substantial expression of genes involved in respiratory denitrification suggests another possible fate for periplasmic nitrite (Figure 3, Supplementary Table S1), raising the possibility that Alviniconcha symbionts may be utilizing nitrate as an alternative electron acceptor to oxygen, potentially reducing competition for oxygen with the host (Hentschel and Felbeck, 1993; Hentschel et al., 1996). Though we did not detect transcripts for the canonical dissimilatory nitrate reductase Nar, in other bacteria, Nap has been shown to catalyze the first step in aerobic denitrification, as the presence of oxygen inhibits activity of Nar (Potter et al., 2001). We posit that Nap has a similar role in Alviniconcha symbionts. All of our samples contained transcripts for the rest of the denitrification pathway, including periplasmic respiratory cytochrome cd1 nitrite reductase NirS, the nitric oxide–reductase complex Nor, and the nitrous oxide–reductase complex Nos. Denitrification genes were more abundantly and consistently expressed in the ɛ-dominated transcriptomes, where they constituted around 2% of all reads. These genes were less abundant (0.3% of reads) in the γ-proteobacterial metatranscriptomes. Notably, NirS, which serves the reduction of nitrite to nitric oxide, and thus may serve to commit nitrite to a dissimilatory pathway, was ∼50-fold more abundant in the ɛ-dominated samples (Figure 3).
Functionally, this assemblage of assimilatory and dissimilatory genes may represent a strategy well suited for life around hydrothermal vents, where fluid mixing leads to ammonium concentrations that are inversely correlated with availability of oxygen as a terminal electron acceptor. The model outlined in Figure 3 would permit purely respiratory reduction of nitrate in holobionts exposed to oxygen-poor and ammonium-rich vent fluid. Conversely, holobionts in more aerobic conditions, with less access to ammonium, could decrease complete denitrification to dinitrogen in favor of assimilating nitrate to meet the needs of biosynthesis. Under this hypothesis, the expression differences we observed between γ- and ɛ-dominated metatranscriptomes would suggest that these symbionts were engaged primarily in either nitrogen assimilation or denitrification in response to variations in water chemistry in their respective habitats.
Despite differences in expression, the complement of nitrogen genes was remarkably consistent between ɛ- and γ-dominated metatranscriptomes. The similarities in nitrogen gene content were especially striking in light of the large differences we observed in sulfur and carbon pathways. Where the latter differences were typical of pathways found in free-living members of their respective proteobacterial classes, the combination of nitrogen-related genes we observed in both classes of Alviniconcha symbionts was fairly unusual, but could potentially catalyze functionally identical pathways in both classes of symbiont. At hydrothermal vents, the capacity to use nitrate for both biosynthesis and respiration is widespread among ɛ-proteobacteria (Sievert and Vetriani, 2012), including the free-living relatives of Alviniconcha ɛ-proteobacterial symbionts, Sulfurimonas (Takai et al., 2006; Sievert et al., 2008; Sikorski et al., 2010). Nitrate respiration is less common among free-living vent γ-proteobacteria (Sievert and Vetriani, 2012), though, curiously, patterns of nitrogen gene expression similar to the Alviniconcha symbionts have been observed in other γ-proteobacterial symbionts, such as those associated with vent tubeworms (Markert et al., 2011; Robidart et al., 2011). Although this model remains to be validated, such functional convergence may reflect similar selective pressures imposed by life in this environment, as well as the symbiotic lifestyle.
Flagellar genes
Flagellar genes showed striking differences in expression between ɛ- and γ-proteobacterial metatranscriptomes (Figure 4, Supplementary Table S1), hinting at potential differences in host–symbiont interactions. The ɛ-dominated samples expressed transcripts for at least 30 flagellum-related genes, including flagellins flaAB and transcripts associated with the flagellar hook, ring, motor, basal body, and biosynthesis (Supplementary Table S1). Only 10 flagellum-related genes were recovered from γ-dominated metatranscriptomes and, in aggregate, were ∼20-fold lower in abundance. While the diversity of roles that flagellar genes have in other symbiotic bacteria complicates interpretation (Anderson et al., 2010), we propose three hypotheses that might explain the observed differences in expression.
First, if flagella are used primarily for motility, their abundant expression in mature-host associations may signal differences in symbiont motility and transmission dynamics between the ɛ- and γ-proteobacterial symbionts. Riftia tubeworm symbionts, while possessing a large number of genes related to motility and chemotaxis, do not appear to possess flagella while inhabiting the host trophosome (Harmer et al., 2008; Robidart et al., 2008). Instead, they may utilize flagella during horizontal transmission (Harmer et al., 2008), when flagellar motility could be important in escaping from parental host tissue and/or chemotaxis towards a new host. Here, the abundant flagellar gene expression in ɛ-symbionts could indicate that they are actively transmitted throughout the lifetime of the host. The lower expression observed in the γ-hosting snails might in turn reflect either a different transmission strategy or, potentially, temporal differences in symbiont transmission.
Second, flagellar proteins are commonly used in host recognition and attachment. It is plausible that the expression of flagellar genes in ɛ-dominated metatranscriptomes relates to host recognition (that is, specificity). Flagellar proteins are critical to symbiont recognition and colonization in other systems: for example, the Euprymna-Vibrio symbioses (Nyholm et al., 2000; Millikan and Ruby, 2004), another highly specific, horizontally transmitted marine symbiosis. A previous study found that one Alviniconcha host type nearly always hosted solely ɛ-proteobacterial symbionts, while others hosted mixed populations of the two γ-proteobacterial symbiont lineages and, occasionally, the ɛ-proteobacterial symbionts (Beinart et al., 2012). The expression of flagellin in ɛ-proteobacterial symbionts may contribute to this specificity.
Finally, flagellar genes may also mediate nutritional export from symbiont to host. Ring- and hook-associated flagellar proteins have been shown to have an important secretory role in the intracellular symbionts of aphids (Maezawa et al., 2006; Toft and Fares, 2008), though they have lost genes for the flagellin tail proteins, which were abundantly expressed in our ɛ-dominated samples. In Alviniconcha, the mode of nutrient transfer in γ-hosting individuals may predominantly be via digestion of symbiont cells, as is thought to be the case for γ-proteobacterial symbionts of other vent animals (Lee et al., 1999). In contrast, translocation of small organic compounds from symbiont to host may have a bigger role in Alviniconcha hosting ɛ-proteobacteria. Compound-specific isotopic observations made by Suzuki et al. (2005) suggest as much, demonstrating that symbiont-associated fatty acids are detected in non-symbiotic host tissues of γ-, but not ɛ-hosting Alviniconcha.
Transposons
Another surprising difference between symbiont types was the increased abundance of transposases in the γ-dominated metatranscriptomes, where predicted transposases accounted for 3–7% of all bacterial transcripts via IMG in MG-RAST (SEED called around 2–5% of the transcripts transposons). Predicted transposases were present in the ɛ-dominated samples, but were much lower in abundance and matched largely to different transposase families (Figure 4, Supplementary Table S1). The presence of transposons in the genomes of other chemoautotrophic symbionts is variable: they are undetected in the genomes of the vertically-transmitted endosymbiont of vent clams (Kuwahara et al., 2007; Newton et al., 2007), while those of the horizontally transmitted vent tubeworm endosymbionts contain at least one (Robidart et al., 2008; Gardebrecht et al., 2012), and nearly 20% of an endosymbiont genome from the oligochaete Olavius algarvensis is composed of transposable elements (Woyke et al., 2006).
Moran and Plague (2004) have proposed that the proliferation of mobile genetic elements occurs early during the transition to an obligate intracellular lifestyle, as genetic bottlenecks during transmission decrease the effective population size of the symbionts, leading to decreased strength of selection. Combined with the lack of a free-living life stage, this is thought to relax purifying selection on the symbiont genome. Although conflicting with the current hypothesis that both γ-proteobacterial and ɛ-proteobacterial symbionts are transmitted via the environment, the dramatic differences in transposase expression observed among these metatranscriptomes hints at the possibility that these two distinct symbiont–host associations either represent different stages of evolutionary development, or exhibit differing transmission modes. When taken in conjunction with the corresponding differences in flagellar gene expression, these data raise the hypothesis that ɛ-proteobacterial symbionts in Alviniconcha, relative to the γ-proteobacterial symbionts, experience more frequent dispersal and fewer genetic bottlenecks.
Conclusions
Our results reveal that Alviniconcha symbionts exhibited marked differences in gene expression related to energy metabolism. The predominance of both hydrogen oxidation and DNR genes in the ɛ-dominated metatranscriptomes would suggest that these holobionts live in more highly reduced and potentially less oxygen-rich fluids. This is consistent with the previously observed patterns of distribution across a regional gradient, wherein ɛ-hosting Alviniconcha were most abundant in the more sulfidic, hydrogen-rich fluids found at the northern vent fields (Beinart et al., 2012). Though these differences in expression do not necessarily imply differences in metabolic capability (this is better addressed via genomic sequencing of each symbiont type), their striking correlation with holobiont biogeography supports the hypothesis that symbiont physiology has an important role in habitat partitioning among the host types in this genus.
Unexpectedly, the observed differences in flagellar genes and transposons between ɛ- and γ-dominated metatranscriptomes hint at differences in symbiont life histories, potentially related to the dynamics of association between host and symbiont, including mode of transmission and nutrient exchange. Though other chemoautotrophic symbioses have been described with widely varying specificities and transmission mechanisms (reviewed in Dubilier et al., 2008), Alviniconcha is thus far unique in that close relatives within the genus host unrelated symbionts, each with apparently variable degrees of specificity (Beinart et al., 2012).
The data presented here illustrate the value of using in situ preservation and shore-based transcriptomics to examine symbiont physiological poise. This technology and methodology allowed us to test a priori hypotheses as well as to identify previously unrecognized differences among these symbionts. Future studies should employ such approaches when studying both host and symbiont gene expression within and among different geochemical habitats to better understand the complex relationships among symbionts, their hosts, and the environment.
Acknowledgments
This material is based upon work supported by the National Science Foundation (OCE-0732369 to PRG and GRF grant no. DGE-1144152 to JGS and RAB), as well as Moore Foundation Investigator and Agouron Institute grants to EFD. We thank the crews of the RV Thomas G Thompson and the ROV JASON II, C DiPerna and P Meneses for assisting with the design and building of the ISMASH and J Dang for assistance with sample processing. We also thank J Bryant, R Barry and T Palden for their help in complementary DNA preparation and pyrosequencing and J Delaney, C Cavanaugh, A Knoll and C Marx for their helpful comments and editing that improved this article.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson JK, Smith TG, Hoover TR. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol. 2010;18:30–37. doi: 10.1016/j.tim.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinart RA, Sanders JG, Faure B, Sylva SP, Lee RW, Becker EL, et al. Evidence for the role of endosymbionts in regional-scale habitat partitioning by hydrothermal vent symbioses. Proc Natl Acad Sci USA. 2012;109:E3241–E3250. doi: 10.1073/pnas.1202690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg I. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks B. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress JJ, Fisher CR, Brooks JM, Kennicutt MC, Bidigare R, Anderson AE. A methanotrophic marine molluscan (bivalvia, mytilidae) symbiosis: mussels fueled by gas. Science. 1986;233:1306–1308. doi: 10.1126/science.233.4770.1306. [DOI] [PubMed] [Google Scholar]
- Childress JJ, Fisher CR, Favuzzi JA, Sanders NK. Sulfide and carbon dioxide uptake by the hydrothermal vent clam, Calyptogena magnifica, and its chemoautotrophic symbionts. Physiol Zool. 1991;64:1444–1470. [Google Scholar]
- Childress JJ, Girguis PR. The metabolic demands of endosymbiotic chemoautotrophic metabolism on host physiological capacities. J Exp Biol. 2011;214:312–325. doi: 10.1242/jeb.049023. [DOI] [PubMed] [Google Scholar]
- Dowd S, Callaway T, Wolcott R, Sun Y, McKeehan T, Hagevoort R, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Micro. 2008;6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felbeck H. Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila jones (Vestimentifera) Science. 1981;213:336–338. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- Fisher CR, Childress JJ.1984Substrate Oxidation by Trophosome Tissue from Riftia pachyptila Jones (Phylum pogonophora vol. 5Elsevier: Amsterdam; PAYS-BAS. [Google Scholar]
- Frigaard N-U, Dahl C. Sulfur Metabolism in phototrophic sulfur bacteria. Adv Microb Physiol. 2009;54:103–200. doi: 10.1016/S0065-2911(08)00002-7. [DOI] [PubMed] [Google Scholar]
- Gardebrecht A, Markert S, Sievert SM, Felbeck H, Thurmer A, Albrecht D, et al. Physiological homogeneity among the endosymbionts of Riftia pachyptila and Tevnia jerichonana revealed by proteogenomics. ISME J. 2012;6:766–776. doi: 10.1038/ismej.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh W, Dam B. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol Rev. 2009;33:999–1043. doi: 10.1111/j.1574-6976.2009.00187.x. [DOI] [PubMed] [Google Scholar]
- Girguis PR, Childress JJ. Metabolite uptake, stoichiometry and chemoautotrophic function of the hydrothermal vent tubeworm Riftia pachyptila: responses to environmental variations in substrate concentrations and temperature. J Exp Biol. 2006;209:3516–3528. doi: 10.1242/jeb.02404. [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Gracey AY. Interpreting physiological responses to environmental change through gene expression profiling. J Exp Biol. 2007;210:1584–1592. doi: 10.1242/jeb.004333. [DOI] [PubMed] [Google Scholar]
- Gracey AY, Chaney ML, Boomhower JP, Tyburczy WR, Connor K, Somero GN. Rhythms of gene expression in a fluctuating intertidal environment. Curr Biol. 2008;18:1501–1507. doi: 10.1016/j.cub.2008.08.049. [DOI] [PubMed] [Google Scholar]
- Grimm F, Franz B, Dahl C.2008Thiosulfate and Sulfur Oxidation in Purple Sulfur Bacteria Microbial Sulfur MetabolismIn: Dahl C, Friedrich CG, (eds). Springer: Berlin Heidelberg; 101–116. [Google Scholar]
- Harada M, Yoshida T, Kuwahara H, Shimamura S, Takaki Y, Kato C, et al. Expression of genes for sulfur oxidation in the intracellular chemoautotrophic symbiont of the deep-sea bivalve Calyptogena okutanii. Extremophiles. 2009;13:895–903. doi: 10.1007/s00792-009-0277-8. [DOI] [PubMed] [Google Scholar]
- Harmer TL, Rotjan RD, Nussbaumer AD, Bright M, Ng AW, DeChaine EG, et al. Free-living tube worm endosymbionts found at deep-sea vents. Appl Environ Microbiol. 2008;74:3895–3898. doi: 10.1128/AEM.02470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Felbeck H. Nitrate respiration in the hydrothermal vent tubeworm Riftia pachyptila. Nature. 1993;366:338–340. [Google Scholar]
- Hentschel U, Hand S, Felbeck H. The contribution of nitrate respiration to the energy budget of the symbiont-containing clam Lucinoma aequizonata: a calorimetric study. J Exp Biol. 1996;199:427–433. doi: 10.1242/jeb.199.2.427. [DOI] [PubMed] [Google Scholar]
- Hugler M, Wirsen CO, Fuchs G, Taylor CD, Sievert SM. Evideince for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the epsilon subdivision of Proteobacteria. J Bacteriol. 2005;187:3020–3027. doi: 10.1128/JB.187.9.3020-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hügler M, Petersen JM, Dubilier N, Imhoff JF, Sievert SM. Pathways of carbon and energy metabolism of the epibiotic community associated with the deep-sea hydrothermal vent shrimp Rimicaris exoculata. PLoS One. 2011;6:e16018. doi: 10.1371/journal.pone.0016018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KS, Beehler CL, Sakamoto-Arnold CM, Childress JJ. In situ measurements of chemical distributions in a deep-sea hydrothermal vent field. Science. 1986;231:1139–1141. doi: 10.1126/science.231.4742.1139. [DOI] [PubMed] [Google Scholar]
- Kuwahara H, Yoshida T, Takaki Y, Shimamura S, Nishi S, Harada M, et al. Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, Calyptogena okutanii. Curr Biol. 2007;17:881–886. doi: 10.1016/j.cub.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Lee RW, Robinson JJ, Cavanaugh CM. Pathways of inorganic nitrogen assimilation in chemoautotrophic bacteria-marine invertebrate symbioses: expression of host and symbiont glutamine synthetase. J Exp Biol. 1999;202:289–300. doi: 10.1242/jeb.202.3.289. [DOI] [PubMed] [Google Scholar]
- Maezawa K, Shigenobu S, Taniguchi H, Kubo T, Aizawa S-i, Morioka M. Hundreds of flagellar basal bodies cover the cell surface of the endosymbiotic bacterium Buchnera aphidicola sp. strain APS. J Bacteriol. 2006;188:6539–6543. doi: 10.1128/JB.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert S, Gardebrecht A, Felbeck H, Sievert SM, Klose J, Becher D, et al. Status quo in physiological proteomics of the uncultured Riftia pachyptila endosymbiont. Proteomics. 2011;11:3106–3117. doi: 10.1002/pmic.201100059. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass E, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikan DS, Ruby EG. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol. 2004;186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Mottl MJ, Seewald JS, Wheat CG, Tivey MK, Michael PJ, Proskurowski G, et al. Chemistry of hot springs along the Eastern Lau Spreading Center. Geochimica Et Cosmochimica Acta. 2011;75:1013–1038. [Google Scholar]
- Nakagawa S, Takai K. Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol Ecol. 2008;65:1–14. doi: 10.1111/j.1574-6941.2008.00502.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takai K, Inagaki F, Hirayama H, Nunoura T, Horikoshi K, et al. Distribution, phylogenetic diversity and physiological characteristics of epsilon- Proteobacteria in a deep-sea hydrothermal field. Environ Microbiol. 2005;7:1619–1632. doi: 10.1111/j.1462-2920.2005.00856.x. [DOI] [PubMed] [Google Scholar]
- Newton ILG, Woyke T, Auchtung TA, Dilly GF, Dutton RJ, Fisher MC, et al. The Calyptogena magnifica chemoautotrophic symbiont genome. Science. 2007;315:998–1000. doi: 10.1126/science.1138438. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–180. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- Potter L, Angove H, Richardson D, Cole J. Nitrate reduction in the periplasm of gram-negative bacteria. Adv Microb Physiol. 2001;45:51–112. doi: 10.1016/s0065-2911(01)45002-8. [DOI] [PubMed] [Google Scholar]
- Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA. Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth. Biochem J. 1999;344:77–84. [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies Jr, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Meth. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu Rev Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- Robidart JC, Bench SR, Feldman RA, Novoradovsky A, Podell SB, Gaasterland T, et al. Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ Microbiol. 2008;10:727–737. doi: 10.1111/j.1462-2920.2007.01496.x. [DOI] [PubMed] [Google Scholar]
- Robidart JC, Roque A, Song P, Girguis PR. Linking hydrothermal geochemistry to organismal physiology: physiological versatility in Riftia pachyptila from sedimented and basalt-hosted vents. PLoS One. 2011;6:e21692. doi: 10.1371/journal.pone.0021692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert SM, Scott KM, Klotz MG, Chain PSG, Hauser LJ, Hemp J, et al. Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl Environ Microbiol. 2008;74:1145–1156. doi: 10.1128/AEM.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert SM, Vetriani C. Chemoautotrophy at deep-sea vents: past, present, and future. Oceanography. 2012;25:218–233. [Google Scholar]
- Sikorski J, Munk C, Lapidus C, Djao ODN, Lucas S, Glavina DelRioT, et al. Complete genome sequence of Sulfurimonas autotrophica type strain (OK10) Stand Genomic Sci. 2010;3:194–202. doi: 10.4056/sigs.1173118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart F, Dmytrenko O, DeLong E, Cavanaugh C. Metatranscriptomic analysis of sulfur oxidation genes in the endosymbiont of Solemya velum. Front Microbiol. 2011;2:134. doi: 10.3389/fmicb.2011.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FJ, Newton ILG, Cavanaugh CM. Chemosynthetic endosymbioses: adaptations to oxic-anoxic interfaces. Trends Microbiol. 2005;13:439–448. doi: 10.1016/j.tim.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Stewart FJ, Ottesen EA, DeLong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4:896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kojima S, Sasaki T, Suzuki M, Utsumi T, Watanabe H, et al. Host-symbiont relationships in hydrothermal vent gastropods of the genus Alviniconcha from the Southwest Pacific. Appl Environ Microbiol. 2006;72:1388–1393. doi: 10.1128/AEM.72.2.1388-1393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Sasaki T, Suzuki M, Nogi Y, Miwa T, Takai K, et al. Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal-vent gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl Environ Microbiol. 2005;71:5440–5450. doi: 10.1128/AEM.71.9.5440-5450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Campbell BJ, Cary SC, Suzuki M, Oida H, Nunoura T, et al. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of epsilonproteobacteria. Appl Environ Microbiol. 2005;71:7310–7320. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Suzuki M, Nakagawa S, Miyazaki M, Suzuki Y, Inagaki F, et al. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int J Syst Evol Micr. 2006;56:1725–1733. doi: 10.1099/ijs.0.64255-0. [DOI] [PubMed] [Google Scholar]
- Tivey MK. Generation of seafloor hydrothermal vent fluids and associated mineral deposits. Oceanography. 2007;20:50–65. [Google Scholar]
- Toft C, Fares MA. The evolution of the flagellar assembly pathway in endosymbiotic bacterial genomes. Mol Biol Evol. 2008;25:2069–2076. doi: 10.1093/molbev/msn153. [DOI] [PubMed] [Google Scholar]
- Vignais PM, Billoud B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- Wendeberg A, Zielinski FU, Borowski C, Dubilier N. Expression patterns of mRNAs for methanotrophy and thiotrophy in symbionts of the hydrothermal vent mussel Bathymodiolus puteoserpentis. ISME J. 2012;6:104–112. doi: 10.1038/ismej.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg JB, Stein JL. Hemoglobin in the symbiont-harboring gill of the marine gastropod Alviniconcha hessleri. Biol Bull. 1995;188:5–7. doi: 10.2307/1542061. [DOI] [PubMed] [Google Scholar]
- Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Takai K. Sulfur metabolisms in epsilon- and gamma-Proteobacteria in deep-sea hydrothermal fields. Front Microbiol. 2011;2192 doi: 10.3389/fmicb.2011.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.