Abstract
High-elevation cold environments are considered ideal places to test hypotheses about mechanisms of bacterial colonization and succession, and about bacterial biogeography. Debris-covered glaciers (glaciers whose ablation area is mainly covered by a continuous layer of rock debris fallen from the surrounding mountains) have never been investigated in this respect so far. We used the Illumina technology to analyse the V5 and V6 hypervariable regions of the bacterial 16S rRNA gene amplified from 38 samples collected in July and September 2009 at different distances from the terminus on two debris-covered glaciers (Miage and Belvedere—Italian Alps). Heterotrophic taxa-dominated communities and bacterial community structure changed according to ice ablation rate, organic carbon content of the debris and distance from the glacier terminus. Bacterial communities therefore change during downwards debris transport, and organic carbon of these recently exposed substrates is probably provided more by allochthonous deposition of organic matter than by primary production by autotrophic organisms. We also investigated whether phylotypes of the genus Polaromonas, which is ubiquitous in cold environments, do present a biogeographical distribution by analysing the sequences retrieved in this study together with others available in the literature. We found that the genetic distance among phylotypes increased with geographic distance; however, more focused analyses using discrete distance classes revealed that both sequences collected at sites <100 km and at sites 9400–13 500 km to each other were more similar than those collected at other distance classes. Evidences of biogeographic distribution of Polaromonas phylotypes were therefore contrasting.
Keywords: biogeography, colonization, Illumina, psychrophiles, soil development, succession
Introduction
Debris-covered glaciers (DCGs) are mountain glaciers (that is, all the glaciers but ice sheets) whose ablation area is covered mostly by debris (Benn and Evans, 2010). DCGs are common on mountain ranges of Asia (Himalaya, Karakoram and Tien Shan), South America (Andes), Alaska, New Zealand and Alps (Smiraglia et al., 2000; Diolaiuti et al., 2003; Mihalcea et al., 2008; Benn and Evans, 2010). Thick supraglacial debris reduces the rate and magnitude of buried ice ablation (Østrem, 1959; Nakawo and Rana, 1999), but the debris surface can be heated by solar radiation to temperatures that can exceed +30 °C (Brock et al., 2010). The debris cover of DCGs is characterized by continuous rock substrate inputs at the highest part of the glacier. The debris is mainly composed of clasts, ranging in size from millimetres to metres, which fall on the glacier surface from surrounding mountains. The debris of DCGs therefore differs from the fine wind-blown particles that constitute the cryoconite, the most abundant debris in the central parts of debris-free glaciers (Laybourn-Parry et al., 2012). The debris is then transported down valley for times that, on some glaciers, can be as long as a century (Pelfini et al., 2007). As a result, the glacier surface is covered by a continuous debris layer, whose thickness generally increases toward the glacier terminus. The long transport on the glacier surface allows debris weathering and alteration, and its colonization not only by microorganisms but also by animals (for example, arthropods) and plants (Pelfini et al., 2007, 2012; Caccianiga et al., 2011; Gobbi et al., 2011). Ecological communities therefore exist on the surface of DCGs, and may be structured according to a chronosequence, with communities increasing in complexity towards the glacier terminus (Gobbi et al., 2011).
The rock coverage of DCGs, by virtue of its ecological features, allows the investigation of ecological hypotheses about microbial colonization and succession of microbial communities in extreme environments, but has been neglected so far in ecological and microbiological studies. Indeed, in Alpine environments, microbial succession studies have typically been conducted only on glacial forefields, where the distance from the receding glacier front served as a proxy for time since organism colonization (Sigler and Zeyer, 2002; Nicol et al., 2005; Nemergut et al., 2007; Schmidt et al., 2008; Sattin et al., 2009; Schutte et al., 2009, 2010; Philippot et al., 2011).
In this study, we collected, at increasing distances from glacier terminus, 38 samples of the debris coverage of two Italian DCGs, the Miage (Valle d' Aosta—Italy) and the Belvedere (Piemonte—Italy) glaciers, and used the Illumina technology to provide a thorough description of the microbial communities, by 16S rRNA gene tag-deep sequencing. The first aim of this study was therefore to verify two hypotheses about microbial communities in the extreme environment of DCGs: (1) that the structure of the microbial communities along the glacier surface changes according to time of exposure of the debris, and (2) that microbial successions of these environments are dominated by autotrophic bacteria in the first stages and by heterotrophic bacteria in later stages.
High-elevation cold environments are considered ideal model systems for microbial biogeographical studies, as they are ‘extreme' ecosystems, are geographically widespread and separated to one another, usually contain low microbial diversity and are linked to one another by the movements of cold dry air masses (Darcy et al., 2011). The bacterial genus Polaromonas is among the dominant bacterial taxa in glacial ice and sediments worldwide, and its presence has been reported particularly in recently deglaciated substrates (Frey et al., 2010; Darcy et al., 2011; Michaud et al., 2012). It is therefore a suitable model taxon for investigating biogeographical patterns of distribution of microorganisms. This study also aims at analysing Polaromonas sequences found on the Miage and Belevedere glaciers together with others published in the literature (Darcy et al., 2011) to investigate whether Polaromonas phylotypes do present a biogeographical distribution at global scale, as suggested recently (Darcy et al., 2011).
Materials and methods
Study sites and environmental data
The ablation zone of Miage glacier (45° 47′N, 06° 52′E, Mont Blanc massif) is debris covered from about 2400 m above sea level (a.s.l.) to the terminus at 1720 m a.s.l. The debris cover is mainly composed by gneisses, schistes and granites (Brock et al., 2010), and ranges in thickness between a few centimetres to more than 1 m at the terminus. Belvedere glacier (45° 57′N, 4° 34′E, Monte Rosa massif) is debris covered from about 2300 m a.s.l. to the terminus at 1750 m a.s.l. Rock debris is mainly composed by gneisses, micaschists and granites, and ranges in thickness between a few centimetres to more than 1 m at the terminus (Diolaiuti et al., 2003).
Debris thickness on both glaciers was estimated by analysing, according to empirical models, kinetic surface temperature maps (pixel size 90 × 90 m2) acquired by the ASTER satellite on 5 August 2009 (Belvedere glacier) and 12 August 2009 (Miage glacier) at 10:40 am local solar time (Taschner and Ranzi, 2002; Mihalcea et al., 2008). Annual surface velocity of both glaciers was measured by the differential global positioning system method (Diolaiuti et al., 2005; Caccianiga et al., 2011). Surface elevation was extracted from 2003 Digital Elevation Models. The organic carbon (OC) content of sampled debris was quantified by the dichromate oxidation and titration (Walkley and Black method; Schumacher, 2002) on the clastic fraction <2 mm, and pH was measured on the same clastic fraction in a distilled water soil suspension (1 : 2.5 soil : water ratio) by a portable pH meter (mod. HI 991300; Hanna Instruments, Woonsocket, RI, USA).
The two glaciers significantly differ in surface temperature, slope at sampling sites and pH values of sampled debris (see Supplementary Information for further details on glaciological features).
Debris sampling and DNA extraction
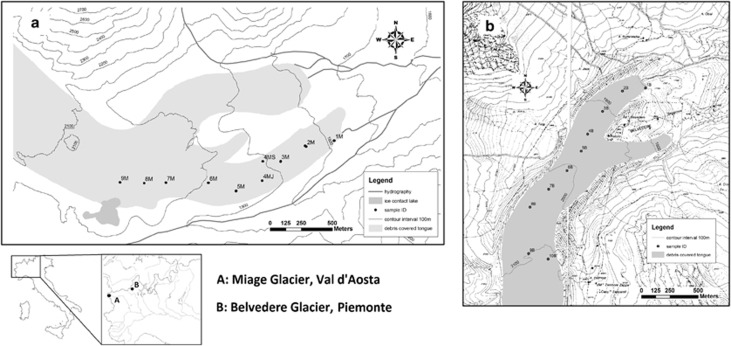
On both glaciers, samples were aseptically collected at the surface of the debris cover at approximately constant intervals (elevation and distance) from 1794 to 2043 m a.s.l. on the Miage glacier and from 1803 to 2085 m a.s.l. on the Belvedere glacier (Supplementary Table S1, Figure 1) both in July 2009 (the 28th on Miage and the 2nd on Belvedere) and September 2009 (the 10th on Miage and the 18th on Belvedere). Large clasts were discarded during sampling. The sampling point position was determined in July by a global positioning system (Oregon 300; Garmin Ltd, Olate, KS, USA). Global positioning system was used to recover the sampling sites in September. In the field, each sampling site was also visually recognized by the aid of a picture of the sampling location taken during the first visit in July. Samples were, therefore, taken at (approximately) the same site, with the only exception of sample 4 on the Miage glacier, which was sampled in different positions in July and September because the sampling point of July could not be reached in September because of a crevasse. In all, 9 samples were aseptically collected on Miage glacier and 10 on Belvedere glacier. Debris samples were kept at +4 °C during transport to the laboratory, which occurred within 12 h. Samples were then divided into two aliquots: one was stored at −20 °C for DNA extraction and the other at +4 °C for the analyses of OC and pH. No replicated samples were taken at each site because multivariate regression-like analyses (see Statistical methods) were used to investigate the change in the structure of microbial communities along an ecological gradient (Lennon, 2011).
Figure 1.
The Miage (a) and Belvedere (b) glaciers, with sampling points.
DNA was extracted from 0.5 g of debris using FastDNA Spin for soil Kit (MP Biomedicals, Solon, OH, USA) following the manufacturer's instructions. Amplification tests were carried out on the extracts and ensured that no inhibition occurred during amplification of the different DNA samples.
Further information on field and laboratory procedures is available in the Supplementary Information.
16S rRNA gene fragment sequencing
The V5 and V6 hypervariable regions of the 16S rRNA gene were polymerase chain reaction-amplified using 783F and 1027R primers (Huber et al., 2007; Wang and Qian, 2009; see also the Supplementary Information), for Genome Analyzer-IIx sequencing. Six to eight purified amplicons with different barcodes at 5′ end of the forward primer (Supplementary Table S2) were pooled in 100 μl samples with a DNA concentration of 50 ng μl−1. Multiplexed sequencing of all the pooled samples was performed on a single Illumina GA-IIx lane, using a paired-end 76 bp protocol and the 4.0 sequencing chemistry. Illumina Real-Time Analysis software, version 1.8.7 (San Diego, CA, USA), was used to perform the cluster extraction and base-calling processing analyses.
Sequence processing and data analysis
Each sequence was assigned to its original sample according to its barcode. After sorting, the reverse read of each paired-end sequence was reverse-complemented and merged with the corresponding forward read, inserting 20 Ns in between (Claesson et al., 2010). A quality cutoff was then applied to remove sequences that did not contain the barcode or with an average base quality value (Q) lower than 30. The barcode was removed from sequences before further processing.
The taxonomic attribution of the filtered sequences was carried out using the stand-alone version of Ribosomal Database Project Bayesian classifier, using 50% confidence cutoff as recently suggested for sequences shorter than 200 bp (Wang et al., 2007; Claesson et al., 2009). Operational taxonomic units (OTUs) were defined on the basis of the Ribosomal Database Project classification, considering the fourth taxonomic level, which for most bacteria corresponds to the Order.
The sequences classified as Polaromonas spp. by Ribosomal Database Project classifier with a confidence level >80% were selected and clustered by UCLUST algorithm through QIIME interface (Caporaso et al., 2010) with the following parameter values: max_accept=0, max_reject=0, sort by abundance and sequence identity 99%. The GenBank Polaromonas sequences (Darcy et al., 2011) were used as reference database. Among the sequences clustered on the seed reference sequences, we selected the unique Polaromonas sequences from the Miage and Belvedere glaciers, respectively. The use of unique sequences at each glacier avoids inflating the effect of samples collected at very short geographical distances. The 43 unique within-glacier Polaromonas sequences obtained were used in the analyses of the biogeography of Polaromonas genus (see Statistical methods) along with 55 sequences from 15 glaciers throughout the world published in Darcy et al. (2011) and obtained from the GenBank (Benson et al., 2010). Polaromonas sequences were aligned using SILVA database as template and the genetic distances between them were computed with MOTHUR (Schloss et al., 2009). The source codes used for sequence processing and data formatting are available upon request from the authors.
Statistical methods
To compare samples that largely differ in the number of sequences (6047–42 864, see also Results), 6000 reads were randomly selected from all libraries and used in the analyses of the structure of microbial communities. Such number of sequences allows an accurate determination of β-diversity among microbial communities (Caporaso et al., 2012). In addition, accumulation curves at each sampling site suggest that α-diversity could be captured with 6000 reads per sample, at least at the fourth taxonomic level (details not shown).
The number of sequences belonging to each OTU was considered an estimate of bacterial abundance at each sampling site. Ecological determinants of the structure of microbial communities were investigated by redundancy analysis (Legendre and Legendre, 1998; Borchard et al., 2011) and hierarchical cluster analysis with the complete linkage method, on Hellinger distances between bacterial communities. This distance was chosen as it is metric, depends on the difference in the proportion of OTUs between samples, decreases the importance of OTU abundance over occurrence and avoids the double-zero problem when comparing OTU composition between samples (Legendre and Legendre, 1998; De Caceres et al., 2010).
We preliminarily investigated the correlations among environmental variables to avoid entering highly correlated predictors in the statistical models (see Supplementary Information for details on variables excluded from the analyses). Predictors entered in models were distance from the glacier terminus, pH, two dichotomous variables indicating the glacier (Belvedere or Miage) and the month (July or September), slope, ablation rate, debris thickness and amount of OC in the sample. Models were simplified by backward selection of non-significant variables. Analyses were performed with the VEGAN package (Oksanen et al., 2009) and the HCLUST procedure in R 2.8.1 (R Development Core Team, 2008).
Difference in OTU abundance among glaciers or months was tested by t-tests, and variation in OTU abundance according to other environmental variables was tested by correlations. P-values were corrected for multiple testing according to the false discovery rate (FDR) procedure (Benjamini and Yekutieli, 2001) using the MULTTEST package in R 2.8.1.
For the analysis of the biogeography of Polaromonas, we calculated the matrix of great-circle distances among glaciers using in-house procedures (see Ambrosini et al., 2009). The matrix of genetic distances among sequences (see above) was then related to the matrix of geographic distances by means of Mantel tests and Mantel correlograms (corrected by the FDR procedure). Sampling sites were unevenly distributed worldwide (Supplementary Table S4 and Supplementary Figure S3). Breaks among distance classes in the Mantel correlogram were therefore arbitrary set to values corresponding to gaps in the distribution of distances among glaciers. The number of genetic distances in each geographical distance class ranged between 315 and 945, thus assuring adequate power to all statistical tests. See also Supplementary Information for why this procedure was preferred over alternative ones.
Data accessibility
Demultiplexed fastq-formatted DNA sequences have been deposited to Sequence Read Archive, study accession number ERP001386 (http://www.ebi.ac.uk/ena/data/view/ERP001386). Fasta-formatted DNA sequences of Polaromonas have been uploaded as Supplementary Information.
Results
After sorting and quality filtering, the number of 2 × 76 bp paired-end sequences obtained from the sequencing was 829 657, while the number of sequences of each library ranged from 6047 to 42 864 (Supplementary Table S3).
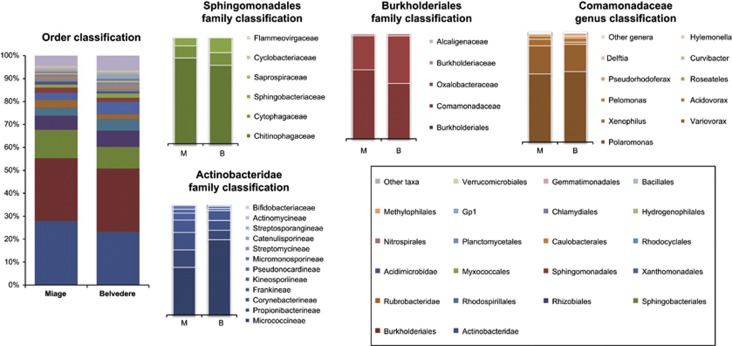
As reported in previous works, the number of sequences classified at 50% confidence level by the Bayesian classifier at a given taxonomic level sharply decreased from Domain to Genus level owing to the detection of unknown bacteria (Roesch et al., 2007; Franzetti et al., 2011). The classification of all the filtered sequences is reported at the fourth taxonomic level (Order; Figure 2 and Supplementary Figure S1) to take into account most of the sequences and to assure a classification accuracy >80% (Claesson et al., 2010). The total number of classified taxa in the sample ranged from 46 to 68 (Supplementary Table S3). At each sample, 33.9–51.7% of sequences were not classified. In all the communities, Actinobacteridae, Sphingobacteriales and Burkholderiales were the dominant taxa and accounted for >50% of the classified sequences. As reported in Figure 2, at lower taxonomic levels most of the Actinobacteridae belonged to suborder Micrococcineae and Propionibacterineae; almost all (74–80%) of the Sphingobacteriales were classified as Chitinophagaceae, while Comamonadaceae family was the dominant taxon within the order Burkholderiales. Furthermore, most of the Comamonadaceae sequences belonged to genera Polaromonas (61% for Miage and 63% for Belvedere) and Variovorax (25% for Miage and 24% for Belvedere).
Figure 2.
Taxonomic classification using a Ribosomal Database Project Bayesian classifier (50% confidence level) of the filtered sequences. Classification of all the filtered sequences is reported at the fourth taxonomic level. Classification at lower taxonomic levels is reported for Actinobacteridae, Burkholderiales and Sphingobacteriales.
Structure of microbial communities
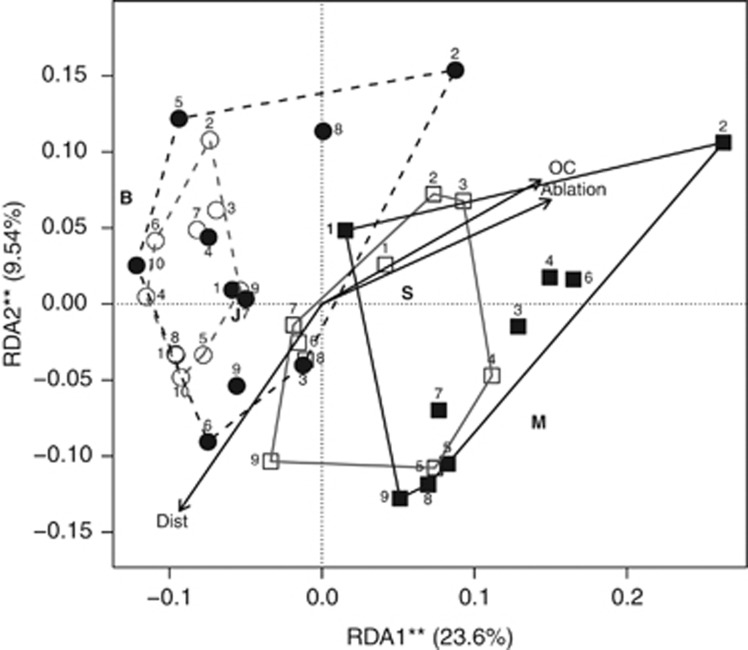
Cluster analysis partitioned samples into three main groups, which however did not correspond to any obvious subdivision according to glacier and/or season (Supplementary Figure S2). Redundancy analysis indicated that the structure of microbial communities differed significantly between glaciers and months, and changed with increasing distance from the glacier terminus, amount of OC in the debris and ablation rate at sampling site (Table 1 and Figure 3).
Table 1. Final redundancy analysis of the structure of the microbial communities in relation to environmental factors.
| Effect | df | Variance | Pseudo-F | P-value |
|---|---|---|---|---|
| Distance from the terminus | 1 | 0.001 | 1.809 | 0.047 |
| Glacier | 1 | 0.003 | 8.289 | 0.005 |
| Month | 1 | 0.001 | 1.916 | 0.035 |
| Organic C | 1 | 0.001 | 2.373 | 0.015 |
| Ablation rate | 1 | 0.001 | 2.247 | 0.010 |
| Residual | 32 | 0.011 |
Abbreviations: d.f., degrees of freedom.
Overall model significance: pseudo-F5,32=4.137, P=0.005.
Significance was assessed by a randomization procedure (Legendre and Legendre, 1998).
Figure 3.
Two-dimensional redundancy analysis plot of microbial communities observed on Belvedere (circles and dashed lines) or Miage (squares and solid lines) glacier in July (open symbols and grey lines) and September (filled symbols and black lines). The proportion of variance explained by each axis is shown. Asterisks indicate statistical significance of axes (**P<0.01). Numbers denote sampling site at each glacier and month. Arrows represent vectors of increasing distance from the glacier terminus (Dist), organic carbon content in the debris (OC) or ablation rate (Ablation). Letters indicate centroids of categorical variables entered in the analyses (B=Belvedere glacier; M=Miage glacier; J=July; S=September).
Relative abundance of Acidimicrobidae, Rubrobacteridae, Sphingobacteriales and Sphingomonadales increased (r⩾0.515, n=38, PFDR⩽0.024) at increasing levels of OC in the debris, and that of Xantomonadales significantly decreased (r=−0.501, n=38, PFDR⩽0.031) at increasing ablation rate. Distance from the terminus did not significantly affect the abundance of any OTU (|r|⩽0.453, n=38, PFDR⩾0.072). Abundance of 16 OTUs significantly differed between Belvedere and Miage glaciers: Acidimicrobidae, Actinobacteridae, Gemmatimonadales, Gp4, Nitrospirales, Rubrobacteridae, Sphingobacteriales and Sphingomonadales were more abundant in the Miage glacier than in the Belvedere glacier (t36⩽−3.390, PFDR⩽0.033), while the reverse held true for Caulobacterales, Chlamydiales, Clostridiales, Gp1, Opitutales, Planctomycetales, Rhodospirillales and Xanthomonadales (t36⩾3.600, PFDR⩽0.024). Finally, abundance of Bacillales differed significantly between months, being larger in July than in September (t36=3.446, PFDR=0.021).
Biogeography of Polaromonas genus
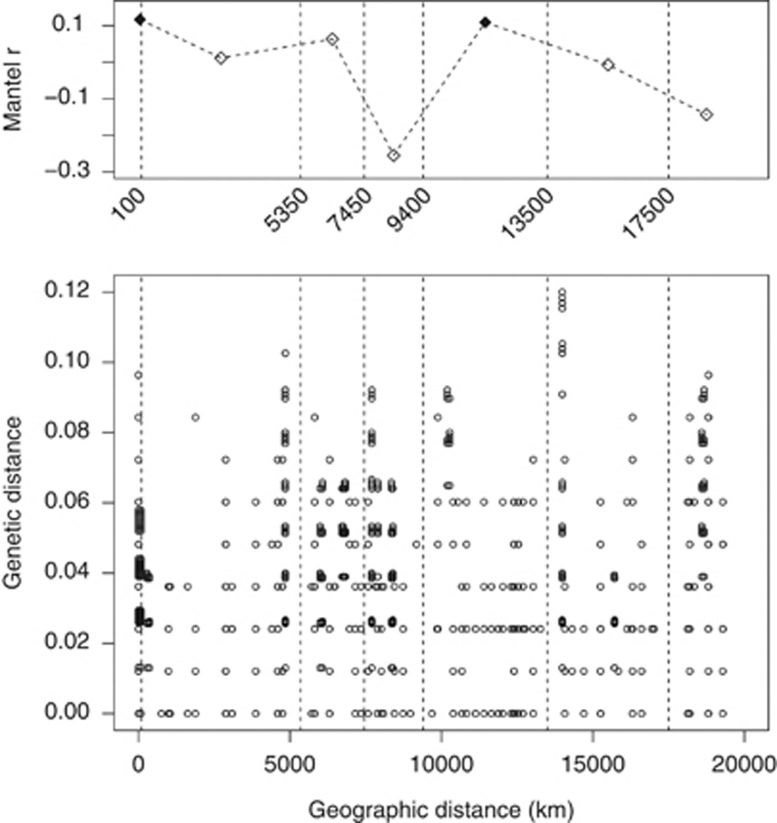
The 43 Polaromonas sequences retrieved in this study were combined with the 55 sequences from Darcy et al. (2011), who reported Polaromonas sequences from 15 sites from the entire world (Supplementary Figure S3). We re-run the analyses carried out by Darcy with this expanded data set. Genetic distance among unique within-site Polaromonas sequences was positively correlated with geographic distance among sites (rM=0.202, P<0.001, one-sided test). Mantel correlogram indicated that sequences sampled at <100 km was significantly more similar than those sampled at larger distance (rM=0.117, PFDR=0.007, one-sided tests). The same held true for sequences retrieved in glaciers between 9400 and 13 500 km to each other (rM=0.109, PFDR=0.007, one-sided tests; Figure 4). Shifting the cutoff limits of these two distance classes did not qualitatively change the results of the correlogram analysis (see Supplementary Information for details).
Figure 4.
Genetic distance among Polaromonas sequences by geographic distance between glaciers. Lower panel shows all pairwise comparisons of genetic distance among Polaromonas sequences (n=4753) and geographic distance among glaciers (n=17). Upper panel shows the Mantel correlogram of genetic distance among the same sequences separated in six distance classes. The midpoint of each distance class is plotted. Shaded diamonds indicate significant tests in the Mantel correlogram. Vertical dashed lines indicate breakpoints between distance classes. The small numbers between the panels indicate the exact position of breakpoints.
Discussion
This work represents the first attempt to characterize the microbial communities on the debris cover of alpine DCGs and to determine possible environmental factors affecting their diversity.
Microbial communities of the debris were dominated by Actinobacteridae, Sphingobacteriales and Burkholderiales. The dominant taxon was the Actinobacteridae that represented 27% and 22% of the classified sequences at Miage and Belvedere glaciers, respectively. Most of these sequences were classified as Micrococcineae and Propionibacterineae, in accordance with recent studies specifically addressing the structure of Actinobacteria in cold environments (Jiang et al., 2010; Sanyika et al., 2012). Moreover, Actinobacteridae are ubiquitous in soils under very different climatic conditions (Madigan et al., 2009) and their high abundance is consistent with the presence of OC in the sediments, despite that the average OC content of debris sampled in Miage and Belvedere glaciers was 0.83 (0.72 s.d.) g kg−1. This value is much lower than that of young soils (1–100 years) in European Alps, which range from 1.9 to 147.8 g kg−1 (Egli et al., 2012).
Sphingobacteriales, which belongs to the phylum Bacteroidetes, is the only major taxon whose relative abundance significantly increased with increasing level of OC in the debris. This confirms the results from Fierer et al. (2007), where Bacteroidetes were positively correlated with C mineralization rates, which is a proxy of OC available to microorganisms. In addition, most of Sphingobacteriales sequences (74–80%) were classified as Chitinophagaceae at the family level (Figure 2). These microorganisms are able to degrade chitin and other complex polymeric organic matter (del Rio et al., 2010). Chitinophagaceae and other chitinolytic bacteria dominated the microbial communities of Antarctic soils and glacier forefields (Brankatschk et al., 2011; Ganzert et al., 2011). Chitin provides both C and N to the microorganisms (Moorhead et al., 2012) and may be supplied by arthropods, whose presence was already documented on the Miage glacier (Gobbi et al., 2011), and by fungi, which occur in recently deglaciated soils (Zumsteg et al., 2012). Interestingly, arthropods' communities showed a turnover along the Miage glacier tongue, and richer and more structured communities occurred close to glacier terminus (Gobbi et al., 2011). Despite the fact that no data on the concentration of chitin in the debris cover of DCGs are available, we speculate that chitin might represent a relevant fraction of the OC in the debris of DCGs.
The majority of the Burkholderiales sequences retrieved in this study belonged to the Comamonadaceae family (65%) and, in particular, to the genus Polaromonas, which represented 5.6% (Miage glacier) and 5.8% (Belvedere glacier) of the total sequences classified at genus level (Figure 2). This confirms the results of Nemergut et al. (2007), who found high abundances of Polaromonas in recently deglaciated areas. According to the genomic and metagenomic data, Polaromonas possesses the dormancy-inducing gene hipA, which might enhance its ability to survive to low temperature and long-range transport (Darcy et al., 2011). Moreover, Polaromonas strains from cold soils are able to oxidize a wide range of substrates (see Darcy et al. (2011) and references therein) and it has been recently demonstrated that a Polaromonas strain is able to weather recently exposed rocks (Frey et al., 2010). Despite the fact that we have no direct evidence of these abilities in the local Polaromonas strains, we can speculate that all these features might provide Polaromonas with selective advantages over other microorganisms in colonizing new substrates and therefore contribute to its high abundances.
It has been postulated that bacterial autotrophs may play a crucial role in the initial stages of microbial colonization and ecosystem development on rock substrates due to the low nutrient content, few available OC and scarce vegetation of fresh substrates (Sigler et al., 2002). Autotrophic bacteria processes may have significant rock-weathering activity. For example, chemolithotrophic iron/sulphur oxidizers may contribute to weather pyrite-bearing rocks by lowering the pH even when present at low abundances in the microbial communities (Borin et al., 2010; Mapelli et al., 2011). Endolithic Cyanobacteria may also contribute to silica dissolution through alkalinization (Mapelli et al., 2012). Interestingly, a red surface patina can be observed on Miage debris, whose lithology shows general similarities with the one described by Borin et al. (2010), but is less widespread on the Belevedere, whose debris has a slightly different lithology and a more acidic substrate (see Supplementary Information). However, no sequence classified as Acidithiobacillales (Order) was found either on Belvedere glacier or Miage glacier, and pH values of the samples (pH>5) suggested a negligible contribution of these bacteria to rock-weathering.
The most described role of autotrophs in early succession stages is the CO2 fixation. Particularly, some works suggested that Cyanobacteria are important sources of C in very early succession stages and some surveys did report high abundances of Cyanobacteria in early colonization stages (Nemergut et al., 2007; Schmidt et al., 2008; Zumsteg et al., 2012). On the contrary, other investigations showed low abundances of Cyanobacteria in glacier forefields without any relation between their abundance and soil age (Sigler et al., 2002; Sattin et al., 2009; Goransson et al., 2011; Knelman et al., 2012). Hence, the role of Cyanobacteria in the very early stages of ecosystem development is currently controversial. Our results add another piece of evidence against the old paradigm stating that these microorganisms are early pioneer organisms. In fact, Cyanobacteria were not among the dominant taxa in the debris, as they represented only 2.1% and 0.9% of the sequences classified at Class level at Belvedere and Miage glaciers, respectively.
Redundancy analysis showed that glacier, month, distance from the terminus, ablation rate and OC content significantly influenced the structure of the bacterial communities. Abundance of 16 taxa significantly differed from Belvedere and Miage glaciers. This is not surprising since the importance of local factors such as climatic conditions, bedrock composition, soil texture and pH in influencing bacterial diversity has been already documented (Mannisto et al., 2007; Lazzaro et al., 2009; Zumsteg et al., 2012). Particularly, the influence of pH was reported in several papers, although with contrasting results (Eskelinen et al., 2009; Lauber et al., 2009; Borin et al., 2010; King et al., 2010; Philippot et al., 2011; Knelman et al., 2012). Despite the fact that the microbial community structure did not show any significant relation with pH, the debris of the Belvedere glacier was on average more acidic than that of the Miage glacier (see Supplementary Information). Hence, the observed difference in microbial community structure between the two glaciers may reflect a difference in average pH of the debris.
On a DCG, distance from glacier terminus, ablation rate and OC content of the debris are intimately related. Indeed, the rock coverage of a DCG is characterized by a continuous input of fresh, poor-in-OC debris from surrounding rocks in the upper part of the glacier. During downwards transport by glacier movements, OC content in the debris increases due to several concomitant mechanisms as a longer exposure of sediments to wind-blown organic matter, increased organic activity at lower altitudes and greater debris colonization by plants and animals. This leads to a gradient in the OC content of debris. Hence, distance from the terminus is a proxy of the time during which debris weathering and bacterial and vegetation colonization occur. Local ablation rate also increases toward the terminus, and is higher in areas where glacier movements determine a finer debris cover and even exposure of ice, for example, in areas with crevasses (Pelfini et al., 2012). Hence, OC content, ablation rate and distance from the terminus may probably be considered proxies of variation in general ecological features of the supraglacial debris that occurs during down valley transport. This interpretation is corroborated by the observations that distance from the terminus is represented by a vector opposite to those representing the other two variables in the redundancy analysis plot (Figure 3). The results from the multivariate analysis therefore suggest that glacier debris cover may host chronosequences similar to those observed in glacier forefields (Sigler and Zeyer, 2002; Nicol et al., 2005; Nemergut et al., 2007; Schmidt et al., 2008; Sattin et al., 2009; Schutte et al., 2009, 2010; Philippot et al., 2011; Knelman et al., 2012; Zumsteg et al., 2012).
Overall, the results of this study of the variation in the structure of microbial communities along DCGs and the detailed analysis of the taxa that predominate in the communities suggest that bacteria with heterotrophic metabolisms dominate the microbial communities of the debris cover of the studied glaciers. Therefore, during the initial stages of soil development in these extreme environments, organic C seems to be mainly provided by allochthonous deposition of organic matter (for example, plant debris, insects, animal excrements) rather than by CO2 incorporation, as already supposed for glacier forefields (Schmidt et al., 2008; Brankatschk et al., 2011). In addition, the nutrient bioavailability in such extreme environments might be enhanced by the activity of specialized rock-weathering bacteria, and chitinolytic bacterial activities might further contribute to enrich supraglacial debris in organic matter.
The study of the biogeographical pattern of Polaromonas distribution revealed that the genetic distance among phylotypes was positively correlated with geographic distance. In addition, phylotypes collected at sites <100 km and between 9400 and 13 500 km to each other were significantly more similar than those collected at other distance classes. The significant similarity of sequences retrieved in glacial areas separated by <100 km and the significance of the Mantel test, which indicates a genetic by distance divergence of Polaromonas strains, suggests that geographic distances contribute to the genetic structuring of Polaromonas phylotypes. However, the Mantel correlogram showed that sequences retrieved at glaciers separated by 9400–13 500 km to each other were significantly more similar to each other than sequences found at glaciers in other distance classes. On the one hand, this result is not surprising, as it is similar to the findings of Darcy et al. (2011), from which most of the Polaromonas sequences used in this paper were obtained, where a significant similarity between sequences at thousands of kilometres to each other was observed. On the other hand, this result is difficult to explain, as we have no clear hypothesis about why glaciers at this distance class should host Polaromonas strains that are significantly more similar to each other than those found on glaciers at other distances. Maybe, Polaromonas strains do are ubiquitous, as suggested by Darcy et al. (2011) and in contrast with the previous results. However, if Polaromonas strains had a really cosmopolitan distribution, the Mantel correlogram would have identified no significant difference in the genetic distance between sequences at any distance class. Further investigation on a larger number of Polaromonas sequences from more glacial areas of the world are needed to assess whether strains of this bacterial genus do show a biogeographical distribution or are cosmopolitan.
In summary, the results of this study on bacterial communities of DCGs add pieces of evidence supporting the hypotheses that (1) the structure of microbial communities changes according to the time of exposure of the sediment; (2) autotrophic organisms are not the only important early pioneers of recently exposed substrates, but heterotrophic and rock-weathering organisms may play a crucial role in soil formation by exploiting allochthonous deposition of organic matter and enhancing nutrient availability. Moreover, contrasting evidences about the hypothesis on the biogeographical distribution of Polaromonas phylotypes were found, and it is still unclear whether strains of this cosmopolitan bacterial genus do show a biogeographical pattern of distribution or they have a worldwide distribution.
Acknowledgments
We gratefully thank Alessia Sacchetti for help during field work, Rocco Piazza and Alessandra Pirola for their support during sequencing and bioinformatic analyses, Chiara Compostella for chemical analysis of debris samples and Daniel Said Pullicino for helpful discussion on OC sources. Comments from two anonymous referees greatly improved the quality of the manuscript. The work was partially funded by the 2008 MIUR PRIN grant (Grant No. 2008723SYJ_001) and by the 2010-2011 MIUR PRIN grant (Grant No. 2010AYKTAB_006) to CS, and by the 2009 FAR grant to RA.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ambrosini R, Møller A, Saino N. A quantitative measure of migratory connectivity. J Theor Biol. 2009;257:203–211. doi: 10.1016/j.jtbi.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- Benn D, Evans J.2010Glaciers and Glaciation2nd edn.Hodder Education: London, UK [Google Scholar]
- Benson D, Karsch-Mizrachi I, Lipman D, Ostell J, Sayers E. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchard D, Gillet F, Legendre F. Numerical Ecology with R. Springer: New York, NY, USA; 2011. [Google Scholar]
- Borin S, Ventura S, Tambone F, Mapelli F, Schubotz F, Brusetti L, et al. Rock weathering creates oases of life in a high Arctic desert. Environ Microbiol. 2010;12:293–303. doi: 10.1111/j.1462-2920.2009.02059.x. [DOI] [PubMed] [Google Scholar]
- Brankatschk R, Towe S, Kleineidam K, Schloter M, Zeyer J. Abundances and potential activities of nitrogen cycling microbial communities along a chronosequence of a glacier forefield. ISME J. 2011;5:1025–1037. doi: 10.1038/ismej.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock BW, Mihalcea C, Kirkbride MP, Diolaiuti G, Cutler MEJ, Smiraglia C. Meteorology and surface energy fluxes in the 2005–2007 ablation seasons at the Miage debris-covered glacier, Mont Blanc Massif, Italian Alps. J Geophys Res. 2010;115:D09106. [Google Scholar]
- Caccianiga M, Andreis C, Diolaiuti G, D'Agata C, Mihalcea C, Smiraglia C. Alpine debris-covered glaciers as a habitat for plant life. Holocene. 2011;21:1011–1020. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, O'Sullivan O, Wang Q, Nikkila J, Marchesi JR, Smidt H, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Wang QO, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcy JL, Lynch RC, King AJ, Robeson MS, Schmidt SK. Global distribution of Polaromonas phylotypes—evidence for a highly successful dispersal capacity. PLoS One. 2011;6:e23742. doi: 10.1371/journal.pone.0023742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Caceres M, Legendre P, Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119:1674–1684. [Google Scholar]
- del Rio TG, Abt B, Spring S, Lapidus A, Nolan M, Tice H, et al. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034T) Stand Genomic Sci. 2010;2:87–95. doi: 10.4056/sigs.661199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diolaiuti G, D'Agata C, Smiraglia C. Belvedere Glacier, Monte Rosa, Italian Alps: tongue thickness and volume variations in the second half of the 20th century. Arct Antarct Alp Res. 2003;35:255–263. [Google Scholar]
- Diolaiuti G, Kirkbride MP, Smiraglia C, Benn DI, D'Agata C, Nicholson L. Calving processes and lake evolution at Miage glacier, Mont Blanc, Italian Alps. Ann Glaciol. 2005;40:207–214. [Google Scholar]
- Egli M, Favilli F, Krebs R, Pichler B, Dahms D. Soil organic carbon and nitrogen accumulation rates in cold and alpine environments over 1 Ma. Geoderma. 2012;183–184:109–123. [Google Scholar]
- Eskelinen A, Stark S, Mannisto M. Links between plant community composition, soil organic matter quality and microbial communities in contrasting tundra habitats. Oecologia. 2009;161:113–123. doi: 10.1007/s00442-009-1362-5. [DOI] [PubMed] [Google Scholar]
- Fierer N, Bradford M, Jackson R. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Franzetti A, Gandolfi I, Gaspari E, Ambrosini R, Bestetti G. Seasonal variability of bacteria in fine and coarse urban air particulate matter. Appl Microbiol Biotechnol. 2011;90:745–753. doi: 10.1007/s00253-010-3048-7. [DOI] [PubMed] [Google Scholar]
- Frey B, Rieder S, Brunner I, Plotze M, Koetzsch S, Lapanje A, et al. Weathering-associated bacteria from the Damma glacier forefield: physiological capabilities and impact on granite dissolution. Appl Environ Microbiol. 2010;76:4788–4796. doi: 10.1128/AEM.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzert L, Lipski A, Hubberten H, Wagner D. The impact of different soil parameters on the community structure of dominant bacteria from nine different soils located on Livingston Island, South Shetland Archipelago, Antarctica. FEMS Microbiol Ecol. 2011;76:476–491. doi: 10.1111/j.1574-6941.2011.01068.x. [DOI] [PubMed] [Google Scholar]
- Gobbi M, Isaia M, De Bernardi F. Arthropod colonisation of a debris-covered glacier. Holocene. 2011;21:343–349. [Google Scholar]
- Goransson H, Venterink HO, Baath L. Soil bacterial growth and nutrient limitation along a chronosequence from a glacier forefield. Soil Biol Biochem. 2011;43:1333–1340. [Google Scholar]
- Huber J, Mark Welch D, Morrison H, Huse S, Neal P, Butterfield D, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Jiang H, Huang Q, Deng S, Dong H, Yu B. Planktonic actinobacterial diversity along a salinity gradient of a river and five lakes on the Tibetan Plateau. Extremophiles. 2010;14:367–376. doi: 10.1007/s00792-010-0316-5. [DOI] [PubMed] [Google Scholar]
- King AJ, Freeman KR, McCormick KF, Lynch RC, Lozupone C, Knight R, et al. Biogeography and habitat modelling of high-alpine bacteria. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1055. [DOI] [PubMed] [Google Scholar]
- Knelman JE, Legg TM, O'Neill SP, Washenberger CL, González A, Cleveland CC, et al. Bacterial community structure and function change in association with colonizer plants during early primary succession in a glacier forefield. Soil Biol Biochem. 2012;46:172–180. [Google Scholar]
- Lauber C, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn-Parry J, Tranter M, Hodson AJ. The Ecology of Snow and Ice Environments. Oxford University Press: Oxford, UK; 2012. [Google Scholar]
- Lazzaro A, Abegg C, Zeyer J. Bacterial community structure of glacier forefields on siliceous and calcareous bedrock. Eur J Soil Sci. 2009;60:860–870. [Google Scholar]
- Legendre P, Legendre L.1998Numerical Ecology2nd English edn.Elsevier: Amsterdam, The Netherlands [Google Scholar]
- Lennon J. Replication, lies and lesser-known truths regarding experimental design in environmental microbiology. Environ Microbiol. 2011;13:1383–1386. doi: 10.1111/j.1462-2920.2011.02445.x. [DOI] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM, Dunlap PV, Clark DP.2009Brock Biology of Microorganisms12th edn.Pearson International Edition: San Francisco, CA, USA [Google Scholar]
- Mannisto M, Tiirola M, Haggblom M. Bacterial communities in Arctic fields of Finnish Lapland are stable but highly pH-dependent. FEMS Microbiol Ecol. 2007;59:452–465. doi: 10.1111/j.1574-6941.2006.00232.x. [DOI] [PubMed] [Google Scholar]
- Mapelli F, Marasco R, Balloi A, Rolli E, Cappitelli F, Daffonchio D, et al. Mineral–microbe interactions: biotechnological potential of bioweathering. J Biotechnol. 2012;157:473–481. doi: 10.1016/j.jbiotec.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Mapelli F, Marasco R, Rizz A, Baldi F, Ventura S, Daffonchio D, et al. Bacterial communities involved in soil formation and plant establishment triggered by pyrite bioweathering on Arctic moraines. Microbial Ecol. 2011;61:438–447. doi: 10.1007/s00248-010-9758-7. [DOI] [PubMed] [Google Scholar]
- Michaud L, Caruso C, Mangano S, Interdonato F, Bruni V, Lo Giudice A. Predominance of Flavobacterium, Pseudomonas, and Polaromonas within the prokaryotic community of freshwater shallow lakes in the northern Victoria Land, East Antarctica. FEMS Microbiol Ecol. 2012;82:391–404. doi: 10.1111/j.1574-6941.2012.01394.x. [DOI] [PubMed] [Google Scholar]
- Mihalcea C, Brock BW, Diolaiuti G, D'Agata C, Citterio M, Kirkbride MP, et al. Using ASTER satellite and ground-based surface temperature measurements to derive supraglacial debris cover and thickness patterns on Miage Glacier (Mont Blanc Massif, Italy) Cold Regions Sci Technol. 2008;52:341–354. [Google Scholar]
- Moorhead DL, Lashermes G, Sinsabaugh RL. A theoretical model of C- and N-acquiring exoenzyme activities, which balances microbial demands during decomposition. Soil Biol Biochem. 2012;53:133–141. [Google Scholar]
- Nakawo M, Rana B. Estimate of ablation rate of glacier ice under a supraglacial debris layer. Geografiska Annaler. 1999;81:695–701. [Google Scholar]
- Nemergut DR, Anderson SP, Cleveland CC, Martin AP, Miller AE, Seimon A, et al. Microbial community succession in an unvegetated, recently deglaciated soil. Microbial Ecol. 2007;53:110–122. doi: 10.1007/s00248-006-9144-7. [DOI] [PubMed] [Google Scholar]
- Nicol GW, Tscherko D, Embley TM, Prosser JI. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ Microbiol. 2005;7:337–347. doi: 10.1111/j.1462-2920.2005.00698.x. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson G, Solymos P, et al. Vegan: Community Ecology Package, R package version 1.15-3. 2009.
- Østrem G. Ice melting under a thin layer of moraine and the existence of ice cores in moraine ridges. Geograf Ann. 1959;41:228–230. [Google Scholar]
- Pelfini M, Santilli M, Leonelli G, Bozzoni M. Investigating surface movements of debris-covered Miage glacier, Western Italian Alps, using dendroglaciological analysis. J Glaciol. 2007;53:141–152. [Google Scholar]
- Pelfini M, Diolaiuti G, Leonelli G, Bozzoni M, Bressan N, Brioschi D, et al. The influence of glacier surface processes on the short-term evolution of supraglacial tree vegetation: a case study of the Miage Glacier, Italian Alps. Holocene. 2012;22:847–856. [Google Scholar]
- Philippot L, Tscherko D, Bru D, Kandeler E. Distribution of high bacterial taxa across the chronosequence of two Alpine glacier forelands. Microbial Ecol. 2011;61:303–312. doi: 10.1007/s00248-010-9754-y. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria; 2008. [Google Scholar]
- Roesch L, Fulthorpe R, Riva A, Casella G, Hadwin A, Kent A, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyika TW, Stafford W, Cowan DA. The soil and plant determinants of community structures of the dominant actinobacteria in Marion Island terrestrial habitats, Sub-Antarctica. Polar Biol. 2012;35:1–13. [Google Scholar]
- Sattin SR, Cleveland CC, Hood E, Reed SC, King AJ, Schmidt SK, et al. Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol. 2009;47:673–681. doi: 10.1007/s12275-009-0194-7. [DOI] [PubMed] [Google Scholar]
- Schloss P, Westcott S, Ryabin T, Hall J, Hartmann M, Hollister E, et al. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SK, Reed SC, Nemergut DR, Grandy AS, Cleveland CC, Weintraub MN, et al. The earliest stages of ecosystem succession in high-elevation (5000 metres above sea level), recently deglaciated soils. Proc R Soc Ser B. 2008;275:2793–2802. doi: 10.1098/rspb.2008.0808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher BA. Methods of the Detemination of Total Organic Corabon (TOC) in Soils and Sediments. NCEA-C-1282. United States Environmental Protection Agency: Las Vegas, NV, USA; 2002. [Google Scholar]
- Schutte U, Abdo Z, Bent S, Williams C, Schneider G, Solheim B, et al. Bacterial succession in a glacier foreland of the high Arctic. ISME J. 2009;3:1258–1268. doi: 10.1038/ismej.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte U, Abdo Z, Foster J, Ravel J, Bunge J, Solheim B, et al. Bacterial diversity in a glacier foreland of the high Arctic. Mol Ecol. 2010;19:54–66. doi: 10.1111/j.1365-294X.2009.04479.x. [DOI] [PubMed] [Google Scholar]
- Sigler W, Crivii S, Zeyer J. Bacterial succession in glacial forefield soils characterized by community structure, activity and opportunistic growth dynamics. Microb Ecol. 2002;44:306–316. doi: 10.1007/s00248-002-2025-9. [DOI] [PubMed] [Google Scholar]
- Sigler WV, Zeyer J. Microbial diversity and activity along the forefields of two receding glaciers. Microb Ecol. 2002;43:397–407. doi: 10.1007/s00248-001-0045-5. [DOI] [PubMed] [Google Scholar]
- Smiraglia C, Diolaiuti G, Casati D, Kirkbride MP.2000Recent areal and altimetric variations of Miage Glacier (Monte Bianco massif, Italian Alps)In Nakawo M, Raymond CF, Fountain A, (eds)Debris Covered Glaciers IAHS: Wallingford, Seattle, WA, USA; 227–233. [Google Scholar]
- Taschner S, Ranzi R.2002Comparing the opportunities of LANDSAT-TM and ASTER data for monitoring a debris covered glacier in the italian alps within the GLIMS project Proceedings of the 22nd International Geoscience and Remote Sensing Symposium (IGRASS 2002) Vol. 224–28 June 2002Toronto, Canada; 1044–1046. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qian P. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One. 2009;4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumsteg A, Luster J, Göransson H, Smittenberg RH, Brunner I, Bernasconi SM, et al. Bacterial, archaeal and fungal succession in the forefield of a receding glacier. Microb Ecol. 2012;63:552–564. doi: 10.1007/s00248-011-9991-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Demultiplexed fastq-formatted DNA sequences have been deposited to Sequence Read Archive, study accession number ERP001386 (http://www.ebi.ac.uk/ena/data/view/ERP001386). Fasta-formatted DNA sequences of Polaromonas have been uploaded as Supplementary Information.