Abstract
Although numerous studies have investigated changes in soil microbial communities across space, questions about the temporal variability in these communities and how this variability compares across soils have received far less attention. We collected soils on a monthly basis (May to November) from replicated plots representing three land-use types (conventional and reduced-input row crop agricultural plots and early successional grasslands) maintained at a research site in Michigan, USA. Using barcoded pyrosequencing of the 16S rRNA gene, we found that the agricultural and early successional land uses harbored unique soil bacterial communities that exhibited distinct temporal patterns. α-Diversity, the numbers of taxa or lineages, was significantly influenced by the sampling month with the temporal variability in α-diversity exceeding the variability between land-use types. In contrast, differences in community composition across land-use types were reasonably constant across the 7-month period, suggesting that the time of sampling is less important when assessing β-diversity patterns. Communities in the agricultural soils were most variable over time and the changes were significantly correlated with soil moisture and temperature. Temporal shifts in bacterial community composition within the successional grassland plots were less predictable and are likely a product of complex interactions between the soil environment and the more diverse plant community. Temporal variability needs to be carefully assessed when comparing microbial diversity across soil types and the temporal patterns in microbial community structure can not necessarily be generalized across land uses, even if those soils are exposed to the same climatic conditions.
Keywords: temporal diversity, 16S rRNA, pyrosequencing, land use, soil moisture, biogeochemistry
Introduction
Soils are dynamic environments and the microorganisms that live in these habitats must contend or respond to changing soil conditions, yet most studies of soil bacterial communities focus on the spatial variation in the diversity and composition of soil microbial communities. In particular, there are a relatively large number of studies documenting how changes in land use alter soil bacterial communities and the biogeochemical processes they carry out. For example, we know that soils in managed agricultural systems often have very different communities from those found in unmanaged systems (Jangid et al., 2008; Wu et al., 2008), with specific management practices, including liming and nitrogen fertilization, often having strong effects on the structure of soil microbial communities (Ramirez et al., 2010; Rousk et al., 2010). We have a more limited understanding of the temporal variability in soil microbial communities, the factors that are responsible for the observed temporal patterns, and how such patterns may vary across different land-use types.
Ecologists have long sought to understand the biotic and abiotic factors that drive temporal changes in the diversity and composition of plant and animal communities. This holds true for microbial ecology as well; there are numerous studies focused on the temporal variability in aquatic microbial communities, identifying the biotic or abiotic factors driving the temporal dynamics (for example, Huber et al., 2002; Gilbert et al., 2009; Shade et al., 2012). In soil systems, there is also a reasonably large body of literature focused on temporal variability in microbially driven biogeochemical processes across time scales ranging from minutes to years, but there are far fewer studies specifically examining the temporal variability in soil microbial communities. As the composition of microbial communities can influence the rates of biogeochemical processes and their responses to changing environmental conditions (Schimel, 2001; Strickland et al., 2009), an improved understanding of the temporal variability in soil microbial communities may help build a more comprehensive understanding of soil function and how soil function changes across land-use types. More generally, temporal analyses of soil microbial communities can provide key insight into the factors influencing the overall diversity of soil microbial communities and the environmental niches inhabited by the large proportion of soil microbes that remain undescribed, just as time series analyses have provided important insight into the ecological attributes of specific plant and animal taxa (Preston, 1960; Ives and Carpenter, 2007).
The few studies that have examined temporal variability in soil microbial communities indicate that the composition of bacterial and fungal communities can vary on the scale of days (Zhang et al., 2011), seasons (Schadt et al., 2003; Kennedy et al., 2006; Lipson, 2007) and years (Buckley and Schmidt, 2003; DeBruyn et al., 2011). In some cases, the changes in these communities can be linked to changes in soil environmental conditions (Rasche et al., 2011). However, we often do not know how the observed temporal variability in microbial diversity and community composition compares in magnitude to the differences between soil types or if soil microbial communities exhibit predictable temporal variation, that is, changes in community structure and diversity that are directly related to measurable changes in soil conditions or plant phenology. Clearly our ability to resolve temporal variability in soil microbial communities will depend on the analytical methods employed and the sampling design (Frostegard et al., 2011), with some DNA-based studies indicating that spatial variability may exceed temporal variability across broader geographic gradients (Fierer and Jackson, 2006; Griffiths et al., 2011). The determination of temporal patterns in soil microbial communities requires, almost by definition, the analysis of a relatively large number of samples as it is necessary to discriminate between the temporal changes in communities and those changes that are related to the inherently high level of spatial heterogeneity observed in soil. As a result, most previous work has relied on methods (such as DNA fingerprinting) that are reasonably high throughput, but quantify community changes with limited phylogenetic or taxonomic resolution. With the availability of high-throughput sequencing-based approaches, it is now feasible to conduct detailed analyses of microbial communities across a relatively large number of samples (Knight et al., 2012), allowing us to describe the temporal variability in soil microbial communities and quantify how temporal variability compares to spatial variability within a given system.
Here we examined how the diversity and composition of soil bacterial communities changed across a 7-month period from replicated, experimental plots representing three land-use types common to the midwestern United States. We used barcoded pyrosequencing of the 16S rRNA gene to compare α-diversity (numbers of taxa or lineages) and β-diversity (differences in community composition) within and between land-use types over time. Given that land use often has a strong effect on soil bacterial communities, we hypothesized that each land use would harbor a unique bacterial community and that changes in α-diversity over time would be lower than differences in α-diversity between land-use types. Likewise, we hypothesized that β-diversity, and the relative abundances of individual taxa, would be more variable across land uses than within individual land use over time. Where we do observe significant temporal shifts in either α-or β-diversity, we hypothesized that these shifts would correspond to measured changes in soil conditions and soil biogeochemical processes.
Materials and methods
Site description and sample collection
The sampling sites are part of the main cropping experiment at W.K. Kellogg Biological Station, a long-term ecological research site located in southern Michigan, USA (42.40 °N, 85.40 °W), which includes several different land-use types common to the region. Each land use is replicated (six plots per land use) in a random block design with all plots located within 2 km of each other (individual plots are ∼1700 m2; http://houghton.kbs.msu.edu/maps/images/2008plotmap.pdf). Average annual precipitation at the site is 890 mm, with approximately half falling as snow, and the mean annual temperature is 9.0 °C. The soils are sandy loam to silt clay loams derived from glacial till and are generally classified as fine-loamy, mixed, mesic Typic Hapludalfs with total soil C and N concentrations of 1.3% and 0.13%, respectively, soil pH of 5.5 and a cation exchange capacity of 5.5 cmol kg−1.
Soil samples were collected from two agricultural land-use types (conventional and reduced-input row crops) and an early successional grassland (noted as T1, T3 and T7 at the KBS Main Cropping site, respectively). Each of the 18 plots were sampled at a∼1-month intervals during the 2008 growing season (May through November excluding September, Figure 1). The conventional till and reduced-input land-uses have an annual rotation of crop plants with corn grown on these plots in 2008. Corn was seeded in early May and the reduced-input plots were also planted with a Trifolium pratense (L.) cover crop. The plant community in the early successional land-use type is dominated by Solidago canadensis (L.), Apocynum cannabinum (L.) and Phleum pratense (L.), and has not been tilled since 1989. The early succession plots are burned annually in April. A nitrogen fertilizer solution of urea, ammonium and nitrate was applied to both conventional and reduced-input plots at planting, but the reduced-input plots received roughly a third of the total fertilizer applied to the conventional plots (156 kg ha−1). The agricultural plots also received a herbicide treatment in the middle of May. Additional details on these plots and their management can be found at http://lter.kbs.msu.edu/datatables/150.
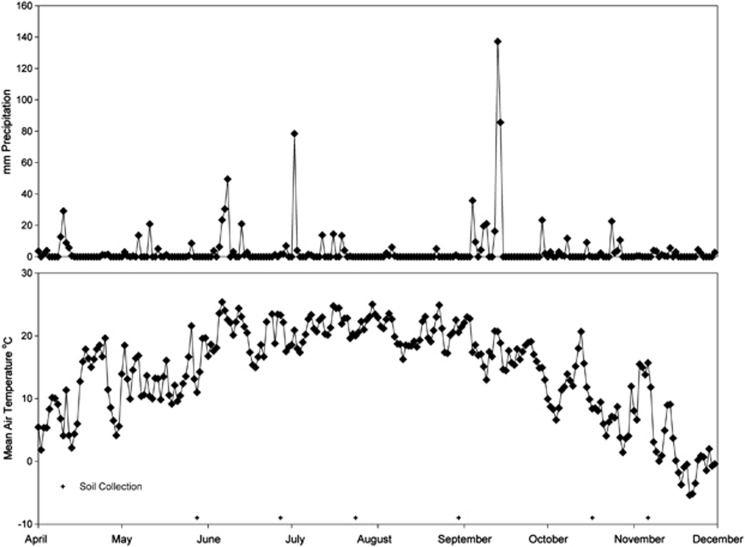
Figure 1.
Precipitation and mean air temperature across the 7-month sampling period. Daily precipitation and mean air temperature were gathered from the KBS weather station (http://lter.kbs.msu.edu/datatables/7). Plus signs (+) indicate time points when soil was collected for the bacterial community analyses.
We collected mineral soil from six locations within each plot to a depth of 5 cm using a standard soil corer (2.5 cm diameter) from each location and combining the cores to make a composite sample representing each plot at each time point. Each composite soil sample was sieved through 2-mm mesh, homogenized and stored at −20 °C before processing/extraction. Together, we analyzed a total of 108 soil samples (3 land-use types × 6 replicate plots per land use × 6 time points per plot).
Biogeochemical and environmental measurements
We evaluated trace gas flux, soil moisture and temperature to identify whether changes in bacterial diversity through time were related to biogeochemical processes. We estimated soil CO2 (kg C-CO2 ha−1 day−1), N2O and CH4 (g N-N2O or C-CH4 ha−1 day−1) fluxes using permanently installed in-situ static chambers on a bimonthly to monthly basis in four locations within each plot following the protocols described in Ambus and Robertson (Ambus and Robertson, 2006). Soil inorganic N concentrations (NH4+ and NO3−) were quantified on a monthly basis using the composited soil samples collected from each plot. Specifically, soils were extracted within 48 h via a 1-M KCl extraction (1:10 w/v), passed through a Whatman no. 1 filter and measured on an OI Analytical Flow Solution IV analyzer (OI Analytical, College Station TX, USA). The air temperature and precipitation data for our study period was based on daily averages from the weather station located at the KBS long-term ecological research (Figure 1).
Molecular analyses
The DNA was extracted with a portion of the 16S rRNA gene amplified and pyrosequenced according to methods described previously (Lauber et al., 2009). Briefly, 0.1–0.2 g of soil was loaded into bead tubes containing solution C1 of the MoBio Power Soil DNA extraction kit (MoBio Laboratories, Inc., Carlsbad, CA, USA), incubated for 10 min at 65 °C and then vortexed for 2 min at maximum speed with the MoBio vortex adapter. The remaining steps of the extraction procedure were performed as directed by the manufacturer. The V4–V5 regions of the 16S rRNA gene were amplified from the extracted DNA using primers designed to amplify bacteria and archaea with few biases against individual taxa (Bates et al., 2011; Bergmann et al., 2011). The primers anneal to positions 515 and 806 (based on Escherichia coli 16S rRNA numbering) and contain the A and B 454 FLX adapters and a 2-bp linker 5′ to the 16S-specific portion of the oligonucleotide. In addition, the forward primer contains a unique 12-bp barcode to allow sequences to be assigned to specific samples. All PCR reactions were performed in triplicate using 1 × 5 Prime Hot Master Mix (5 PRIME Inc., Gaithersburg, MD, USA), 5pmol forward and reverse primers, and 2 μl template DNA (Bates et al., 2011). Amplicons from replicate PCR reactions were pooled and cleaned using the MoBio PCR clean up kit and quantified using a Picogreen dsDNA assay (Life Technologies, Grand Island, NY, USA). An equal amount of amplified DNA from each sample was combined into a single tube and sequenced at Engencore (University of South Carolina) on a Roche 454 GS-FLX-Titanium sequencer.
Quality filtering and sequence analyses
Sequences were processed using QIIME (Caporaso et al., 2010b), an analysis pipeline available at http://qiime.sourceforge.net/, using the default parameters to eliminate sequences of poor quality (quality score <25, homopolymer runs of >6 nt and length <200 nt). Phylotypes were determined at the ⩾97% sequence similarity level using the UCLUST algorithm (Edgar, 2010). A representative sequence for each phylotype was aligned against the Greengenes coreset (DeSantis et al., 2006) using PyNAST (Caporaso et al., 2010a), with sequences classified using the Hugenholtz taxonomy via BLAST (Altschul et al., 1990). The alignment was filtered to remove common gaps with a phylogenetic tree constructed de novo using FastTree (Price et al., 2009). α-diversity (diversity of microbial communities found within individual samples) was estimated using Faith's phylogenetic diversity metric (Faith's PD (Faith, 1992) and a taxonomic metric, the number of phylotypes per sample. We relied on Faith's phylogenetic diversity metric for correlation analyses instead of the more commonly used taxonomic metrics as phylogenetic diversity levels are less sensitive to sequencing errors and/or errors introduced during de novo identification of unique phylotypes (Kuczynski et al., 2010). All downstream analyses, including α- and β-diversity analyses, as well as estimates of taxon abundances were determined at a set sequencing depth per sample (1000 randomly selected sequences per sample). This per-sample rarefaction of the data set is essential to ensure that differences in sequencing depth do not bias our estimates of α-or β-diversity (Lozupone et al., 2011; Lundin et al., 2012).
Data analysis
The overall effect of land use on α- and β-diversity was determined using ANOVAs (implemented in R, http://www.r-project.org/) and permutational MANOVAs (PerMANOVAs (Clarke and Gorley, 2006)), respectively. Temporal variability in β-diversity were assessed using the PerMANOVA procedure with month as the main factor, allowing for full permutation of the raw data with Monte Carlo tests accounting for Type III error, where the fixed effects sum to zero with 9999 permutations. Changes in β-diversity across time (using May as the baseline) were calculated for each land use as the percent difference between average monthly UniFrac distances, a phylogenetic metric of pairwise differences between communities (Lozupone and Knight, 2005). Mantel tests were conducted in PRIMER (Clarke and Gorley, 2006) to identify relationships between weighted UniFrac distances and euclidean distances of Julian date, soil gas flux and the measured soil properties. We used Pearson correlations to identify relationships between soil properties, Faith's PD and taxon abundances.
Results and discussion
Sequencing effort
The barcoded pyrosequencing of our soils resulted in 278 408 quality sequences averaging 2600 sequences per sample (ranging from 118 to 6003) with a mean read length of 304 bp (the full length of the amplified gene region). Of the initial 108 samples, one failed to sequence with an additional 9 samples having <1000 sequences, yielding a total of 98 samples that were used for downstream analyses. The sequences are deposited in EBI under the accession number ERP002214.
α-Diversity is highly variable across time
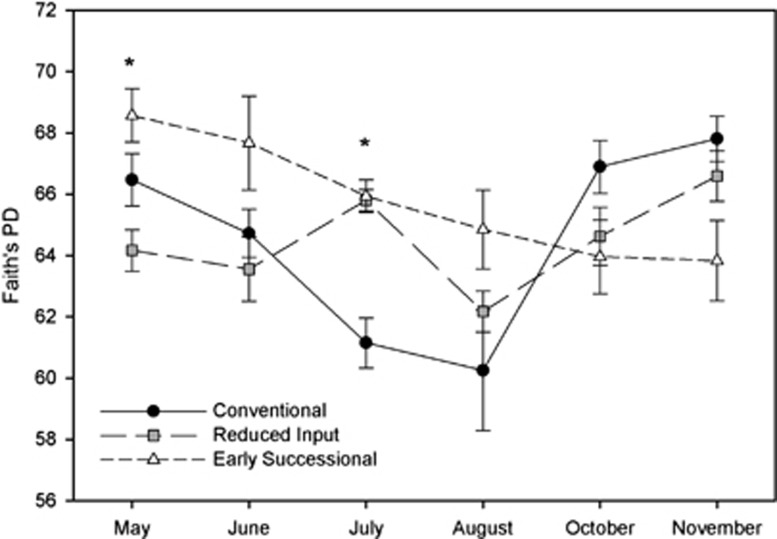
Contrary to our hypothesis, the temporal variability in α-diversity within a given land use was greater than the differences in α-diversity between land-use types (Figure 2). The α-diversity patterns between the land-use types varied across the sampling period and no single land-use type consistently harbored the most diverse bacterial communities across all time points (Figure 2). Furthermore, α-diversity on the agricultural land use was more variable than was observed in the early successional soils. These results suggest that comparisons of α-diversity levels that are based on a single time point should be considered with caution as the patterns may change over time, that is, patterns evident at a single time point may not hold across multiple time points. Spatial variability may exceed temporal variability in situations where the soils being examined represent broad gradients in soil properties (Fierer and Jackson, 2006), but this may not always be the case and, as shown here, plots with high levels of diversity in 1 month may have far lower levels of diversity if sampled in a different month.
Figure 2.
Changes in α-diversity measured using Faith's phylogenetic diversity (PD) (Faith, 1992). The average PD for each land use on each sampling date is plotted with ±1 s.e. of the mean. Asterisks indicate a significant land-use effect on Faith's PD (ANOVA P<0.05) for a given sampling date.
Relationship of α-diversity to soil properties
The distinct seasonal patterns in α-diversity within the agricultural and early successional land uses (Figure 2) suggest that a unique set of factors may be linked to the temporal dynamics of soil bacterial communities in these plots. α-Diversity was significantly correlated with soil moisture and temperature in the conventional (Pearson r=0.47 and −0.63, respectively, P<0.05 in both cases) and reduced-input land uses (r=0.72 and −0.55, respectively; P<0.05 in both cases) while only soil moisture was correlated with Faith's PD in the soils from the early successional land-use type (r=0.51, P=0.03) (Supplementary Table 1). A similar pattern of correlation was also noted between the number of phylotypes and the same edaphic factors (Supplementary Table 1). Soil inorganic N levels and fluxes of C-CH4, C-CO2 and N-N2O were poor predictors of the α-diversity patterns (Supplementary Table 1), suggesting that temporal changes in α-diversity were unrelated to changes in biogeochemical conditions over the time scale of this study.
The relationship of α-diversity to soil moisture and temperature varied across the land-use types. Whereas Faith's PD was negatively correlated with soil moisture in the agricultural land uses, it was positively correlated with diversity in the early successional plots even though the temporal patterns in soil moisture levels were nearly identical across all plots (Supplementary Table 2). These distinct diversity patterns may be related to differences in plant phenologies between the land-use types that could directly or indirectly influence the temporal patterns of diversity. In particular, we suspect that temporal differences in rhizodeposition rates may be contributing to shifts in diversity, as we know that the inputs of microbially available organic C from rhizodeposition can vary over time (Butler et al., 2003; Kaiser et al., 2010) and that microbial communities can be sensitive to these sources of organic C (Grayston et al., 2001; Macdonald et al., 2004; Paterson et al., 2007).
β-Diversity varies more across land uses than across time
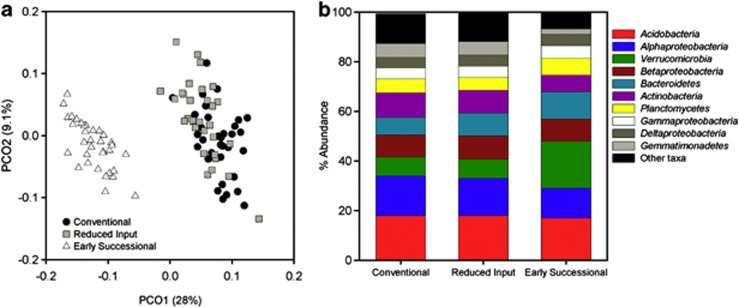
As predicted, land-use had a significant effect on bacterial β-diversity with the agricultural and early successional land-use types harboring soil bacterial communities that were distinct from one another regardless of the time the soils were collected (Figure 3a; PerMANOVA, pseudo-F=18, P<0.001, Supplementary Figure 1), and are in contrast with previous work indicating temporal variability exceeded treatment effects in a Californian grassland (Cruz-Martinez et al., 2009).The unique clustering of the early successional land use apart from the other land-use types was due to higher relative abundances of taxa belonging to the Verrucomicrobia, Bacteroidetes and Planctomycetes phyla in these soils compared with the agricultural soils (Figure 3b). In particular, Verrucomicrobia were 2.5 times more abundant in the early successional soils compared with agricultural soils, a pattern that is consistent with previous work showing this phylum to be less prevalent in cultivated soils than in other land uses (Buckley and Schmidt, 2001b). Though the bacterial communities in the conventional and reduced-input agricultural soils were significantly different from one another (PerMANOVA pseudo-F =3.6, P=0.001), the dominant phyla and proteobacterial sub-phyla shared similar relative abundances across these agricultural land-use types (Figure 3). The differences between the cultivated and early successional grassland plots (Figure 3a) mirror those reported previously where agricultural and non-agricultural soils (even those located in close proximity) often harbor distinct communities (Buckley and Schmidt, 2001a; Bissett et al., 2011; Jangid et al., 2011). However, the specific taxa driving the separation in communities across different land-use types at this site (Figure 3b) are not necessarily the same as those driving the land-use patterns observed in other studies; the taxa responses are likely variable across studies due to study-specific differences in land use, soil or site characteristics. Nevertheless, this work not only highlights that agricultural practices can have strong effects on soil biota, effects that were larger in magnitude than the temporal variability in community composition within land-use types at KBS, but also suggests that the magnitude of change over time may be specific for individual soil types or climatic regions (Cruz-Martinez et al., 2009).
Figure 3.
Weighted UniFrac and taxonomic composition between land uses. (a) Pairwise, weighted UniFrac distances visualized on principle coordinates plot (PCoA) with the percent of variation explained by each axis noted in parentheses. PerMANOVA pseudo-F and P-value for land-use effect on β-diversity are reported in the text. (b) Mean relative abundances of taxa within each land use. The abundance of each taxon was calculated as the percentage of sequences per land use for a given bacterial group.
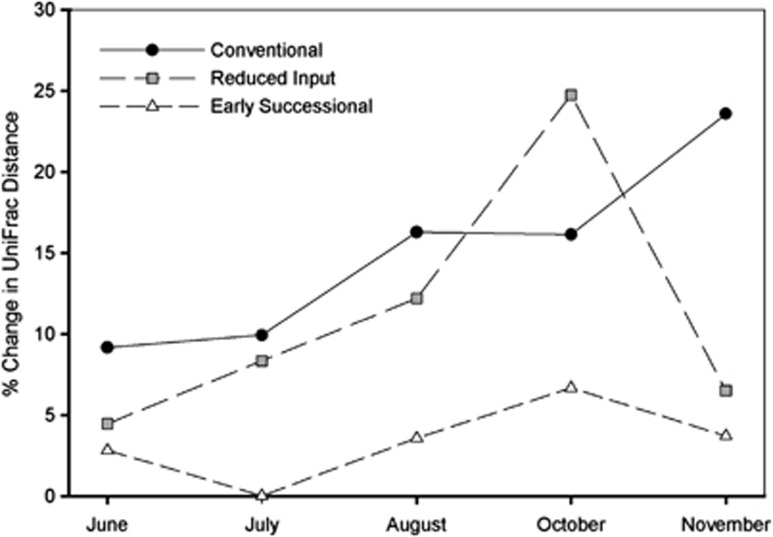
Even though the agricultural soils always had communities that were distinct from those found in the early successional soils, bacterial communities within individual land-use types did vary significantly across the growing season (pseudo-F>1.5, P<0.01 in all cases, Supplementary Figure 1). UniFrac distances changed, on average, by more than 10% from the initial time point for both the conventional and reduced-input land uses (Figure 4), while average UniFrac distances differed by only 5% in the early successional land-use plots across the time series. These patterns are reflected in Table 1 showing that the relative abundances of the dominant phyla and sub-phyla were more variable in the agricultural soils than in the early successional soils. The differences in the magnitude of the temporal variability could be related to the land management practices (for example, fertilizer applications) or differences in plant community composition and phenology. Likewise, differences in the life history strategies of the dominant bacterial taxa found in these land-use types, and their relationship to soil properties (discussed below) likely contribute to the temporal variability observed in these soils.
Figure 4.
Percent difference in UniFrac distances within land-use types over time. Changes in UniFrac distance were calculated as the percent difference from May using the average pairwise distance at each of the proceeding time points.
Table 1. Relative abundances of taxa across the sampling period in each land-use type.
|
Conventional |
Reduced input |
Early successional |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| May | June | July | August | October | November | May | June | July | August | October | November | May | June | July | August | October | November | |
| Acidobacteria | 17 | 20 | 20 | 23 | 15 | 15 | 17 | 19 | 19 | 20 | 15 | 16 | 15 | 15 | 17 | 19 | 16 | 18 |
| Alphaproteobacteria | 17 | 16 | 18 | 17 | 16 | 15 | 14 | 15 | 17 | 15 | 14 | 16 | 12 | 13 | 13 | 12 | 13 | 13 |
| Verrucomicrobia | 7.2 | 8.0 | 8.1 | 7.5 | 8.0 | 7.3 | 7.7 | 7.7 | 8.6 | 8.0 | 7.0 | 7.0 | 18 | 18 | 18 | 20 | 19 | 20 |
| Betaproteobacteria | 10 | 8.7 | 8.6 | 8.5 | 8.2 | 9.7 | 11 | 9.0 | 9.0 | 8.8 | 8.7 | 10 | 9.5 | 8.3 | 10 | 7.6 | 8.9 | 9.1 |
| Bacteroidetes | 6.8 | 6.2 | 6.2 | 7.6 | 5.6 | 8.5 | 9.6 | 10 | 8.0 | 11 | 6.5 | 8.1 | 14 | 9.7 | 9.3 | 11 | 9.3 | 10 |
| Actinobacteria | 8.6 | 9.2 | 6.6 | 7.5 | 12 | 14 | 9.3 | 7.9 | 5.6 | 7.3 | 16 | 12 | 6.7 | 8.5 | 5.0 | 5.7 | 9.3 | 5.0 |
| Planctomycetes | 5.7 | 5.5 | 5.7 | 5.9 | 5.7 | 5.3 | 4.6 | 5.2 | 5.9 | 5.5 | 5.0 | 5.1 | 6.6 | 7.2 | 6.7 | 8.3 | 6.0 | 5.8 |
| Gammaproteobacteria | 4.7 | 5.3 | 4.7 | 2.6 | 4.0 | 5.2 | 6.2 | 4.9 | 4.6 | 3.2 | 3.2 | 3.9 | 5.0 | 5.3 | 5.5 | 4.0 | 5.0 | 5.3 |
| Deltaproteobacteria | 5.3 | 4.5 | 3.8 | 3.3 | 4.9 | 4.2 | 3.8 | 4.7 | 5.1 | 4.0 | 4.8 | 5.4 | 4.5 | 5.6 | 5.6 | 3.9 | 4.2 | 4.2 |
| Gemmatimonadetes | 7.3 | 5.6 | 5.6 | 5.1 | 5.6 | 4.0 | 5.7 | 5.0 | 6.7 | 5.1 | 4.7 | 5.2 | 2.1 | 2.4 | 2.4 | 2.0 | 1.8 | 1.9 |
| Chloroflexi | 4.0 | 3.9 | 4.3 | 3.9 | 5.4 | 5.0 | 3.1 | 3.3 | 2.9 | 3.2 | 5.5 | 4.3 | 2.2 | 2.6 | 2.8 | 2.4 | 3.6 | 2.2 |
| Firmicutes | 0.78 | 1.9 | 1.1 | 1.3 | 1.3 | 1.0 | 3.3 | 1.5 | 1.1 | 1.8 | 1.3 | 1.3 | 0.35 | 0.33 | 0.45 | 0.45 | 0.60 | 1.02 |
| OP10 | 1.2 | 1.6 | 1.8 | 2.0 | 1.1 | 0.93 | 1.1 | 1.5 | 1.8 | 2.0 | 0.93 | 0.88 | 0.45 | 0.47 | 0.55 | 1.0 | 0.64 | 0.64 |
| Crenarchaeota | 1.1 | 1.0 | 1.8 | 1.2 | 1.1 | 1.1 | 1.2 | 1.6 | 1.4 | 1.5 | 1.2 | 1.6 | 0.52 | 0.53 | 0.38 | 0.47 | 0.48 | 0.92 |
| Cyanobacteria | 0.55 | 0.80 | 1.2 | 1.6 | 2.4 | 1.8 | 0.5 | 1.2 | 0.9 | 1.5 | 4.5 | 1.1 | 0.27 | 0.27 | 0.28 | 0.42 | 0.26 | 0.32 |
| Nitrospirae | 0.60 | 0.60 | 0.80 | 0.60 | 0.66 | 0.33 | 0.73 | 0.75 | 0.58 | 0.36 | 0.60 | 0.88 | 0.50 | 0.37 | 0.28 | 0.25 | 0.44 | 0.38 |
Values represent the mean abundance of each taxon for each sampling point for taxa with greater than 0.5% abundance. A color version of this table is available at the ISME journal online.
β-diversity and temporal changes in soil properties
We analyzed relationships between β-diversity, time and soil properties within each land use using Mantel tests. Within each of the land-use types, Julian date was significantly correlated with UniFrac distances across the sampling period (Table 2), but sampling date was a far better predictor of community composition in the agricultural soils than in the soils from the early successional land use (Table 2). Of the remaining factors, soil moisture and temperature were significantly correlated with community similarity in both agricultural land-use types (Global R>0.27, P<0.05 in both cases), but not in the early successional plots even though all soils shared similar moisture and temperature regimes (Supplementary Table 2). We did not find any significant relationships between measured soil gas fluxes and inorganic soil N concentrations within any of the land-use types, suggesting that temporal changes in biogeochemical processes do not correspond to temporal changes in the composition of the soil bacterial communities. The temporal variability in soil processes may be more related to changes in the relative abundances of active members of the soil community, which could be documented using RNA-based approaches (Buckley and Schmidt, 2001a; Baldrian et al., 2012), not the DNA-based approach used here. Alternatively, we would expect these processes to be controlled, in part, by the fungal communities (which were not characterized here) or only a small portion of the bacterial community that changes in membership over time, thus, obscuring any relationships between overall community composition and the measured process rates.
Table 2. Mantel correlation coefficients between soil properties and UniFrac distance.
| Conventional | Reduced input | Early successional | |
|---|---|---|---|
| C-CH4 (g ha−1 d−1) | −0.13 | −0.05 | −0.03 |
| C-CO2 (kg ha−1 d−1) | 0.004 | −0.01 | −0.07 |
| N-N2O (g ha−1 d−1) | 0.154 | 0.03 | −0.04 |
| Julian date | 0.51 | 0.38 | 0.18 |
| Percent soil moisture | 0.43 | 0.42 | 0.08 |
| Soil temperature (°C) | 0.54 | 0.27 | 0.14 |
| Inorganic N (mg kg−1) | 0.07 | 0.04 | 0.04 |
Normalized soil data were used to calculate pairwise euclidean distances before performing Mantel correlations. Bold text indicates P-values <0.05.
Temporal shifts in the relative abundances of specific bacterial taxa
Regardless of the land-use type, acidobacterial abundances typically peaked in August (19–23%) and decreased to seasonal lows (16%) during the end of the sampling period, while Actinobacteria were most abundant in October (9–16%) and November (Table 1; further summarized in Supplementary Table 3). The observed changes in acidobacterial and actinobacterial abundances were significantly correlated with soil moisture and temperature in all of the land-use types with Acidobacteria becoming less abundant when the soils were moister and cooler, and Actinobacteria exhibiting the opposite pattern (Table 1; Supplementary Table 2; Smit et al., 2001). These patterns could be related to the moisture or temperature preferences of these taxa (Placella et al., 2012) or, alternatively, these patterns may be related to a presumed late season peak in plant litter and root exudates that may favor Actinobacteria over the more oligotrophic Acidobacteria (Fierer et al., 2007). The relative abundances of Gammaproteobacteria, a group known to be sensitive to soil moisture (Lipson, 2007), were typically lowest in August when the soils were driest (Table 1, Supplementary Table 2), though relative abundances were only significantly correlated with soil moisture in the conventional land-use type (Table 1, Supplementary Table 1). Alternatively, since Gammaproteobacteria is often considered to be a copiotrophic group (Fierer et al., 2007), thriving when carbon availability is highest, the observed changes in relative abundances may be driven by changes in the quantity and quality of organic carbon available from plant litter inputs or root exudation. As carbon availability is likely to be closely tied to soil moisture and temperature conditions (Knorr et al., 2005), it is difficult to identify the specific in-situ factors driving the observed changes in relative abundances of these diverse taxonomic groups over time.
Conclusions
Not only does land use has a significant role in determining the composition of soil bacterial communities, land use also influences how these communities change over time. Both α- and β-diversity varied over time but the soils from the different land-use types did not exhibit identical temporal dynamics even though all the soils were located in close proximity and exposed to the same climatic conditions. This finding suggests that land-use type and, in particular, differences in vegetation dynamics may have a large role in modulating the temporal variability in soil bacterial communities.
α-Diversity was highly variable over time with the land uses harboring the most diverse soil bacterial communities changing on a month-to-month basis. In contrast, the agricultural and early successional land-use types always had communities that were distinct from one another regardless of the month the soils were collected, suggesting that β-diversity patterns are relatively constant over time, a pattern that has been observed elsewhere (Krave et al., 2002; Fierer and Jackson, 2006; Wallenstein et al., 2007). We observed significant differences in β-diversity over time, however, these differences were generally lower in magnitude than the differences between the agricultural and early successional land-use types. Within land-use types, shifts in community composition were often correlated with soil moisture and temperature conditions, suggesting that these factors directly or indirectly regulate the structure of soil bacterial communities. Additional research is required to identify the specific drivers of the temporal dynamics exhibited by soil bacterial communities and to determine whether the observed shifts in bacterial community composition parallel changes in the functional attributes of these communities over time.
Acknowledgments
We thank Donna Lyons, Stacey VanderWulp, Kevin Kahmark, and Cathy McMinn for assistance with the sample collection and the laboratory analyses. In addition, we thank members of the Fierer lab group for their help with manuscript preparation. This work was funded by grants awarded to NF from the National Science Foundation (DEB-0953331) and the US Department of Agriculture (2008-34158-04713). Grants to JTL from National Science Foundation (DEB 0842441) and the US Department of Agriculture (2008-35107-04481 and 2011-67019-30225).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Bio. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ambus P, Robertson GP. The effect of increased n deposition on nitrous oxide, methane and carbon dioxide fluxes from unmanaged forest and grassland communities in Michigan. Biogeochemistry. 2006;79:315–337. [Google Scholar]
- Baldrian P, Kolarik M, Stursova M, Kopecky J, Valaskova V, Vetrovsky T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6:248–258. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissett A, Richardson AE, Baker G, Thrall PH. Long-term land use effects on soil microbial community structure and function. Appl Soil Ecol. 2011;51:66–78. [Google Scholar]
- Buckley DH, Schmidt TM. The structure of microbial communities in soil and the lasting impact of cultivation. Microb Ecol. 2001a;42:11–21. doi: 10.1007/s002480000108. [DOI] [PubMed] [Google Scholar]
- Buckley DH, Schmidt TM. Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soil. FEMS Microbiol Ecol. 2001b;35:105–112. doi: 10.1111/j.1574-6941.2001.tb00793.x. [DOI] [PubMed] [Google Scholar]
- Buckley DH, Schmidt TM. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ Microbiol. 2003;5:441–452. doi: 10.1046/j.1462-2920.2003.00404.x. [DOI] [PubMed] [Google Scholar]
- Butler JL, Williams MA, Bottomley PJ, Myrold DD. Microbial community dynamics associated with rhizosphere carbon flow. Appl Environ Microb. 2003;69:6793–6800. doi: 10.1128/AEM.69.11.6793-6800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010a;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010b;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Gorley RN.2006PRIMER6th ednPRIMER-E Ltd.: Plymouth, UK [Google Scholar]
- Cruz-Martinez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF. Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J. 2009;3:738–744. doi: 10.1038/ismej.2009.16. [DOI] [PubMed] [Google Scholar]
- DeBruyn JM, Nixon LT, Fawaz MN, Johnson AM, Radosevich M. Global biogeography and quantitative seasonal dynamics of gemmatimonadetes in soil. Appl Environ Microb. 2011;77:6295–6300. doi: 10.1128/AEM.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Frostegard A, Tunlid A, Baath E. Use and misuse of PLFA measurements in soils. Soil Biol Biochem. 2011;43:1621–1625. [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, et al. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- Grayston SJ, Griffith GS, Mawdsley JL, Campbell CD, Bardgett RD. Accounting for variability in soil microbial communities of temperate upland grassland ecosystems. Soil Biol Biochem. 2001;33:533–551. [Google Scholar]
- Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS. The bacterial biogeography of British soils. Environ Microbiol. 2011;13:1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- Huber JA, Butterfield DA, Baross JA. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl Environ Microb. 2002;68:1585–1594. doi: 10.1128/AEM.68.4.1585-1594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives AR, Carpenter SR. Stability and diversity of ecosystems. Science. 2007;317:58–62. doi: 10.1126/science.1133258. [DOI] [PubMed] [Google Scholar]
- Jangid K, Williams MA, Franzluebbers AJ, Sanderlin JS, Reeves JH, Jenkins MB, et al. Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol Biochem. 2008;40:2843–2853. [Google Scholar]
- Jangid K, Williams MA, Franzluebbers AJ, Schmidt TM, Coleman DC, Whitman WB. Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Biol Biochem. 2011;43:2184–2193. [Google Scholar]
- Kaiser C, Koranda M, Kitzler B, Fuchslueger L, Schnecker J, Schweiger P, et al. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 2010;187:843–858. doi: 10.1111/j.1469-8137.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N, Brodie E, Connolly J, Clipson N. Seasonal influences on fungal community structure in unimproved and improved upland grassland soils. Can J Microbiol. 2006;52:689–694. doi: 10.1139/w06-015. [DOI] [PubMed] [Google Scholar]
- Knight R, Jansson J, Field D, Fierer N, Desai N, Fuhrman JA, et al. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol. 2012;30:513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr W, Prentice IC, House JI, Holland EA. Long-term sensitivity of soil carbon turnover to warming. Nature. 2005;433:298–301. doi: 10.1038/nature03226. [DOI] [PubMed] [Google Scholar]
- Krave AS, Lin B, Braster M, Laverman AM, van Straalen NM, Roling WFM, et al. Stratification and seasonal stability of diverse bacterial communities in a Pinus merkusii (pine) forest soil in central Java, Indonesia. Environ Microbiol. 2002;4:361–373. doi: 10.1046/j.1462-2920.2002.00304.x. [DOI] [PubMed] [Google Scholar]
- Kuczynski J, Liu ZZ, Lozupone C, McDonald D, Fierer N, Knight R. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods. 2010;7:813–U867. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil ph as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microb. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson DA. Relationships between temperature responses and bacterial community structure along seasonal and altitudinal gradients. FEMS Microbiol Ecol. 2007;59:418–427. doi: 10.1111/j.1574-6941.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin D, Severin I, Logue JB, Östman Ö, Andersson AF, Lindström ES. Which sequencing depth is sufficient to describe patterns in bacterial α- and β-diversity. Environ Microbiol Rep. 2012;4:367–372. doi: 10.1111/j.1758-2229.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- Macdonald LM, Paterson E, Dawson LA, McDonald AJS. Short-term effects of defoliation on the soil microbial community associated with two contrasting Lolium perenne cultivars. Soil Biol Biochem. 2004;36:489–498. [Google Scholar]
- Paterson E, Gebbing T, Abel C, Sim A, Telfer G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 2007;173:600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- Placella SA, Brodie EL, Firestone MK. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc Natl Acad Sci USA. 2012;109:10931–10936. doi: 10.1073/pnas.1204306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston FW. Time and space and the variation of species. Ecology. 1960;41:611–627. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez KS, Lauber CL, Knight R, Bradford MA, Fierer N. Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology. 2010;91:3463–3470. doi: 10.1890/10-0426.1. [DOI] [PubMed] [Google Scholar]
- Rasche F, Knapp D, Kaiser C, Koranda M, Kitzler B, Zechmeister-Boltenstern S, et al. Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME J. 2011;5:389–402. doi: 10.1038/ismej.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- Schadt CW, Martin AP, Lipson DA, Schmidt SK. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science. 2003;301:1359–1361. doi: 10.1126/science.1086940. [DOI] [PubMed] [Google Scholar]
- Schimel J.2001Biogeochemical models: implicit vs. explicit microbiologyIn: Schulze E, Heimann M, Harrison S, Holland E, Lloyd J, Prentice IC, et al (eds).Global Biogeochemical Cycles in the Climate System Academic Press: New York, NY, USA; 177–183. [Google Scholar]
- Shade A, Read JS, Youngblut ND, Fierer N, Knight R, Kratz TK, et al. Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J. 2012;6:2153–2167. doi: 10.1038/ismej.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit E, Leeflang P, Gommans S, van den Broek J, van Mil S, Wernars K. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl Environ Microb. 2001;67:2284–2291. doi: 10.1128/AEM.67.5.2284-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland MS, Lauber C, Fierer N, Bradford MA. Testing the functional significance of microbial community composition. Ecology. 2009;90:441–451. doi: 10.1890/08-0296.1. [DOI] [PubMed] [Google Scholar]
- Wallenstein MD, McMahon S, Schimel J. Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol Ecol. 2007;59:428–435. doi: 10.1111/j.1574-6941.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- Wu T, Chellemi DO, Graham JH, Martin KJ, Rosskopf EN. Comparison of soil bacterial communities under diverse agricultural land management and crop production practices. Microb Ecol. 2008;55:293–310. doi: 10.1007/s00248-007-9276-4. [DOI] [PubMed] [Google Scholar]
- Zhang NL, Xia JY, Yu XJ, Ma KP, Wan SQ. Soil microbial community changes and their linkages with ecosystem carbon exchange under asymmetrically diurnal warming. Soil Biol Biochem. 2011;43:2053–2059. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.