Abstract
Different bacterial strains can have different value as food for heterotrophic nanoflagellates (HNF), thus modulating HNF growth and community composition. We examined the influence of prey food quality using four Limnohabitans strains, one Polynucleobacter strain and one freshwater actinobacterial strain on growth (growth rate, length of lag phase and growth efficiency) and community composition of a natural HNF community from a freshwater reservoir. Pyrosequencing of eukaryotic small subunit rRNA amplicons was used to assess time-course changes in HNF community composition. All four Limnohabitans strains and the Polynucleobacter strain yielded significant HNF community growth while the actinobacterial strain did not although it was detected in HNF food vacuoles. Notably, even within the Limnohabitans strains we found significant prey-related differences in HNF growth parameters, which could not be related only to size of the bacterial prey. Sequence data characterizing the HNF communities showed also that different bacterial prey items induced highly significant differences in community composition of flagellates. Generally, Stramenopiles dominated the communities and phylotypes closely related to Pedospumella (Chrysophyceae) were most abundant bacterivorous flagellates rapidly reacting to addition of the bacterial prey of high food quality.
Keywords: flagellate community composition, freshwater, flagellate growth, food quality of bacteria, Limnohabitans, 454 pyrosequencing
Introduction
A unique role of pelagic bacterial communities is their ability to transform dissolved organic material into particulate material making it available to higher trophic levels. Small protists, largely heterotrophic nanoflagellates (HNF) are generally considered to be the major link connecting dissolved organic material, bacteria and the grazer food chain (Jürgens and Matz, 2002; Sherr and Sherr, 2002). However, knowledge of which species or taxa of bacterioplankton are actually consumed by small protists is still quite rudimentary (Boenigk and Arndt, 2002) and available for only a few freshwater habitats with a rather limited taxonomic resolution (Jezbera et al., 2005; Pernthaler, 2005; Salcher et al., 2008). Thus, the important questions of which bacterioplankton taxa represent the major link in carbon flow to the grazer food chain (Šimek et al., 2006; Salcher et al., 2007), and which grazer taxa mediate this process in different aquatic environments remain unanswered (Montagnes et al., 2008; Nolte et al., 2010; Weber et al., 2012).
In a broad variety of freshwater ecosystems, bacterioplankton communities are frequently dominated by representatives of only a few phylogenetic clusters of Betaproteobacteria and Actinobacteria (Newton et al., 2011). Among Betaproteobacteria, two groups of microbes, differing in many aspects of their lifestyles (Jezbera et al., 2012), are globally distributed and abundant in a wide array of habitats — the genus Limnohabitans (mostly affiliated with the R-BT065 cluster, Kasalický et al., 2010, Jezbera et al., 2013) and Polynucleobacter necessarius subsp. asymbioticus (Hahn et al., 2012).
Limnohabitans bacteria are abundant in circum neutral or alkaline lakes (Šimek et al., 2010), they display high growth rates and metabolic flexibility, with a notably tight relationship to algal-derived organic substances and algal exudates (Peréz and Sommaruga, 2006; Šimek et al., 2011). Their high growth potential and a significant contribution to bulk bacterioplankton biomass are counterbalanced by a marked vulnerability to protist grazing (Jezbera et al., 2005; Šimek et al., 2006; Salcher et al., 2008). These ecological traits, together with the fact that strains representing different lineages of the genus Limnohabitans have been recently isolated (Kasalický et al., 2013), make this bacterial group an invaluable model for testing their role in carbon flow to higher trophic levels.
One can postulate that certain bacterial taxa, those with high growth and grazing induced mortality rates, should have a prominent role in carbon flow (acting as ‘link', Sherr et al., 1987) to higher trophic levels in a given environment. Thus, the growth parameters and a biomass increase of natural HNF communities feeding on such taxa can be suggested as a measure of carbon flow from a particular bacterial group to the predator and, moreover, of the food quality of a particular bacterial prey type for HNF.
Many different methods to study bacterivory by flagellates have been proposed; however, none are appropriate for assessing the dynamics of a particular prey item abundant and present in natural bacterioplankton (Montagnes et al., 2008). Most of the approaches build on ingestion of labeled bacterial prey surrogates (for example, Sherr and Sherr (1993); Montagnes et al., 2008), recently improved by detection using, for example, fluorescence in situ hybridization (FISH)-probes targeting bacteria directly in food vacuoles (Jezbera et al., 2005; Šimek et al., 2007). However, ingestion of prey need not yield population growth of HNF (Boenigk et al., 2004; Tarao et al., 2009). Novel methodical approaches are consequently needed to examine the role of particular bacterioplankton taxa in carbon transfer to HNF. We assume that prey quality and its availability can influence the community composition of HNF (Pernthaler, 2005) and this community composition can be now unveiled by using pyrosequencing of eukaryotic small subunit (SSU) rRNA amplicons (Massana et al., 2004; Nolte et al., 2010).
In this study, we exploited an innovative experimental design to address the following aims: (i) to examine growth community parameters (growth rate, lag phase and flagellate growth efficiency) of freshwater HNF assemblages feeding on bacterial strains of different food quality with focus on closely related Limnohabitans strains of diverse cell size and morphology compared with strains from Actinobacteria and Polynucleobacter lineages and (ii) to examine how the identity of the bacterial prey can modulate HNF community composition using pyrosequencing of SSU rRNA amplicons.
Materials and methods
Experimental organisms
Six different bacterial strains (Table 1), from three important freshwater clades (Newton et al., 2011), were used as prey items for flagellate predators. Four bacterial strains affiliated with the genus Limnohabitans (from the R-BT065 subcluster of Betaproteobacteria, Šimek et al., 2001; Kasalický et al., 2010), of different sizes and shape, were selected: L. parvus (strain II-B4T) and L. planktonicus (strain II-D5T, Kasalický et al., 2010), both isolated from Římov reservoir (South Bohemia, Czech Republic), and further two undescribed Limnohabitans strains—2KL-27 and 2KL-1 isolated from Klíčava reservoir (Central Bohemia, Czech Republic). Other two strains represented phylogenetically distinct prey items, one strain of the genus Polynucleobacter (P. cosmopolitanus, strain MWH-MoIso2, Betaproteobacteria, Hahn, 2003; Boenigk et al., 2004) and the other (MWH-Wo1, Hahn and Pöckl, 2005) from the Luna 2 cluster of Actinobacteria.
Table 1. Characteristics of bacterial strains used as prey for the natural heterotrophic nanoflagellate community from the Římov reservoir.
| Species | Strain | Affiliation | Cell volume (in μm3) | Cell length (in μm) | Cell shape | Origin | Reference |
|---|---|---|---|---|---|---|---|
| Limnohabitans planktonicus | II-D5T | R-BT065 cluster, Betaproteobacteria | 0.135±0.079 | 1.075±0.276 | Large rod | Římov reservoir, Czech Republic | Kasalický et al., 2010 |
| Limnohabitans parvus | II-B4T | R-BT065 cluster, Betaproteobacteria | 0.052±0.035 | 0.670±0.185 | Short rod | Římov reservoir, Czech Republic | Kasalický et al., 2010 |
| Limnohabitans sp. | 2KL-27 | R-BT065 cluster, Betaproteobacteria | 0.067±0.038 | 0.748±0.185 | Coccoid | Klíčava reservoir Czech Republic | Kasalický et al., 2013 |
| Limnohabitans sp. | 2KL-1 | R-BT065 cluster, Betaproteobacteria | 0.204±0.110 | 1.164±0.294 | Large solenoid | Klíčava reservoir Czech Republic | Kasalický et al., 2013 |
| Polynucleobacter cosmopolitanus | MWH-MoIso2T | D-subcluster Betaproteobacteria | 0.049±0.023 | 0.625±0.115 | Short curved rods | Lake Mondsee, Austria | Boenigk et al., 2004 |
| Undescribed Actinobacterium | MWH-Wo1 | Luna 2 cluster Actinobacteria | 0.071±0.023 | 0.765±0.132 | Small solenoid | Lake Wolfgangsee, Austria | Hahn and Pöckl, 2005 |
Data on cell dimension of each tested strain represent mean value±s.d. of >200 cells measured.
Experimental design and sampling
Before the experiment, the bacterial cultures (Table 1) were pre-grown in liquid 3 g l−1 NSY medium (Hahn et al., 2004). Cells from 50 ml were concentrated by centrifugation at 5.000 g and subsequently re-suspended into 50 ml of <0.2 μm filtered and sterilized water from Římov reservoir (South Bohemia, Czech Republic). The cultures were kept on a shaker overnight to permit even re-suspension of cells and adaptation to the reservoir water. Subsequently, bacteria were enumerated as described below.
We used a natural HNF community to examine the food prey quality of the different bacteria (Table 1) and their effects on HNF community composition. The experiment was scheduled for the onset of the spring maximum of HNF in the reservoir (∼4.5 × 103 cells ml−1). A 10-liter water sample from Římov reservoir was collected on 18 April 2011 and then gravity filtered through 5-μm pore-size, 147-mm diameter filters. HNF in the filtered water were thus released from zooplankton grazing. After 30 h there was an approximate two-fold HNF abundance increase and a decrease in cell numbers of free-living bacteria (from 2.5 to ∼1 × 106 cells ml−1). The vast majority of remaining bacteria was composed of small, for HNF inedible flocks or filaments.
After the 30 h pre-incubation, 250 ml of the 5-μm filtrates were further manipulated by addition of the respective bacterial strains. Triplicate experiment setup is illustrated in Figure 1. As the tested prey bacteria possessed markedly different mean cell volume (MCV, Table 1), the initial cell numbers of each bacterial strain added into experimental treatments was set to yield approximately the same initial biovolume for all six strains. Notably, the biovolume of the bacterial prey added into the HNF community represented ∼25-fold the background bacterial biomass (mostly composed of grazing-resistant morphotypes) present in the pre-incubated HNF solution. A 5-μm filtrate containing the same starting HNF community but with no bacteria added was used as control. All treatments were incubated in the dark at 18 °C and subsamples were taken aseptically at 12–24 h intervals. At selected time points bacterial samples were collected for: (i) FISH and (ii) DNA extraction for pyrosequencing.
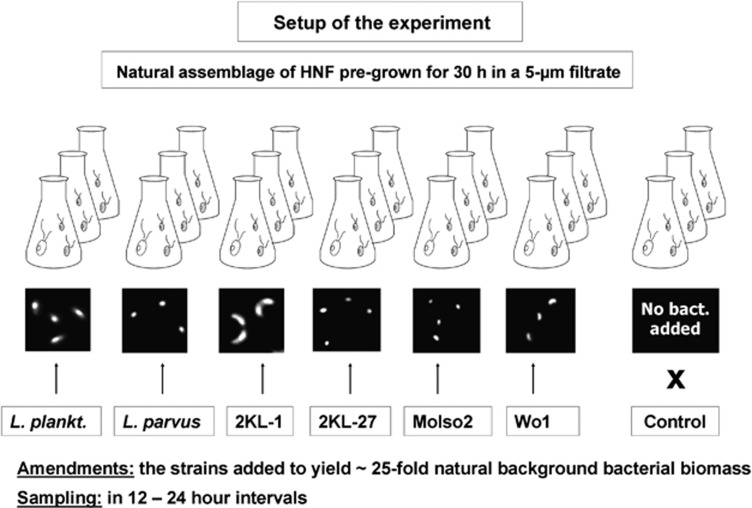
Figure 1.
Experimental design: a natural HNF community in 5-μm filtered water from Římov reservoir was pre-incubated for 30 h and then subjected to additions of different bacterial food items as the major HNF food source (for details see Methods). Note that the initial concentration of natural background bacteria was in all treatments 1.09±0.10 × 106 cells ml−1 (Mean±s.d.). The bacterial strains L. parvus, L. planktonicus, 2KL-27, 2KL-1, MoIso2 and Wo1 (for cell size and morphology see the inserted microphotograph, for further details see Table 1) were added to yield ∼25-fold natural background of bacterial biovolume present in the non-amended 5-μm filtrate used as control. Subsamples were collected in 12–24 h intervals.
Bacterial abundance and sizing, flow cytometry
Bacterial abundance was measured via flow cytometry in samples stained with the fluorochrome Syto13 (Molecular Probes, Eugene, OR, USA) using the FACS-Calibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) as detailed in Gasol and Del Giorgio (2000). Only at t63 and t87 hours, bacteria were counted microscopically (for details see Šimek et al. (2007)) to quantify accurately also bacterial cells in small grazing-resistant flocks or filaments resulting from HNF grazing pressure. Bacteria (>200 cells per sample) were sized by using the semiautomatic image analysis systems (NIS-Elements 3.0, Laboratory Imaging, Prague, Czech Republic).
Catalyzed reporter deposition fluorescence in situ hybridization
Group-specific oligonucleotide probes (ThermoHybaid, Ulm, Germany) and the catalyzed reporter deposition fluorescence in situ hybridization protocol (Pernthaler et al., 2002) were used to track: (a) time-course changes in proportions of the added prey bacteria, and (b) presence of prey bacteria in HNF food vacuoles (Jezbera et al., 2005). Subsamples from control (no bacteria added) and from all bacterial prey-amended treatments were collected at times t0, t37 and t63 hours of the experiment. Oligonucleotide probes were employed to target: the R-BT065 cluster (probe R-BT065, Šimek et al., 2001), which covers all four Limnohabitans strains used (Kasalický et al., 2013); the whole Polynucleobacter cluster (probe PnecABCD-445, Hahn et al., 2005); and the entire Actinobacteria phylum (probe HGC69a).
Heterotrophic nanoflagellate enumeration, biovolume and growth efficiency
Subsamples (1–5 ml) were stained with DAPI and HNF abundance was determined via epifluorescence microscopy as described elsewhere (Šimek et al., 2001). To calculate mean volumes of HNF cells (approximated to prolate spheroids), lengths and widths of 100 cells in triplicate treatments were measured manually on-screen with a built-in tool of a PC-based image analysis system (NIS-Elements 3.0, LIM, Prague, Czech Republic). Estimates of HNF growth efficiency as % based on cellular biovolume were calculated as follows:
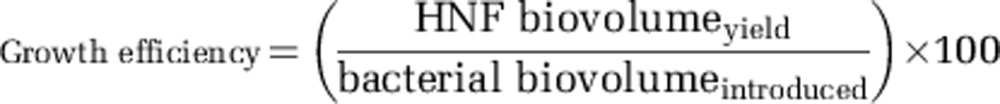 |
Where HNF 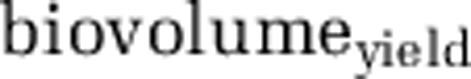 is the HNF biovolume increment (that is, maximum HNF biovolume reached minus HNF biovolume present at t0) divided by bacterial strain-specific biovolume introduced into the treatment at t0. The maximum HNF growth rate was calculated using log-transformed data on HNF abundance with linear regression as the slope of the best-fit line. The three consecutive time points on the HNF growth curve yielding the largest r2 were selected for calculation. The lag was calculated as the period from the time zero to the intercept between the best-fit line of HNF growth and the zero-time level of HNF abundance.
is the HNF biovolume increment (that is, maximum HNF biovolume reached minus HNF biovolume present at t0) divided by bacterial strain-specific biovolume introduced into the treatment at t0. The maximum HNF growth rate was calculated using log-transformed data on HNF abundance with linear regression as the slope of the best-fit line. The three consecutive time points on the HNF growth curve yielding the largest r2 were selected for calculation. The lag was calculated as the period from the time zero to the intercept between the best-fit line of HNF growth and the zero-time level of HNF abundance.
Pyrosequencing of eukaryotic communities and data analysis
Extraction and purification of DNA from triplicate subsamples (50–200 ml, see Figure 1) collected at times t0, t37 and t63 hours of the experiment were performed as described previously (Jezberová et al., 2010).
We used slightly modified broad eukaryotic PCR primers targeting the SSU rRNA genes. The primers were tagged with a 5′-tail adapter for the 454 sequencing (Table 2). The forward primer also contained a 6–10 bp tag for each of the samples inserted between the 454-adapter A and the SSU-specific part, the reverse primer was modified with a 5′ BioTEG modification. The primers amplify a fragment of the SSU rRNA gene including the variable V9 region. PCR was carried out in a 20-μl reaction with 0.2 U Phusion High-Fidelity DNA Polymerase (Finnzymes Oy, Espoo, Finland), 200 μℳ dNTPs and 0.25 pmol of each primer. The cycling profile consisted of 1 min denaturation at 95 °C, followed by 30 PCR cycles (95 °C for 30 s, 60 °C for 45 s, 72 °C for 60 s) with a final extension step of 10 min at 72 °C. DNA from each triplicate subsample was subjected to PCR separately. PCR was carried out eight times per sample in order to generate more products and the PCR products were pooled upon completion of the reaction.
Table 2. HPLC-purified PCR primers used, which carry sequences specific for the SSU of the rRNA gene.
| Sequence (5′-3′) | References | |
|---|---|---|
| Adapter A | CCATCTCATCCCTGCG TGTCTCCGACTCAG | |
| SSU forward primer | GTACACACCGCCCGTC | Lane, 1991; Stoeck et al., 2010 |
| SSU reverse primer | TGATCCTTCTGCAGGTTCACCTAC | Medlin et al., 1988; Stoeck et al., 2010 |
| Adapter B | BioTEG-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG CTGAGACTGCCAAGGCACACAGGGGATAGG |
Abbreviation: SSU, small subunit.
The pooled PCR products of each sample were gel-purified using QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and quantified on an agarose gel. We pooled 525 ng from each sample and sequenced the pooled DNA on a 454 Roche (Durham, NC, USA) FLX sequencer (titanium chemistry). Read sequences will be deposited in the NCBI Short Read Archive (deposition in progress).
CANGS DB (Pandey et al., 2011) was used for processing, adapter and primer clipping, quality filtering and grouping of sequences according to barcodes. Briefly, sequences that did not fit the following quality criteria were removed: (i) no ambiguous nucleotides (that is, no Ns in the sequence reads); (ii) quality score >20, when averaged across the read after clipping adapters and primers; (iii) minimum sequence length of 130 bp (including PCR primers); and (iv) at least two copies of the read present in two different samples in the entire data set before clipping primers (Medinger et al., 2010).
Flagellate taxonomic classification
Reads obtained from each sample were assigned to taxa defined by the NCBI taxonomy by using the software MEGAN 4 (Huson et al., 2007). The assignment to taxa is based on BLAST results. Reads were compared by BLAST (Altschul et al., 1990) against the NCBI-nr database, and the resulting data sets were loaded in MEGAN. As sequence sets obtained for the different samples varied in read numbers, each sample was analyzed by using an adjusted LCA value. A basic LCA parameter of 200 was selected and LCA parameters for individual samples were normalized according to sample size. For each sample, only the number of reads assigned in a pre-run (LCA parameter set as 200) to protist taxa was considered for normalizing LCA parameters. Matching between taxon assignment by MEGAN 4 and phylogeny derived classification of reads was examined by comparing MEGAN results with a phylogenetic tree constructed with reads of a representative subset of reads. Both methods resulted in a comparable assignment of reads to taxa or phylogenetic clusters, respectively.
Results
Flagellate growth responses to bacterial prey
We tested the effects of food quality of six bacterial strains of different size, morphology and taxonomic affiliation (Table 1) on the growth and community composition of a natural HNF community from a freshwater reservoir. The bacterial strains were added in numbers (Figure 2) that compensated for their different MCVs to yield approximately the same initial total prey biovolume in all treatments (Figure 3).
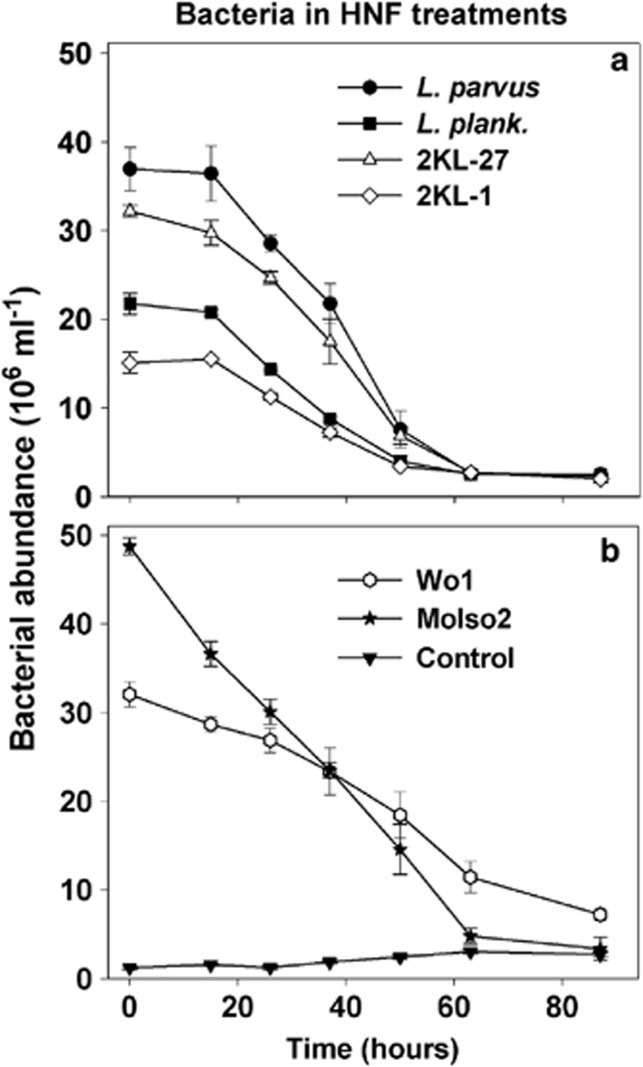
Figure 2.
Time-course changes in bacterial abundance in 5-μm treatments amended by bacterial strains of the genus Limnohabitans (a), that is, L. parvus, L. planktonicus, 2KL-27 and 2KL-1, and (b) with the strains MoIso2 and Wo1 compared to control with no bacteria added. Values are means for triplicates; error bars show s.d..
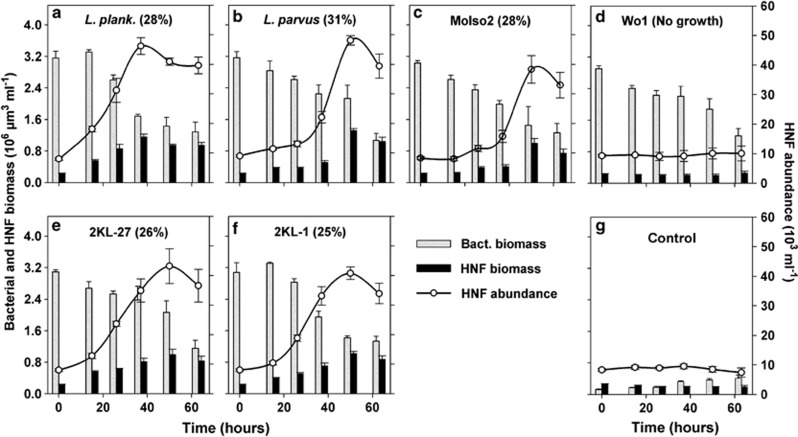
Figure 3.
Time-course changes in HNF abundance and biovolume compared with bacterial biovolume in the treatments amended with respective bacterial strains (a–f, for further details see text to Figure 1 and Table 1) compared with control (g) with no bacteria added. Values are means for triplicates; error bars show s.d.. Values of growth efficiency, as % of bacterial biovolume introduced into the treatment at t0, are listed in parenthesis for the treatments where positive HNF growth was detected (for details see Methods).
HNF grazing decimated all prey types within 63 h to the abundance level of control treatment, except for the strain Wo1 (Actinobacteria) with a markedly smaller rate of cell decline (Figure 2). All six strains were clearly detected in HNF food vacuoles using FISH probes (Supplementary Figure 1) and all but the Wo1 strain, yielded significant growth of HNF communities, although with different growth dynamics (Figures 3 and 4). For instance, HNF numbers and biovolume peaked in the treatment amended with L. planktonicus at t37 hours, while in most other cases the HNF peak abundance appeared later at t50 hours. However, although cell abundance of the Wo1 strain decreased (Figure 2), no HNF growth was detected compared with control treatment (Figures 3 and 4). Bacterial cellular biovolume added into treatments compared with the net HNF biovolume increments allowed for estimates of growth efficiency of HNF on different food items (Figure 3), ranging from 25% (strain 2KL-1) to 31% (L. parvus).
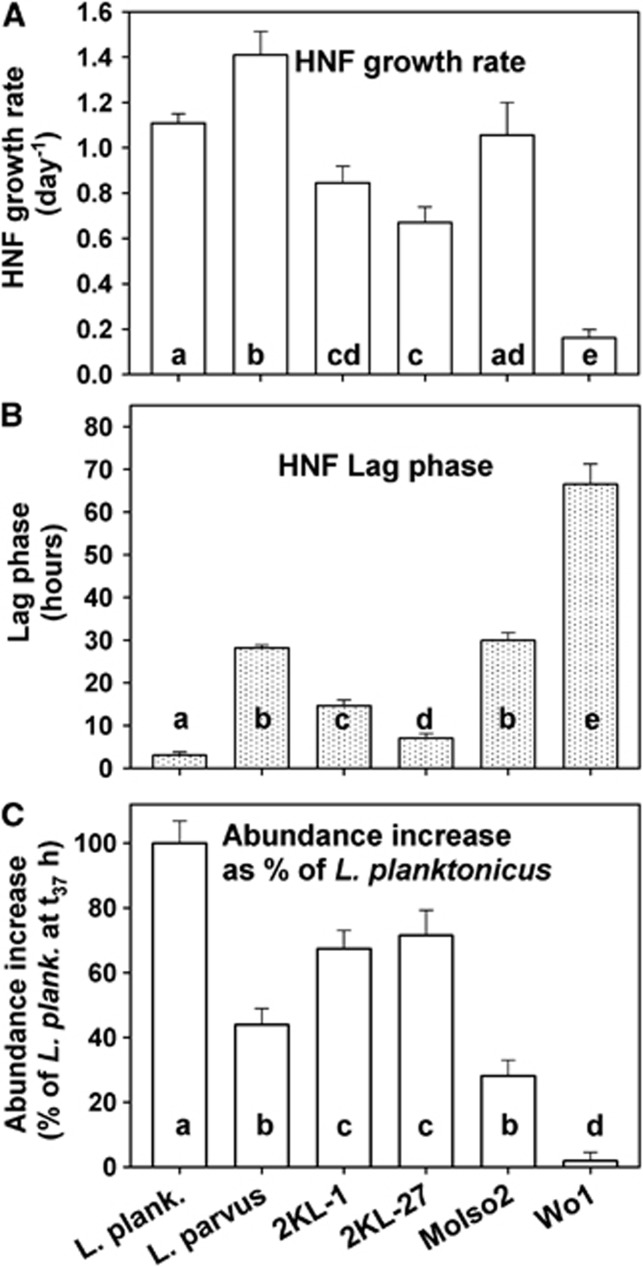
Figure 4.
Comparison of HNF maximum growth rates (A), length of their lag phase after the treatment amendment (B) and the rate of HNF abundance increase growing on different bacterial strains (for further details see text to Figure 1 and Table 1) as related to the treatment with L. planktonicus (C) where the HNF abundance and biovolume peaked already at t37 hours (compare Figure 3). Values are means for triplicates; error bars show s.d. Different letters indicate a significant difference (P<0.05, ANOVA, followed by Tukey's multiple comparison tests) between treatments amended with different bacterial strains.
The similar initial biovolumes of the distinct bacterial prey yielded different HNF growth dynamics (for significance see Figure 4). For instance, HNF in the L. planktonicus treatment achieved a population peak significantly faster (Figure 4) than in other treatments with almost no lag phase. In contrast, HNF in the L. parvus treatment grew even faster but after relatively long lag phase, resembling rather growth parameters of HNF growing on strain MoIso2 of a similar cell size (Table 1). In contrast, even the relatively closely related and similarly sized Limnohabitans strains, that is, L. parvus and KL-27 (Table 1 and Figure 4), gave distinct HNF growth dynamics, while the similarly sized gram-positive strain Wo1 did not support detectable HNF community growth. Note that MCVs of all the tested bacterial strains were relatively stable during the first 50 h of the experiment; that is, the coefficient of variation of MCV for each strain was within 12% of the strains' MCVs listed in Table 1 (values for t0 hours). However, the starting prevalence of the target bacteria (tracked by FISH probes) in the treatments diminished (Figure 2) and MCVs of all bacteria present in the treatments almost doubled towards the experimental end (data not shown). It resulted from increasing proportions of larger grazing-resistant morphotypes (small flocks and filaments) that developed from the background bacteria present in the original sample as a response to increasing HNF abundance (Figure 3) and bacterivory.
Effects of bacterial prey quality on HNF community dynamics and composition
Prey-specific differences in HNF growth parameters were also reflected in the analysis of pyrosequenced eukaryotic SSU rRNA amplicon (in total 102 239 reads were analyzed). Among reads representing protistan taxa, Stramenopiles clearly dominated with 55–82% of the total numbers of reads over all treatments and the course of the study, while other groups such as Katablepharidaceae (0–12%), Choanoflagellida (6–13%) and sum of Opisthokonta and Rhizaria (6–18%) appeared either only irregularly or did not show any clear trend in the samples (data not shown). Groups of Cryptophyta, Chlorophyta and Telonema-related reads composed generally a negligible proportion (mostly<1%), or such reads were detectable only sporadically in control samples (data not shown).
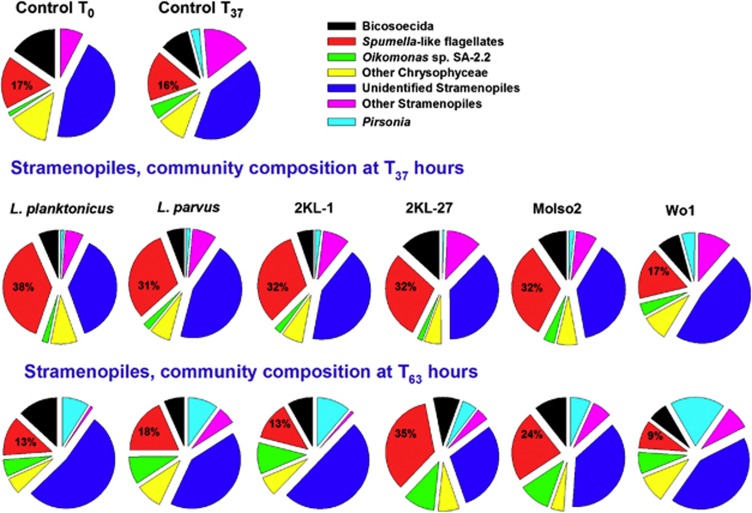
Thus, Stramenopiles, including also typical bacterivorous groups of Spumella-like flagellates, represented the most abundant protistan groups of interest for further analysis of the bacterial prey-related responses of the HNF community. Generally, all prey types supporting HNF growth (Figure 3) induced an approximately two-fold increase in the relative proportions of the Spumella-like HNF (from 16 to 31–38% of total Stramenopiles; Figure 5) towards exponential growth phase of HNF (t37 hours). No such community shift was observed in Wo1 and control treatments (without HNF growth, Figure 3), in which the overall Stramenopiles composition developed similarly and differed significantly from other treatments supporting HNF growth (P<0.01, two-way ANOVA, followed by Bonferroni's post-test). Notably, the relative proportions of Oikomonas sp. (SA-2.2) and Pirsonia clusters significantly increased (P<0.01) between t37–t63 hours in all treatments (Figure 5).
Figure 5.
A pie chart of the relative proportion (as %) of different subgroups of Stramenopiles within total Stramenopiles (accounting for 58–82% of the potentially bacterivorous flagellate groups) in treatments amended with different bacterial strains at times t39 and t63 compared with control treatments at t0 and t39 hours. For further details regarding the bacterial strains see text to Figure 1 and Table 1.
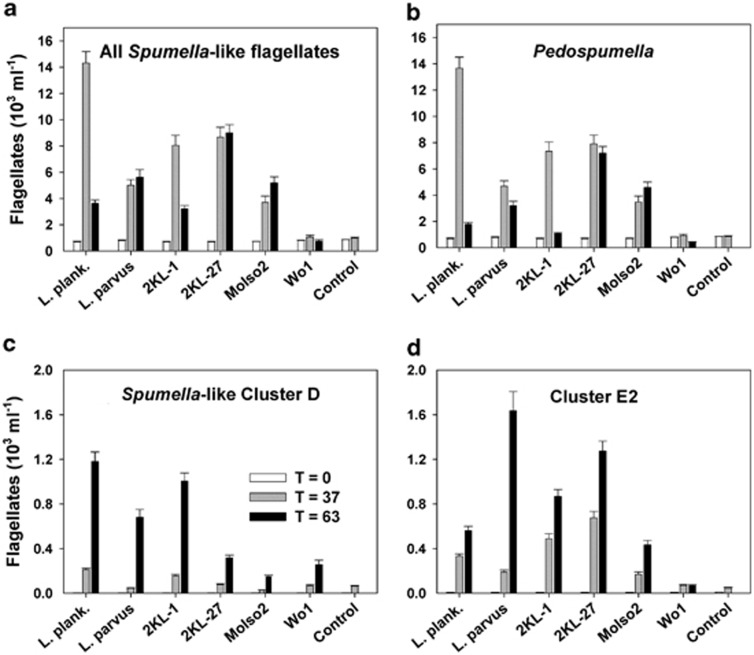
An increase in proportions of the Spumella-like HNF paralleled a steep increase in total HNF abundance between t0 and t37 hours (compare Figures 3 and 5) indicating high net growth rates of the Spumella-like HNF in all but Wo1 and control treatments. Overall, highly significant differences in growth rates of Spumella-like HNF were detected in the different treatments (P<0.0001, ANOVA, Table 3). For instance, the Spumella-like HNF grew significantly faster (Table 3) in L. planktonicus >2KL-1≅2KL-27>L. parvus ≅ MoIso2 >> Wo1 treatment (≅, comparable growth). In t37 hours, the vast majority the Spumella-like reads was affiliated with Pedospumella sp. (see phylogenetic tree, Supplementary Figure 2), with the highest proportion in L. planktonicus (>95%) and lowest in Wo1 treatment (86%, Supplementary Figure 3). Overall, L. planktonicus treatment yielded 2–3-fold higher abundance of Pedospumella-like HNF at t37 hours than other bacteria, however, followed by their dramatic ∼7-fold decrease at t63 hours observed also in 2KL-1 treatment (Figure 6). In contrast, no such dramatic changes in abundance of Pedospumella sp. between t37–63 hours were observed in L. parvus, 2KL-27 and MoIso2 treatments.
Table 3. Estimates of maximum growth rates (mean value based on triplicate treatments) of the whole group of Spumella-like HNF and of three most abundant subgroups of the Spumella-like HNF detected in sequence analysis (compare absolute numbers and relative proportions of the target HNF groups in Figures 3, 5 and 6 and Supplementary Figure 3) between t 0 and t 37 of the experiment.
| Flagellate group |
Flagellate growth rate on different bacterial strains (per day, mean±s.d.) |
|||||
|---|---|---|---|---|---|---|
| L. planktonicus | L. parvus | 2KL-1 | 2KL-27 | MoIso2 | Wo1 | |
| Whole Spumella-like group | 1.70±0.05A | 1.01±0.10B | 1.37±0.08C | 1.40±0.08C | 0.86±0.11B | 0.07±0.04D |
| Pedospumella sp. | 1.69±0.08A | 0.98±0.09B | 1.33±0.08C | 1.36±0.04C | 0.84±0.13B | −0.01±0.03D |
| Spumella-like Cluster D | 2.88±0.09A | 1.88±0.10B | 2.73±0.08A | 2.27±0.08C | 1.59±0.11D | 2.20±0.09C |
| Cluster E2 | 2.28±0.07A | 1.91±0.10B | 2.57±0.10C | 2.77±0.13C | 1.88±0.11B | 1.32±0.07D |
Different superscripted letters indicate a significant difference between flagellate growth rates in different treatments (P<0.0001, one-way ANOVA, followed by Tukey's multiple comparison tests).
Figure 6.
Absolute numbers of all Spumella-like flagellates (a) and of their three most abundant subclusters—Pedospumella, Spumella-like cluster D and Cluster E2 (b–d, respectively) in treatments amended with different bacterial strains at times t37 and t63 compared with control treatment. These data are based on the relative proportion of these subclusters within the target protistan groups (Figure 5 and Supplementary Figure 3) related to total HNF numbers (Figure 3). Note different y axis scaling for c, d. Values are means for triplicates; error bars show s.d.. For further details regarding the bacterial strains see text to Figure 1 and Table 1.
Another two groups related to Spumella-like HNF, that is, cluster D (related to the so far undescribed Spumella-like flagellate strain JBC27) and E2 (related to the undescribed Spumella-like flagellate strains JBM08 and JBM18), rapidly increased both their relative and absolute numbers in some treatments (compare Figure 6 and Supplementary Figure 3), however, in distinct prey-specific fashions. Their growth rates differed significantly with the prey type (P<0.0001, ANOVA, Table 3). Overall, in L. planktonicus, L. parvus and 2KL-1 treatments similar relative proportions of the major Spumella-like flagellate groups were found at t63 hours, whereas significantly lower proportions (P<0.01, ANOVA, followed by Bonferroni's post-test) and absolute numbers of the Spumella-like Cluster D (related to the JBC27 strain) were detected in 2KL-27 and MoIso2 treatments. Interestingly, while no growth of total HNF can be seen in the Wo1 treatment (similar to control in this aspect, Figure 3), this prey addition induced some increase in relative proportions and abundance of Spumella-like Cluster D at t63 hours (Figure 6 and Supplementary Figure 3).
Discussion
Major findings of the study
This study demonstrated the suitability of Limnohabitans bacteria as prey for flagellates and clear prey-specific community growth responses of HNF communities to the different food quality of the tested bacteria. To the best of our knowledge, this the first study clearly documenting strong prey-specific effects of even closely related bacteria on HNF community composition. This is an ecological aspect that has been long under debate (for example, Boenigk et al., 2004; Pernthaler, 2005; Montagnes et al., 2008) but without any direct evidence concerning natural HNF assemblages.
Notably, the fast growing Limnohabitans bacteria (Šimek et al., 2006; Jezbera et al., 2012) likely represent a high quality resource supporting rapid growth of natural HNF communities. We are confident in this conclusion as recently four more tested strains from other Limnohabitans lineages (Kasalický et al., 2013) supported fairly rapid growth of HNF communities in another two freshwater ecosystems (Šimek, unpublished data). Our results lend solid support to previous speculations based on preliminary indirect evidence on the key role of these bacteria in carbon flow from alga-derived substrates (Šimek et al., 2010, 2011) to the plankton grazer food chain (Jezbera et al., 2005). The important role of the Limnohabitans sharply contrasts to that of the Wo1 actinobacterial strain. Although cells of Wo1 were ingested (Supplementary Figure 1) this prey did not support any HNF community growth, likely because of limited digestibility of the bacterium for flagellates (Tarao et al., 2009).
Methodical aspects—examining growth parameters of flagellates
In this study, an innovative combination of methods such pyrosequencing and FISH-targeting of prey bacteria directly in HNF food vacuoles were exploited, with the experimental design building on prey-depletion approach (Montagnes et al., 2008) in ∼3-day incubations with the HNF community established after 30-h in 5-μm filtrate. Thus, in pelagic environments even more profound shifts in HNF communities (Figure 5 and Supplementary Figure 3) could be expected as a response to, for example, phytoplankton succession related bacterial blooms (Eiler and Bertilsson, 2004) lasting for few days or weeks.
In our experimental design exploited, the HNF growth responses could be directly attributed to a particular prey item dominating the prey assemblage. Moreover, due to enhanced grazing pressure in 5-μm treatments the less abundant naturally occurring bacteria, present after 30-h incubation were mostly composed of grazing-resistant flocks and filaments (Jürgens and Matz, 2002), that is, of bacterial biomass that was likely irrelevant to the prey-specific growth responses of HNF.
Most importantly, our approach yielded ecologically meaningful prey-specific flagellate growth parameters. For instance, the combination of a high growth rate and short lag phase of the HNF communities indicates a marked suitability of a bacterial prey for flagellates present in the assemblage, that is, demonstrating a minimum acclimatization time to the prey offered. Notably, L. planktonicus yielded almost immediate exponential HNF growth with minimum lag allowing for the most rapid accumulation of an HNF population peak among all the strains (Figure 3). Such results indicate that the strain represents a high quality prey. In contrast, for all the other strains a significantly prolonged lag phase were observed (Figure 3), thus indicating much longer acclimatization time and likely selection of flagellates capable to exploit more efficiently the prey offered.
Prey-specific aspects of HNF growth responses
The prey-specific HNF growth responses were reflected in different patterns and timing of the HNF community shifts (Figures 3 and 5 and Supplementary Figure 3). Among Stramenopiles, mainly the typical bacterivorous Spumella-like flagellate taxa (Boenigk et al., 2004, 2005) approximately doubled their relative proportions in the community and dramatically increased absolute numbers (Figure 6) during the exponential HNF growth (t37 hours) in all, but Wo1 treatment. This clearly points to the key role of bacterial food quality characteristics as the Wo1 strain was the only one that did not support HNF community growth (see also Tarao et al., 2009). Interestingly, throughout the course of the study and among experimental treatments, there was no bloom of choanoflagellates observed (considered as bacterivores, Boenigk and Arndt, 2002) as this group generally comprised relatively stable low proportions of total protistan reads (data not shown).
During the stationary HNF growth phase (t63 hours) the bulk Spumella-like cluster dropped in both relative and absolute numbers in all but 2KL-27 and MoIso2 treatments, for which quite unique trends were observed (Figures 5 and 6). Interestingly, cell size and morphology of 2KL-27 resembled closely L. parvus or MoIso2 Polynucleobacter strains (compare Figure 1 and Table 1), but induced significantly different HNF growth responses (Figure 4). It indicates that even within closely related Limnohabitans bacteria non-morphology related traits have an important role in regulating exploitation of particular bacteria (Boenigk and Arndt, 2002; Jürgens and Matz, 2002). Interestingly, the very small cells of the ultramicrobacterial (<0.1 μm3) strain MoIso2, supported only slow growth of several Poteriospumella spp. genotypes in laboratory experiments (Boenigk et al., 2004). However, in this study after longer lag phase, indicating ‘HNF community acclimatization time', this prey yielded rapid doubling times of different members of the Spumella-like cluster (Table 3). This together with highly prey-specific HNF community shift in MoIso2 treatment points to the role of predator–prey species-specific interactions supporting the idea of clear niche partitioning even among closely related flagellate species (Boenigk et al., 2004).
Our study documents, at different taxonomic levels, the strong impact of prey on the resulting HNF community dynamics over different phases of population growth. Aside from a common trend of decreasing HNF numbers at t63 hours (Figure 3), their MCVs increased (data not shown) in parallel with significantly higher proportions (P<0.01, ANOVA, Bonferroni's post-test) of sequences matching with the Oikomonas sp. SA-2.2 cluster or those loosely related to Pirsonia (Figure 5). Although the Oikomonas is considered a bacterivore (Boenigk and Arndt, 2002), the Pirsonia genus has been reported as a diatom feeder in brackish waters (Schnepf and Schweikert, 1997; Kühn et al., 1996). This overall community shift may reflect an increasing proportion of larger flagellates within Stramenopiles (Figure 5) preying upon smaller and highly abundant Spumella-like HNF that become food limited at t63 hours (Figure 2). However, this shift to larger flagellates, in contrast with the bulk decline in HNF abundance (Figure 3), could partially be related to zooplankton removal involved in our experimental design. Generally, in plankton environments larger HNF are efficiently limited by zooplankton grazing (Jürgens and Matz, 2002) and thus smaller HNF, such that Spumella-like bacterivorous chrysomonads frequently dominate freshwater plankton (Šimek et al., 1997; Nolte et al., 2010).
The bacterial prey of high quality (except for Wo1 strain) supported growth of the bulk HNF community with doubling time of 11–23 h (Figure 3), but in the range of 5.8–16 h for the representatives of rapidly growing flagellates (Table 3) affiliated with the Spumella-like subclusters D (related to the undescribed flagellate strain JBC27) and E2 (related to the undescribed Spumella-like flagellate strains JBM08 and JBM18). The rapid flagellate growth mainly on L. planktonicus, 2KL-1 and 2Kl-27 strains is comparable to that found for typical Spumella-like flagellates cultured under optimal food conditions (Boenigk et al., 2007). This together with high HNF growth efficiency (Figure 3, compare Fenchel (1986)) is evidence of the food quality of most Betaproterobacteria strains used in this study.
Although the Spumella-like flagellates from the clusters D and E2 were practically absent in the control at t0 hours, due the their high growth potential (compare Pedospumella sp. in Table 3) they significantly increased in relative and absolute numbers toward the end of the experiment, mainly in L. planktonicus, L. parvus and 2KL-1 treatments. Moreover, the flagellates affiliated with the Spumella-like cluster D were the only predator group, which grew in the presence of the Wo1 strain. Although being considered as poor quality prey (Tarao et al., 2009), Wo1 did yield a certain prey-specific HNF community shift.
Concluding remarks
This study clearly demonstrated that different food properties of even closely related bacterial prey can modulate differently growth dynamics of flagellates and in turn also the overall HNF community composition. During the exponential growth phase the driving force of such a shift was the Pedospumella cluster (likely due to the ‘inoculum size' at t0) that overgrew other bacterivorous flagellate groups and comprised ∼15–40% of total HNF numbers. Notably, our approach facilitated determining bulk parameters of HNF community growth while revealing many significant differences related to bacterial prey quality (Figure 3). Moreover, short lag phase and rapid growth responses of major bacterivorous HNF taxa to a particular prey type might indicate its high food quality. Our experimental design combined with pyrosequencing of the grazer community could provide important insights regarding the question which bacterial strains are active in carbon transfer to the grazer food chain in a particular aquatic system and which flagellate groups are the key players in the trophic transfer.
Acknowledgments
This study was largely supported by the Grant Agency of the Czech Republic under research grant 13-00243S awarded to K.Š and by project FWF P 21151 awarded to J.B. Partial support was provided by the project Postdok_BIOGLOBE (contracting V. K., CZ.1.07/2.3.00/30.0032) co-financed by the European Social Fund and the state budget of the Czech Republic. We thank Ulrike Koll for DNA extraction and contributions to analysis of pyrosequencing reads and John Dolan for English text corrections. We thank to two anonymous reviewers for their constructive criticism of the original submission.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
Microphotographs of bacterial populations.
Maximum likelihood (ML) phylogenetic tree of Chrysophyceae.
A pie chart of the relative proportion (as %) of different subgroups of Spumella-like bacterivorous flagellates.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Boenigk J, Arndt H. Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Anton Leeuw Int J G. 2002;81:465–480. doi: 10.1023/a:1020509305868. [DOI] [PubMed] [Google Scholar]
- Boenigk J, Jost S, Stoeck T, Garstecki T. Differential thermal adaptation of clonal strains of a protist morphospecies originating from different climatic zones. Environ Microbiol. 2007;9:593–602. doi: 10.1111/j.1462-2920.2006.01175.x. [DOI] [PubMed] [Google Scholar]
- Boenigk J, Pfandl K, Stadler P, Chatzinotas A. High diversity of the ‘Spumella-like' flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ Microbiol. 2005;7:685–697. doi: 10.1111/j.1462-2920.2005.00743.x. [DOI] [PubMed] [Google Scholar]
- Boenigk J, Stadler P, Wiedlroither A, Hahn MW. Strain-specific differences in the grazing sensitivities of closely related ultramicrobacteria affiliated with the Polynucleobacter cluster. Appl Environ Microbiol. 2004;70:5787–5793. doi: 10.1128/AEM.70.10.5787-5793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boenigk J. Some remarks on strain specifity and general patterns in the ecology of Spumella (Chrysophyceae) Nova Hedwigia Beih. 2005;128:167–178. [Google Scholar]
- Eiler A, Bertilsson S. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ Microbiol. 2004;6:1228–1243. doi: 10.1111/j.1462-2920.2004.00657.x. [DOI] [PubMed] [Google Scholar]
- Fenchel T. The ecology of heterotrophic microflagellates. Adv Microb Ecol. 1986;9:57–97. [Google Scholar]
- Gasol JM, Del Giorgio PA. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar. 2000;64:197–224. [Google Scholar]
- Hahn MW, Pöckl M. Ecotypes of planktonic Actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Appl Environ Microbiol. 2005;71:766–773. doi: 10.1128/AEM.71.2.766-773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Pöckl M, Wu QL. Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. Appl Environ Microbiol. 2005;71:4539–4547. doi: 10.1128/AEM.71.8.4539-4547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Scheuerl T, Jezberová J, Koll U, Jezbera J, Šimek K, et al. The passive yet successful way of planktonic life: genomic and experimental analysis of the ecology of a planktonic Polynucleobacter population. PLoS ONE. 2012;7:e32772. doi: 10.1371/journal.pone.0032772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Stadler P, Wu QL, Pöckl M. The filtration–acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J Microbiol Methods. 2004;57:379–390. doi: 10.1016/j.mimet.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hahn MW. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl Environ Microbiol. 2003;69:5248–5254. doi: 10.1128/AEM.69.9.5248-5254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezbera J, Horňák K, Šimek K. Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microb Ecol. 2005;52:351–363. doi: 10.1016/j.femsec.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Jezbera J, Jezberová J, Koll U, Horňák K, Šimek K, Hahn MW. Major freshwater bacterioplankton groups: Contrasting trends in distribution of Limnohabitans and Polynucleobacter lineages along a pH gradient of 72 habitats. FEMS Microb Ecol. 2012;81:467–479. doi: 10.1111/j.1574-6941.2012.01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezbera J, Jezberová J, Šimek K, Kasalický V, Hahn MW. Patterns of Limnohabitans microdiversity across a large set of freshwater habitats as revealed by reverse line blot hybridization. PLoS ONE. 2013;8:e58527. doi: 10.1371/journal.pone.0058527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezberová J, Jezbera J, Brandt U, Lindström ES, Langenheder S, Hahn MW. Ubiquity of Polynucleobacter necessarius subsp asymbioticus in lentic freshwater habitats of a heterogenous 2000 km2 area. Environ Microbiol. 2010;12:658–669. doi: 10.1111/j.1462-2920.2009.02106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens K, Matz C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Anton Leeuw Int J G. 2002;81:413–434. doi: 10.1023/a:1020505204959. [DOI] [PubMed] [Google Scholar]
- Kasalický V, Jezbera J, Šimek K, Hahn MW. Limnohabitans planktonicus sp. nov., and Limnohabitans parvus sp. nov., two novel planktonic Betaproteobacteria isolated from a freshwater reservoir. Int J Syst Evol Microbiol. 2010;60:2710–2714. doi: 10.1099/ijs.0.018952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasalický V, Jezbera J, Šimek K, Hahn MW. The diversity of the Limnohabitans genus, an important group of freshwater bacterioplankton, by characterization of 35 isolated strains. PLoS ONE. 2013;8:e58205. doi: 10.1371/journal.pone.0058209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn SF, Drebes G, Schnepf E. Five new species of the nanoflagellate Pirsonia in the German Bight, North Sea, feeding on planktonic diatoms. Helgol Meeresunters. 1996;50:205–222. [Google Scholar]
- Lane DJ.199116S/23S sequencingIn: Stackebrandt E, Goodfellow M (eds)Nucleic Acid Technologies in Bacterial Systematic Wiley: NY, USA; 115–175. [Google Scholar]
- Massana R, Balague V, Guillou L, Pedros-Alio C. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microb Ecol. 2004;50:231–243. doi: 10.1016/j.femsec.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Medinger R, Nolte V, Pandey RV, Jost S, Ottenwälder B, Schlötterer C, et al. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol. 2010;19:32–40. doi: 10.1111/j.1365-294X.2009.04478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Montagnes DJS, Barbosa AB, Boenigk J, Davidson K, Jürgens K, Macek M, et al. Selective feeding behaviour of key free-living protists: avenues for continued study. Aquat Microbiol Ecol. 2008;53:83–98. [Google Scholar]
- Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A Guide to the natural history of freshwater lake bacteria. Microb Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte V, Pandey RV, Jost S, Medinger R, Ottenwälder B, Boenigk J, et al. Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol Ecol. 2010;19:2908–2915. doi: 10.1111/j.1365-294X.2010.04669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RV, Nolte V, Boenigk J, Schlötterer C. CANGS DB: a stand-alone web-based database tool for processing, managing and analyzing 454 data in biodiversity studies. BMC Res Notes. 2011;4:227. doi: 10.1186/1756-0500-4-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol. 2002;68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Peréz MT, Sommaruga R. Differential effect of algal- and soil-derived dissolved organic matter on alpine lake bacterial community composition and activity. Limnol Oceanogr. 2006;51:2527–2537. [Google Scholar]
- Salcher MM, Hofer J, Horňák K, Jezbera J, Sonntag B, Vrba J, et al. Modulation of microbial predator-prey dynamics by phosphorus availability: growth patterns and survival strategies of bacterial phynogenetic clades. FEMS Microb Ecol. 2007;60:40–50. doi: 10.1111/j.1574-6941.2006.00274.x. [DOI] [PubMed] [Google Scholar]
- Salcher MM, Pernthaler J, Zeder M, Psenner R, Posch T. Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ Microbiol. 2008;10:2074–2086. doi: 10.1111/j.1462-2920.2008.01628.x. [DOI] [PubMed] [Google Scholar]
- Schnepf E, Schweikert M. Pirsonia, phagotrophic nanoflagellates incertae sedis, feeding on marine diatoms: attachment, fine structure and taxonomy. Arch Protistenkd. 1997;147:361–371. [Google Scholar]
- Sherr EB, Sherr BF, Albright L. Bacteria: link or sink. Science. 1987;235:88. doi: 10.1126/science.235.4784.88a. [DOI] [PubMed] [Google Scholar]
- Sherr EB, Sherr BF.1993Protistan grazing rates via uptake to fluorescently labelled preyIn: Kemp P, Sherr BF, Sherr EB, Cole J (eds)Handbook of methods in aquatic microbial ecology Lewis: Boca Raton, FL, USA; 695–701. [Google Scholar]
- Sherr EB, Sherr BF. Significance of predation by protists in aquatic microbial food webs. Anton Leeuw Int J G. 2002;81:293–308. doi: 10.1023/a:1020591307260. [DOI] [PubMed] [Google Scholar]
- Šimek K, Hartman P, Nedoma J, Pernthaler J, Vrba J, Springmann D, et al. Community structure, picoplankton grazing and zooplankton control of heterotrophic nanoflagellates in a eutrophic reservoir during the summer phytoplankton maximum. Aquat Microb Ecol. 1997;12:49–63. [Google Scholar]
- Šimek K, Horňák K, Jezbera J, Nedoma J, Vrba J, Straškrabová V, et al. Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environ Microbiol. 2006;8:1613–1624. doi: 10.1111/j.1462-2920.2006.01053.x. [DOI] [PubMed] [Google Scholar]
- Šimek K, Kasalický V, Jezbera J, Jezberová J, Hejzlar J, Hahn MW. Broad habitat range of the phylogenetically narrow R-BT065 cluster, representing a core group of the betaproteobacterial genus Limnohabitans. Appl Environ Microbiol. 2010;76:631–639. doi: 10.1128/AEM.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Kasalický V, Zapomělová E, Horňák K. Algal-derived substrates select for distinct betaproteobacterial lineages and contribute to niche separation in Limnohabitans strains. Appl Environ Microbiol. 2011;77:7307–7315. doi: 10.1128/AEM.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Pernthaler J, Weinbauer MG, Horňák K, Dolan JR, Nedoma J, et al. Changes in bacterial community composition, dynamics, and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl Environ Microbiol. 2001;67:2723–2733. doi: 10.1128/AEM.67.6.2723-2733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimek K, Weinbauer MG, Horňák K, Jezbera J, Nedoma J, Dolan J. Grazer and virus-induced mortality of bacterioplankton accelerates development of Flectobacillus populations in a freshwater community. Environ Microbiol. 2007;9:789–800. doi: 10.1111/j.1462-2920.2006.01201.x. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Bass D, Nebel M, Christe R, Jones MDH, Breiner H-W, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Tarao M, Jezbera J, Hahn MW. Involvement of cell surface structures in size-independent grazing resistance of freshwater Actinobacteria. Appl Environ Microbiol. 2009;75:4720–4726. doi: 10.1128/AEM.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, del Campo J, Wylezich C, Massana R, Jürgens K. Unveiling trophic functions of uncultured protist taxa by incubation experiments in the brackish Baltic Sea. PLoS ONE. 2012;7:e41970. doi: 10.1371/journal.pone.0041970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microphotographs of bacterial populations.
Maximum likelihood (ML) phylogenetic tree of Chrysophyceae.
A pie chart of the relative proportion (as %) of different subgroups of Spumella-like bacterivorous flagellates.