Abstract
Electronic health records (EHRs) are increasingly useful for health services research. For relatively uncommon conditions, such as multiple myeloma (MM) and its treatment-related complications, a combination of multiple EHR sources is essential for such research. The Shared Health Research Information Network (SHRINE) enables queries for aggregate results across participating institutions. Development of a rational search strategy in SHRINE may be augmented through analysis of pre-existing databases. We developed a SHRINE query for likely non-infectious treatment-related complications of MM, based upon an analysis of the Multiparameter Intelligent Monitoring in Intensive Care (MIMIC II) database. Using this query strategy, we found that the rate of likely treatment-related complications significantly increased from 2001 to 2007, by an average of 6% a year (p=0.01), across the participating SHRINE institutions. This finding is in keeping with increasingly aggressive strategies in the treatment of MM. This proof of concept demonstrates that a staged approach to federated queries, using external EHR data, can yield potentially clinically meaningful results.
Background
The advent of electronic health records (EHRs) has created the possibility of phenome-driven research.1–3 However, such research is often hampered by small sample sizes at individual institutions, and limited granularity of large electronic databases. For example, the study of complication rates of multiple myeloma (MM) has been hampered by the lack of detailed information in large databases such as the National Cancer Institute's Surveillance, Epidemiology and End Results (SEER, http://seer.cancer.gov). New tools such as Informatics for Integrating Biology and the Bedside (i2b2), which is a common architecture for EHR-derived databases, and the Shared Health Research Information Network (SHRINE), a federated query tool which extracts information from multiple individual i2b2 instances, may significantly enable phenome-driven research.4–8
In this pilot study, we explored the degree to which SHRINE, which is enabled at five Harvard Medical School institutions (Beth Israel Deaconess Medical Center, Brigham and Women's Hospital, Children's Hospital Boston, Dana-Farber Cancer Institute, and Massachusetts General Hospital), could be employed to find statistically significant patterns of changing rates of serious complications in patients with MM. To develop a rational search strategy, we used a separate publicly available database to generate a list of encoded complications associated with the treatment of MM. The treatment of MM has changed significantly over the past decade, and we hypothesized that resultant patterns of treatment-related complications might also have changed.
Methods
We first ran a general SHRINE query for all patients diagnosed with MM, using any of the eight International Classification of Disease, Ninth Revision, Clinical Modification (ICD-9-CM) codes in the Agency for Healthcare Research and Quality's Clinical Classifications Software (CCS) Group 40: MM, as well as the codes for plasma cell leukemia (203.1*) and neoplasm of uncertain behavior of plasma cells (238.6).9 We considered a patient to have ‘established care’ at one of the SHRINE institutions if they had five or more occurrences of one or more of these primary diagnosis codes.
To identify codes pertaining to serious MM-associated complications, we conducted an analysis of patients with MM, identified as above, in the Multiparameter Intelligent Monitoring in Intensive Care (MIMIC II) database.10 MIMIC II is a large, publicly available, database of hospital admissions occurring between 2001 and 2007, where at least part of the admission was spent in the intensive care unit. The ICD-9-CM codes and derived CCS groupings generated at the end of each identified admission were used as a proxy for phenotype. The one-sided exact binomial test was used to calculate whether the observed rate of an ICD-9-CM code and/or CCS grouping in the MM subgroup was significantly greater than the expected rate of the code/grouping in the overall MIMIC II cohort.11 Statistical significance was defined as p=0.05 divided by the total number of binomial tests performed (the Bonferroni correction).12
Significant ICD-9-CM codes and CCS groups were then manually categorized as (1) infectious complications of MM or its treatment; (2) non-infectious complications of MM; (3) non-infectious treatment-related complications; or (4) non-specific/unrelated. Using these categories, we queried SHRINE for rates of encoding for non-infectious treatment-related complications for patients diagnosed with MM between January 1, 2001 and December 31, 2007. Queries were performed in 1-year periods for the MM diagnosis, with 60 months allowed for complications, in order to capture late complications. Individual hospital results of ‘10 or fewer patients’ were considered to be zero entries, and the mean results of the remainder were combined to form an aggregate estimate. Simple linear regression was used to evaluate for a trend in complications over time, with p<0.05 considered statistically significant; R2 was calculated to determine goodness of fit.11 The SHRINE query topic was approved by the SHRINE data steward; both SHRINE and MIMIC II are institutional review board exempt. All investigators completed the appropriate human subjects training.
Results
Eighty-five patients were diagnosed with MM in the MIMIC II dataset, accounting for 102 total hospital admissions. In aggregate, there were 1203 ICD-9-CM codes from these hospitalizations, corresponding to 407 unique ICD-9-CM diagnoses and 137 unique CCS groupings. Excluding the ICD-9-CM codes and CCS group corresponding to MM itself, there were 24 significant diagnosis codes and CCS groupings (table 1). Five CCS groupings (CCS 133, CCS 259, CCS 58, CCS 63, and CCS 3) were felt to be too non-specific to be included in the query strategy. ICD-9-CM codes 733.13, 117.3, and 584.9 were subsumed by CCS groups 207, 4, and 157, respectively. Of the remainder, eight were determined to be non-infectious and treatment-related (V42.81, CCS 2617, 996.85, V42.82, CCS 81, CCS 95, CCS 237, and 288); five were non-infectious complications of MM (CCS 207, 285.22, CCS 157, CCS 209, and 275.42) and three were infectious complications of MM or its treatment (CCS 2, CCS 122, CCS 4).
Table 1.
Significant (p value≤9.19×10−5) ICD-9-CM codes and CCS groups from the MIMIC II analysis, with associated p values
| ICD-9-CM or CCS | Description | O:E Ratio | p Value |
|---|---|---|---|
| CCS 207 | Pathological fracture | 30.7 | 6.02×10−17 |
| V42.81 | Bone marrow replaced by transplant | 53.4 | 1.20×10−14 |
| 285.22 | Anemia in neoplastic disease | 24.8 | 2.10×10−14 |
| CCS 259 | Residual codes; unclassified | 5.8 | 4.82×10−13 |
| 733.13 | Pathologic fracture of vertebrae | 23.4 | 3.90×10−12 |
| CCS 2617 | E codes: adverse effects of medical drugs | 8.2 | 5.67×10−10 |
| 996.85 | Complications of transplanted; bone marrow, graft-versus-host disease (acute) (chronic) | 24.6 | 2.30×10−09 |
| CCS 58 | Other nutritional; endocrine; and metabolic disorders | 15.5 | 3.42×10−08 |
| V42.82 | Transplant; peripheral stem cells | 54.5 | 5.00×10−08 |
| CCS 63 | Diseases of white blood cells | 10.3 | 2.51×10−07 |
| CCS 81 | Other hereditary and degenerative nervous system conditions | 17.3 | 5.60×10−07 |
| CCS 2 | Septicemia (except in labor) | 2.9 | 9.85×10−07 |
| CCS 3 | Bacterial infection; unspecified site | 4.1 | 1.66×10−06 |
| CCS 133 | Other lower respiratory disease | 8.8 | 3.78×10−06 |
| 117.3 | Aspergillosis, Infection by Aspergillus species, mainly A fumigatus, A flavus group, A terreus group | 22.2 | 4.00×10−06 |
| CCS 122 | Pneumonia (except that caused by tuberculosis or sexually transmitted disease) | 4.3 | 4.64×10−06 |
| CCS 4 | Mycoses | 6.9 | 8.77×10−06 |
| CCS 157 | Acute and unspecified renal failure | 2.4 | 1.11×10−05 |
| CCS 209 | Other acquired deformities | 68.4 | 1.97×10−05 |
| CCS 95 | Other nervous system disorders | 3.9 | 2.83×10−05 |
| 584.9 | Acute kidney failure, unspecified | 2.2 | 3.00×10−05 |
| CCS 237 | Complication of device; implant or graft | 3.3 | 3.58×10−05 |
| 275.42 | Hypercalcemia | 13.9 | 3.70×10−05 |
| 288 | Neutropenia | 21.4 | 4.40×10−05 |
Entries shown in italic were considered to be a marker for treatment-related complications and were used for subsequent SHRINE queries. O:E, observed:expected occurrence of code/group, patients with MM versus general MIMIC II population.
CCS, Clinical Classifications Software; ICD-9-CM, International Classification of Disease, Ninth Revision, Clinical Modification; MIMIC, Multiparameter Intelligent Monitoring in Intensive Care; MM, multiple myeloma; SHRINE, Shared Health Research Information Network.
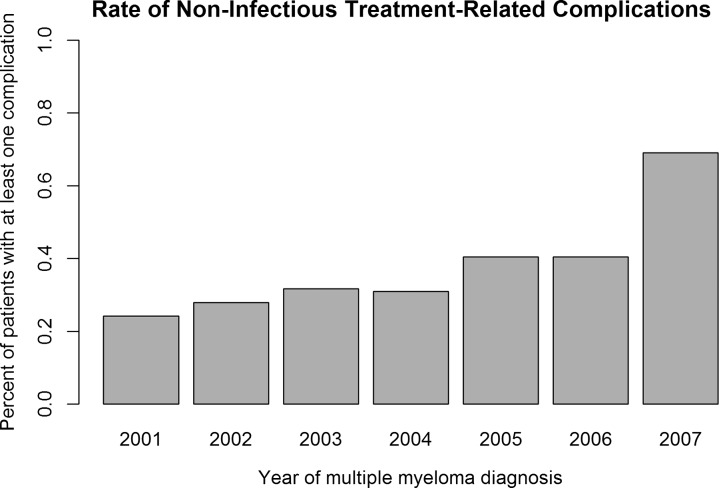
We identified a total of 3307 patients in SHRINE with five or more occurrences of any of the primary terms; representation was across the participating institutions, with no majority (<50%) from any single institution. A total of 1235 patients had one or more occurrences of any of the selected condition codes, giving an overall rate of 37% for non-infectious treatment-related complications. The rate of these codes increased significantly, by an average of 6.0% a year (p value 0.01, R2=0.74). Much of this increase was seen between the years 2006 and 2007, although the trend towards increasing complications remained significant even with exclusion of the 2007 data (3.4% a year, p value 0.003, R2=0.91). These results are summarized in figure 1.
Figure 1.
Rate of non-infectious treatment-related complications. Cases identified in Shared Health Research Information Network, in yearly intervals, with one or more non-infectious treatment-related complications, as defined in table 1, occurring up to 60 months after the time of case definition.
Discussion
This pilot project has demonstrated that SHRINE can be used to obtain aggregate information about patients diagnosed with MM across multiple institutions. Furthermore, we demonstrated a significant increase in the rate of condition codes for non-infectious treatment-related complications in this population, using encoded definitions identified through external analysis. At least three significant developments during the evaluated time period may account for these findings, including the apparent discrepancy seen between 2006 and 2007: (1) autologous, and to a lesser degree allogeneic, stem cell transplants became widespread in the treatment of MM13 14; (2) bortezomib, thalidomide, and lenalidomide became frequently used in the treatment of MM15–17; (3) universal healthcare was introduced in Massachusetts.
The apparent discontinuity between 2006 and 2007 was seen across multiple institutions. One possible explanation is that the seminal studies establishing lenalidomide as a treatment for MM were published in 200718 19; long-term follow-up of the Assessment of Proteasome Inhibition for Extending Remissions trial, which established bortezomib as a treatment for MM, was also published in 2007.20 Bortezomib and lenalidomide both commonly cause neuropathy, and thus individual codes within CCS groups 81 and 95 may account for the jump in the complication rate in 2007; confirmation of this hypothesis will be sought in future work. Universal healthcare, which was started mid-2006 might have resulted in more patients entering treatment over time. Finally, although changes is coding practices might also account for a portion of the observed changes, this would be difficult to demonstrate, without patient-level chart abstraction. MM is considered incurable, although the recent advances in treatment described above have improved disease-free survival and, possibly, overall survival. Whether the increase in treatment-related complications seen in this particular population has been accompanied by an increase in disease-free survival or by a decrease in overall mortality could not be determined with the current version of SHRINE.
Given that there are 17 000 ICD-9-CM codes, the initial evaluation for complications using the MIMIC II database was essential for the rational development of SHRINE queries. While MIMIC II represents a generally sicker population than that in SHRINE, its ease of use and comprehensiveness make it a useful database. Analysis of the results from MIMIC II makes it clear that human experts (including several of the authors of this paper) could generate only a partial list of the complication codes found, and this problem will become even more acute with future coding systems such as ICD-10. Future work will include validation of the results from MIMIC II on more general electronic medical record databases, possibly with the aid of phenotype visualization tools that are being developed.21
This general approach has several notable weaknesses. As mentioned, since MIMIC II represents a critically ill cohort, the patterns of complications, as defined by ICD-9-CM codes and CCS groupings, may not reflect a more typical outpatient population. However, ICD-9-CM phenotype is better defined in the inpatient setting owing to the increased use of multiple codes, on average about 10 for each patient in the case of MIMIC II. Even with this increased level of phenotypic detail, causality is often difficult to determine with the ICD-9-CM coding system. For instance, MM is often associated with infections due to frequent concomitant hypogammaglobulinemia; the treatment for MM is also associated with infections through other immunosuppressive mechanisms. Because ICD-9-CM does not distinguish the underlying cause of infection, we were unable to reliably assess infectious treatment-related complications. Thus, we probably underestimated the rate of overall treatment-related complications. Additionally, it must be acknowledged that phenome-wide analysis does not directly account for biases and confounding variables. It is possible that we also misestimated the study population in SHRINE, as a result of our definition of ‘established care.’ The cut-off point of five or more primary diagnosis code occurrences was chosen as a compromise between including patients who might have been only seen on a few occasions (eg, for a second opinion) and excluding patients who succumbed rapidly to their disease (eg, critically ill patients not surviving an initial hospitalization). There is no distinct cut-off point that can reliably differentiate over-exclusions and over-inclusions. On a related note, one important general caveat about SHRINE queries is that the same patient may be seen across multiple institutions, and thus be counted twice.
Subject to these limitations, SHRINE is a promising application which will become increasingly powerful with the participation of additional institutions. In conclusion, we have shown that SHRINE, when informed by data derived from an external database, can be a powerful tool to help answer specific health services research questions. For MM, in particular, this study demonstrates that treatment-related complications appear to have increased over time. Future work will focus on replicating this finding over an increased number of institutions, as SHRINE becomes more comprehensive, and investigating whether outcomes also changed over the observed time period. Whether complications which by their nature could be secondary to disease or to treatment (such as infectious complications in the case of MM) can be reliably incorporated through further algorithmic refinement will also be a focus of future work. Finally, the generalizability of the process will be examined by evaluating several other disease phenotypes.
Acknowledgments
We acknowledge Dr Charles Safran, whose advice and support during this process was invaluable; Katia Zilber-Izhar, Shared Health Research Information Network (SHRINE) project coordinator; and Andy McMurry, whose efforts in spreading awareness about SHRINE have been much appreciated.
Correction notice: This article has been corrected since it was published Online First. Robin M Joyce was incorrectly listed as Robin A Joyce.
Contributors: JLW, KB and RAJ conceived the study design; JLW performed the experiments; JLW and GA analyzed the data; JLW and RAJ contributed to the manuscript writing; all authors approved the final manuscript.
Funding: This work was supported, in part, by grants 5R21DA025168-02, 1R01HG004836-01, and 4R00LM009826-03 to GA. The funder had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Data from Multiparameter Intelligent Monitoring in Intensive Care (MIMIC II) are publicly accessible. Shared Health Research Information Network (SHRINE) aggregates data from individual institutions, which are the owners of such data. Any SHRINE data beyond aggregated reports requires institutional review board review and approval from each participating institution.
References
- 1.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet 2010;11:855–66 [DOI] [PubMed] [Google Scholar]
- 2.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010;26:1205–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res 2010;62:1120–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:624–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res 2009;19:1675–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet 2011;12:417–28 [DOI] [PubMed] [Google Scholar]
- 8.Kohane IS, Churchill SE, Murphy SN. A translational engine at the national scale: informatics for integrating biology and the bedside. J Am Med Inform Assoc 2012;19:181–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cusack CM, Shah S. Web-based tools from AHRQ's National Resource Center. AMIA Annu Symp Proc 2008:1221. [PubMed] [Google Scholar]
- 10.Saeed M, Villarroel M, Reisner AT, et al. Multiparameter Intelligent Monitoring in Intensive Care II: a public-access intensive care unit database. Crit Care Med 2011;39:952–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson B, Trapp RG. Basic & clinical biostatistics. 4th edn. New York: Lange Medical Books/McGraw-Hill, Medical Pub. Division, 2004 [Google Scholar]
- 12.Rice TK, Schork NJ, Rao DC. Methods for handling multiple testing. Adv Genet 2008;60:293–308 [DOI] [PubMed] [Google Scholar]
- 13.Attal M, Harousseau JL, Stoppa AM, et al. A prospective, randomized trial of autologous bone marrow transplantation and chemotherapy in multiple myeloma. Intergroupe Francais du Myelome. N Engl J Med 1996;335:91–7 [DOI] [PubMed] [Google Scholar]
- 14.Child JA, Morgan GJ, Davies FE, et al. High-dose chemotherapy with hematopoietic stem-cell rescue for multiple myeloma. N Engl J Med 2003;348:1875–83 [DOI] [PubMed] [Google Scholar]
- 15.San Miguel JF, Schlag R, Khuageva NK, et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med 2008;359:906–17 [DOI] [PubMed] [Google Scholar]
- 16.Palumbo A, Bringhen S, Caravita T, et al. Oral melphalan and prednisone chemotherapy plus thalidomide compared with melphalan and prednisone alone in elderly patients with multiple myeloma: randomised controlled trial. Lancet 2006;367:825–31 [DOI] [PubMed] [Google Scholar]
- 17.Rajkumar SV, Hayman SR, Lacy MQ, et al. Combination therapy with lenalidomide plus dexamethasone (Rev/Dex) for newly diagnosed myeloma. Blood 2005;106:4050–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimopoulos M, Spencer A, Attal M, et al. Lenalidomide plus dexamethasone for relapsed or refractory multiple myeloma. N Engl J Med 2007;357:2123–32 [DOI] [PubMed] [Google Scholar]
- 19.Weber DM, Chen C, Niesvizky R, et al. Lenalidomide plus dexamethasone for relapsed multiple myeloma in North America. N Engl J Med 2007;357:2133–42 [DOI] [PubMed] [Google Scholar]
- 20.Richardson PG, Sonneveld P, Schuster M, et al. Extended follow-up of a phase 3 trial in relapsed multiple myeloma: final time-to-event results of the APEX trial. Blood 2007;110:3557–60 [DOI] [PubMed] [Google Scholar]
- 21.Warner JL, Alterovitz G. Phenome-based analysis as a means for discovering context-dependent clinical reference ranges. AMIA Annu Symp Proc 2012: 1441–9 [PMC free article] [PubMed] [Google Scholar]