Abstract
The hereditary sensory and autonomic neuropathies (HSAN) are rare inherited neuropathies presenting with sensory loss and complications, including ulcers, infections, osteomyelitis and amputations. Usually, sensory symptoms predominate although motor involvement can occur. Autonomic features may be minimal (then hereditary sensory neuropathy, HSN, is preferred). HSAN has been classified into five subtypes depending on clinical presentation.1
Hereditary sensory and autonomic neuropathy II (HSANII or HSNII) is an early onset, autosomal recessive sensory neuropathy with ulcero-mutilating complications due to mutations in the HSN2 isoform of the WNK1 gene.2 Recently, a similar phenotype was described in a Saudi-Arabian family, and a homozygous nonsense mutation found in a new gene, FAM134B (family with sequence similarity 134, member B), encoding a newly identified Golgi protein. The index case in this family was initially thought to have leprosy. Three additional families (out of 75 patients) with similar phenotypes were found to have homozygous loss of function mutations in FAM134B.3
Here, we report the clinical and pathological findings in a further patient with HSNII due to a homozygous mutation in FAM134B.
Case Report
The patient (III-2) is from a consanguineous Somalian family (figure 1A) and had a normal birth and developmental milestones. She had recurrent foot infections and ulcers from the age of 5 years, and a right forefoot amputation aged 10 years. She started using a wheelchair aged 11 years. She described feeling pain and sweating normally. A cousin (III-7) had similar symptoms of recurrent infections and amputations.
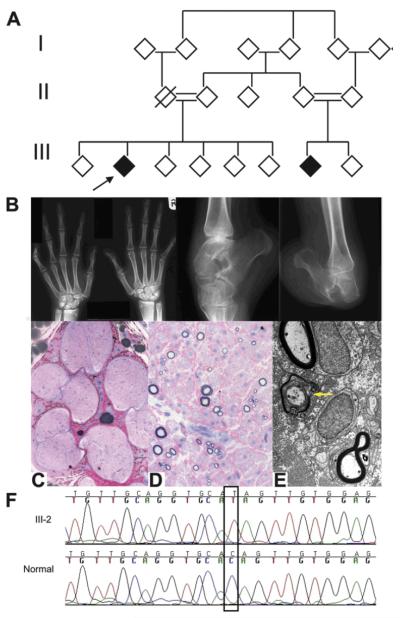
Figure 1.
(A) Filled symbols=affected; empty symbols=unaffected; cross-slashed symbols=deceased; double line=consanguinity. The proband is indicated by the arrow. (B) Radiographs of the hands and ankles. (C–E) Sural nerve biopsy. (C, D) Semi-thin resin sections, stained with MBABF. (C) Seven fascicles with severe depletion of myelinated fibres. Higher magnification (D) shows that both large and small myelinated fibres are affected. There is no inflammation. Electron microscopy (E) shows remaining myelinated fibres, with occasional demyelination (yellow arrow) and occasional bands of Büngner (not shown). Scale bar: (A) 500 μm, (B) 65 μm and (C) 1.1 μm. (F) Chromatograms of FAM134B in affected patient (top) and control (bottom). The homozygous nonsense mutation c.471 C→T is outlined.
At 16 years of age she was admitted to hospital with a discharging ulcer on the right tibia. Split-skin smears were negative for leprosy. The tibial ulcer had underlying chronic osteomyelitis and, despite intravenous antibiotics, required excision. A sural nerve biopsy was performed using standard procedures.
Neuropsychological tests were limited by language and cultural barriers but she performed adequately. Pupils were normal, pulse was regular and there was no postural hypotension. There was scoliosis, widespread scarring and hypopigmentation of the skin, and a painful callus on the palm. There was wasting of small hand muscles, fingernails were dystrophic and toenails absent. Fixed deformities were present at the fingers, knees and ankles. There was an amputation of the right forefoot. In the upper limbs there was weakness of intrinsic hand muscles to MRC grade 4. In the lower limbs there was marked weakness of all muscle groups, in a length dependent pattern, with no movement at the ankles. Proprioception and vibration were normal; there was patchy loss of pinprick in a glove and stocking distribution. Only triceps and supinator reflexes were present. Plantar responses were mute.
Plain radiographs (figure 1B) showed acroosteolysis and resorption of the terminal phalanges of the thumb and fingers, with joint destruction and fixed deformities of the little fingers. There was disruption of the right talocalcaneal joint with resorption of the anterior calcaneus. All bones were osteopenic.
Nerve conduction studies demonstrated an axonal, sensory more than motor, length dependent large fibre neuropathy (see supplementary table available online). Thermal thresholds were abnormal in the lower limbs, suggesting additional small fibre dysfunction. Autonomic function tests showed no fall in blood pressure after 4 min of sitting after supine testing (she could not stand). There was a blunted blood pressure response to isometric exercise, no response to cutaneous cold stimulation and a normal rise in heart rate during hyperventilation.
Sural nerve biopsy (figure 1C–E) demonstrated a severely reduced density of large myelinated fibres with occasional regenerative clusters. There was loss of both myelinated and unmyelinated fibres with no evidence of inflammation. Sequencing the FAM134B gene (primers available on request) revealed a homozygous nonsense mutation p.Q145X (figure 1F). This change was absent in 168 chromosomes from 84 European controls.
Sequencing of NGFB4, HSN22 and SPTLC15 was normal. Other members of the family were not available for examination or genetic testing.
Comment
This patient had an early onset, autosomal recessive, ulcero-mutilating neuropathy with amputations. There was considerable motor involvement but no significant autonomic involvement. Neurophysiology and nerve biopsy demonstrated an axonal neuropathy.
This report expands the phenotype of HSNII due to FAM134B mutations. In contrast to the previously reported cases,3 this patient had no evidence of significant autonomic involvement. Motor weakness was not described previously; our patient had significant length dependent weakness although this was complicated by severe joint deformities. Our patient was initially thought to have leprosy, similar to the first reported case.3
The nerve biopsy findings showed severe depletion of small and large myelinated and unmyelinated fibres. Nerve biopsy was previously reported in two of the original families with FAM134B mutations, also showing an axonal neuropathy but with a preference for small myelinated fibres.3
The nonsense mutation found in this patient, c.471C→T (p.Q145X), was described in the original paper in a Turkish family, is predicted to lead to nonsense mediated decay resulting in absence of the protein or a non-functional gene product.3 This family had onset in the first decade, impaired nociception, ulcerations of hands and feet, and chronic osteomyelitis; however, weakness was not described. Nerve conduction studies showed an axonal sensory neuropathy with some motor involvement, similar to our patient. It is of interest that although FAM134B is predominantly expressed in dorsal root ganglia, motor neurons do show some weak staining which may explain why our patient had motor involvement. As we confirmed the mutation by direct sequencing, we cannot exclude the possibility that the patient may have had a large deletion of one allele rather than being homozygous for the mutation; however, large deletions have not been described in this gene to date and a homozygous mutation is more likely given the consanguineous pedigree.
FAM134B belongs to a family of genes of unknown function. In mice, FAM134B is predominantly expressed in sensory and autonomic ganglia and is a component of the Golgi matrix.3 The FAM134B protein overlaps with the cis-Golgi marker giantin and has a similar response to brefeldin A as Golgi resident matrix proteins.3 The Golgi apparatus is made of stacks of flattened cisternae (from cis to trans); newly synthesised lipids and proteins enter the cis-Golgi network from the endoplasmic reticulum, are modified, and leave via the trans-Golgi network to be distributed to their final destinations. The mechanism by which mutations in FAM134B cause HSNII is unknown. It is possible that a structural change in the Golgi apparatus may occur, affecting trafficking of neurotrophins which are critical for survival of sensory and autonomic ganglia neurons.
In conclusion, this report expands the phenotype of HSNII due to mutations in FAM134B emphasising that motor involvement may occur and providing further evidence for the pathogenicity of mutations in FAM134B.
Supplementary Material
Acknowledgements
We are grateful to the patients and families who support this research.
Funding: MMR has received funding/grant support from the Medical Research Council (MRC) and the Muscular Dystrophy Campaign. HH has received funding/grant support from the NIHR UCLH/UCL Comprehensive Biomedical Research Centre (CBRC), the MRC (MRC Centre and HH fellowship, G108/638 and G0802760) and the Muscular Dystrophy Association (MDA). This work was undertaken at University College London Hospitals/University College London, which received a proportion of funding from the Department of Health's National Institute for Health Research Biomedical Research Centres funding scheme.
Footnotes
Competing interests: None.
Ethics approval: This study was conducted with the approval of the Joint Medical and Ethics Committee at The National Hospital for Neurology and Neurosurgery.
Provenance and peer review: Not commissioned; externally peer reviewed.
REFERENCES
- 1.Houlden H, Blake J, Reilly MM. Hereditary sensory neuropathies. Curr Opin Neurol. 2004;17:569–77. doi: 10.1097/00019052-200410000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Lafreniere RG, MacDonald ML, Dube MP, et al. Identification of a novel gene (HSN2) causing hereditary sensory and autonomic neuropathy type II through the Study of Canadian Genetic Isolates. Am J Hum Genet. 2004;74:1064–73. doi: 10.1086/420795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurth I, Pamminger T, Hennings JC, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. 2009;41:1179–81. doi: 10.1038/ng.464. [DOI] [PubMed] [Google Scholar]
- 4.Einarsdottir E, Carlsson A, Minde J, et al. A mutation in the nerve growth factor beta gene (NGFB) causes loss of pain perception. Hum Mol Genet. 2004;13:799–805. doi: 10.1093/hmg/ddh096. [DOI] [PubMed] [Google Scholar]
- 5.Bejaoui K, Wu C, Scheffler MD, et al. SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat Genet. 2001;27:261–2. doi: 10.1038/85817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.