Abstract
I investigated the potential contribution of Y-linked genes by analyzing 16 Y-consomic strains that had been established on a DH-strain background. The results provided evidence that only the Y chromosome from the C3H/HeJ strain was different from most other inbred strains. The CBA strain has the lightest testis and the DDD strain has the heaviest testis among mouse strains; however, Y-consomic analysis revealed that there were no significant differences in testis weight among DH, DH-Chr YDDD, and DH-Chr YCBA strains, suggesting that YDDD and YCBA themselves do not influence testis weight. QTL analysis in DDD × DH F2 mice identified significant testis weight QTLs on chromosomes 9, 14, and 17, and the DDD allele at all these loci was associated with an increase in testis weight. Contribution of Y chromosome itself to testis weight was thus rather modest, and therefore, major testis weight determinants are autosomal. However, it was uncertain whether there would be any effects by interactions between Y chromosomal and autosomal genes.
Keywords: mouse, QTL, testis weight, Y-consomic strain
Introduction
Normal development of the testis is crucially important for ensuring reproductive success. Testis weight has a direct connection with male fertility; that is, spermatogenic ability. The rate of sperm production largely depends on the total length and/or diameter of the seminiferous tubes, which, in turn, are the primary determinants of testis weight.1) In fact, testis weight depends on the rate of sperm production; for example, the testis weight in polygamous males is heavier than that in monogamous males in primates.2) A seasonal change in testis weight is reported in wild animals (seasonal breeders); they tend to have a greater testis weight in their breeding season.3) On the other hand, there is an apparent genetic aspect to the control of testis weight. Indeed, in laboratory mice, strain differences in testis weight have been apparent to many biologists and geneticists who work with divergent strains.4),5)
Testis weight is probably determined by the action of multiple genes under the influence of non-heritable environmental effects. In addition to genes on autosomes and the X chromosome, the relevance of Y-chromosome-linked (hereafter called Y-linked) genes has been suggested, because the testis develops only in males, and the testis-determining Sry gene is Y-linked. The effect of Y-linked genes on testis weight has been debated, but the results are conflicting.1),5)–8) For a definitive evaluation of the effect of the Y chromosome, it is crucial to synchronize the genetic background other than the Y chromosome. Analyzing Y-chromosome consomic strains (hereafter called Y-consomic strains) is the best way of accomplishing this. Accordingly, I established a series of Y-consomic strains, and I addressed whether there was any contribution of Y-linked genes to testis weight. In addition, involvement of genes on autosomes and/or on the X chromosome has been suggested.7),8) Another line of evidence also supports the probability of an autosomal contribution; that is, in mouse lines selected for testis weight in males, the ovulation rate in females increased.9),10) Because marked variations in testis weight are observed among mouse inbred strains in terms of either absolute weight or weight relative to body weight, the genetic basis for this variability can be investigated genetically with the aid of QTL analysis. To date, several QTL analyses have addressed the issue of the genetics of testis weight.5),11)–14) Although they demonstrated the presence of testis weight genes on several chromosomes, it is expected that novel genes or loci underlying testis weight will be identified in different genetic crosses. In this study, I performed QTL analysis on testis weight in the inbred DDD/Sgn mouse strain. The DDD/Sgn is one of the mouse strains that have an extremely large testis. The testis weight in DDD/Sgn is about one and a half times greater than that in common inbred mouse strains, such as C57BL/6J.
Materials and methods
Mice and genetic cross.
Inbred mouse strains DDD/Sgn (hereafter called DDD for convenience), DH/Sgn (DH), CF1/Sgn (CF1), RR/Sgn (RR), and SS/Sgn (SS) were maintained in the National Institute of Agrobiological Sciences (Tsukuba, Japan). A/J (A), CAST/EiJ (CAST), AKR/J (AKR), RF/J (RF), SJL/J (SJL), and SWR/J (SWR) strains were purchased from the Jackson Laboratory (Bar Harbor, ME). BALB/cA (BALB), C3H/HeJ (C3H), C57BL/6J (B6), and KK/Ta (KK) were purchased from CLEA Japan (Tokyo). CBA/N (CBA) was purchased from Japan SLC (Hamamatsu, Japan). I would like to mention the origin and characteristics of the DDD strain briefly on the basis of information from Mouse Genome Informatics (MGI, http://www.jax.org). In 1928, the original colony of dd mice was introduced from Germany into the Kitasato institute, Tokyo. Their descendants were shipped to the Health Institute of Manchuria Railway, Tailen, China in 1934. Two males and eight females from the Tailen colony were shipped back to the Institute for Infectious Diseases (Denken), Tokyo. Inbreeding of dd mice maintained at Denken was commenced in 1957, and the resulting inbred strain was named DDD after dd at Denken. As described below, F1- Dh/+ male mice resulting from a cross between DDD females and DH-Dh/+ are essentially lethal during neonatal period; however, this does not occur in the reciprocal cross.15)
For analysis of the effect of Y-linked genes by use of a series of Y-consomic strains, a Y-consomic strain, which has a Y chromosome from DDD (hereafter called YDDD), onto a DH background (hereafter called DH-Chr YDDD) has been produced by successive backcrossing (backcross generation: N30, sample size: n = 41). In a similar way, Y-consomic strains DH-Chr YA (N21, n = 27), DH-Chr YB6 (N29, n = 32), DH-Chr YBALB (N29, n = 24), DH-Chr YCAST (N30, n = 26), DH-Chr YCBA (N11, n = 11), DH-Chr YCF1 (N19, n = 21), DH-Chr YC3H (N28, n = 40), DH-Chr YKK (N12, n = 5), DH-Chr YRR (N11, n = 18), DH-Chr YSS (N11, n = 10), DH-Chr YAKR (N30, n = 37), DH-Chr YRF (N30, n = 32), DH-Chr YSJL (N31, n = 29), and DH-Chr YSWR (N34, n = 27), were established and used in this study.
Structurally, two kinds of Mus musculus Y chromosomes are known to coexist among the inbred mouse strains, that is, M. m. musculus Y (YMus) and M. m. domesticus Y (YDom), on the basis of nucleotide sequences of the Sry gene.16),17) Y chromosomes, YA, YB6, YBALB, YCAST, YCF1, YC3H, YKK, and YRR belong to YMus, YAKR, YRF, YSJL, and YSWR belong to YDom, and YCBA and YSS are unknown.
For QTL analysis, reciprocal F1 males were produced between DDD and DH. For F2 analysis, DDD females were crossed with DH males to produce F1, and F1 males and females were intercrossed to produce F2 males. Hereafter, I defined the DDD as having D alleles, and the DH as having H alleles, throughout the genome.
All mice were maintained in a specific-pathogen-free facility with a regular light cycle of 12 hr light: 12 hr dark, with controlled temperature and humidity. They had free access to food (CE-2, CLEA Japan) and water. Experiments were approved by the Institutional Animal Care and Use Committee of the National Institute of Agrobiological Sciences.
Phenotype measurements.
At the age of 80 ± 1 days after birth, mice were weighed with an electric balance to the nearest 0.01 g. Then the mice were killed, and the testis on both sides was removed and placed in physiologic saline. After they were rinsed, I wiped excessive moisture with wet chromatography paper, and the paired testes weight was determined with an electric balance to the nearest 1 mg. The spleen weight was determined in the same way. The weight of the spleen was analyzed as a reference for a parenchymatous organ. Trait names have been abbreviated as follows: Bw for body weight (g), Tw for absolute paired testis weight (mg), rTw for relative testis weight [Tw (mg)/Bw (g)], Sw for absolute spleen weight (mg), and rSw for relative spleen weight [Sw (mg)/Bw (g)]. Although rTw (and rSw) has been used to minimize the inevitable effects by Bw traditionally, Tw and rTw were analyzed separately in the study.
Genotyping.
Genomic DNA was isolated from the tails of mice with a commercial DNA extraction kit (Wizard Genomic DNA Purification Kit, Promega, Madison, WI). Microsatellite sequence length polymorphism was detected by electrophoresis subsequent to PCR. Most microsatellite primers were purchased as MapPairs (Research Genetics, Huntsville, AL), whereas others were synthesized on the basis of information from MGI. PCR amplification was carried out by use of a Takara PCR thermal cycler MP (TaKaRa Biomedicals, Tokyo) under the following conditions: 1 cycle at 94 °C for 5 min; 35 cycles at 94 °C for 30 s, 55 °C for 1 min, and 72 °C for 45 s; 1 cycle at 72 °C for 7 min. All PCR products were electrophoresed on 10% polyacrylamide gels for 70 min and visualized by ethidium bromide staining.
QTL analysis.
For identifying putative testis weight QTLs, a total of 72 F2 mice (which were selected out of 143 F2 mice), including 24 mice showing the highest Tw and 24 mice showing the lowest Tw. These 72 F2 mice also included 20 mice showing the highest rTw and 22 mice showing the lowest rTw. Furthermore, these 72 F2 mice were shown to include 19 out of 24 mice having the highest Bw and 18 out of 24 mice having the lowest Bw. The 72 F2 mice were genotyped for a total of 92 microsatellite marker loci distributed on all autosomes and the X chromosome. Initially, the QTL analysis was performed in 143 F2 mice with the Mapmaker/EXP version 3.0b and the Mapmaker/QTL 1.1b computer program.18) Of these 143 F2, 72 mice were genotyped completely, and the genotypes of most of the remaining mice were labeled as missing data. In general, once a log logarithm of odds (LOD) score of more than 2.8 (threshold LOD score for suggestive linkage as recommended by Lander and Kruglyak19)) was obtained, the remaining 71 F2 mice, and newly produced 30 F2 were genotyped for all microsatellite markers located on relevant chromosomes. At this stage, QTL analysis was again performed in all 173 F2 mice with the Map Manager QTX b20 software.20) The interval mapping was performed with this program. Because the interval mapping function of Map Manager QTX is most accurate when the phenotypic data are normally distributed, the normality was assessed by Kolmogorov-Smirnov test. As a result, because distribution of Bw in F2 mice did not follow a normal distribution, log-transformed Bw data was analyzed. Significant threshold values at genome-wide 5% level were calculated by performing 1,000 permutations of the phenotypic data. Although it depends on the traits, approximate threshold LOD scores (because QTL calculates a LRS, it is converted to a LOD score by dividing by 4.605) for five phenotypic traits are as follows: 2.1 for suggestive linkage, 3.6–3.7 for significant linkage, and 5.1–5.7 for highly significant linkage. Once a significant QTL was identified, the 95% confidence interval (CI) for the QTL was defined as a 1.5-LOD score support interval. Potential interaction between marker loci was evaluated pairwise. For this analysis, the threshold LOD score for significance at genome-wide 5% level was obtained for all traits by performing 1,000 permutations on the interaction model of Map Manager QTX b20, and then the significance of the total effect of the two loci was tested. Significant threshold LOD scores for total effects are as follows: 8.3 for Bw, 8.6 for Tw, and 8.4 for rTw. For a pair of loci showing the significant total effect, interaction testing was performed according to the User Manual for QTX (by Chmielewicz KM and Manly KF, http://www.mapmanager.org/mmQTX.html).
Statistics.
A statistical analysis between two groups was performed by use of Student’s or Welch’s t-test, and the statistical analysis among mouse groups with three possible genotypes in QTL analysis was performed by one-way ANOVA. Statistical analysis on Y-consomic strains was performed by use of Tukey’s multiple comparison tests with SPSS software (SPS for Windows Release 7.5.1J, SPSS Inc., Chicago, IL). P < 0.05 was considered to indicate significant difference.
Results
Analyses of Bw, Tw, and rTw in Y-consomic strains.
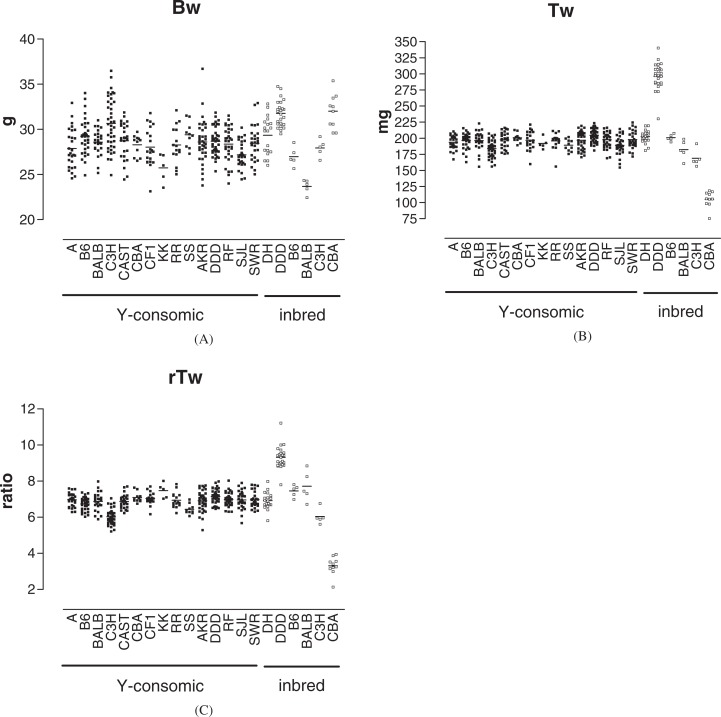
Scatter plots of trait values (Bw, Tw, and rTw) in 16 Y-consomic strains (including DH) and 6 inbred strains are shown in Fig. 1(A–C), and the results of Tukey’s multiple comparison tests are listed in Tables 1–6. DH was listed in plots as one of the inbred strains, but was also included in the statistical analysis in Y-consomic strains as DH-Chr YDH. B6, BALB, and C3H are progenitor strains for the DH. CBA is known as having the smallest testis among mouse strains on the basis of some publications1),5),7),21) and information from the commercial breeder (Japan SLC Inc., http://www.jslc.co.jp).
Fig. 1.
Scatter plots of trait values in 15 Y-consomic strains (▪) and 6 inbred strains (□). Each point represents the trait value of an individual mouse. As to the Y-consomic strains, only the donor strain symbols are presented at X-axis. Each horizontal bar indicates the mean of the trait value. A: Bw (body weight, g), B: Tw (absolute paired testis weight, mg), C: rTw [relative testis weight is expressed as Tw (mg)/Bw (g)]. All measurements were done at the age of 80 ± 1 days after birth.
Table 1.
Result of Tukey’s multiple comparison tests for Bw in Y consomic strains
| B6 | BALB | C3H | CAST | CBA | CF1 | KK | RR | SS | AKR | DDD | RF | SJL | SWR | DH | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | — | — | *** | — | — | — | — | — | — | — | — | — | — | — | — |
|
| |||||||||||||||
| B6 | — | — | — | — | — | * | — | — | — | — | — | ** | — | — | |
|
|
|
||||||||||||||
| BALB | ** | — | — | — | — | — | — | — | — | — | — | — | — | ||
|
|
|
||||||||||||||
| C3H | * | * | *** | *** | ** | — | *** | *** | *** | *** | *** | — | |||
|
|
|
||||||||||||||
| CAST | — | — | — | — | — | — | — | — | — | — | — | ||||
|
|
|
||||||||||||||
| CBA | — | — | — | — | — | — | — | — | — | — | |||||
|
|
|
||||||||||||||
| CF1 | — | — | — | — | — | — | — | — | — | ||||||
|
|
|
||||||||||||||
| KK | — | — | — | — | — | — | — | * | |||||||
|
|
|
||||||||||||||
| RR | — | — | — | — | — | — | — | ||||||||
|
|
|
||||||||||||||
| SS | — | — | — | — | — | — | |||||||||
|
|
|
||||||||||||||
| AKR | — | — | — | — | — | ||||||||||
|
|
|
||||||||||||||
| DDD | — | — | — | — | |||||||||||
|
|
|
||||||||||||||
| RF | — | — | — | ||||||||||||
|
|
|
||||||||||||||
| SJL | — | * | |||||||||||||
|
|
|
||||||||||||||
| SWR | — | ||||||||||||||
P < 0.05,
P < 0.01,
P < 0.001,
Not Significant
Table 6.
Result of tukey’s multiple comparison tests for rTw in six inbred strains
P < 0.05,
P < 0.01,
P < 0.001,
Not Significant
The Bw of a DH-Chr YC3H was greater than that of other Y-consomic strains except for DH-Chr YB6, DH-Chr YSS, and DH-Chr YDH (Fig. 1A and Table 1), suggesting that YC3H was different from the Y chromosome of most inbred strains in the ability to control Bw. In inbred strains, the Bw of the BALB was significantly smaller than that of all other strains (Fig. 1A and Table 2). Although there was no significant difference in Bw between DDD and CBA, these were significantly heavier than other strains. Because there was no significant difference in Bw among DH-Chr YBALB, DH-Chr YDDD, and DH-Chr YCBA (Table 1), this suggested that YBALB, YDDD, and YCBA themselves had no significant effects on Bw.
Table 2.
Result of Tukey’s multiple comparison tests for Bw in six inbred strains
P < 0.05,
P < 0.01,
P < 0.001,
Not Significant
The DH-Chr YC3H had a significantly lower Tw than did other Y-consomic strains except for DH-Chr YA, DH-Chr YKK, DH-Chr YRR, DH-Chr YSS, and DH-Chr YSJL (Fig. 1B and Table 3), suggesting that YC3H was different from the Y chromosomes of most inbred strains in the ability to control Tw. Among inbred strains, DDD had a significantly higher Tw than did all other strains, whereas CBA had a significantly lower Tw than did all other strains (Table 4). Because there was no significant difference in Tw among DH-Chr YDDD, DH-Chr YCBA, and DH-Chr YDH (Table 3), this suggested that YDDD and YCBA themselves had no significant effects on Tw. In contrast, C3H had a significantly lower Tw than did other inbred strains except for BALB (Table 4); thus, it was possible that the lower Tw in C3H is partly due to the effect of YC3H.
Table 3.
Result of Tukey’s multiple comparison tests for Tw in Y consomic strains
| B6 | BALB | C3H | CAST | CBA | CF1 | KK | RR | SS | AKR | DDD | RF | SJL | SWR | DH | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | — | — | — | — | — | — | — | — | — | — | * | — | — | — | — |
|
| |||||||||||||||
| B6 | — | ** | — | — | — | — | — | — | — | — | — | — | — | — | |
|
|
|
||||||||||||||
| BALB | ** | — | — | — | — | — | — | — | — | — | — | — | — | ||
|
|
|
||||||||||||||
| C3H | ** | ** | * | — | — | — | *** | *** | *** | — | *** | *** | |||
|
|
|
||||||||||||||
| CAST | — | — | — | — | — | — | — | — | — | — | — | ||||
|
|
|
||||||||||||||
| CBA | — | — | — | — | — | — | — | — | — | — | |||||
|
|
|
||||||||||||||
| CF1 | — | — | — | — | — | — | — | — | — | ||||||
|
|
|
||||||||||||||
| KK | — | — | — | — | — | — | — | — | |||||||
|
|
|
||||||||||||||
| RR | — | — | — | — | — | — | — | ||||||||
|
|
|
||||||||||||||
| SS | — | * | — | — | — | — | |||||||||
|
|
|
||||||||||||||
| AKR | — | — | — | — | — | ||||||||||
|
|
|
||||||||||||||
| DDD | — | *** | — | — | |||||||||||
|
|
|
||||||||||||||
| RF | — | — | — | ||||||||||||
|
|
|
||||||||||||||
| SJL | — | * | |||||||||||||
|
|
|
||||||||||||||
| SWR | — | ||||||||||||||
P < 0.05,
P < 0.01,
P < 0.001,
Not Significant
Table 4.
Result of Tukey’s multiple comparison tests for Tw in six inbred strains
P < 0.05,
P < 0.01,
P < 0.001,
Not Significant
Regarding rTw, DH-Chr YC3H had a significantly lower rTw than did other Y-consomic strains except for DH-Chr YSS (Fig. 1C and Table 5), suggesting that YC3H was different from the Y chromosomes of most inbred strains in the ability to control rTw. Among inbred strains, DDD had a significantly higher rTw than did all other strains, whereas CBA had a significantly lower rTw than did all other strains (Table 6). The finding that there was no significant difference in rTw among DH-Chr YDDD, DH-Chr YCBA, and DH-Chr YDH (Table 5) suggested that YDDD and YCBA themselves had no significant effects on rTw. Again, C3H had a significantly lower rTw than did the other inbred strains (Table 6); thus, it was possible that the lower rTw in C3H is partly due to the effect of YC3H. In all traits examined, there were also significant differences among several Y-consomic strains (Tables 1, 3, and 5), but, as can be seen, DH-YC3H was stood out among Y-consomic strains. The singularity of the YC3H was expressed most prominently in rTw, probably because the Bw-increasing effect and the Tw-decreasing effect of YC3H were combined.
Table 5.
Result of Tukey’s multiple comparison tests for rTw in Y consomic strains
| B6 | BALB | C3H | CAST | CBA | CF1 | KK | RR | SS | AKR | DDD | RF | SJL | SWR | DH | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | — | — | *** | — | — | — | — | — | — | — | — | — | — | — | — |
|
| |||||||||||||||
| B6 | — | *** | — | — | — | * | — | — | — | ** | — | — | — | — | |
|
|
|
||||||||||||||
| BALB | *** | — | — | — | — | — | — | — | — | — | — | — | — | ||
|
|
|
||||||||||||||
| C3H | *** | *** | *** | *** | *** | — | *** | *** | *** | *** | *** | *** | |||
|
|
|
||||||||||||||
| CAST | — | — | — | — | — | — | — | — | — | — | — | ||||
|
|
|
||||||||||||||
| CBA | — | — | — | * | — | — | — | — | — | — | |||||
|
|
|
||||||||||||||
| CF1 | — | — | * | — | — | — | — | — | — | ||||||
|
|
|
||||||||||||||
| KK | — | *** | — | — | — | — | — | — | |||||||
|
|
|
||||||||||||||
| RR | — | — | — | — | — | — | — | ||||||||
|
|
|
||||||||||||||
| SS | — | *** | * | — | — | — | |||||||||
|
|
|
||||||||||||||
| AKR | — | — | — | — | — | ||||||||||
|
|
|
||||||||||||||
| DDD | — | — | — | — | |||||||||||
|
|
|
||||||||||||||
| RF | — | — | — | ||||||||||||
|
|
|
||||||||||||||
| SJL | — | — | |||||||||||||
|
|
|
||||||||||||||
| SWR | — | ||||||||||||||
P < 0.05,
P < 0.01,
P < 0.001,
Not Significant
These results suggest that some Y chromosomes have effects on Bw, Tw, and rTw, but the Y chromosomes themselves of most inbred mouse strains had only modest effects on these traits. Instead, it is suggested that these traits are controlled mainly by autosomal and/or X-linked genes. Therefore, to search for such genes, I performed a subsequent QTL analysis.
QTL analysis.
The 72 F2 mice were genotyped for a total of 92 microsatellite marker loci distributed on all autosomes and on the X chromosome; the average distance between markers was approximately 17.4 cM (1,600 cM/92). A list of microsatellite markers used in the present study with their chromosomal location from the information retrieved from the MGI (February 5, 2008) is available upon request.
In the initial screening of 72 selected from 143 F2 mice, a significant linkage (LOD score ≥ 4.3 was applied to an initial analysis) was identified on chromosome 17 (D17Mit164 for rTw), and suggestive linkages (LOD score ≥ 2.8 was applied to an initial analysis) were identified on chromosomes 4 (near D4Mit214 for Tw), 9 (D9Mit229 for Tw), 11 (D11Mit236 for Bw and Sw), and 14 (D14Mit165 for rTw). The remaining 71 F2 mice and 30 newly produced F2 mice were genotyped for all of the loci on chromosomes 9, 11, 14, and 17, and for D4Mit214. In addition, D1Mit293, D5Mit240, D7Mit250, D10Mit188, D12Mit141, D13Mit139, D18Mit60, and D18Mit123 were genotyped, because these loci showed LOD scores with a near-suggestive threshold for some traits.
Pearson’s correlation coefficient between Bw and Tw was 0.46 (P = 9.68 × 10−11), that between Bw and Sw was 0.42 (P = 6.76 × 10−9), and that between Tw and Sw was 0.06 (P = 0.42). It was thus suggested that both of Tw and Sw are influenced by Bw to a certain extent, but distinct genetic bases may underlie in the control of Tw and Sw.
Bw QTLs.
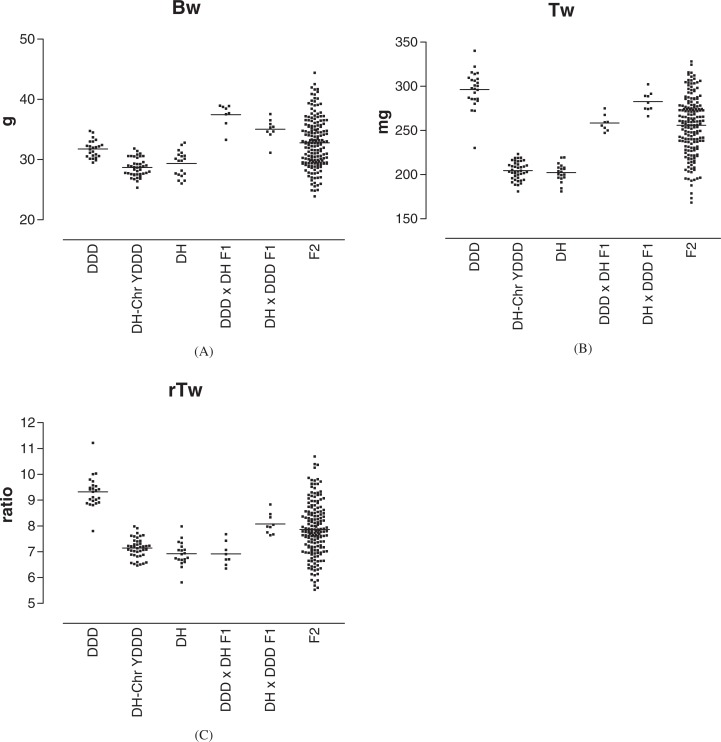
Scatter plots of Bw for DDD, DH-Chr YDDD, DH, DDD × DH F1, DH × DDD F1, and F2 are shown in Fig. 2A. In the comparison of Bw among DDD, DH-Chr YDDD, and DH, DDD mice were significantly heavier than DH and DH-Chr YDDD, but no significant difference was identified between DH and DH-Chr YDDD. The results suggested that YDDD itself had no significant effect on Bw in the DH background, and the larger Bw is attributed to autosomal, X-linked, and/or mitochondrial genes. Therefore, it was anticipated that some of these gene loci would be revealed by QTL analysis.
Fig. 2.
Scatter plots of trait values in DDD (n = 24–25), DH-Chr YDDD (n = 41), DH (n = 19), ♀DDD × ♂DH F1 (n = 8), ♀DH × ♂DDD F1 (n = 9), and ♀DDD × ♂DH F2 (n = 171–173). Each point represents the trait value of an individual mouse. Each horizontal bar indicates mean of the trait value. A: Bw (body weight, g), B: Tw (absolute paired testis weight, mg), C: rTw [relative testis weight is expressed as Tw (mg)/Bw (g)]. All measurements were done at the age of 80 ± 1 days after birth.
Interestingly, DH F1 were significantly heavier than HD F1 mice (P < 0.02). Because YDDD did not differ from YDH in its effect on Bw, this reciprocal cross effect should again be attributed to the effect of X-linked genes, mitochondrial genes, or imprinted genes.
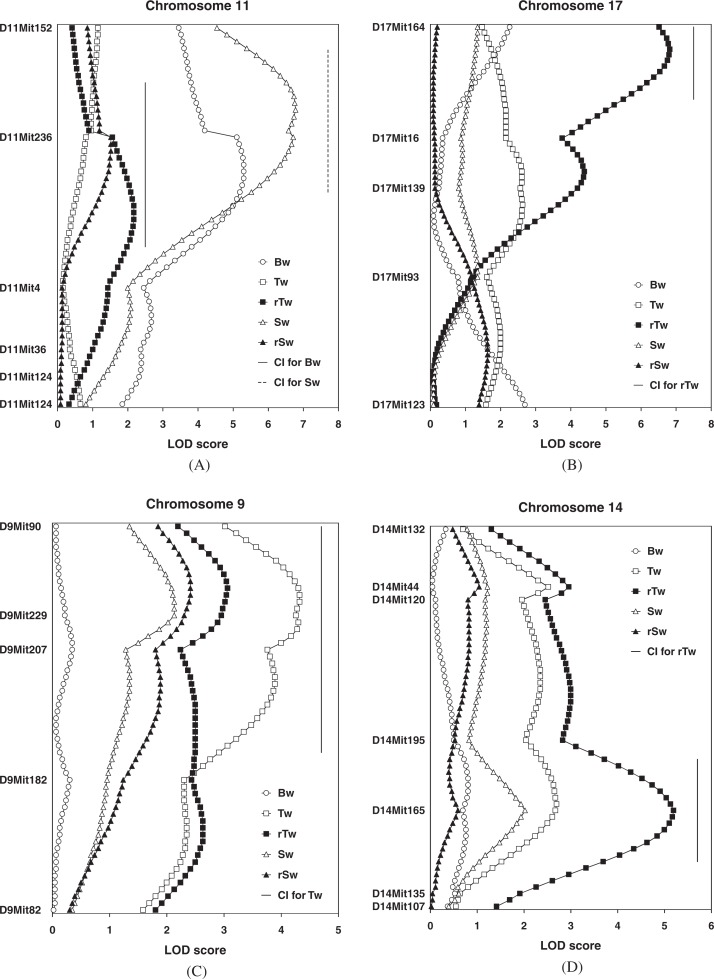
One significant Bw QTL was identified on proximal chromosome 11, near D11Mit236, with a peak LOD score of 5.3 (Table 7, Fig. 3A). I named this locus Bwdq1 (body weight in DDD male QTL 1). The D allele at Bwdq1 increased the Bw in an additive manner (Tables 7, 8). In addition, two separate suggestive Bw QTLs were identified on proximal- and distal-part of chromosome 17 (Table 7, Fig. 3B). At the proximal locus (near D17Mit164), the D allele was associated with a decrease in Bw, while the D allele was associated with an increase in Bw at the distal locus (near D17Mit123).
Table 7.
QTLs identified in this study
| Traitsa | Chr (Closest marker) | Peak positionb (CIc) | LOD (% Varianced) | Adde | Domf | QTL symbolg |
|---|---|---|---|---|---|---|
| Bw | 11 (D11Mit236) | 23 (17–32) | 5.3 (13)** | 0.03 | −0.01 | Bwdq1 |
| 17 (D!7Mit164) | 4 | 2.3 (6)* | −0.02 | −0.00 | ||
| 17 (D17Mit123) | 57 | 2.7 (7)* | 0.02 | −0.00 | ||
| Tw | 4 (D4Mit214) | 18 | 2.9* | |||
| 5 (D5Mit240) | 59 | 2.6* | ||||
| 9 (D9Mit229) | 24 (h–50) | 4.3 (11)** | 15.96 | −6.30 | Twdq1 | |
| 14 (D14Mit165) | 51 | 2.7 (7)* | 12.50 | −5.64 | ||
| 17 (D17Mit139) | 33 | 2.6 (7)* | 13.15 | −0.21 | ||
| rTw | 9 (D9Mit229) | 22 | 3.1 (8)* | 0.45 | −0.08 | |
| 11 (D11Mit236) | 28 | 2.2 (6)* | −0.39 | 0.21 | ||
| 14 (D14Mit165) | 52 (46–57) | 5.2 (13)*** | 0.45 | −0.46 | Rtwdq1 | |
| 17 (D17Mit164) | 7 (h–13) | 6.8 (17)*** | 0.64 | −0.28 | Rtwdq2 | |
| Sw | 9 (D9Mit229) | 25 | 2.1 (6)* | −4.60 | −4.11 | |
| 11 (D11Mit236) | 18 (14–26) | 6.8 (17)*** | 9.76 | 3.83 | Swdq1 | |
| rSw | 9 (D9Mit229) | 22 | 2.4 (6)* | −0.17 | −0.08 |
Bw, log-transformed Bw (g); Tw, paired testis weight in mg; rTw, paired testis weight relative to Bw [Tw (mg)/Bw (g)]; Sw, spleen weight in mg; rSw, spleen weight relative to Bw [Sw (mg)/Bw (g)].
Peak position of the LOD score plot curve is expressed as distance from the centromere in cM.
95% confidence interval (CI) is defined by 1.5-LOD support interval. CI is given only to significant QTLs.
Total variance explained by QTL at this locus is expressed as percent. When only one or two markers are fully genotyped in all 173 F2 as to the relevant chromosome, this value is not given.
suggestive,
significant, and
highly significant.
The additive component of the QTL D allele effect. Positive value indicates that the D allele is associated with increased trait values, and negative value indicates that the D allele is associated with decreased trait values. When only one or two markers are fully genotyped in all 173 F2 as to the relevant chromosome, this value is not given.
The dominance component of the QTL D allele effect. When only single marker is fully genotyped in all 173 F2 as to the relevant chromosome, this value is not given.
Assignment of the QTL symbol is limited to significant and highly significant QTLs.
Proximal end of CI cannot be defined because it extends proximally.
Fig. 3.
LOD score plots of Bw (body weight), Tw (absolute paired testis weight), rTw (relative testis weight), Sw (absolute spleen weight), and rSw (relative spleen weight) on chromosomes 11 (A), 17 (B), 9 (C), and 14 (D). Each vertical line indicates 95% confidence interval (CI) for a defined trait. X-axis represents LOD score. Y-axis represents microsatellite localization; the upper part of the graph is in the direction of the centromere, and the lower part is in the direction of the telomere.
Table 8.
Effect of significant QTLs on the basis of single point statistics
| QTL (Closest marker) | Trait | Mean ± S.E.M. trait values by marker genotype
|
Nominal P value | ||
|---|---|---|---|---|---|
| H/H | H/D | D/D | |||
| Bwdq1 (D11Mit236) | Bw (g) | 30.93 ± 0.58 | 32.64 ± 0.41 | 35.34 ± 0.68 | 0.0000087 |
| Tw (mg) | 249.83 ± 5.32 | 255.27 ± 3.36 | 263.95 ± 5.23 | 0.16 | |
| rTw | 8.14 ± 0.18 | 7.87 ± 0.11 | 7.51 ± 0.14 | 0.029 | |
| Twdq1 (D9Mit229) | Bw (g) | 32.86 ± 0.61 | 32.48 ± 0.43 | 33.37 ± 0.74 | 0.539 |
| Tw (mg) | 244.61 ± 4.78 | 253.12 ± 3.44 | 274.28 ± 4.42 | 0.000067 | |
| rTw | 7.49 ± 0.14 | 7.84 ± 0.12 | 8.31 ± 0.16 | 0.0013 | |
| Rtwdq1 (D14Mit165) | Bw (g) | 32.36 ± 0.70 | 33.29 ± 0.44 | 31.96 ± 0.59 | 0.19 |
| Tw (mg) | 246.93 ± 4.94 | 253.02 ± 3.36 | 271.13 ± 4.90 | 0.0023 | |
| rTw | 7.67 ± 0.15 | 7.65 ± 0.10 | 8.56 ± 0.18 | 0.0000081 | |
| Rtwdq2 (D17Mit164) | Bw (g) | 34.02 ± 0.55 | 32.46 ± 0.45 | 31.25 ± 0.67 | 0.0056 |
| Tw (mg) | 252.40 ± 3.87 | 253.01 ± 3.87 | 269.26 ± 5.46 | 0.036 | |
| rTw | 7.49 ± 0.13 | 7.81 ± 0.10 | 8.69 ± 0.19 | 0.00000040 | |
Tw QTLs.
In a similar way as for the Bw, scatter plots of the Tw for DDD, DH-Chr YDDD, DH, DDD × DH F1, DH × DDD F1, and F2 are shown in Fig. 1B. In the comparison of Tw among DDD, DH-Chr YDDD, and DH, DDD mice had a significantly larger Tw than did DH and DH-Chr YDDD, but no significant difference was identified between DH and DH-Chr YDDD. The results suggest that YDDD itself had no significant effect on Tw in the DH background.
A reciprocal cross effect was observed; that is, DH × DDD F1 had a significantly larger Tw than did DDD × DH F1 (P < 0.0003).
One significant Tw QTL was identified on chromosome 9, near D9Mit229, with a LOD score of 4.3 (Table 7, Fig. 3C). I named this locus Twdq1 (testis weight in DDD male QTL 1). The D allele at Twdq1 increased the Tw in an additive manner (Tables 7, 8). In addition, four suggestive Tw QTLs were identified on chromosomes 4, 5, 14, and 17 (Table 7). At all these loci, the D allele was associated with an increase in Tw.
rTw QTLs.
Scatter plots of rTw for DDD, DH-Chr YDDD, DH, DDD × DH F1, DH × DDD F1, and F2 are shown in Fig. 1C. In the comparison of rTw among DDD, DH-Chr YDDD, and DH, DDD mice had significantly larger rTw than did DH and DH-Chr YDDD, but no significant difference was identified between DH and DH-Chr YDDD. The results suggest that YDDD itself had no significant effect on rTw in the DH background.
A reciprocal cross effect was again observed; that is, DH × DDD F1 had a significantly larger rTw than did DDD × DH F1 (P < 0.00006).
Two highly significant rTw QTLs were identified on chromosomes 14 (near D14Mit165) and 17 (near D17Mit164) (Table 7, Figs. 3B and D). I assigned the locus symbols Rtwdq1 (relative testis weight in DDD male QTL 1) and Rtwdq2 (relative testis weight in DDD male QTL 2) to these QTLs, respectively. The peak LOD score for Rtwdq1 was 5.2, and this locus explained 13% of the F2 variance. The D allele at Rtwdq1 was recessive to the H allele, and it increased the rTw (Tables 7, 8). On the other hand, the peak LOD score for Rtwdq2 was 6.8, and this locus explained 17% of the F2 variance. The D allele at Rtwdq2 increased rTw in an additive manner (Tables 7, 8). In addition, two suggestive QTLs were identified on chromosomes 9 and 11. Of these, a locus on chromosome 9 was mapped to near D9Mit229, with a peak LOD score of 3.1 (Fig. 3C). At this locus, the D allele was associated with an increase in rTw. This locus was suggested to be allelic with Twdq1.
It was thus revealed that YDDD itself had no significant effects on Bw, Tw, and rTw, and significant QTLs for these traits were confirmed on DDD autosomes; however, it was uncertain whether there would be any effects by interactions between Y chromosomal and autosomal genes.
Sw and rSw QTLs.
Although a data plot is not shown, the DDD strain had very large spleen among the inbred strains, and F2 mice showed a spectrum of Sw and rSw.
One highly significant Sw QTL was identified on chromosome 11 (near D11Mit236) (Table 7, Fig. 3A). I assigned the locus symbol Swdq1 (spleen weight in DDD male QTL 1) to this QTL. The peak LOD score for Swdq1 was 6.8, and this locus explained 17% of the F2 variance. The D allele at Swdq1 increased the Sw in an additive manner. Swdq1 was suggested to be allelic with Bwdq1, and probably for this reason, only one suggestive rSw locus was identified on chromosome 9. This locus was also suggestive QTL for Sw.
Next, potential interaction between marker loci was evaluated pairwise for Bw, Tw, and rTw. One significant interaction was detected for rTw. A marker pair of D1Mit293 and D12Mit141 showed LOD score of 8.9 as a total LOD score for association, and LOD score of 4.1 as an interaction LOD score. Interaction LOD score was higher than that for significant threshold LOD score obtained for interval mapping, and the P value for interaction effect was 0.002 (recommended P value was less than 0.01); these results satisfied a criterion for declaring significant pairwise interaction. Because both loci had not been genotyped completely till then, I genotyped both loci in 173 F2 mice. As a result, the interaction of this marker pair was revealed to be not statistically significant.
Finally, the effect of a significant QTL on other traits was examined. Because Bw and Tw are mutually interrelated traits, it is appropriate to assess whether the QTL has any effect on other related traits by using a point-wise, rather than a genome-wide, significance threshold of P = 0.05.22)Table 8 summarizes the results. It can be seen that Bwdq1 had a significant effect on the rTw, but the D allele was associated with a decreased rTw. Twdq1 also had a significant effect on the rTw, and the D allele was associated with an increased rTw. Rtwdq1 had no significant effect on Bw, but had significant effect on Tw. In contrast to Rtwdq1, Rtwdq2 had significant effect on the Bw and Tw. The D allele at the Rtwdq2 was associated with a decreased Bw, but was associated with an increased Tw. These results suggested the presence of a complex interrelation between Bw and Tw, and therefore rTw.
Discussion
Effect of Y-linked genes on testis weight is rather modest.
The effect of the Y-linked genes on testis weight has been argued about for many years, and the results are still conflicting.1),5)–8) Hayward and Shire,1) Le Roy et al.,5) and Hunt and Mittwoch,7) and reported results that support the presence of a Y chromosomal effect. In contrast, Herrick and Wolfe6) and Chubb8) claimed that it is unlikely that the Y chromosome has a significant effect. A problem is that the former three studies were done on some CBA substrains, whereas the latter two did not use the CBA strain. This is why I analyzed Y-consomic strains by incorporating the CBA strain in this study. Nevertheless, there was still a major discrepancy among the results of the preceding three studies1),5),7) and those of the present one with regard to the effect of YCBA. Hayward and Shire1) produced several genetic crosses including F1, F2, F3, F4, and N2 backcross progeny between CBA/FaCam and SF/Cam strains, and they showed that rTw is clearly segregated with the type of Y chromosome; that is, YCBA is associated with a lower rTw. Although they admitted that there was an autosomal contribution, they estimated that 41% of the difference in rTw was due to the Y chromosome. Hunt and Mittwoch7) produced F1, F2, and N2 backcross progeny between CBA/Gr and BALB/c, and they showed that the Tw is affected by factors on the Y chromosome as well as those on autosomes and the X chromosome. They estimated that approximately 46% of the difference in Tw was attributable to the effect of the Y chromosome. These two studies are similar in presenting the argument that a relatively large portion of the difference in testis weight is due to the Y chromosome. Le Roy et al.5) produced F1 and F2 progeny between CBA/H and NZB/BINJ, performed a QTL analysis on Tw, and identified testis weight determinants on several autosomes and the X chromosome. They also showed evidence that a Y-consomic strain carrying YCBA (to be exact, the non-recombining part of the YCBA) on an NZB background had a significantly lower Tw than did the NZB strain. The most plausible explanation for the inconsistency between Le Roy’s study5) and this study is the difference in the background strain. YCBA reduced the Tw on the NZB background, but not on the DH background. A CBA substrain difference was unlikely to be suspected, because all CBA substrains, including the present strain CBA/N, had very small Tw and rTw. The combination of the Y chromosome and the background genome may be crucially important for controlling testis weight, and native YNZB may be essential for sustaining a relatively larger Tw in the NZB strain, because the Tw was not changed in the CBA strain when its Y chromosome was replaced by that from the NZB strain.5) In this regard, it is interesting to replace the Y chromosome in the DDD strain by the Y chromosome of a different strain (i.e. DH-Chr YDH, see below), but this cannot be tested immediately.
In addition, although I did not measure trait values in the inbred AKR strain, several studies reported that the AKR strain has a smaller testis than do the other inbred strains.4),23) However, YAKR itself had no significant effects on any of the traits examined in the present study (Tables 1, 3, and 5). This result also suggested the absence of a testis weight determinant on the Y chromosome in the AKR strain. Overall, the effect of the Y chromosome itself on testis weight was surely present, but it was generally rather modest. However, I cannot rule out a possibility that there would be any effects by interactions between Y chromosomal and autosomal genes. It seems to be crucially important to establish and analyze DDD-Chr YDH for verifying the possibility.
Concerning the singularity of YC3H, colleagues and I have previously reported that YC3H was different from YB6, YBAL, and YDH24) in the ability to cause neonatal lethality in (♀DDD × ♂DH-Dh/+) F1-Dh/+ males.15),24) Colleagues and I previously reported that F1-Dh/+ male mice resulting from a cross between DDD females and DH-Dh/+ males were essentially lethal during neonatal period; however, this did not occur in the reciprocal cross.15) Subsequent genetic mapping analysis revealed that the lethality was caused by a combination of three independent gene loci; that is the Dh locus on chromosome 1, Grdhq1 locus on the X chromosome, and the Y-linked gene locus from some inbred strains.24) As to the Y-linked gene, YB6, YBAL, and YDH caused lethality, but YC3H did not. The singularity of YC3H among these four strains was supported by the nucleotide polymorphisms of the Sry gene.24) On the basis of the results of statistical comparison presented in Tables 2, 4, and 6, YDH was suggested to be the same as YB6 rather than as YBALB.
Major testis weight determinants are autosomal.
Several coincidental QTLs or candidate genes can be postulated for the present QTLs. In particular, there are numerous potential genes or loci within a 95% CI for Bwdq1. Among nearly ten candidate genes picked up in an MGI search, colony stimulating factor 2 (granulocyte-macrophage) (Csf2, 29.5 cM)25),26) and glycine receptor, alpha 1 subunit (Glra1, 30.0 cM),27) are known to have effects on spleen weight; therefore, taking the coincidental occurrence of Bwdq1 and Swdq1 into consideration, these two genes are the best candidates for these QTLs. Several overlapping QTLs are also known. Among them, weight gain in high growth mice 7 (Wg7)28) and body weight, 10 weeks, QTL 3 (Wt10q3),29) can be regarded as coincidental QTLs.
Although I do not enumerate them in detail, many candidate genes can be postulated for Twdq1; this is partly because of a very large CI for this locus (Table 7). According to the mutant phenotypes provided by MGI, many of the candidate genes may have crucial roles in spermatogenesis. It is impossible to point out which is the best candidate gene for Twdq1 at the moment.
In contrast to Twdq1 on chromosome 9, only a few candidate genes can be postulated for Rtwdq1 on chromosome 14 and Rtwdq2 on chromosome 17. Indeed, the fibronectin type III domain containing 3a (Fndc3a)30) is the only plausible candidate gene for Rtwdq1. For Rtwdq2, there are three possible candidate genes; of these, one is a gene and the remaining two are QTLs. The first candidate is a high mobility group AT-hook 1 (Hmga1).31) The second candidate is a male hybrid sterility QTL 1 (Mhstq1).12)Mhstq1 causes male infertility as well as a small testis. Because proximal chromosome 17 holds five hybrid sterility gene loci (Hst1, Hst4-7), an apparent association of Rtwdq2 with male fertility is suggested. Third, an association between the H2 haplotype and testis weight has been reported.22),32) Iványi et al.32) assigned the gene symbol Hom1 (hormone metabolism 1) to this locus. Shukri and Shire33) also reported that the segregation pattern for Tw was identical to that for the H2 locus. However, H2 is located at 23 cM, which is outside the CI for Rtwdq2; therefore, H2 is clearly excluded from being a candidate. However, assuming that the gene causative of Hom1 is not H2, and that Hom1 is allelic to Rtwdq2, there are no discrepancies between the present result and previously reported results, because there was a point-wise significant linkage for rTw at 23 cM position in the present study (Fig. 3B). Interestingly, Gregorová and Iványi23) produced and analyzed C57BL/10ScSnPh × AKR/J F2 mice and found that the AKR allele at the H2 locus was associated with an increased Tw (not significant) and rTw (significant), but was associated with a decreased Bw (significant). With regard to the effect of a locus on proximal chromosome 17, the inverse correlation between Bw and rTw (Tw) was also observed in this study. Finally, a suggestive QTL on chromosome 11 for rTw has an overlapping QTL, low testis weight 1 (Lstw1), which was identified in an interspecific recombinant congenic mice between B6 and Mus spretus.14)
Because of the limitation of the experimental cross design, I could not investigate the effects of mitochondrial and imprinted genes as well as the effects by interactions between Y chromosomal and autosomal genes, although the presence of these effects was suggested in this study. Nevertheless, I identified one suggestive and one significant QTLs for Tw, and two highly significant QTLs for rTW. The identification of the genes underlying these QTLs will provide insight into the genetic control of testis weight.
Acknowledgments
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported. This study was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Nos. 15500305 and 19500373).
Non-standard abbreviation
- QTL
quantitative trait locus
- LRS
likelihood ratio statistics
- LOD
logarithm of odds
References
- 1).Hayward, P and Shire, J.G.M. (1974) Nature 250, 499–500 [Google Scholar]
- 2).Harcourt, A.H., Harvey, P.H., Larson, S.G. and Short, R.V. (1981) Nature 293, 55–57 [DOI] [PubMed] [Google Scholar]
- 3).Leader-Williams, N. (1979) J. Reprod. Fert. 57, 117–126 [DOI] [PubMed] [Google Scholar]
- 4).Shire, J.G.M. andBartke, A. (1972) J. Endocrinol. 55, 163–171 [DOI] [PubMed] [Google Scholar]
- 5).Le Roy, I., Tordjman, S., Migliore-Samour, D., Degrelle, H and Roubertoux, P.L. (2001) Genetics 158, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Herrick, C.S. and Wolfe, H.G. (1977) Genetics 86, s27 [Google Scholar]
- 7).Hunt, S.E. and Mittwoch, U. (1987) Genet. Res. Camb. 50, 205–211 [DOI] [PubMed] [Google Scholar]
- 8).Chubb, C. (1992) Biol. Reprod. 47, 29–36 [DOI] [PubMed] [Google Scholar]
- 9).Land, R.B. (1973) Nature 241, 208–209 [DOI] [PubMed] [Google Scholar]
- 10).Mafizul Islam, A.B.M., Hill, W.G. and Land, R.B. (1976) Genet. Res. Camb. 27, 23–32 [DOI] [PubMed] [Google Scholar]
- 11).Zídek, V., Musilová, A., Pintíř, J., Šimákovaá, M. and Pravenec, M. (1998) Mamm. Genome 9, 503–505 [DOI] [PubMed] [Google Scholar]
- 12).Elliott, R.W., Poslinski, D., Tabaczynski, D., Hohman, C. and Pazik, J. (2004) Mamm. Genome 15, 704–710 [DOI] [PubMed] [Google Scholar]
- 13).Oka, A., Mita, A., Sakurai-Yamatani, N., Yamamoto, H., Takagi, N., Takano-Shimizu, T., Toshimori, K., Moriwaki, K. and Shiroishi, T. (2004) Genetics 166, 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).L’Hôte,D., Serres, C., Laissue, P., Oulmouden, A., Rogel-Gaillard, C., Montagutelli, X and Vaiman, D. (2007) Genetics 176, 1907–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Suto, J., Wakayama, T., Imamura, K., Goto, S. and Fukuta, K. (1996) Exp. Anim. 45, 99–101 [DOI] [PubMed] [Google Scholar]
- 16).Kunieda, T. and Toyoda, Y. (1993) Genomics 13, 236–237 [DOI] [PubMed] [Google Scholar]
- 17).Coward, P., Nagai, K., Chen, D., Thomas, H.D., Nagamine, C.M. and Lau, Y.F.C. (1994) Nat. Genet. 6, 245–250 [DOI] [PubMed] [Google Scholar]
- 18).Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E., Newburg, L. (1987) Genomics 1, 174–181 [DOI] [PubMed] [Google Scholar]
- 19).Lander, E. and Kruglyak, L. (1995) Nat. Genet. 11, 241–247 [DOI] [PubMed] [Google Scholar]
- 20).Manly, K.F., Cudmore, R.H.Jr. and Meer, J.M. (2001) Mamm. Genome 12, 930–932 [DOI] [PubMed] [Google Scholar]
- 21).Bartke, A. and Krzanowska, H. (1972) J. Hered. 63, 172–174 [DOI] [PubMed] [Google Scholar]
- 22).Galli, J., Li, L.-S., Glaser, A., Östenson, C.-G., Jiao, H., Fakhrai-Rad, H., Jacob, H.J., Lander, E.S. and Luthman, H. (1996) Nat. Genet. 12, 31–37 [DOI] [PubMed] [Google Scholar]
- 23).Gregorová, S. and Iványi, P. (1976) Folia Biol. (Praha) 22, 82–86 [PubMed] [Google Scholar]
- 24).Suto, J., Yamanaka, H. and Sekikawa, K. (1999) Mamm. Genome 10, 777–783 [DOI] [PubMed] [Google Scholar]
- 25).Robertson, S.A., Roberts, C.T., Farr, K.L., Dunn, A.R. and Seamark, R.F. (1999) Biol. Reprod. 60, 251–261 [DOI] [PubMed] [Google Scholar]
- 26).Riopel, J., Tam, M., Mohan, K., Marino, M.W. and Stevenson, M.M. (2001) Infect. Immun. 69, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Hirzel, K., Muller, U., Latal, A.T., Hulsmann, S., Grudzinska, J., Seeliger, M.W., Betz, H. and Laube, B. (2006) Neuron 52, 679–690 [DOI] [PubMed] [Google Scholar]
- 28).Farber, C.R. and Medrano, J.F. (2007) Genetics 175, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Moody, D.E., Pomp, D., Nielsen, M.K. and Van Vleck, L.D. (1999) Genetics 152, 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).MacGregor, G.R., Russel, L.D., Van Beek, M.E., Hanten, G.R., Kovac, M.J., Kozak, C.A., Meistrich, M.L. and Overbeek, P.A. (1990) Proc. Natl. Acad. Sci. USA 87, 5016–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Liu, J., Schiltz, J.F., Ashar, H.R. and Chada, K.K. (2003) Mol. Reprod. Dev. 66, 81–89 [DOI] [PubMed] [Google Scholar]
- 32).Iványi, P., Gregorová, S. and Micková, M. (1972) Folia Biol. (Praha) 18, 81–97 [PubMed] [Google Scholar]
- 33).Shukri, N.M. and Shire, J.G.M. (1989) J. Reprod. Fert. 87, 587–592 [DOI] [PubMed] [Google Scholar]