Abstract
One new phenylalanine derivative 4′-OMe-asperphenamate (1), along with one known phenylalanine derivative (2) and two new cytochalasins, aspochalasin A1 (3) and cytochalasin Z24 (4), as well as eight known cytochalasin analogues (5–12) were isolated from the fermentation broth of Aspergillus elegans ZJ-2008010, a fungus obtained from a soft coral Sarcophyton sp. collected from the South China Sea. Their structures and the relative configurations were elucidated using comprehensive spectroscopic methods. The absolute configuration of 1 was determined by chemical synthesis and Marfey’s method. All isolated metabolites (1–12) were evaluated for their antifouling and antibacterial activities. Cytochalasins 5, 6, 8 and 9 showed strong antifouling activity against the larval settlement of the barnacle Balanus amphitrite, with the EC50 values ranging from 6.2 to 37 μM. This is the first report of antifouling activity for this class of metabolites. Additionally, 8 exhibited a broad spectrum of antibacterial activity, especially against four pathogenic bacteria Staphylococcus albus, S. aureus, Escherichia coli and Bacillus cereus.
Keywords: soft coral-derived fungus, Aspergillus elegans, phenylalanine derivative, cytochalasin, antifouling
1. Introduction
Marine-derived fungi have proven to be a promising source of structurally novel and biologically active secondary metabolites that have become interesting and significant resources for drug discovery [1]. Especially, the genus Aspergillus has been known to be a major contributor to the bioactive secondary metabolites of marine fungal origin, for example, antibacterial bisabolene-type sesquiterpenoids from sponge-derived fungus, Aspergillus sp. [2], cytotoxic hetero-spirocyclic γ-lactams from A. sydowii [3], 14-membered macrolides from A. ostianus [4], anthraquinone derivatives with naphtho [1,2,3-de] chromene-2,7-dione skeleton from A. glaucus [5] and cytochalasins from A. flavipes [6].
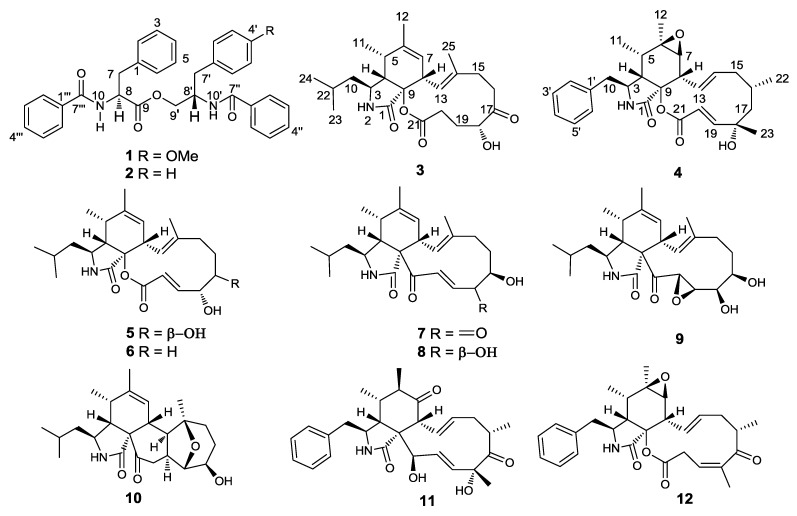
In our search for new antibacterial, cytotoxic and antifouling natural products from marine fungi in the South China Sea, we have found several bioactive compounds, including sesquiterpenoids, quinolinone alkaloids, azaphilone derivatives, resorcylic acid lactones and anthraquinone derivatives, from marine fungi through the bioassay-guided isolation [2,7,8,9,10,11,12]. In our ongoing investigations on the marine-derived fungi isolated from marine invertebrates, a fungus, Aspergillus elegans ZJ-2008010, obtained from a soft coral, Sarcophyton sp., attracted our attention. The EtOAc extract of a fermentation broth of the fungus exhibited antifouling activity against the larval settlement of the barnacle, Balanus amphitrite. Bioassay-guided fractionation of the bioactive extract led to the isolation of one new phenylalanine derivative, 4′-OMe-asperphenamate (1), together with one known phenylalanine derivative asperphenamate (2) [13,14] and two new cytochalasins, aspochalasin A1 (3) and cytochalasin Z24 (4), as well as eight known cytochalasins (5–12), determined as aspochalasins I (5) and J (6) [15], aspochalasins B (7) and D (8) [16,17], aspochalasin H (9) [18], aspergillin PZ (10) [19], zygosporin D (11) [20] and rosellichalasin (12) [21] (Figure 1). All isolated metabolites (1–12) were evaluated for their antifouling and antibacterial activities. Herein, we report the isolation, structure elucidation and biological activities of these compounds.
Figure 1.
Structures of compounds 1–12.
2. Results and Discussion
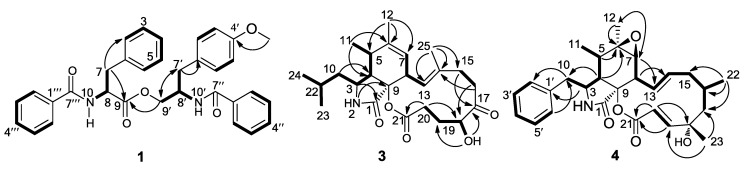
4′-OMe-asperphenamate (1) was obtained as a white powder. Its molecular formula was established as C33H32N2O5 (19 degrees of unsaturation) from HRESIMS, combined with 1H and 13C NMR spectroscopic data (Table 1). The 1H NMR spectrum of 1 showed the presence of 19 aromatic protons in the downfield, including three monosubstituted phenyls at δH [7.38 (2H, m), 7.24 (1H, m) and 7.23 (2H, m); 7.65 (2H, m), 7.50 (1H, m) and 7.30 (2H, m); and 7.70 (2H, m), 7.42 (1H, m) and 7.30 (2H, m)] and one disubstituted phenyl at δH [7.12 (1H, d, J = 8.4 Hz), 7.10 (1H, d, J = 8.4 Hz), 6.82 (1H, d, J = 8.4 Hz) and 6.81 (1H, d, J = 8.4 Hz)]. In addition, two secondary amino protons at δH 6.71 (1H, d, J = 8.4 Hz) and 6.59 (1H, d, J = 6.6 Hz) were also observed in the downfield. In the highfield, two methine groups at δH 4.88 (1H, m) and 4.62 (1H, m), one oxygenated methylene group at δH [4.52 (1H, dd, J = 10.8, 4.2 Hz) and 4.05 (1H, dd, J = 10.8, 3.0 Hz)], one methoxyl group at δH 3.74 (3H, s), two methylene groups at δH [3.22 (1H, dd, J = 14.4, 6.6 Hz) and 3.16 (1H, dd, J = 14.4, 6.6 Hz)] and at δH [3.00 (1H, dd, J = 13.2, 6.6 Hz) and 2.89 (1H, dd, J = 13.2, 8.4 Hz)] were observed. These spectroscopic features combined with the 13C NMR spectrum suggested that 1 very closely resembled asperphenamate (2) [13,14]. The only difference in the 1H NMR spectrum was the presence of a singlet methyl signal at δH 3.74 (3H, s) in 1 instead of an aromatic proton signal at δH 7.30 (1H, m) in 2. Furthermore, in the 13C NMR spectrum, the C-4′ signal moved downfield significantly [δC 158.9 (C) in 1 vs. 126.8 (CH) in 2], indicating a methoxy group was located at C-4′. The location of the methoxy group at C-4′ was confirmed by the heteronuclear multiple bond correlation (HMBC) correlation from 4′-OCH3 at δH 3.74 (3H, s) to C-4′ at δC 158.9 (C). The planar structure of 1 was further confirmed by the 1H-1H COSY and HMBC experiments (Figure 2).
Table 1.
NMR spectroscopic data (600/150 MHz, DMSO-d6) for compound 1.
| Position | 1H (J in Hz) | 13C | Position | 1H (J in Hz) | 13C |
|---|---|---|---|---|---|
| 1 | - | 134.2, C | 8′ | 4.62, m | 50.4, CH |
| 2 | 7.23, m | 129.4, CH | 9′ | 4.52, dd (10.8, 4.2) | 65.4, CH2 |
| 3 | 7.38, m | 128.8, CH | 4.05, dd (10.8, 3.0) | ||
| 4 | 7.24, m | 127.2, CH | 10′ | 6.71, d (8.4) | - |
| 5 | 7.23, m | 128.8, CH | 1″ | - | 133.4, C |
| 6 | 7.38, m | 129.4, CH | 2″ | 7.65, m | 127.2, CH |
| 7 | 3.22, dd (14.4, 6.6) | 37.4, CH2 | 3″ | 7.30, m | 128.5, CH |
| 3.16, dd (14.4, 6.6) | 4″ | 7.50, m | 132.1, CH | ||
| 8 | 4.88, m | 55.2, CH | 5″ | 7.30, m | 128.5, CH |
| 9 | - | 172.1, C | 6″ | 7.65, m | 126.9, CH |
| 10 | 6.59, d (6.6) | - | 7″ | - | 167.5, C |
| 1′ | - | 137.3, C | 1′″ | - | 137.2, C |
| 2′ | 7.10, d (8.4) | 130.3, CH | 2′″ | 7.70, m | 127.1, CH |
| 3′ | 6.81, d (8.4) | 114.3, CH | 3′″ | 7.30, m | 128.5, CH |
| 4′ | - | 158.9, C | 4′″ | 7.42, m | 131.5, CH |
| 5′ | 6.82, d (8.4) | 114.3, CH | 5′″ | 7.30, m | 128.5, CH |
| 6′ | 7.12, d (8.4) | 130.3, CH | 6′″ | 7.70, m | 127.1, CH |
| 7′ | 3.00, dd (13.2, 6.6) | 36.7, CH2 | 7′″ | - | 166.7, C |
| 2.89, dd (13.2, 8.4) | 4′-OCH3 | 3.74, s | 54.6, CH3 |
Figure 2.
1H-1H COSY correlations and key heteronuclear multiple bond correlation (HMBC) correlations of compounds 1, 3 and 4.
The absolute configurations of C-8 and C-8′ in 1 were determined by chemical synthesis and Marfey’s method [22]. The major acid hydrolysis products of 1 with 6 N HCl at 105 °C for 19 h were Phe and 2-amino-3-(4-methoxyphenyl)-1-propanol. The hydrolysates of 1, as well as the standard l-Phe and dl-Phe, were derivatized with Marfey’s reagent, 1-fluoro-2,4-dinitrophenyl-5-l-alanine amide (FDAA). Analysis of the FDAA derivatives of the hydrolysates from 1 by HPLC, compared with the FDAA derivatives from l-Phe and dl-Phe, revealed the presence of an l-Phe moiety in 1 (Figure S32). Therefore, the absolute configuration at C-8 was assigned as S.
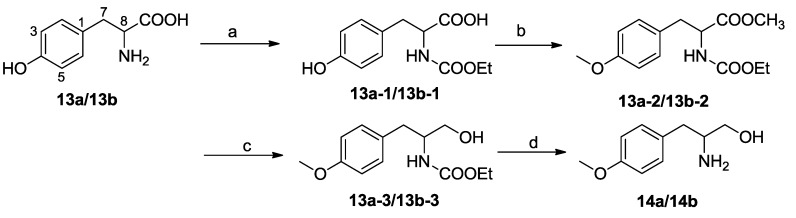
To determine the absolute configuration at C-8′, (R/S)-2-amino-3-(4-methoxyphenyl)-1-propanol (14a) and (S)-2-amino-3-(4-methoxyphenyl)-1-propanol (14b) were synthesized. Compounds 14a and 14b were prepared from dl-Tyr (13a) and l-Tyr (13b) (Figure 3), respectively, according to the literature method [23]. Also, 14a and 14b were derivatized with Marfey’s reagent (FDAA). Analysis of the FDAA derivatives of the hydrolysates from 1 by HPLC, compared with the FDAA derivatives from 14a and 14b, revealed the presence of a 14b moiety in 1 (Figure S32). Accordingly, the absolute configuration at C-8′ was assigned as S. The 8S8′S configurations in 1 were consistent with those in asperphenamate (2), confirming that these two compounds share the same biogenetic path way.
Figure 3.
The preparation of compounds 14a and 14b.
Aspochalasin A1 (3) was isolated as a white powder. Its molecular formula was established as C24H35NO5 (eight degrees of unsaturation) by HRESIMS, combined with 1H and 13C NMR spectroscopic data (Table 2). The 1H NMR spectrum recorded in CDCl3 intuitively revealed five methyl groups, two olefinic protons [δH 6.00, 1H, d, J = 11.4 Hz, H-13; δH 5.37, 1H, br s, H-7], an amide group (δH 5.93, 1H, br s, NH), six methine protons, five methylene units, along with one hydroxy group (Table 2). These features characteristically revealed the structure of 3 as possessing a (2-methylpropyl) isoindolone moiety, consistent with a cytochalasin skeleton [16]. These structural features were also confirmed by the 13C NMR and DEPT spectra (Table 2). In the 13C NMR spectrum, except for the carbon signals of the (2-methylpropyl) isoindolone moiety, there were also ten resonance signals revealed that 3 possessed a 12-membered lactone macrocycle, containing a methyl group (δC 15.9, CH3, C-25), a double bond [δC 125.6 (CH), C-13 and δC 136.2 (C), C-14] and two carbonyl groups [δC 212.7 (C), C-17 and δC 171.8 (C), C-21]. The 1H and 13C NMR spectra of 3 were closely related to those of aspochalasin M [24]. The obvious difference was the absence of the macrocyclic ketone moiety as in aspochalasin M at δC 214.5 (C-21) and the presence of a lactone at δC 171.8 (C-21) in 3. The differences were also observed for the chemical shifts of H-18/C-18 [δH 5.00 (1H, ddd, J = 10.8, 3.6, 3.6 Hz) and δC 74.0 (CH) in 3 vs. δH 4.15 (1H, dd, J = 5.0, 5.0 Hz) and δC 77.4 (CH) in aspochalasin M] and the large coupling constant J = 10.8 Hz between H-18 (δH 5.00) and H-19 (δH 1.53) in 3, instead of the small coupling constant J = 5.0 Hz between H-18 (δH 4.15) and H-19 (δH 2.00) in aspochalasin M. These features established an axial H-18 in 3 in contrast to an equatorial H-18 in aspochalasin M. This suggested that the OH at C-18 in 3 has a α-orientation different from the β-orientation in aspochalasin M. The 1H-1H COSY and HMBC allowed the deduction of the gross structure of 3 as shown in Figure 2.
Table 2.
NMR spectroscopic data (600/150 MHz, CDCl3) for compounds 3 and 4.
| Position | 3 | 4 | ||
|---|---|---|---|---|
| 1H (J in Hz) | 13C | 1H (J in Hz) | 13C | |
| 1 | - | 172.0, C | - | 166.9, C |
| 2 | 5.93, br s | - | 6.30, br s | - |
| 3 | 3.00, m | 52.6, CH | 3.68, m | 53.6, CH |
| 4 | 2.49, dd (4.2, 3.6) | 52.6, CH | 3.04, br d (5.4) | 49.2, CH |
| 5 | 2.74, m | 35.1, CH | 2.22, m | 35.9, CH |
| 6 | - | 141.2, C | - | 57.2, C |
| 7 | 5.37, br s | 122.5, CH | 2.74, d (5.4) | 60.8, CH |
| 8 | 3.43, d (10.8) | 42.1, CH | 2.99, d (10.8, 5.4) | 47.0, CH |
| 9 | - | 86.8, C | - | 85.2, C |
| 10 | 1.96, m | 46.4, CH2 | 2.86, dd (12.6, 8.4) | 45.1, CH2 |
| 1.57, m | 2.77, dd (12.6, 6.6) | |||
| 11 | 1.21, d (6.6) | 14.3, CH3 | 0.93, d (6.6) | 12.7, CH3 |
| 12 | 1.78, br s | 20.3, CH3 | 1.17, br s | 19.5, CH3 |
| 13 | 6.00, d (10.8) | 125.6, CH | 6.03, dd (13.2, 10.8) | 124.2, CH |
| 14 | - | 136.2, C | 5.21, ddd (13.2, 10.8, 3.6) | 138.8, CH |
| 15 | 2.68, m | 37.2, CH2 | 2.11, m | 44.1, CH2 |
| 2.39, m | 2.08, m | |||
| 16 | 2.69, m | 35.5, CH2 | 1.27, m | 29.3, CH |
| 2.45, m | ||||
| 17 | - | 212.7, C | 1.76, dd (13.2, 1.8) | 53.8, CH2 |
| 1.68, dd (13.2, 5.4) | ||||
| 18 | 5.00, ddd (10.8, 3.6, 3.6) | 74.0, CH | - | 72.8, C |
| 19 | 2.10, m | 29.1, CH2 | 7.11, d (15.6) | 159.1, CH |
| 1.53, m | ||||
| 20 | 2.65, m | 31.4, CH2 | 5.69, d (15.6) | 119.1, CH |
| 2.44, m | ||||
| 21 | - | 171.8, C | - | 172.8, C |
| 22 | 1.19, m | 25.6, CH | 1.03, d (6.6) | 27.1, CH3 |
| 23 | 0.93, d (6.6) | 21.0, CH3 | 1.33, s | 22.0, CH3 |
| 24 | 0.90, d (6.6) | 23.9, CH3 | - | - |
| 25 | 1.80, br s | 15.9, CH3 | - | - |
| 1′ | - | - | - | 137.0, C |
| 2′/6′ | - | - | 7.15, br d (7.2) | 129.4, CH |
| 3′/5′ | - | - | 7.30, m | 129.0, CH |
| 4′ | - | - | 7.25, m | 127.1, CH |
| 18-OH | 3.56, d (3.6) | |||
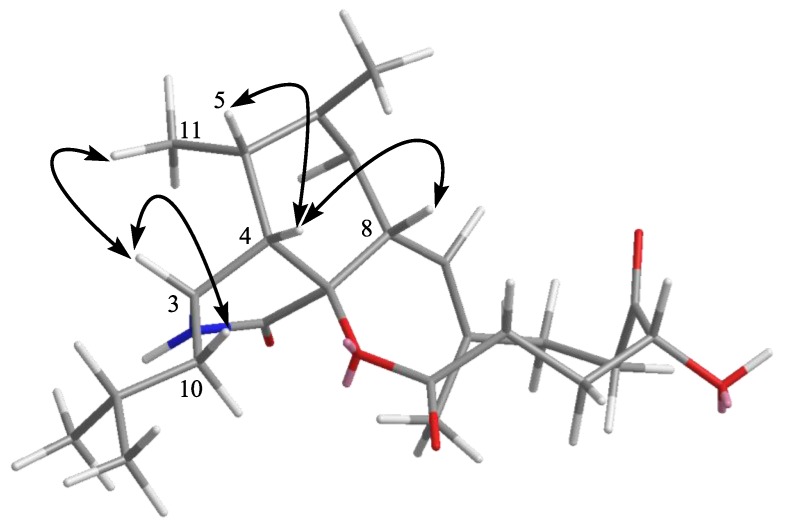
The relative stereochemistry of 3 was determined by NOESY experiments and comparison of NMR data with those of aspochalasin M [24]. In the NOESY experiments (Figure 4), the signal of H-3 showed correlations with H-10 and H-11 and H-4 with H-5 and H-8, indicating that the relative configurations of the perhydroisoindol-1-one moiety in 3 were in accord with those of reported cytochalasins. The modified Mosher’s method [25] was tried to determine the absolute configuration of C-18 in 3; unfortunately, the reactions failed. A literature search revealed that the stereochemistry of the cyclohexane and isoindole moieties in all isolated cytochalasins, so far, are the same [26]. Therefore, based on the above data and the biogenesis consideration, 3 was determined as aspochalasin A1.
Figure 4.
Key NOESY correlations of compound 3.
Cytochalasin Z24 (4) was also obtained as a white power. Its molecular formula was established as C28H35NO5 by HRESIMS. The general features of its NMR spectroscopic data (Table 2) were markedly similar to those of cytochalasin Z22 [27]. Detailed comparison of NMR data of these two compounds suggested that they had the same 10-phenyl-substituted 6,7-epoxyperhydroisoindol-1-one skeleton. The only significant difference in the 1H NMR spectrum was the presence of a methylene group at δH [1.76 (dd, J = 13.2, 1.8 Hz) and 1.68 (dd, J = 13.2, 5.4 Hz)] for H-17 in 4, and in the 13C-NMR spectrum, a methylene group at δC 53.8 (CH2) for C-17 was observed in 4, instead of a carbonyl carbon at δC 212.3 (C) for C-17 in cytochalasin Z22. The gross structure of 4 was further confirmed by 1H-1H COSY and HMBC spectra (Figure 2).
The configuration of the core skeleton, 10-phenyl-substituted 6,7-epoxyperhydroisoindol-1-one in 4 could be assigned based on biogenetic origin with those of cytochalasin Z22 [27]. In the 1H and 13C NMR spectra of 4, the chemical shifts and coupling constants of the H-atoms of C-3, C-4, C-5, C-7, C-8, 11-Me and 12-Me and the chemical shifts of the C-atoms of C-3–C-8, 11-Me and 12-Me were similar to those of cytochalasin Z22, suggesting that C-3–C-8 in 4 possess the same configurations as in Z22. The configurations of C-16 and C-18 of the macrocyclic moiety in 4 were assigned by comparison of the NMR data with those of [11]-cytochalasa-6(12),13,19-triene-1,21-dione-7,18-dihydroxy-16,18-dimethyl-10-phenyl-(7S*,13E,16S*,18S*,19E) [28], [δH 1.27 (1H, m) and δC 29.3 (CH) for C-16, δC 72.8 (C) for C-18 in 4] vs. [δH 1.33 (1H, m) and δC 28.8 (CH) for C-16, δC 72.8 (C) for C-18 in [11]-cytochalasa-6(12),13,19-triene-1,21-dione-7,18-dihydroxy-16,18-dimethyl-10-phenyl-(7S*,13E,16S*,18S*,19E)]. These results indicated that the two compounds shared the same macrocyclic moiety, and these were also confirmed by other known cytochalasins based on the shared biogenesis. The absolute configuration of 4 was then deduced as 3S,4S,5S,6R,7S,8S,9S,13E,16S,18S,19E. Therefore, the structure of 4 was established and named cytochalasin Z24.
The structures of known compounds (2, 5–12) were identified by comparison of their spectroscopic data with those in the literature [13,14,15,16,17,18,19,20,21]. Among them, asperphenamate (2), an uncommon phenylalanine derivative, has already been isolated from some bioactive natural sources, such as fungi, Aspergillus flavipes [29], Penicillium megasporum [30], P. brevicompactum [31] and P. canadense [32], and plants, Anaphalis subumbellata [33], Artemisia anomala [34] and Croton hieronymi [13]. In the present paper, this is the first report of isolated asperphenamate from marine-derived fungus.
Cytochalasins are a large group of fungal alkaloids with a wide range of biological activities targeting cytoskeletal processes [35]. Cytochalasin biosynthesis has been revealed by the formation of an acetate- and methionine-derived octa- or nonaketide chain and the attachment of an amino acid. The type of cytochalasins depends on the incorporated amino acids as structural subunits [36,37]. Compounds 3–9, 11 and 12 are a class of cytochalasins with 12-membered or 11-membered carbocyclic (or oxygen-containing) rings connecting the C-8 and C-9 positions of a perhydroisoindol-1-one moiety. The substituents at C-3 in compounds 3 and 5–9 is a 2-methylpropyl group; and in compounds 4, 11 and 12 is a phenyl group, belonging to the class of 10-phenyl-[11]-cytochalasin. Compound 10 belongs to an unusual type of aspochalasins with a pentacyclic system, of which only three examples have been reported.
All the isolated compounds were evaluated for antifouling activity against the larval settlement of the barnacle, Balanus amphitrite. Compounds 5, 6, 8 and 9 showed strong antifouling activity with the EC50 values of 34, 14, 6.2 and 37 μM, respectively. Despite the slight structural differences in their macrocycles, 8, bearing an α,β-unsaturated ketone moiety, was found to be considerably more active than 6, with an α,β-unsaturated lactone moiety, suggesting the importance of an electrophilic α,β-unsaturated carbonyl moiety for the antifouling activity of these cytochalasins. Compound 8 displayed more activity than 9, indicating that the double-bond at C-19 and C-20 might be essential for the antifouling activity of cytochalasins. This is the first report of antifouling activity for this class of metabolites.
The antibacterial activity of all isolated compounds were also assessed against six terrestrialpathogenic bacteria and two marine pathogenic bacteria. Compounds 1, 2, 5, 8 and 10 exhibited selective antibacterial activity (Table 3). Compounds 1 and 2 showed selective antibacterial activity against S. epidermidis, with the MIC values of 10 μM for each. Compound 5 showed moderate activity against S. epidermidis and S. aureus with the MIC values of 20 and 10 μM, respectively, while 8 showed a broad spectrum of antibacterial activity, especially towards four pathogenic bacteria, S. epidermidis, S. aureus, E. coli and B. cereus.
Table 3.
Antibacterial activity for compounds 1, 2, 5, 8 and 10.
| Compound | MIC (μM) | |||
|---|---|---|---|---|
| S. epidermidis | S. aureus | E. coli | B. cereus | |
| 1 | 10 | >20 | >20 | >20 |
| 2 | 10 | >20 | >20 | >20 |
| 5 | 20 | 10 | >20 | >20 |
| 8 | 10 | 10 | 10 | 10 |
| 10 | 20 | >20 | >20 | >20 |
| Ciprofloxacin a | 0.30 | 0.30 | 0.60 | 1.20 |
a Ciprofloxacin was used as a positive control.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were measured on a JASCO P-1020 digital polarimeter. IR spectra were recorded on a Nicolet Nexus 470 spectrophotometer. 1H and 13C NMR spectra were recorded on a JEOL Eclips-600 spectrometer at 600 MHz for 1H and 150 MHz for 13C in DMSO-d6 or CDCl3. Chemical shifts δ are reported in ppm, using TMS as internal standard, and coupling constants (J) are in Hz. ESIMS and HRESIMS spectra were measured on a Q-TOF Ultima Global GAA076 LC mass spectrometer. HPLC separation was performed using a Hitachi L-2000 prep-HPLC system coupled with a Hitachi L-2455 photodiode array detector. A Kromasil C18 preparative HPLC column (10 × 250 mm, 5 μm and 4.6 × 250 mm, 5 μm) was used. Analysis of FDAA derivatives by HPLC was performed using Waters 2695 prep-HPLC system coupled with a Waters 2489 UV detector. A Waters C18 analytical HPLC column (4.6 × 250 mm, 5 μm) was used. Silica gel (Qing Dao Hai Yang Chemical Group Co.; Qing Dao, China; 200–300 mesh), octadecylsilyl silica gel (Unicorn, Merck KGaA, Darmstadt, Germany; 45–60 μm) and Sephadex LH-20 (Amersham Biosciences Inc., Piscataway, NJ, USA) were used for column chromatography (CC). Precoated silica gel plates (Yan Tai Zi Fu Chemical Group Co.; Yan Tai, China; G60, F-254) were used for thin layer chromatography (TLC).
3.2. Fungal Materials
The fungal strain, Aspergillus elegans ZJ-2008010, was isolated from a piece of tissue from the inner part of the fresh soft coral, Sarcophyton sp. (GX-WZ-20080011), which was collected from the Weizhou coral reef in the South China Sea in September, 2008. The strain was deposited in the Key Laboratory of Marine Drugs, the Ministry of Education of China, School of Medicine and Pharmacy, Ocean University of China, Qingdao, China, with the access code ZJ-2008010. The fungal strain was cultivated in 50 L potato glucose liquid medium (15 g of glucose and 30 g of sea salt in 1 L of potato infusion, in 1 L Erlenmeyer flasks, each containing 300 mL of culture broth) at 25 °C without shaking for 4 weeks.
3.3. Identification of Fungus
The Fungus was identified according to its morphological characteristics and a molecular biological protocol by 16S rRNA amplification and sequencing of the ITS region. The sequence data have been submitted to GenBank, with an accession number JF694928, and the fungal strain was identified as Aspergillus elegans.
3.4. Extraction and Isolation
The fungal cultures were filtered through cheesecloth, and the filtrate was extracted with EtOAc (3 × 50 L, 12 h each). The organic extracts were concentrated in vacuo to yield an oily residue (20.2 g), which was subjected to silica gel column chromatography (CC) (petroleum ether, EtOAc v/v, gradient 100:0–0:100) to generate nine fractions (Fr. 1–Fr. 9). Fr. 2 was isolated by CC on silica gel eluted with petroleum ether-EtOAc (4:1) and then subjected to Sephadex LH-20 CC eluting with mixtures of petroleum ether-CHCl3-MeOH (2:1:1) to obtain compounds 1 (3.0 mg) and 2 (20.0 mg). Fr. 3 was subjected to repeated Sephadex LH-20 CC (CHCl3/MeOH, v/v, 1:1) and further purified by using HPLC on an ODS semi-preparative column (Kromasil C18, 10 × 250 mm, 5 μm, 2 mL/min) eluted with 85% MeOH/H2O to obtain compounds 3 (3.6 mg), 4 (4.0 mg), 7 (10.0 mg), 11 (7.0 mg) and 12 (5.0 mg). Fr. 4 was subjected to repeated Sephadex LH-20 CC (CHCl3/MeOH, v/v, 1:1) and further purified on HPLC (82% MeOH/H2O) to afford compounds 5 (10.8 mg), 6 (8.2 mg) and 8 (11.0 mg). Fr. 5 was subjected to repeated Sephadex LH-20 CC (MeOH) and further purified on HPLC (60% MeOH/H2O) to afford compounds 9 (9.0 mg) and 10 (12.0 mg).
4′-OMe-asperphenamate (1): White powder; [α]24D −38.0 (c 0.20, MeOH); UV (MeOH) λmax (log ε) 214(2.90), 280(2.50), 314(1.20) nm; IR (KBr) νmax 3448, 1712, 1624 cm−1; 1H and 13C NMR: see Table 1; HRESIMS m/z 537.2215 [M + H]+ (calcd. for C33H33N2O5, 537.2209).
Aspochalasin A1 (3): White powder; [α]24D +12.0 (c 0.15, CHCl3); UV (MeOH) λmax (log ε) 214(2.90); IR (KBr) νmax 3462, 1700, 1448 cm−1; 1H and 13C NMR: see Table 2; HRESIMS m/z 418.2582 [M + H]+ (calcd. for C24H36NO5, 418.2593).
Cytochalasin Z24 (4): White powder; [α]24D −25.0 (c 0.12, CHCl3); UV (MeOH) λmax (log ε) 214(4.1), 232(3.1), 265(3.0), 289(2.9); IR (KBr) νmax 3310, 1710, 1446 cm−1; 1H and 13C NMR: see Table 2; HRESIMS m/z 466.2407 [M + H]+ (calcd. for C28H36NO5, 466.2410).
3.5. Synthesis of Compounds 14a and 14b
(S/R)-3-(4-Hydroxyphenyl)-2-[(ethoxycarbonyl)amino] propionic acid (13a-1) or (S)-3-(4-hydroxyphenyl)-2-[(ethoxycarbonyl)amino] propionic acid (13b-1): Ethyl chloroformate (255 μL, 3.3 mmol) was added to a solution of dl-Tyr or l-Tyr (13a or 13b , 543 mg, 3 mmol, respectively) and NaHCO3 (750 mg, 9 mmol, respectively) in a mixture of H2O/THF (15 mL/15 mL, respectively). After stirring at room temperature (rt) overnight, the mixture was diluted with H2O and washed with Et2O, successively. The aqueous layer was acidified and extracted with EtOAc (3 × 18 mL). The combined extracts were washed (H2O), dried (Na2SO4) and concentrated to afford 500 mg (92%) of 13a-1 or 13b-1, respectively, colorless oil.
Methyl (S/R)-2-[(ethoxycarbonyl)amino]-3-(4-methoxyphenyl) propanoate (13a-2) or methyl (S)-2-[(ethoxycarbonyl)amino]-3-(4-methoxyphenyl) propanoate (13b-2): CH3I (370 μL, 6 mmol) was added to a solution of 13a-1 or 13b-1 (500 mg, 1.97 mmol, respectively) in dry acetone (5 mL, respectively) containing suspended K2CO3 (822 mg, 6 mmol, respectively). The mixture was refluxed at rt overnight, then diluted with H2O and extracted with EtOAc (3 × 50 mL, respectively). The combined extracts were washed (H2O), dried (Na2SO4) and concentrated, afforded 410 mg (82%) of 13a-2 or 13b-2, respectively, colorless oil.
(R/S)-2-[(Ethoxycarbonyl)amino]-3-(4-methoxyphenyl) propan-1-ol (13a-3) and (S)-2-[(ethoxycarbonyl)amino]-3-(4-methoxyphenyl) propan-1-ol (13b-3): A solution of 13a-2 or 13b-2 (200 mg, 0.71 mmol, respectively) in THF (10 mL, respectively) was added dropwise over 2 h to a cold (0 °C) solution of LiAlH4 (48 mg, 1.26 mmol, respectively) in THF (10 mL, respectively) maintained under Ar. The mixture was stirred overnight at rt; then, it was cooled to 0 °C and extracted with EtOAc. The combined extracts were washed (H2O), dried (Na2SO4) and concentrated. The crude product over silica gel (petroleum ether/EtOAc: 4/1) afforded 160 mg (80%) of 13a-3 or 13b-3, respectively, colorless oil.
(S/R)-2-Amino-3-(4-methoxyphenyl)-1-propanol (14a) and (S)-2-amino-3-(4-methoxyphenyl)-1-propanol (14b): A solution of 13a-3 or 13b-3 (160 mg, 0.63 mmol) and KOH (900 mg, 16.3 mmol, respectively) in CH3OH/H2O (2:1, 7 mL, respectively) was stirred at 80 °C 4 h. The mixture was then extracted with EtOAc (3 × 50 mL). The combined extracts were washed (H2O), dried (Na2SO4) and concentrated. The crude product was over silica gel (petroleum ether/EtOAc: 3/1) to yield 120 g (75%) of 14a or 14b, respectively, white solid.
3.6. Absolute Configuration Determination of 1 by Marfey’s Method [22]
A solution of 1 (1.5 mg) in 6 M HCl (1 mL) was heated to 105 °C for 19 h. The solution was then evaporated to dryness and the residue redissolved in H2O (250 μL). A 50 μL portion of the acid hydrolysate solution was then placed in a 1 mL reaction vial and treated with a 1% solution of FDAA (200 μL) in acetone followed by 1.0 M NaHCO3 (40 μL). The reaction mixture was heated at 45 °C for 1 h, cooled to room temperature and then acidified with 2.0 M HCl (20 μL). In a similar fashion, 14a and 14b and the standard dl- and l-Phe were derivatized with FDAA separately. The FDAA derivatives of the hydrolysates, 14a and 14b and standard amino acids were subjected to HPLC analysis (Waters C18 column; 5 μm, 4.6 × 250 mm; 1.0 mL/min) at 30 °C using the following gradient program: solvent A, H2O + 0.1% TFA; solvent B, MeCN; linear gradient: 0 min, 25% B, 40 min, 60% B, 45 min, 100% B; UV detection at 340 nm. The retention times for the FDAA derivatives of hydrolysates of 1 were 22.3 and 23.6 min, respectively; standard l-Phe, d-Phe, (S)-2-amino-3-(4-methoxyphenyl)-1-propanol (14b) and (R)-2-amino-3-(4-methoxyphenyl)-1-propanol were 23.6, 25.7, 22.3 and 24.1 min, respectively (Figure S28, Supporting Information).
3.7. Biological Assays
Antifouling activity against the larval-attachment was determined using cyprids of the barnacle, B. amphitrite Darwin, according to literature procedures [38]. Adults of B. amphitrite exposed to air for more than 6 h were collected from the intertidal zone in Hong Kong and then were placed in a container filled with 0.22 μm of filtered seawater (FSW) to release nauplii. The collected nauplii were reared to the cyprid stage according to the method described by Thiyagarajan et al. [38]. When kept at 26–28 °C and fed with Chaetoceros gracilis, larvae developed to cyprids on the fourth day. Fresh cyprids were used in the tests. Larval settlement assays were performed using 24-well polystyrene plates (Becton Dickinson 353047 [39]).
Antibacterial activity was determined against six terrestrial pathogenic bacteria, including Staphylococcus epidermidis, S. aureus, Escherichia coli, Bacillus subtilis, B. cereus and Micrococcus luteus, and two marine pathogenic bacteria, Vibrio parahaemolyticus and Listonella anguillarum, by the microplate assay method [40].
4. Conclusions
Twelve secondary metabolites, including two phenylalanine derivatives (1,2) and ten cytochalasins (3–12), have been isolated from the fermentation broth of a soft coral-derived fungus, Aspergillus elegans ZJ-2008010. Compound 1 is a new phenylalanine derivative, and 3 and 4 are new cytochalasin analogues. Their structures and the relative configurations were elucidated using comprehensive spectroscopic methods. The absolute configuration of 1 was determined by chemical synthesis and Marfey’s method. Asperphenamate (2) is the first report of isolated asperphenamate from marine-derived fungus. Compound 10 belongs to an unusual type of aspochalasins with a pentacyclic system, of which only three examples have been reported. Cytochalasins showed strong antifouling activity against the larval settlement of the barnacle, B. amphitrite. This is the first report of antifouling activity for this class of metabolites.
Acknowledgements
We appreciate Pei-Yuan Qian and his coworkers from Hong Kong University of Science and Technology for evaluation of the antifouling activity. This work was supported by the National Natural Science Foundation of China (No. 41130858; 41176121), the National High Technology Research and Development Program of China (863 Program) (No. 2013AA093001), the Natural Science Foundation of Shandong Province (ZR2011HM085), the Program for New Century Excellent Talents in University, Ministry of Education of China (No. NCET-11-0472) and the Natural Science Foundation of Hainan Province (213021).
Supplementary Files
Supplementary (PDF, 1846 KB)
References
- 1.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H.G., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2012;29:144–222. doi: 10.1039/c2np00090c. [DOI] [PubMed] [Google Scholar]
- 2.Li D., Xu Y., Shao C.L., Yang R.Y., Zheng C.J., Chen Y.Y., Fu X.M., Qian P.Y., She Z.G., de Voogd N.J., Wang C.Y. Antibacterial bisabolane-type sesquiterpenoids from the sponge-derived fungus Aspergillus sp. Mar. Drugs. 2012;10:234–241. doi: 10.3390/md10010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren H., Liu R., Chen L., Zhu T.J., Zhu W.M., Gu Q.Q. Two new hetero-spirocyclic γ-lactam derivatives from marine sediment-derived fungus Aspergillus sydowi D 2–6. Arch. Pharmacal Res. 2010;33:499–502. doi: 10.1007/s12272-010-0401-4. [DOI] [PubMed] [Google Scholar]
- 4.Kito K., Ookura R., Yoshida S., Namikoshi M., Ooi T., Kusumi T. New cytotoxic 14-membered macrolides from marine-derived fungus Aspergillus ostianus. Org. Lett. 2008;10:225–228. doi: 10.1021/ol702598q. [DOI] [PubMed] [Google Scholar]
- 5.Du L., Zhu T.J., Fang Y.C., Liu H.B., Gu Q.Q., Zhu W.M. Aspergiolide A, a novel anthraquinone derivative with naphtho [1,2,3-de] chromene-2,7-dione skeleton isolated from a marine-derived fungus Aspergillus glaucus. Tetrahedron. 2008;64 doi: 10.1016/j.tet.2008.03.012. [DOI] [Google Scholar]
- 6.Lin Z.J., Zhang G.J., Zhu T.J., Liu R., Wei H.J., Gu Q.Q. Bioactive cytochalasins from Aspergillus flavipes, an endophytic fungus associated with the mangrove plant Acanthus ilicifolius. Helv. Chim. Acta. 2009;92:1538–1544. doi: 10.1002/hlca.200800455. [DOI] [Google Scholar]
- 7.Wei M.Y., Wang C.Y., Liu Q.A., Shao C.L., She Z.G., Lin Y.C. Five sesquiterpenoids from a marine-derived fungus Aspergillus sp. isolated from a gorgonian Dichotella gemmacea. Mar. Drugs. 2010;8:941–949. doi: 10.3390/md8040941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao C.L., Wang C.Y., Gu Y.C., Wei M.Y., Pan J.H., Deng D.S., She Z.G., Lin Y.C. Penicinoline, a new pyrrolyl 4-quinolinone alkaloid with an unprecedented ring system from an endophytic fungus Penicillium sp. Bioorg. Med. Chem. Lett. 2010;20:3284–3286. doi: 10.1016/j.bmcl.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 9.Shao C.L., Wang C.Y., Wei M.Y., Gu Y.C., She Z.G., Qian P.Y., Lin Y.C. Aspergilones A and B, two benzylazaphilones with an unprecedented carbon skeleton from the gorgonian-derived fungus Aspergillus sp. Bioorg. Med. Chem. Lett. 2011;21:690–693. doi: 10.1016/j.bmcl.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Shao C.L., Wu H.X., Wang C.Y., Liu Q.A., Xu Y., Wei M.Y., Qian P.Y., Gu Y.C., Zheng C.J., She Z.G., Lin Y.C. Potent antifouling resorcylic acid lactones from the gorgonian-derived fungus Cochliobolus lunatus. J. Nat. Prod. 2011;74:629–633. doi: 10.1021/np100641b. [DOI] [PubMed] [Google Scholar]
- 11.Zheng C.J., Shao C.L., Guo Z.Y., Chen J.F., Deng D.S., Yang K.L., Chen Y.Y., Fu X.M., She Z.G., Lin Y.C., Wang C.Y. Bioactive hydroanthraquinones and anthraquinone dimers from a soft coral-derived Alternaria sp. J. Nat. Prod. 2012;75:189–197. doi: 10.1021/np200766d. [DOI] [PubMed] [Google Scholar]
- 12.Yang K.L., Wei M.Y., Shao C.L., Fu X.M., Guo Z.Y., Xu R.F., Zheng C.J., She Z.G., Lin Y.C., Wang C.Y. Antibacterial anthraquinone derivatives from a sea anemone-derived fungus Nigrospora sp. J.Nat. Prod. 2012;75:935–941. doi: 10.1021/np300103w. [DOI] [PubMed] [Google Scholar]
- 13.Catalan C.A., Heluani C.S., Kotowicz C., Gedris T.E., Herz W. A linear sesterterpene, two squalene derivatives and two peptide derivatives from Croton hieronymi. Phytochemistry. 2003;64:625–629. doi: 10.1016/S0031-9422(03)00202-4. [DOI] [PubMed] [Google Scholar]
- 14.Iapatra S.K., Pal M.K., Mallik A.K., Talapatra B. Structure and synthesis of (−)-anabellamide. A new phenylalanine derived ester amide from Anaphalis subumbellata: Occurrence of 4′-hydroxydehydrokawain. J.Nat. Prod. 1983;46:1401–1403. [Google Scholar]
- 15.Zhou G.X., Wijeratne E.M.K., Bigelow D., Pierson L.S.I., VanEtten H.D., Gunatilaka A.A.L. Aspochalasins I, J, and K: Three new cytotoxic cytochalasans of Aspergillus flavipes from the rhizosphere of Ericameria laricifolia of the Sonoran desert. J. Nat. Prod. 2004;67:328–332. doi: 10.1021/np030353m. [DOI] [PubMed] [Google Scholar]
- 16.Keller-Schierlein W., Kupfer E. Metabolites of microorganisms. The aspochalasins A, B, C, and D. Helv. Chim. Acta. 1979;62:1501–1524. doi: 10.1002/hlca.19790620516. [DOI] [Google Scholar]
- 17.Gebhardt K., Schimana J., Hoeltzel A., Dettner K., Draeger S., Beil W., Rheinheimer J., Fiedler H.P. Aspochalamins A–D and aspochalasin Z produced by the endosymbiotic fungus Aspergillus niveus LU 9575. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2004;57:707–714. doi: 10.7164/antibiotics.57.707. [DOI] [PubMed] [Google Scholar]
- 18.Tomikawa T., Shin-Ya K., Seto H., Okusa N., Kajiura T., Hayakawa Y. Structure of aspochalasin H, a new member of the aspochalasin family. J. Antibiot. 2002;55:666–668. doi: 10.7164/antibiotics.55.666. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Wang T., Pei Y.H., Hua H.M., Feng B.M. Aspergillin PZ, a novel isoindole-alkaloid from Aspergillus awamori. J. Antibiot. 2002;55:693–695. doi: 10.7164/antibiotics.55.693. [DOI] [PubMed] [Google Scholar]
- 20.Fujii Y., Tani H., Ichinoe M., Nakajima H. Zygosporin D and two new cytochalasins produced by the fungus Metarrhizium anisopliae. J. Nat. Prod. 2000;63:132–135. doi: 10.1021/np990331t. [DOI] [PubMed] [Google Scholar]
- 21.Kimura Y., Nakajima H., Hamasaki T. Structure of rosellichalasin, a new metabolite produced by Rosellinia necatrix. Agric. Biol. Chem. 1989;53:1699–1701. doi: 10.1271/bbb1961.53.1699. [DOI] [Google Scholar]
- 22.Marfey P. Determination of d-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg. Res. Commun. 1984;49:591–596. doi: 10.1007/BF02908688. [DOI] [Google Scholar]
- 23.Ousmer M., Braun N.A., Bavoux C., Perrin M., Ciufolini M.A. Total synthesis of tricyclic azaspirane derivatives of tyrosine: FR901483 and TAN1251C. J. Am. Chem. Soc. 2001;123:7534–7538. doi: 10.1021/ja016030z. [DOI] [PubMed] [Google Scholar]
- 24.Lin Z.J., Zhu T.J., Wei H.J., Zhang G.J., Wang H., Gu Q.Q. Spicochalasin A and new aspochalasins from the marine-derived fungus Spicaria elegans. Eur. J. Org. Chem. 2009;18:3045–3051. [Google Scholar]
- 25.Kusumi T., Fujita Y., Ohtani I., Kakisawa H. Anomaly in the modified Mosher’s method: Absolute configurations of some marine cembranolides. Tetrahedron Lett. 1991;32:2923–2926. doi: 10.1016/0040-4039(91)80650-U. [DOI] [Google Scholar]
- 26.Naruse N., Yamamoto H., Murata S., Sawada Y., Fukagawa Y., Oki T. Aspochalasin E, a new antibiotic isolated from a fungus. J. Antibiot. 1993;46:679–681. doi: 10.7164/antibiotics.46.679. [DOI] [PubMed] [Google Scholar]
- 27.Wang F.Z., Wei H.J., Zhu T.J., Li D.H., Lin Z.J., Gu Q.Q. Three new cytochalasins from the marine-derived fungus Spicaria elegans kla03 by supplementing the cultures with l- and d-tryptophan. Chem. Biodivers. 2011;8:887–894. doi: 10.1002/cbdv.201000133. [DOI] [PubMed] [Google Scholar]
- 28.Buchanan M., Hashimoto T., Asakawa Y. Five 10-phenyl-[11]-cytochalasans from a Daldinia fungal species. Phytochemistry. 1995;40:135–140. [Google Scholar]
- 29.Clark A.M., Hufford C.D., Robertson L.W. Two metabolites from Aspergillus flavipes. Lloydia. 1977;40:146–151. [PubMed] [Google Scholar]
- 30.Nozawa K., Udagawa S., Nakajima S., Kawai K. Studies on fungal products. Part 25. A dioxopiperazine derivative from Penicillium megasporum. Phytochemistry. 1989;28:929–931. doi: 10.1016/0031-9422(89)80145-1. [DOI] [Google Scholar]
- 31.Bird B.A., Campbell I.M. Occurrence and biosynthesis of asperphenamate in solid cultures of Penicillium brevicompactum. Phytochemistry. 1982;21:2405–2406. doi: 10.1016/0031-9422(82)85217-5. [DOI] [Google Scholar]
- 32.McCorkindale N.J., Baxter R.L., Roy T.P., Shields H.S., Stewart R.M., Hutchinson S.A. Synthesis and chemistry of N-benzoyl-O-[N′-benzoyl-l-Phenylalanyl]-l-Phenylalaninol, the major mycelial metabolite of Penicillium canadense. Tetrahedron. 1978;34:2791–2795. doi: 10.1016/0040-4020(78)88421-X. [DOI] [Google Scholar]
- 33.Poi R., Adityachoudhury N. Occurrence of two rare amides in Medicago polymorpha. Indian J. Chem. Sec. A. 1986;25:1245–1246. [Google Scholar]
- 34.Jakupovic J., Chen Z.L., Bohlmann F. Artanomaloide, a dimeric guaianolide and phenylalanine derivatives from Artemisia anomala. Phytochemistry. 1987;26:2777–2779. doi: 10.1016/S0031-9422(00)83589-X. [DOI] [Google Scholar]
- 35.Binder M., Tamm C. The cytochalasans: A new class of biologically active microbial metabolites. Angew. Chem. Int. Ed. Engl. 1973;12:370–380. doi: 10.1002/anie.197303701. [DOI] [PubMed] [Google Scholar]
- 36.Schuemann J., Hertweck C. Molecular basis of cytochalasan biosynthesis in fungi: Gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. J. Am. Chem. Soc. 2007;129:9564–9565. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y.G., Tian R.R., Liu S.C., Chen X.L., Liu X.Z., Che Y.S. Alachalasins A–G, new cytochalasins from the fungus Stachybotrys chartarum. Bioorg. Med. Chem. 2008;16:2627–2634. doi: 10.1016/j.bmc.2007.11.042. [DOI] [PubMed] [Google Scholar]
- 38.Thiyagarajan V., Harder T., Qiu J.W., Qian P.Y. Energy content at metamorphosis and growth rate of the early juvenile barnacle Balanus amphitrite. Mar. Biol. (Berl.) 2003;143:543–554. doi: 10.1007/s00227-003-1077-9. [DOI] [Google Scholar]
- 39.Li Y.X., Zhang F.Y., Xu Y., Matsumura K., Han Z., Liu L.L., Lin W.H., Jia Y.X., Qian P.Y. Structural optimization and evaluation of butenolides as potent antifouling agents: modification of the side chain affects the biological activities of compounds. Biofouling. 2012;28:857–864. doi: 10.1080/08927014.2012.717071. [DOI] [PubMed] [Google Scholar]
- 40.Pierce C.G., Uppuluri P., Teistan A.R., Wormley F.L.J., Mowat E., Ramage G., Lopez-Ribot J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008;3:1494–1500. doi: 10.1038/nport.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary (PDF, 1846 KB)