Abstract
The MYC proto-oncogene is frequently activated in human cancers through a variety of mechanisms. Its deregulated expression, unconstrained by inactivation of key checkpoints, such as p53, contributes to tumorigenesis. Unlike its normal counterpart, which is restrained by negative regulators, the unleashed MYC oncogene produces a transcription factor that alters global gene expression through transcriptional regulation, resulting in tumorigenesis. Key genes involved in ribosomal and mitochondrial biogenesis, glucose and glutamine metabolism, lipid synthesis, and cell-cycle progression are robustly activated by MYC, contributing to the acquisition of bioenergetics substrates for the cancer cell to grow and proliferate.
MYC is a master regulator of cell metabolism. In cells with deregulated MYC expression, key genes involved in cell growth and proliferation are robustly activated, resulting in tumorigenesis.
The MYC proto-oncogene, which belongs to the MYC family of genes (MYC, MYCN, MYCL), is downstream from many signal transduction pathways that are responsive to growth factors or the cellular microenvironment, such as cellular engagement with the extracellular matrix and nutrient or growth factor availability (Eilers and Eisenman 2008; Dang 2012b). For example, stimulation of receptor tyrosine kinases triggers activation of MEK-ERK and PI3K pathways, which in turn induces MYC expression. TGF-β can down-regulate MYC expression in a SMAD-dependent manner. The MYC proto-oncogene is, therefore, tightly linked to growth signals, cellular adherence, and the availability of nutrients. In this regard, depletion of MYC is detrimental for embryogenesis. However, the reliance of adult tissues on MYC depends on the tissue type and the role of other MYC family members for the renewal of specific adult tissues.
The inference that overexpressed MYC contributes to the genesis of certain human cancers is compatible with the tumorigenic activity of retroviral v-Myc in naturally occurring chicken tumors (see Connacci-Sorrell 2013). Hence, it is not surprising that ectopic expression of MYC in many transgenic murine tissues results in their oncogenic transformation, often associated with other genetic alterations such as loss of p53 or ARF (Zindy et al. 1998; Eischen et al. 1999). Intriguingly, the context in which MYC is activated could significantly affect tumorigenesis and the tumor type. For example, in a MYC-inducible system restricted to liver cells, activation of MYC in utero results in an aggressive postnatal hepatoblastoma-like diffuse liver cancer that kills the animals within several weeks after birth (Beer et al. 2004). Activation of MYC several weeks after birth resulted in multifocal, less aggressive liver tumors in adult animals.
Deregulated expression of MYC in cancers must be inherently different from regulated MYC expression in normal stem cell compartments. Deregulated MYC expression is surmised to induce a transcriptional response network that is different from the response triggered by endogenous normal MYC, which is fully restrained by feedback loops. In this regard, deregulated MYC expression predisposes cells to growth factor or nutrient deprivation-induced cell death, while deprived normal cells withdraw into the cell-cycle G1 phase (Askew et al. 1991; Evan et al. 1992; Shim et al. 1998; Yuneva et al. 2007). In addition, the MYC transcriptional program is likely to be different in the background of tumor suppressor mutations versus the program induced with normal cellular checkpoints still intact.
MYC FUNCTION
MYC produces a transcription factor MYC, which dimerizes with MAX to bind DNA and regulates gene expression (Eilers and Eisenman 2008). Global mapping of MYC-binding sites and MYC’s effects on gene expression have revealed that MYC is widely associated with chromatin from a variety of cell types. High-quality MYC-binding sites are on the order of 10,000 or more per human genome, although MYC binding does not necessarily correlate with gene expression changes, particularly in systems in which MYC is inducible (Zeller et al. 2006; Kim et al. 2010; Lee et al. 2012; Lin et al. 2012; Nie et al. 2012). In general, the tighter the binding of MYC and the closer the binding is to the proximal promoter of a gene, the higher is the change in gene expression on MYC induction. In fact, several studies provide evidence suggesting that MYC’s major role is its ability to recruit P-TEFb and relieve RNA polymerase pausing, thereby activating transcription (Eberhardy and Farnham 2001; Rahl et al. 2010).
Activation of MYC in many experimental systems is associated with cellular growth and proliferation programs. Indeed, loss-of-function analysis of MYC documents its central role in the growth programs of T lymphocytes, keratinocytes, and intestinal crypt cells (Zanet et al. 2005; Sansom et al. 2007; Wang et al. 2011). The transcriptional program triggered by MYC must be qualitatively and quantitatively different from that of a resting cell, in which expression of cyclin-dependent kinase inhibitors are generally elevated and expression of cyclin-dependent kinases are repressed (Hermeking et al. 2000; Seoane et al. 2001; Staller et al. 2001). The findings by two key papers documenting that widespread association of MYC with almost all genes suggest that MYC functions simply by amplifying gene expression (Lin et al. 2012; Nie et al. 2012; see Young 2013). These findings, however, do not account for the groups of genes that are repressed by MYC (Seoane et al. 2001; Staller et al. 2001). If MYC simply amplifies genes that are already expressed, then the relative ratios of growth-promoting versus growth-arresting gene expression would not be altered after MYC is activated. If MYC, however, were activated in slowly proliferating cells, then MYC-mediated amplification of growth-promoting genes would further drive tumor progression. As such, it is surmised that MYC alters the transcriptional program in favor of cell growth rather than just amplifying genes that are already expressed in resting cells, particularly genes that restrain resting cells from proliferating.
BIOENERGETICS OF GROWING CELLS
If the model of MYC function is that it activates transcriptional programs that favor cell growth and proliferation, and suppress programs that cause cell growth arrest, then MYC must favor the induction of key programs involved in the bioenergetics of growing cells.
The growth of a normal mammalian cell depends on growth factor signaling and nutrient availability (Cantor and Sabatini 2012; Dang 2012a). In the case of yeast, the availability of nutrients is sufficient to trigger a transcriptional program of growth without the need for growth factors (Lippman and Broach 2009). There are, however, no clear yeast orthologs for MYC:MAX. In mammalian cells, it is envisioned that growth factor receptor signaling, in the presence of adequate nutrients, would induce cascades of gene expression responses that should result in (1) import of key nutrients, (2) generation of ATP and NTPs, (3) generation of key building blocks for the synthesis of macromolecules, (4) stimulation of protein synthesis and the synthesis of macromolecules, (5) biogenesis of key cellular organelles, (6) orchestration of the cell-cycle machinery and stimulation of cell growth and division, and (7) regulation of cell- type-specific differentiation from the stem cell compartments. With this picture in mind, MYC is surmised to affect different genes quantitatively in a fashion that allows for the stoichiometric production of key components of the growing cell, supported by an adequate supply of energy and building blocks. Metabolism is up-regulated as cell growth increases and is therefore not just along for the ride.
In certain cell models, resting cells use fatty acid oxidation as a means to generate ATP, maintain membrane potentials, and turn over organelles (Wang et al. 2011). As the cell is induced to grow, glycolysis and glutaminolysis are induced through increased expression of glucose and amino acid transporters (Vander Heiden 2011; Cantor and Sabatini 2012; Dang 2012a). Glucose is imported through transporters into the cell and is subsequently phosphorylated by hexokinase to form glucose-6-phosphate, which is subsequently split into 3-carbon molecules, releasing ATP and culminating in the production of pyruvate. The glycolytic intermediate 3-phosphoglycerate is a substrate for the synthesis of lipids, as well as a precursor of serine and glycine (Chaneton et al. 2012). High metabolic flux through glycolysis hence provides growing cells with building blocks for macromolecular synthesis from glucose as well as ATP. Glucose could also be shunted into the pentose phosphate pathway for the production of NADPH and ribose for reductive biosynthetic reactions and nucleotide synthesis, respectively (Cantor and Sabatini 2012).
The final product of glycolysis, pyruvate, could be further oxidized in the mitochondrion through its conversion to acetyl-CoA, which combines with oxaloacetate to produce citrate, for subsequent degradation in the tricarboxylic acid (TCA) cycle (Cantor and Sabatini 2012). Citrate can also be exported to the cytosol and converted to acetyl-CoA by acetyl-CoA citrate lyase (ACLY) for fatty acid synthesis. In growing cells, a significant amount of pyruvate is converted to lactate, a phenomenon known as aerobic glycolysis or the Warburg effect (Cantor and Sabatini 2012; Zaidi et al. 2012). The shunting of pyruvate to lactate, catalyzed by lactate dehydrogenase (LDH), is necessary for the rapid regeneration of NAD+ from NADH. This NAD+ could be reused by glyceraldehyde 3-phosphate dehydrogenase (GADPH), one of the intermediate steps of the glycolytic pathway, to maintain glycolytic flux (Le et al. 2010). The flow of glucose to pyruvate is maintained partly by recycling of NAD+, whereas its shunting into the mitochondrion is limited by the activity of pyruvate dehydrogenase (PDH), which converts pyruvate to acetyl-CoA. PDH is potently regulated by pyruvate dehydrogenase kinases (PDKs), which phosphorylate and inhibit PDH activity, particularly under hypoxia (Kim et al. 2006; Papandreou et al. 2006).
Glutamine is imported into growing cells through the activation of transporters such as ASCT2 (DeBerardinis and Cheng 2010; Vander Heiden 2011). Once in the cytosol, glutamine can be imported into the mitochondrion, converted to glutamate by glutaminase, and subsequently metabolized to α-ketoglutarate by either glutamate dehydrogenase or glutamate-oxaloacetate transaminase. α-Ketoglutarate is further oxidized in the TCA cycle to generate ATP or provide the carbon skeleton for the production of aspartate, which is a key substrate for nucleotide biosynthesis. Glutamine in the cytosol could also be converted to glutamate by cytosolic glutaminase or exported through the antiporter SLC7A5, which extrudes glutamine in exchange for import of branched-chain amino acids such as leucine (Nicklin et al. 2009).
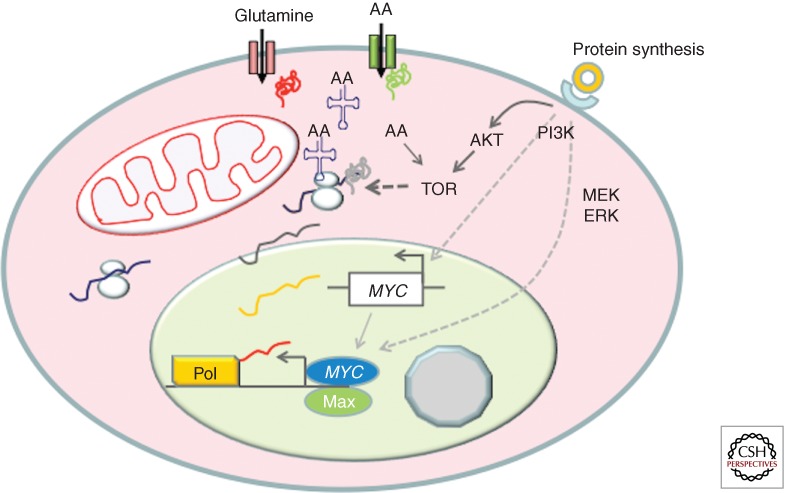
The influx of amino acids, particularly branched-chain amino acids, further activates mTOR downstream from receptor-mediated PI3K activation in a cell stimulated with growth factors (Fig. 1) (Laplante and Sabatini 2012). Protein synthesis is activated downstream from mTOR, resulting in increased ribosomal protein production, which requires concurrent increased mRNAs involved in all aspects of protein synthesis and ribosome biogenesis. Meanwhile, components necessary for nucleotide biosynthesis must also increase accordingly for the production of rRNA, mRNA, and noncoding RNAs. Specifically, the coordinated production of stoichiometric amounts of rRNAs and ribosomal proteins is required for the orderly assembly of ribosomes, such that an imbalance in the production of these components could result in excess levels of specific ribosomal proteins (RPL5, RPL11, RPL23, RPL26, or RPS7) that binds and inhibits Mdm2 ubiquitin ligase activity, releasing p53 to activate a checkpoint-mediated growth arrest (Deisenroth and Zhang 2010).
Figure 1.
Growth factor (doughnut) receptor engagement signals TOR activation through PI3K and MYC activation through the MEK-ERK pathway. MYC in turn stimulates genes involved in amino acid import and protein synthesis, particularly the aminoacyl-tRNA synthetases. The imported amino acids, such as leucine, further activate mTOR activity. The timing of events is only for heuristic purposes and is not based on evidence.
Further, nutrient-sensing mechanisms have been linked to the regulation of rRNA synthesis (Drygin et al. 2010). Glucose deprivation could cause an imbalanced synthesis of rRNA and ribosomal proteins, resulting in the activation of p53 and growth arrest. These complex mechanisms could have evolved from simpler signal transduction systems in yeast that sense nutrients and regulate ribosome biosynthesis. In the case of yeast, glucose and glutamine are sensed and signaled through Ras and TOR, which inactivate repressors of ribosome biogenesis genes (Lippman and Broach 2009). Mutational inactivation of these repressors renders mutant yeast constitutive for ribosomal protein production and addicted to the availability of glucose and glutamine. Hence, production of the central cellular machinery for cell growth, the ribosomes, is tightly linked to nutrient availability.
Not only do glucose and glutamine contribute to the synthesis of glycoproteins and complex carbohydrates of a growing cell, they are also involved in the production of lipids for membrane production. Breakdown of glucose generates phosphoglycerate that is used as the glycerol backbone for lipid synthesis and citrate that contributes to fatty acid synthesis (Cantor and Sabatini 2012). Glutamine is converted to α-ketoglutarate and further broken down by the TCA cycle to produce oxaloacetate, which combines with acetyl-CoA to form citrate that contributes to fatty acid synthesis (Zaidi et al. 2012). Reverse TCA cycling has also been documented, particularly in some hypoxic cells, providing a pathway for the generation of citrate from reductive carboxylation of α-ketoglutarate mediated by isocitrate dehydrogenase in the reverse direction (Yoo et al. 2008; Mullen et al. 2012).
In aggregate, the observations discussed above show that the growing cell is a DNA-instructed complex bioreactor that imports nutrients and oxygen to assemble macromolecules and cellular organelles, replicate DNA, and subsequently produce two daughter cells. The growing cell requires the production of ribosomes, which accounts for most of the cellular dry mass, for the synthesis of all proteins and enzymes that are involved in the synthesis of the other components of the cell. Ribosomes support the translation of many mRNAs that are induced in the growing cell. What then regulates transcription and the construction of two new cells from one original copy?
MYC IS THE MASTER REGULATOR OF TRANSCRIPTION IN GROWING CELLS
The fact that MYC is required for cell growth and could induce the expression of so many genes suggests that it is the master regulator of cell growth. But MYC is not expected to activate gene expression in a haphazard fashion without discrimination; it must stimulate genes involved in cell growth coordinately so that the mixture of transcribed mRNAs is translated into stoichiometric amounts of proteins for the balanced production of many complex macromolecules and cellular structures. This view is compatible with the observations that MYC is tightly linked to the generation of ribosomes through its ability to activate genes involved in ribosome biogenesis (van Riggelen et al. 2010). In fact, MYC is a unique transcription factor with its ability to activate transcription mediated by all three RNA polymerases, I, II, and III (Gomez-Roman et al. 2003; Grandori et al. 2005). The different classes of RNAs are essential for the synthesis of rRNAs, 5S RNA, and ribosomal proteins, which are all required for ribosome biogenesis. The link between MYC and ribosome biogenesis is further underscored by genetic evidence in Drosophila (Johnston et al. 1999; Grewal et al. 2005; Orian et al. 2005).
In mammalian cells, loss and gain of MYC function have been associated with diminished and enhanced cell growth, respectively. The importance of MYC for cell growth has been well documented in lymphocytes, particularly those with floxed allele of MYC that could be deleted by Cre recombinase (de Alboran et al. 2001; Wang et al. 2011). T lymphocytes without MYC failed to mount a growth response on stimulation of the T-cell receptor pathway. Loss of MYC also inhibits WNT-induced colonic crypt cell growth and proliferation (Sansom et al. 2007). Overexpression of MYC in B lymphocytes was shown to be sufficient to increase cell growth that could be unlinked from cell proliferation (Schuhmacher et al. 1999). Acute expression of MYC by adenoviral gene transfer into liver cells in vivo also resulted in significant cell growth without a high level of cell proliferation (Kim et al. 2000). All of these studies underscore MYC as a master transcriptional regulator of cell size increase or cell growth. mTOR, on the other hand, is the master posttranscriptional regulator of cell growth, and these two pathways are interdependent in growing cells. As such, everolimus, a TOR inhibitor, restored oncogene-induced senescence and diminished tumorigenesis in the Eμ-myc transgenic mouse lymphoma model (Wall et al. 2013).
MYC REGULATION OF GLYCOLYSIS AND GLUTAMINOLYSIS
In contrast to the false belief that metabolism is along for the ride as cells grow and divide, the finding that MYC directly regulates genes involved in glucose metabolism as well as those in ribosome biogenesis suggest that increased production of metabolic enzymes as a cell grows is far from being a passive phenomenon. Rather, enhanced metabolic capacity of a growing cell is essential for biomass accumulation and high-fidelity DNA replication. This enhanced capacity requires rewiring of the metabolic capacity of a resting cell into that of a growing and proliferating cell.
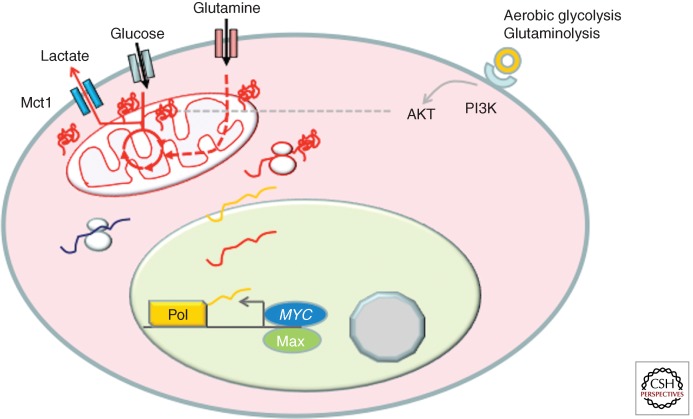
The normal hepatocyte, for example, can undergo gluconeogenesis as part of its normal role to store glucose when nutrient is ample. However, the proliferating hepatocellular carcinoma cell tends to drive glucose down the glycolytic pathway so that glucose could be used to generate building blocks and ATP for the growing cell (Morrish et al. 2009; Hu et al. 2011; Yuneva et al. 2012). In this regard, MYC stimulates virtually all genes involved in glycolysis relative to its induction of genes involved in gluconeogenesis, which is diminished in liver cancer cells (Fig. 2) (Osthus et al. 2000; Hu et al. 2011). For many glycolytic genes, MYC binds phylogenetically conserved MYC consensus E boxes (CACGTG) to activate these genes (Kim et al. 2004). The fact that these sites are conserved suggests that there must be evolutionary pressure to maintain them during evolution.
Figure 2.
MYC is depicted to induce genes involved in glycolysis and glutaminolysis. MYC induction of glucose and glutaminase transporters allow for import of these nutrients, which are depicted to be metabolized and enter the tricarboxylic acid (TCA) cycle (circle in the mitochondrion) to form hybrid intermediates with carbons derived from glucose and glutamine. Glucose is also shown converted to lactate, which is exported by a monocarboxylate transporter Mct1 that is potently activated by MYC. AKT is shown to also increase the glucose-transporting activity.
In addition to the ability of MYC to induce glycolytic genes, MYC has also been implicated in promoting RNA splicing for the expression of pyruvate kinase M2 (PKM2), which has been associated with proliferating cells, versus the alternative form, PKM1 (David et al. 2010). MYC is thought to induce the expression of splicing factors (PTB, hnRNPA1, and hnRNPA2) that favor PKM2 splicing over PKM1. PKM2 is subjected to allosteric regulation that slows the catalytic conversion of phosphoenolpyruvate to pyruvate, allowing for glycolytic intermediates to be channeled into biosynthetic pathways, such as the synthesis of serine and glycine (Chaneton and Gottlieb 2012; Chaneton et al. 2012).
MYC also stimulates genes involved in glutamine metabolism at the transcriptional and posttranscriptional levels (Wise et al. 2008; Gao et al. 2009). Mitochondrial glutaminase protein levels were elevated by MYC induction in the P493-6 B-cell model of human Burkitt’s lymphoma; however, changes in glutaminase (GLS) mRNA levels were very modest relative to changes in protein levels. This observation led to the finding that MYC could repress microRNAs, miR23a, and miR23b, which in turn suppress the synthesis of GLS (Chang et al. 2008; Gao et al. 2009). It appears that MYC could also transcriptionally increase GLS mRNA levels in other systems (Wise et al. 2008; Gao et al. 2009). In addition, MYC was found to induce the expression of other genes involved in glutamine metabolism, such as glutamine transporters (ASCT2 [SLC1A5] and SLC7A5). Glutaminase converts glutamine to glutamate for its oxidation in the TCA cycle and also for protein synthesis or glutathione synthesis. Thus, in addition to its ability to stimulate glucose uptake and glycolysis, MYC also stimulates glutamine consumption and metabolism. In fact, the growing B cell uses a TCA cycle comprised of hybrid intermediates containing carbons from both glucose and glutamine to generate carbon skeletons for macromolecular synthesis (DeBerardinis and Cheng 2010; Le et al. 2012).
Intriguingly, MYC induction of glutamine metabolism appears to be important for cell survival under glucose or oxygen-deprived conditions (Le et al. 2012). Metabolomic tracking studies using 13C-labeled glucose or glutamine in a MYC-inducible P493-6 B-cell lymphoma model document the ability of MYC to drive a glucose-independent cycle using glutamine as the substrate. Further, this study showed a significant contribution of glutamine to glutathione synthesis, particularly under hypoxic conditions. In a parallel metabolomics study, these P493-6 B cells were studied in two states. P493-6 cells are driven by an Epstein–Barr viral EBNA2 protein-dependent nontumorigenic program with endogenous levels of (low) MYC or driven by a tumorigenic program with ectopic (high) MYC alone (Murphy et al. 2013). In both states, these proliferating B cells used a TCA cycle with hybrid intermediates containing both glucose and glutamine carbons. The major difference between these two states, however, is that high MYC increased glutaminolysis by fourfold, whereas glycolysis only increased by 1.2-fold, suggesting that high levels of MYC drive glutaminolysis in the tumorigenic state. In aerobic conditions, a significant amount of glutamine is also converted to proline through MYC’s induction of genes involved in proline biosynthesis (Liu et al. 2012b). Inhibition of glutaminase has been documented to inhibit progression of P493 lymphoma in vivo, indicating that glutamine metabolism is required for full tumorigenic potential (Le et al. 2012).
Studies of a transgenic MYC-inducible mouse liver cancer model indicate that imported glutamine contributes to the production of alanine from pyruvate through transaminases in the premalignant livers (Hu et al. 2011). In frank liver tumor cells, glutaminase levels and glutamine metabolism are increased by MYC. Glycolysis is also heightened with an increased rate of pyruvate to lactate conversion in these liver tumors. Similarly, the MYC-inducible lung cancer model displays both glycolysis and glutaminolysis induced by MYC; however, in contrast to the liver cancer model, glutamine synthetase is also expressed in the lung cancer model (Yuneva et al. 2012). In this regard, these lung cancer cells can produce glutamine from glutamate in addition to their ability to import glutamine.
MYC REGULATION OF NUCLEOTIDE BIOSYNTHESIS AND LIPID SYNTHESIS
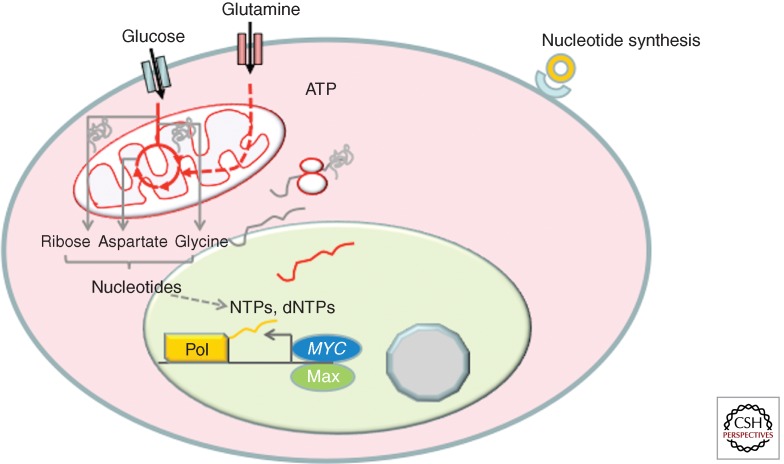
The ability of MYC to induce glycolysis and glutaminolysis supports the cell’s need for ATP and building blocks. Both glucose and glutamine are required for nucleotide biosynthesis (Fig. 3). MYC has been shown to activate many, but not all, genes involved in nucleotide metabolism (Liu et al. 2008; Mannava et al. 2008).
Figure 3.
MYC is shown to induce genes involved in nucleotide synthesis. Both glucose and glutamine provide the carbon skeleton and nitrogen for the de novo synthesis of pyrimidines and purines. Pools of nucleotides, NTPs and dNTPs, support nucleic acid synthesis for transcription and DNA replication.
Recently, it has become more apparent that MYC can induce glycolytic flux from 3-phosphoglycerate (3-PG) for the synthesis of serine and glycine, which are essential for nucleotide biosynthesis (Vazquez et al. 2011, 2013). For example, phosphoglycerate dehydrogenase (PHGDH), which converts 3-PG to 3-phosphopyruvate (3-PP), and PSAT, which converts 3-PP to phosphoserine, are both induced by MYC. SHMT2, which converts serine to glycine coupled with transfer of a carbon for folate synthesis, is one of the earlier established MYC targets, and was shown to partially rescue the growth of MYC null rat fibroblasts (Nikiforov et al. 2002). These studies, taken together, indicate that MYC coordination of glutamine metabolism with nucleotide synthesis is essential for cell growth and proliferation.
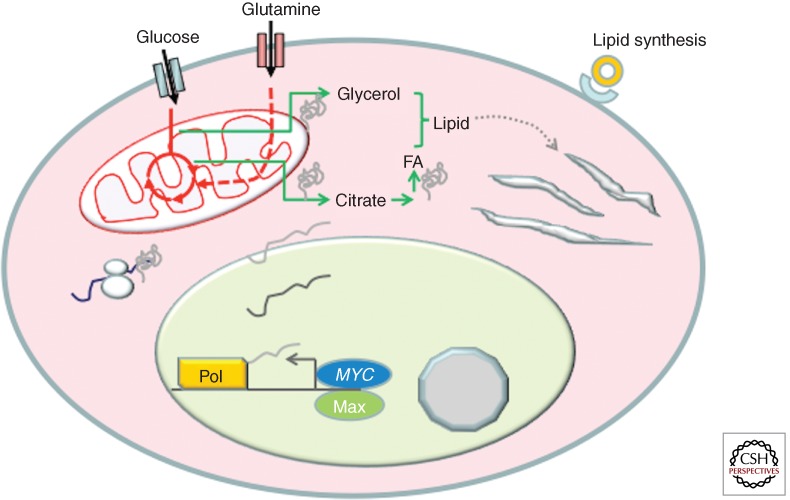
Glucose and glutamine are also essential for fatty acid synthesis (Fig. 4). Citrate generated from glucose and/or glutamine through the TCA cycle is exported into the cytosol and converted by ACLY to acetyl-CoA. Although ACLY is not highly induced by MYC, ACACA (acetyl-CoA carboxylase; ACACA converts acetyl-CoA to malonyl-CoA), FASN (fatty acid synthetase; FASN adds two carbons from acetyl-CoA to the growing fatty acid chain to form longer chains), and SCD (stearoyl-CoA desaturase; SCD introduces a double bond into longer chain fatty acids such as palmitate to form the unsaturated fatty acid oleate) are highly responsive to MYC as documented in the MYC target gene database and more recent studies (Zeller et al. 2003; Loven et al. 2012). Functionally, MYC induces fatty acid synthesis partially through the use of glucose carbons (Morrish et al. 2010). Genes encoding mevalonate and cholesterol synthetic enzymes are also induced but the magnitude is modest. Whether SREBP1 or SREBP2 is involved in the MYC-mediated increase in these mRNAs remains to be established. It is notable, however, that both FASN and SCD have conserved E boxes to which MYC may bind, suggesting that MYC could be sufficient to induce genes involved in fatty acid synthesis without requiring SREBP. Whether MYC elevates genes involved in cholesterol synthesis indirectly through SREBP is not currently known. From these observations and metabolomics studies documenting the incorporation of glucose carbons into fatty acids in response to MYC, it could be surmised that MYC induces lipid synthesis through specific sets of target genes.
Figure 4.
MYC is shown to stimulate genes involved in lipid synthesis. Citrate generated from glucose or glutamine is converted to acetyl-CoA for fatty acid synthesis. Glycerol coming from glucose provides the glyceride backbone for lipid synthesis. Not shown is the pathway for cholesterol synthesis, which is likely to be dependent on SREBP.
MYC REGULATION OF MITOCHONDRIAL BIOGENESIS
As cells grow, MYC induces processes that enable synthesis of new cellular components such as ribosomes, nucleotides, and lipids. These components are used to produce numerous cellular organelles including mitochondria, which are essential for high levels of ATP production in support of a growing cell. In addition to respiration, mitochondria mediate many biosynthetic pathways (Wallace 2012). The TCA cycle is housed in the mitochondrial matrix, and its production of intermediates such as aspartate is essential for de novo nucleotide biosynthesis. Its production of citrate is involved in fatty acid synthesis. Succinyl-CoA from the TCA cycle is an essential substrate for the synthesis of heme, a prosthetic element of many enzymes and respiratory proteins. Dihydroorotate dehydrogenase is a respiratory chain-coupled mitochondrial enzyme that is involved in pyrimidine biosynthesis. These examples illustrate the importance of the mitochondrion to support the biosynthetic functions in cancer cells in addition to its canonical function as an ATP generator.
MYC was shown to induce mitochondrial biogenesis apparently through the direct activation of many genes involved in mitochondrial biogenesis, specifically PGC-1β (Morrish et al. 2003; Li et al. 2005; Zhang et al. 2007; Kim et al. 2008; see Hockenbery 2013). Induction of MYC expression is accompanied by increased mitochondrial mass, which is diminished by siRNA-mediated knockdown of PGC-1β expression. Analyses of both loss and gain of MYC function have provided support for the idea that MYC functions in mitochondrial biogenesis in a number of cell systems. Intriguingly, iron is essential for mitochondrial biogenesis particularly because components of mitochondria contain iron, such as sulfur–iron cluster and heme-containing proteins. In this regard, the transferrin receptor TFRC, which imports iron, is a critical MYC target gene (O’Donnell et al. 2006). A recent study confirms the role of MYC in mitochondrial biogenesis and also implicates MYC in regulation of mitochondrial fusion and fission (Graves et al. 2012). These functional studies have been corroborated by independent genomic studies that link MYC to the induction of mitochondrial genes (Morrish et al. 2003; Li et al. 2005; Kim et al. 2008).
Overexpression of MYC also increases the expression of its target C1QBP or p32, which is involved in mitochondrial protein synthesis (Fogal et al. 2010). Knockdown of p32 diminishes mitochondrial function and respiration, and results in decreased cellular proliferation and remarkably diminished tumorigenesis. This study highlights the importance of mitochondrial function in cancer cells, which had been inaccurately characterized as being driven by the Warburg effect independent of mitochondrial function (Koppenol et al. 2011). ARK5, an AMPK family member, was identified in a synthetic lethality screen for targets in MYC-overexpressing cells (Liu et al. 2012a). Knockdown of ARK5 is accompanied by decreased mitochondrial function, implying that ARK5 is involved in maintenance of mitochondrial integrity and bioenergetic homeostasis. These studies collectively indicate that MYC has many connections to mitochondrial biogenesis and function (Fig. 5).
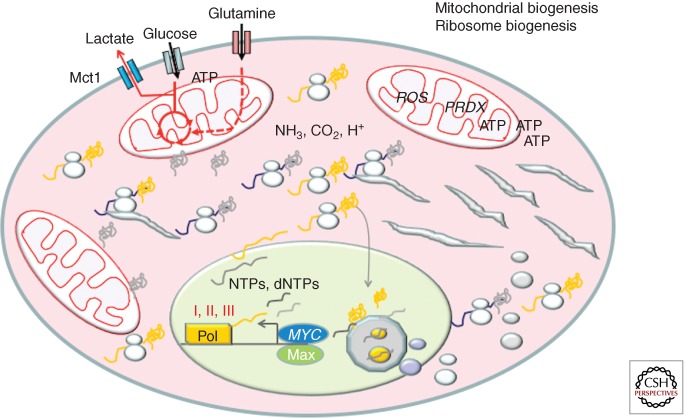
Figure 5.
MYC activates genes involved in mitochondrial and ribosomal biogenesis. The figure depicts the synthesis of rRNAs and ribosomal proteins, which are imported into the nucleolus for ribosome assembly. Assembled ribosomes are exported to support the synthesis of proteins for the growing cell. Mitochondria are duplicated along with mtDNA replication.
In vivo studies have also connected MYC with increased mitochondrial biogenesis when MYC was overexpressed in adult murine myocardium. MYC overexpression increased mitochondrial content, respiration, and glycolytic flux. Conversely, conditional deletion of MYC in stressed mouse myocardium resulted in decreased glycolysis and reduced mitochondrial biogenesis (Ahuja et al. 2010). Hence, both in vitro and in vivo studies link MYC to the genesis of mitochondria, which are essential organelles for the growing cell.
MYC, METABOLISM, AND THE ENTRY OF CELLS INTO S PHASE
As MYC and other factors push growing cells to a critical mass at the restriction point, cells are catapulted into S phase in which DNA replication begins with the nucleotide pools that have accumulated and continued to be synthesized. MYC’s role at the G1–S-phase junction involves its direct activation of E2F and CDK4, which inactivates Rb to enable the transcription factors E2F to activate genes involved in DNA replication (Rempel et al. 2009). Intriguingly, MYC together with E2F binds to numerous genes involved in DNA replication, suggesting coordination between these two transcription factors (Zeller et al. 2006). MYC also attenuates E2F function through activation of the miR-17 cluster of six microRNAs (O’Donnell et al. 2005). One of these microRNAs, miR-92, targets E2F in a feed-forward negative regulatory loop (Aguda et al. 2008). This loop ensures that MYC induction of E2F1 expression is attenuated as cells initiate DNA replication (Pickering et al. 2009). Knockdown of miR-92 levels in serum-stimulated fibroblasts is associated with increased double-strand DNA breaks, suggesting that unattenuated E2F function could diminish the fidelity of DNA synthesis resulting in defects and cell-cycle arrest. The mechanism for this DNA replication stress is unclear, but it could be related to an imbalance in the expression of genes that regulate nucleotide pools versus those that drive the DNA replication machinery when E2F1 is not attenuated by miR-92.
Overexpression of MYC can also induce endoreplication in normal cells and cancer cells treated with microtubule inhibitors or irradiation (Li and Dang 1999; Sheen and Dickson 2002; Pierce et al. 2004; Zanet et al. 2005). In these cases, there appears to be an uncoupling of MYC-induced DNA replication and cell division. This activity of MYC appears to have a physiological role, in that MYC is required for endoreplication, which normally occurs in keratinocytes (Zanet et al. 2005). It could be surmised that MYC induction of endoreplication needs to be coordinated with the availability of nutrients for nucleotide synthesis.
MYC-DEREGULATED METABOLISM IN CANCERS AND THERAPEUTIC OPPORTUNITIES
Could MYC regulation of metabolism be exploited for therapeutic purposes? We have hypothesized that the drive of cell growth by deregulated MYC, unrestrained by negative-feedback loops, renders MYC-overexpressing cells dependent on a continual supply of nutrients and thereby causing them to be nutrient addicted (Dang 2011). Indeed many cell types that overexpress MYC were sensitive to glucose-deprivation-induced cell death. Knockdown of lactate dehydrogenase A (LDHA), which converts pyruvate to lactate, significantly curbed the clonogenicity of lymphoma cells and inhibited tumorigenicity of mammary cancer, lung cancer, and neuroblastomas with amplified N-MYC in mice (Shim et al. 1997; Fantin et al. 2006; Xie et al. 2009; Le et al. 2010; Qing et al. 2010; Wang et al. 2012). These observations suggest that deregulated MYC causes cancer cells to be more vulnerable to glucose deprivation, which would cause the withdrawal of normal cells from the cell cycle into a G1-like state.
Surprisingly, diminished LDHA function owing to specific mutations of LDHA did not decrease the ability of the λ-MYC transgene to induce murine lymphomas (Nilsson et al. 2012). The observation that MYC could also induce LDHB suggests that LDHB could substitute for LDHA, indicating that redundancies may pose challenges in targeting metabolism for therapeutic purposes (Menssen and Hermeking 2002). It is also notable that p32, which is involved in mitochondrial biogenesis and respiration, is required for tumorigenesis, suggesting that mitochondrial function may play key roles for the genesis and maintenance of specific types of cancers.
MYC-overexpressing cells are similarly addicted to glutamine, such that withdrawal of glutamine caused apoptosis of human cells with deregulated expression of MYC (Yuneva et al. 2007). This was confirmed in subsequent studies, which also implicated key enzymes in glutamine metabolism (Wise et al. 2008; Gao et al. 2009). Knockdown of GLS expression remarkably decreased the growth of lymphoma and prostate cancer cells (Gao et al. 2009). Further, a recent study of lung cancer cell lines documents a correlation between the expression of the shorter alternatively spliced form of GLS, termed GAC, with glutamine dependence and sensitivity to a glutaminase inhibitor, BPTES (van den Heuvel et al. 2012). Two studies showed that inhibitors of glutaminase, compound 968 and BPTES, also inhibited growth of MYC-induced tumor xenografts (Wang et al. 2010; Le et al. 2012). Another study of N-MYC-amplified neuroblastoma indicates that these tumors are dependent on glutamine and could be inhibited with a nonspecific transaminase inhibitor, aminooxyacetate (Qing et al. 2012). In aggregate, these studies suggest the feasibility of using glutaminase inhibitors for therapeutic purposes.
CONCLUDING REMARKS
There is overwhelming experimental evidence, from both gain- and loss-of-function studies, implicating MYC in the regulation of cell growth. Hence, activation of MYC must orchestrate the expression of genes that promote cell growth and suppress those that inhibit growth. It stands to reason then, that genes that generate more bioenergetic substrates for this cell growth would be induced by MYC. Both in vitro and in vivo models have provided substantial evidence that MYC induces many genes involved in ribosome biogenesis in growing cells. In concert with these, genes involved in glucose and glutamine metabolism are also induced to support ATP production and macromolecular synthesis for the replication of organelles. In contrast to normal cells, in which MYC is under the tight control of negative regulatory pathways and feedback loops, cells with deregulated expression of MYC escape this control, resulting in an uncontrolled gene expression program. Uncontrolled expression of genes involved in building up cell mass through ribosome biogenesis, for example, would force a demand for a continual supply of nutrients to be available for cells with MYC overexpression. This demand renders MYC-transformed cells addicted to nutrients, and hence they could be killed through nutrient deprivation or interference with key enzymes involved in key metabolic pathways such as glycolysis or glutaminolysis. In contrast, nutrient deprivation of normal cells would trigger negative regulatory pathways that diminish MYC expression, causing cells to withdraw in the G1 phase of the cell cycle. We surmise that this difference could be exploited for therapeutic targeting of cancer cells with metabolic inhibitors.
ACKNOWLEDGMENTS
Our work is supported by National Cancer Institute, National Institutes of Health Leukemia and Lymphoma Society, and AACR-Stand-up-to-Cancer grants and the Abramson Family Cancer Research Institute.
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB 2008. MicroRNA regulation of a cancer network: Consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci 105: 19678–19683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja P, Zhao P, Angelis E, Ruan H, Korge P, Olson A, Wang Y, Jin ES, Jeffrey FM, Portman M, et al. 2010. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J Clin Invest 120: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew DS, Ashmun RA, Simmons BC, Cleveland JL 1991. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 6: 1915–1922 [PubMed] [Google Scholar]
- Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, Arvanitis C, Attardi LD, Feng S, Ruebner B, et al. 2004. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol 2: e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR, Sabatini DM 2012. Cancer cell metabolism: One hallmark, many faces. Cancer Discov 2: 881–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B, Gottlieb E 2012. Rocking cell metabolism: Revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci 37: 309–316 [DOI] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. 2012. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Connacci-Sorrell M. Cold Spring Harb Perspect Med. 2013 doi: 10.1101/cshperspect.a014357. (to be published) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2011. Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb Symp Quant Biol 76: 369–374 [DOI] [PubMed] [Google Scholar]
- Dang CV 2012a. Links between metabolism and cancer. Genes Dev 26: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2012b. MYC on the path to cancer. Cell 149: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL 2010. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463: 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14: 45–55 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T 2010. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y 2010. Ribosome biogenesis surveillance: Probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 29: 4253–4260 [DOI] [PubMed] [Google Scholar]
- Drygin D, Rice WG, Grummt I 2010. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol 50: 131–156 [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J Biol Chem 276: 48562–48571 [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc’s broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 13: 2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69: 119–128 [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P 2006. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9: 425–434 [DOI] [PubMed] [Google Scholar]
- Fogal V, Richardson AD, Karmali PP, Scheffler IE, Smith JW, Ruoslahti E 2010. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol 30: 1303–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, Elster J, Beer-Stolz D, Van Houten B, Vockley J, et al. 2012. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS ONE 7: e37699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7: 295–302 [DOI] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O’Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. 2000. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci 97: 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hockenbery D 2013. Myc and mitochondrial biogenesis and function. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PE, Nelson SJ, Kurhanewicz J, Vigneron DB, Goga A 2011. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab 14: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA 2000. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci 97: 11198–11202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV 2004. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol 24: 5923–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185 [DOI] [PubMed] [Google Scholar]
- Kim J, Lee JH, Iyer VR 2008. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS ONE 3: e1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV 2011. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer 11: 325–337 [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM 2012. mTOR signaling in growth control and disease. Cell 149: 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV 2010. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci 107: 2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. 2012. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab 15: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BK, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, et al. 2012. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res 22: 9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Dang CV 1999. c-Myc overexpression uncouples DNA replication from mitosis. Mol Cell Biol 19: 5339–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 25: 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SI, Broach JR 2009. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc Natl Acad Sci 106: 19928–19933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N, Sedivy JM, Zeller KI, Dang CV 2008. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE 3: e2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ulbrich J, Muller J, Wustefeld T, Aeberhard L, Kress TR, Muthalagu N, Rycak L, Rudalska R, Moll R, et al. 2012a. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature 483: 608–612 [DOI] [PubMed] [Google Scholar]
- Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM 2012b. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci 109: 8983–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA 2012. Revisiting global gene expression analysis. Cell 151: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA 2008. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle 7: 2392–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hermeking H 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc Natl Acad Sci 99: 6274–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Giedt C, Hockenbery D 2003. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev 17: 240–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM 2009. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene 28: 2485–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Noonan J, Perez-Olsen C, Gafken PR, Fitzgibbon M, Kelleher J, VanGilst M, Hockenbery D 2010. Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J Biol Chem 285: 36267–36274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ 2012. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481: 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TA, Dang CV, Young JD 2013. Isotopically nonstationary 13C flux analysis of Myc-induced metabolic reprogramming in B-cells. Metab Eng 15: 206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD 2002. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol 22: 5793–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson LM, Forshell TZ, Rimpi S, Kreutzer C, Pretsch W, Bornkamm GW, Nilsson JA 2012. Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis. PLoS Genet 8: e1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Yu D, Zeller KI, Kim JW, Racke F, Thomas-Tikhonenko A, Dang CV 2006. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol Cell Biol 26: 2373–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, Grewal SS, Knoepfler PS, Edgar BA, Parkhurst SM, Eisenman RN 2005. Genomic binding and transcriptional regulation by the Drosophila Myc and Mnt transcription factors. Cold Spring Harb Symp Quant Biol 70: 299–307 [DOI] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV 2000. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 275: 21797–21800 [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC 2006. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3: 187–197 [DOI] [PubMed] [Google Scholar]
- Pickering MT, Stadler BM, Kowalik TF 2009. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene 28: 140–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Yost C, Britton JS, Loo LW, Flynn EM, Edgar BA, Eisenman RN 2004. dMyc is required for larval growth and endoreplication in Drosophila. Development 131: 2317–2327 [DOI] [PubMed] [Google Scholar]
- Qing G, Skuli N, Mayes PA, Pawel B, Martinez D, Maris JM, Simon MC 2010. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1α. Cancer Res 70: 10351–10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G, Li B, Vu A, Skuli N, Walton ZE, Liu X, Mayes PA, Wise DR, Thompson CB, Maris JM, et al. 2012. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 22: 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rahl PB, Young RA 2013. Myc and transcription elongation. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel RE, Mori S, Gasparetto M, Glozak MA, Andrechek ER, Adler SB, Laakso NM, Lagoo AS, Storms R, Smith C, et al. 2009. A role for E2F activities in determining the fate of Myc-induced lymphomagenesis. PLoS Genet 5: e1000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F 1999. Control of cell growth by c-Myc in the absence of cell division. Curr Biol 9: 1255–1258 [DOI] [PubMed] [Google Scholar]
- Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J 2001. TGF-β influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nature Cell Biol 3: 400–408 [DOI] [PubMed] [Google Scholar]
- Sheen JH, Dickson RB 2002. Overexpression of c-Myc alters G1/S arrest following ionizing radiation. Mol Cell Biol 22: 1819–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV 1997. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci 94: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Chun YS, Lewis BC, Dang CV 1998. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci 95: 1511–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel F, et al. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nature Cell Biol 3: 392–399 [DOI] [PubMed] [Google Scholar]
- van den Heuvel AP, Jing J, Wooster RF, Bachman KE 2012. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Ther 13: 1185–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG 2011. Targeting cancer metabolism: A therapeutic window opens. Nat Rev Drug Discov 10: 671–684 [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- Vazquez A, Markert EK, Oltvai ZN 2011. Serine biosynthesis with one carbon catabolism and the glycine cleavage system represents a novel pathway for ATP generation. PLoS ONE 6: e25881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, Tedeschi PM, Bertino JR 2013. Overexpression of the mitochondrial folate and glycine-serine pathway: A new determinant of methotrexate selectivity in tumors. Cancer Res 73: 478–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M, Poortinga G, Stanley KL, Lindemann RK, Bots M, Chan CJ, Bywater MJ, Kinross KM, Astle MV, Waldeck K, et al. 2013. The mTORC1 inhibitor everolimus prevents and treats Emu-Myc lymphoma by restoring oncogene-induced senescence. Cancer Discov 3: 82–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC 2012. Mitochondria and cancer. Nat Rev Cancer 12: 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, et al. 2010. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18: 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Loo TY, Shen JG, Wang N, Wang DM, Yang DP, Mo SL, Guan XY, Chen JP 2012. LDH-A silencing suppresses breast cancer tumorigenicity through induction of oxidative stress mediated mitochondrial pathway apoptosis. Breast Cancer Res Treat 131: 791–800 [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Valera VA, Merino MJ, Amato AM, Signoretti S, Linehan WM, Sukhatme VP, Seth P 2009. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol Cancer Ther 8: 626–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK 2008. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem 283: 20621–20627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y 2007. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 178: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. 2012. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 15: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N, Swinnen JV, Smans K 2012. ATP-citrate lyase: A key player in cancer metabolism. Cancer Res 72: 3709–3714 [DOI] [PubMed] [Google Scholar]
- Zanet J, Pibre S, Jacquet C, Ramirez A, de Alboran IM, Gandarillas A 2005. Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J Cell Sci 118: 1693–1704 [DOI] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol 4: pR69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci 103: 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL 2007. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11: 407–420 [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 12: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]