Abstract
Caspases are proteases at the heart of networks that govern apoptosis and inflammation. The past decade has seen huge leaps in understanding the biology and chemistry of the caspases, largely through the development of synthetic substrates and inhibitors. Such agents are used to define the role of caspases in transmitting life and death signals, in imaging caspases in situ and in vivo, and in deconvoluting the networks that govern cell behavior. Additionally, focused proteomics methods have begun to reveal the natural substrates of caspases in the thousands. Together, these chemical and proteomics technologies are setting the scene for designing and implementing control of caspase activity as appropriate targets for disease therapy.
Synthetic substrates and inhibitors are used to define fundamental properties of caspases and their roles in promoting apoptosis and inflammation. They may also be used to design therapies that control caspase activity.
WHAT IT TAKES TO BE A CASPASE
The need to recycle scarce amino acids in early biotic environments required that proteases were among the first proteins to appear in the most primitive replicating organisms. Since that distant origin, proteases have diversified into every biological niche, occupied every cellular and extracellular compartment, and are encoded in all organisms on the planet with the exception of the most primitive viruses (Lopez-Otin and Overall 2002; Rawlings et al. 2008). Diverging from their primeval role in protein degradation and recycling, most of the 600 or so mammalian proteases are involved in very limited and specific proteolytic events—initiating and transmitting signals—and a good example of this is the caspase family (Timmer and Salvesen 2007; Crawford and Wells 2011). The caspases belong to a clan, or structurally related group, known as cysteine protease clan CD that also contains the plant metacaspases (Vercammen et al. 2004), and mammalian and plant proteases of the legumain family (involved in specific processing events), the eukaryotic protease separase (required for sister chromatid separation during mitosis), and the bacterial proteases clostripain and gingipain (Chen et al. 1998). Common to all of these proteases is a shared stringent specificity for one type of amino acid side chain at the primary (P1) specificity position in substrates (Arg or Lys for metacaspases, clostripain, and gingipains; Asn for legumains; and Arg for separase). Thus, the caspase fold—the clan CD fold—supports unusually stringent specificity at the primary cleavage site. Distinguishing the caspases from each other are different requirements at the extended substrate recognition positions.
The name “caspase” derives from cysteine-dependent aspartate-specific protease (Alnemri et al. 1996): Catalysis is governed by a critical conserved Cys side chain of the enzyme and by a highly stringent selectivity for cleaving peptides and proteins on the carboxy-terminal side of Asp residues. The use of a Cys side chain as a nucleophile during peptide-bond hydrolysis is common to several protease families. But the primary specificity for Asp is very rare among proteases. Of the currently known human and mouse proteases, only the caspase activator Granzyme B, a serine protease, shares this primary specificity (Odake et al. 1991; Harris et al. 2000). The primary specificity pockets of caspases are almost identical, being formed by the side chains of the strictly conserved residues Arg-179, Arg-341, and Gln-283 (caspase-1 numbering convention). This deep, highly basic pocket (Fig. 1) is perfectly shaped to accommodate an Asp side chain, accounting for the up to four orders of magnitude lower catalytic efficiency for cleavage of peptides with a P1 Glu residue in most caspases (Stennicke et al. 2000), with the notable exception of the Drosophila caspase Dronc, which seems to have some specificity for Glu in addition to Asp (Hawkins et al. 2000; Snipas et al. 2008).
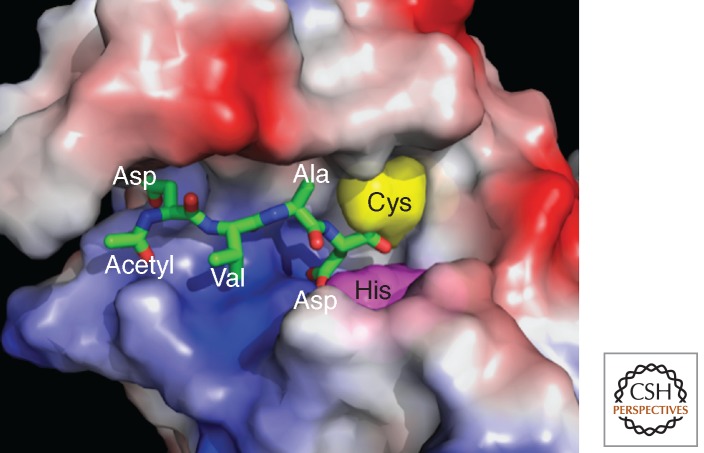
Figure 1.
A caspase catalytic cleft. The active-site cleft of caspase-3 bound to the tetrapeptide inhibitor acetyl-Asp-Val-Ala-Asp-fluoromethylketone (Ac-DVAD-FMK), taken from PDB code 1CP3 (Mittl et al. 1997). The caspase is shown as an electrostatic potential surface, where blue hues denote positive potential and red hues negative potential. The deep basic cleft next to the catalytic Cys (yellow) and His (purple) residues of the enzyme occupies the P1 Asp residue and is conserved in all active caspase structures. To the left of this are the P2–P4 residues, some occupying clearly visible specific pockets. The location of residues to the right (the P′ side) are unavailable in this or similar caspase structures because all common peptidyl inhibitors of caspases are designed without P′ substituents.
CASPASE SUBSTRATES
Synthetic Substrates
A very large number of reports of the design, construction, and application of synthetic substrates have led to some pretty clear insights into the catalytic mechanisms and specificity preferences of individual caspases. There are principally two ways to investigate caspase substrates: (1) the inherent preferences in the active-site cleft that distinguish them from each other, and (2) their natural biological substrates. The first tells us a lot about caspase catalysis and is much easier to investigate than the second. This, in turn, has led to a consensus of the “inherent subsite preference” defining the preference of caspases for individual peptide sequences, and to the development of peptide-based and genetically encoded probes that have been used to follow caspase activity in cells and even whole organisms. Importantly, understanding the inherent subsite preference of caspases has not really achieved the expected predictive power of defining optimal in vivo protein substrates, as described below.
Peptide-Based Substrates
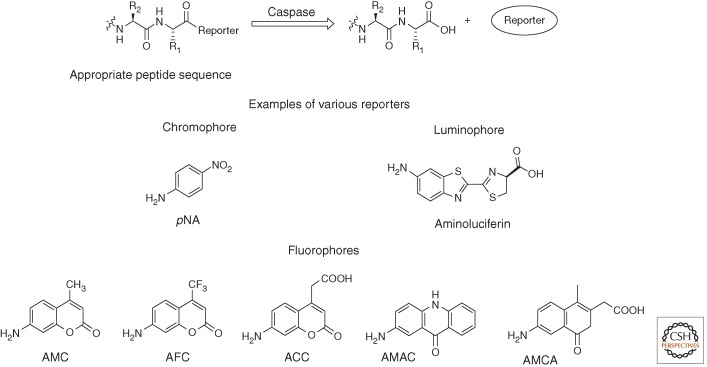
Several strategies to identify inherent subsite preference have been applied to caspases, generally incorporating peptides attached to reporter groups (Fig. 2). This allows for definition of preferred sequences on the “unprimed” side—the residues amino-terminal—of the scissile bond. Positional scanning substrate combinatorial library (PS-SCL) methods, first introduced for caspases in 1997 (Rano et al. 1997), are based on synthesis of libraries of substrate mixtures, which are linked with highly fluorescent molecules. These fluorophores, which are called reporter groups, occupy the P1′ position, and after enzyme cleavage, they are released and emit fluorescence when excited by an appropriate wavelength (Fig. 2).
Figure 2.
Caspase cleavage of reporter group from substrate. Examples of reporter groups of the three main categories (fluorophores, chromophores, and luminophores).
Measurements of liberated fluorescence yield data on reaction kinetics, allow for identification of optimal inherent subsite preference, and provide a powerful method for rapid and reliable analysis of protease specificity. However, it is worth noting that PS-SCL approaches are only helpful for identification of nonprime residues of a substrate (Poreba and Drag 2010). Interestingly, such studies can yield somewhat unexpected results, as exemplified by scanning of caspase-1. Thus, the WEHD sequence is the most favorable tetrapeptide recognition motif for caspase-1 (Rano et al. 1997), inconsistent with the original optimal sequence YVAD for the enzyme (Thornberry and Molineaux 1995) and divergent from the sequence YVHD found to be cleaved in caspase-1’s natural substrate pro-IL-1β. Nevertheless, further investigation with several individual substrates confirmed that the WEHD tetrapeptide was better than the YVAD tetrapeptide because the kcat/KM value was ∼50-fold higher (Table 1).
Table 1.
kcat/KM and Ki values for selected synthetic substrates cleaved by caspase-1
| Substrate | kcat/KM × 105 (m−1 sec−1) |
|---|---|
| Ac-WEHD-AMC | 33.4 |
| Ac-YVHD-AMC | 2.81 |
| Ac-YVAD-AMC | 0.66 |
| Pro-IL-1β | 1.5 |
From Rano et al. (1997).
The pioneering work of Thornberry et al. (1997) using a tetrapeptide PS-SCL approach and Talanian et al. (1997) using sets of individual peptide substrates, subsequently refined by others, led to our current understanding of the inherent subsite preferences summarized in Figure 3. Importantly, caspase-2 seems to be an outlier because its activity on tetrapeptides was very low, and cleaved substrates efficiently only when an S5 residue was provided, with hydrophobic residues favored in the this position (Talanian et al. 1997). A newer strategy that substantially simplified the synthesis of fluorogenic libraries, using the bifunctional fluorogenic group 7-amino-4-carbamoylmethylcoumarin (ACC), was subsequently introduced (Harris et al. 2000), allowing solid-phase synthesis of PS-SCLs (Maly et al. 2002). The advantage of this technique is that any amino acid may be placed at any position, and the approach was applied to examine caspase-3 substrate specificity (Walters et al. 2009). The results were consistent with those obtained from AMC-based substrate libraries (Thornberry et al. 1997), confirming that the type of fluorophore used does influence substrate specificity. Microarray techniques using peptide nucleic acid–encoded libraries of rhodamine-based fluorogenic substrates enable identification of individual subsite preferences via hybridization to complementary DNA oligonucleotides. Using this technique for defining optimal caspase-3 substrates, and a library of 192 peptide sequences, revealed agreement with the known enzyme specificity (Winssinger et al. 2004), validating the approach and confirming that inherent subsite occupancy of caspases is independent of the technique used.
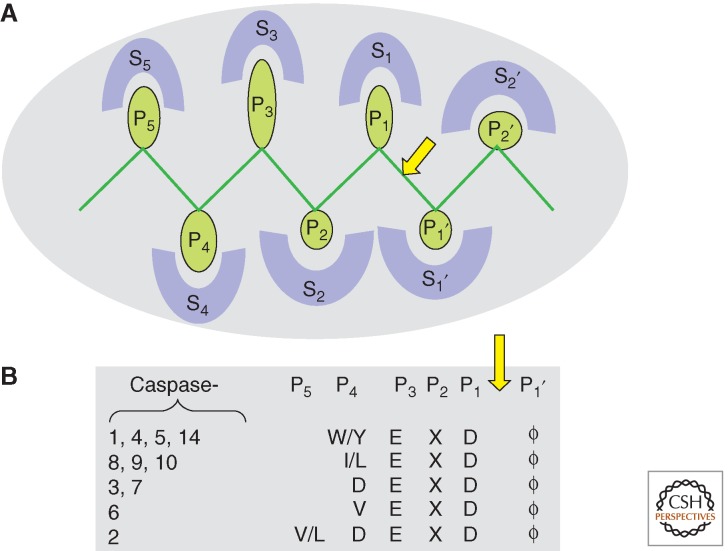
Figure 3.
Inherent substrate specificity. The convention in naming substrate residues in protease active sites is to name the residue to the amino terminal of the scissile bond (arrow) P1 and the one to the carboxy-terminal side P1′. (A) Other residues (green) are numbered consecutively from these origins. Many of these residues will occupy specific pockets on the enzyme, numbered with the matching S designation (blue), each of which may be constructed of several enzyme residues. The objective of this interaction mode, which almost always binds the cleavage region in an extended peptide conformation, is to align the substrate accurately into register with the catalytic machinery. (B) Caspases bind substrates through typical interactions of the active-site cleft with amino acids in the cleavage site. Studies with synthetic peptides and peptide libraries have shown that substrate residues P4–P1′ contribute to binding through association with S4 through S1′ pockets in the active site. All caspases show exquisite selectivity for Asp in the P1 position, a preference for Glu in P3, and intolerance for charged residues at P1′, where Gly, Ala, Thr, Ser, and Asn are preferred (denoted by the ϕ symbol). Distinguishing between them is primarily the nature of the P4 residue, and the inherent substrate specificities are generally well understood at the atomic level. Importantly, caspase-2 has an additional interaction at S5 that enhances catalysis at least 30-fold.
The most striking differences among caspases-1, -3, and -8 can be observed in the S4 pocket. Caspase-1 prefers bulky, hydrophobic residues (tyrosine and tryptophan) (Rano et al. 1997; Thornberry et al. 1997), caspase-3 has a near absolute requirement for aspartic acid (Ganesan et al. 2006a,b), and caspase-8 can liberally accommodate residues, but branched leucine and valine are most preferable (Fig. 3) (Blanchard et al. 1999; Wei et al. 2000).
All methods described above that use fluorogenic peptide substrates are based on measurements of increasing fluorescence after caspase cleavage. However, this is not the only way. 2-Aminoacridone (AMAC) has been used as a reporting group for a caspase substrate (Ac-DEVD-AMAC) and by measuring the decrease of fluorescence after enzyme cleavage (Lozanov et al. 2009). In this approach, because the products of hydrolysis are not fluorescent, background correction is not necessary; thus, there are some advantages in this strategy over the commonly used ones (Table 2). Luciferase-based substrates incorporate the same specificity design, but use aminoluciferin as the leaving group. Like all luciferase assays, the luciferin is oxidized by luciferase and light is emitted. This method has lower background levels and higher sensitivity than fluorescent assays and is suitable for examining cell systems (O’Brien et al. 2005), cell lysates (Kindermann et al. 2010), and in vivo assays (Hickson et al. 2010).
Table 2.
Comparison of kinetic parameters of substrate DEVD for caspase-3 linked with different reporter groups
| Substrate | kcat (sec−1) | KM (µm) | kcat/KM × 106 (m−1 sec−1) | References |
|---|---|---|---|---|
| Ac-DEVD-AMAC | 9.95 | 4.68 | 2.13 | Lozanov et al. 2009 |
| Ac-DEVD-AMCA | 5.86 | 13.65 | 0.42 | Lien et al. 2004 |
| Ac-DEVD-AMC | 9.1 | 10 | 1.4 | Garcia-Calvo et al. 1999 |
| Ac-DEVD-AFC | — | 16.8 | 1.3 | Deveraux et al. 1997; Sun et al. 1997 |
| Cbz-DEVD-aminoluciferin | — | 7.66 | — | O’Brien et al. 2005 |
| Ac-DEVD-pNA | 2.4 | 11 | 0.22 | Talanian et al. 1997 |
| Cbz-DEVD-SiR600 | 3.8 | 1.2 | 3.1 | Kushida et al. 2012 |
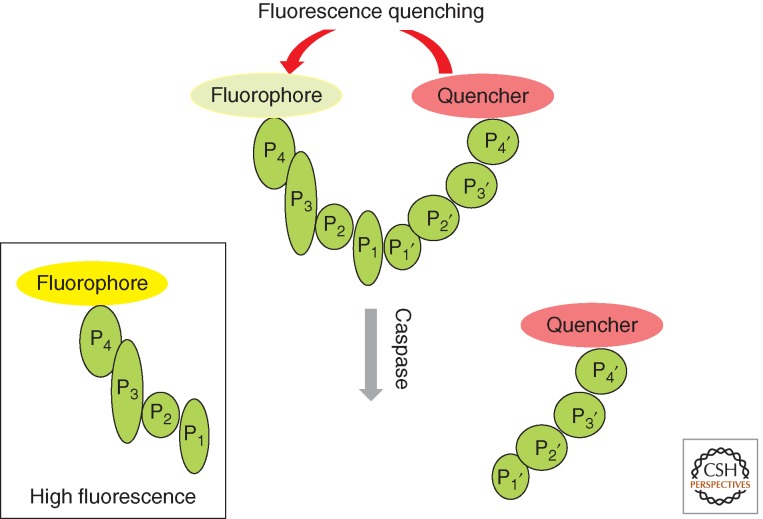
Importantly, the methods to determine substrate specificity described above can only be used to identify nonprime substrate specificity; however, they fail to determine the prime site of a substrate. To establish prime and nonprime substrate specificity, other approaches are required. Internally quenched fluorescence peptide substrates overcome this problem (Stennicke et al. 2000). In this technique, a donor (fluorogenic group) and an acceptor (quenching group) are located on the opposite sides of the substrate. Intact substrates do not show a fluorescence signal (or a very weak one), and after protease cleavage, the quenching group is separated from the fluorophore and increased fluorescence emission is observed (Fig. 4). Thus, it was revealed that in the position P1′, small residues, like Gly, Ser, and Ala, are favored. Interestingly, substrates with large aromatic residues such as Phe and Tyr were cleaved efficiently as well. However, peptides with charged residues, branched aliphatic residues, and Pro were poorly tolerated (Stennicke et al. 2000; Petrassi et al. 2005).
Figure 4.
Schematic representation of fluorescence-quenched substrates. Peptide selectivity elements (green) are bracketed by a fluorophore and a cognate quencher. Cleavage between the fluorophore and quencher results in enhanced fluorescence.
Nonchemical synthesis-based techniques have been applied to defining caspase specificity, revealing essentially the same answer as the peptide-based approaches (Boulware and Daugherty 2006). Thus, cellular libraries of peptide substrates (CLiPS) is a biologically based technique that displays peptide sequences genetically encoded in a sacrificial protein on the surface of bacteria. Fluorescence-activated cell sorting (FACS) is used to find optimal substrates and allows for quantitative measurement of whole-cell fluorescence and determination of kinetic parameters. The investigation of caspase-3 substrates using this technique disclosed the DXVD↓G sequence as an optimal sequence, consistent with previous findings. Proteomic identification of protease cleavage sites (PICS) uses mass spectroscopy to interrogate a proteome-derived peptide library treated with a protease to establish the substrate specificity (Schilling and Overall 2008). Both CLiPS and PICS, with caveats, are capable of disclosing the prime-side preference of caspases, aligning well with the fluorescence-quenching chemical synthesis described above.
Overlapping Substrate Specificity
With all the work on optimal subsite preferences of caspases, it might seem that the defined substrates constitute the selective ones. However, there is a problem of cross-reactivity among caspases. It is extremely important to realize that simple peptide substrates, developed to examine the activity of caspases, lack selectivity for individual caspases (McStay et al. 2008). The majority of the substrates are only useful for studies on individual purified caspases but are ineffective for research on complex biological samples such as lysates of apoptotic cells. The consensus sequences of caspases are overlapping; thus, one caspase may cleave a substrate that was designed for other caspases. These findings indicate that caution is needed, especially while studying cell extracts (Timmer and Salvesen 2007; McStay et al. 2008).
Apoptotic caspases-2, -3, -6, -7, -8, -9, and -10 were analyzed with commercially available peptide-based substrates conjugated to the fluorophore AFC (McStay et al. 2008). Interestingly, only caspase-2 and caspase-9 cleaved their putative preferred substrates (VDVAD and LEHD, respectively) the most efficiently and did not recognize other substrates. The findings are in line with reports describing that for efficient cleavage, caspase-2 requires the P5 residue in the substrate (Talanian et al. 1997; Tang et al. 2011). Caspases-8 and -10 preferred the substrate sequence of caspase-9 (LEHD) rather than their assumed substrate of IETD sequence, although both substrates were hydrolyzed well. Interestingly, caspase-6 apart, from cleaving its experimentally determined preferred substrate VEID, also cleaved these two substrates efficiently. Caspase-7 showed activity for its expected DEVD substrate and additionally for the caspase-2 substrate (VDVAD). The most promiscuous turned out to be caspase-3, which cleaved all substrates, albeit with vastly different rates, and often with higher rates than caspases for which the substrates were designed (McStay et al. 2008). Parallel studies using a colorimetric assay to examine the substrate specificity of caspases-1, -2, -3, -6, -7, -8, and -9 revealed similar overlapping specificity (Pereira and Song 2008).
An investigation of the specificity of 10 caspases (1–10) using substrates linked to a 2-aminocridone fluorophore and analyzed by HPLC to establish enzyme activity allowed simultaneous evaluation of a set of substrates in a single sample containing a chosen caspase (Benkova et al. 2009). The substrates used were based on YVAD, VDVAD, DEVD, LEVD, WEHD, VEID, IETD, LEHD, and AEVD, intended for caspases-1, -2, -3/7, -4, -5/1, -6, -8, -9, -10, respectively. As revealed in previous analyses, only caspase-2 cleaved its putative substrate, owing to its strong requirement for P5 residue for cleavage. But, as revealed previously, the caspase-2 substrate was efficiently cleaved by other caspases, caspase-3 being most dominant. Thus, several groups, using different methods, clearly showed that a problem of overlapping substrate specificity exists among caspases. None of the investigated substrates showed high specificity toward an individual caspase. However, for each of the three groups of caspases (inflammatory caspases, initiator caspases, and effector caspases), there was some sequence selectivity observed.
These studies clearly show that the currently available peptide substrates for caspases show high cross-reactivity and are not appropriate for investigations of individual caspases in complex mixtures. This is further complicated when once considers that some caspases have higher intrinsic catalytic efficiency and some have higher concentrations than the others. When the activity is measured in cell lysates, the high caspase-3 concentration masks the activity of other caspases, even if a so-called preferred artificial substrate is used. Therefore, it constitutes a complicated task. However, the commercially available substrates are still valuable for examination of individual purified caspases (Timmer and Salvesen 2007; McStay et al. 2008; Benkova et al. 2009). This somewhat complex scenario could be simplified in knockdown cells, in which depletion of specific caspases could be used to draw conclusions of the participation of other caspases in selective substrate cleavage. A caveat here would take into account the hierarchical organization of caspase pathways, and thus experiments would be restricted to knockdown of effector, rather than apical, caspases.
Substrates for Cell and In Vivo Imaging
Although there is no real hope of defining the contribution of individual caspase activation in complex mixtures by using currently available simple peptide substrates, there has been reasonable progress in showing overall caspase activity in situ. Single-cell imaging techniques using caspase reporters have been very useful in observing the kinetics of activation in cell populations, but, of course, the same caveats apply as noted above regarding specificity. Rhodamine-based dyes coupled to tetrapeptides have shown utility in uncovering caspase activity, primarily in apoptotic models in cell culture. Thus, compounds such as Ac-DEVD-N′-octyloxycarbonyl-R 110 (Cai et al. 2001), N-Ac-DEVD-N′-(poly-fluorobenzoyl)-R 110 (Zhang et al. 2003), and Ac-DEVD-N′-morpholinecarbonyl-R 110 and the bis-peptide substrate (Cbz-DEVD)2-R 110 have been used for biological studies in living cells (Liu et al. 1999). Another fluorescent dye based on rhodamine, 2-Me SiR600, where the oxygen in the xanthene moiety was replaced by silicon, resulted in red fluorescence of instead of green. Such probes will be valuable in multicolor imaging, because they provide an additional color window. Furthermore, they appear to be more suitable for in vivo imaging because tissues are more transparent to red than to green light (Kushida et al. 2012). NucView488 may also be an excellent reporter group for biological studies on cells, because the substrate DEVD-NucView488 is reported to be highly cell-permeable, efficiently cleaved by caspases, and is very sensitive (Cen et al. 2008).
In attempts to enhance cell penetration of chemical probes, a cell penetrating Tat peptide can be coupled to a far-red quencher, QSY 21, fluorophore Alexa Fluor 647, joined to caspase recognition sequence DEVD (Bullok and Piwnica-Worms 2005). Upon caspase cleavage, the quencher and fluorophore are liberated. The main advantage of this probe is its far-red fluorescence, so there is a little interference from autofluorescence of cells. Subsequently, the same group developed a second-generation apoptosis imaging probe, KcapQ, in which the penetrating peptide was replaced by insertion of d-amino acids to prevent adventitious cleavage and reduce toxicity (Maxwell et al. 2009).
Genetically encoded probes based on Förster (fluorescence) resonance energy transfer (FRET) have been applied to analyze caspase activity in single cells. Using a CFP–YFP pair connected by a flexible linker that contains the caspase cleavage sequence DEVDG, it is possible to monitor the rate of caspase activation following apoptotic induction (Rehm et al. 2002). When the linker is cleaved, energy transfer diminishes and the CFP signal increases, an event that can be monitored by live-cell microscopy. Thus, the time to caspase activation varies among individual cells, depending on the type of treatment and concentration of apoptosis inducer used. Once initiated, disruption of the FRET signal tends to be rapid (<15 min) and largely independent of the apoptotic inducer, attesting to the fast nature of caspase activation. Importantly, FRET probe cleavage is substantially slower in caspase-3-deficient cells, revealing the importance of caspase-3 cleavage of the reporter. As a means to partly distinguish between apical and effector caspases, the DEVD/G sequence can be replaced by DEVD/R (Albeck et al. 2008), based on the differential preference for Arg in P1′ of caspase-3 and caspase-8 (Stennicke et al. 2000). An additional attempt to provide for simultaneous imaging of differential caspase specificities led to the construction of a tripartite FRET probe with the general architecture CFP-YFP-mRFP (mRFP, monomeric red fluorescent protein). The sequence DEVD was inserted between CFP and YFP, whereas VEID was inserted between YFP and mRFP (CFP-C3-YFP-C6-mRFP). After protease cleavage, flow cytometry was used to distinguish activities of DEVDase and VEIDase, allowing for separate detection of two FRET signals (Wu et al. 2006).
Natural (Endogenous) Substrates
Several hundred proteins have been reported to be cleaved in a caspase-dependent manner during apoptosis in vivo (or at least in cellulo), and the list is still growing. It has been proposed that the sum total of these proteolytic events defines the complex cellular morphology known as apoptosis (Martin and Green 1995; Salvesen and Dixit 1997). To test this prediction, it would be necessary to separate the cleavage events that cause apoptotic function/morphology from those innocent victims that are inevitable given the complexity of animal proteomes. It is not so easy to do this in a scientifically rigorous manner, especially given the continual proliferation of reported proteins that are cleaved by caspases, deriving from the sensitive and high-throughput focused proteomics techniques that have recently become available.
Only a small group of these proteins is likely to be biologically significant, whereas the others are just cleaved as bystanders (Fuentes-Prior and Salvesen 2004; Crawford and Wells 2011). To validate whether a suspected protein is an important caspase substrate, one could consider whether it obeys the following criteria, which are related to the confidence in their assignment as naturally occurring events (Timmer and Salvesen 2007):
Proteins shown to be cleaved by recombinant caspases in a purified system (preferably including rate—kcat/KM—calculations)
Cleavage products identified in cells
Cleavage-site mutants generated, showing abrogation of cleavage
Cleavage-site mutants introduced into deficient cells
Cleavage-site mutants introduced transgenically into whole animal
So far, only a handful of the hundreds of reported events have been confirmed to be of significance in vivo, and readers interested in more detailed discussion of this topic are referred to reviews (Timmer and Salvesen 2007; Crawford and Wells 2011). Just as for any posttranslational modification, caspase cleavage can have a gain-of-function or loss-of-function consequence for the protein. For loss-of-function events, one would expect a substantial amount of the protein to be cleaved, but for a gain-of-function event, there need only be a small portion of the total molecules cleaved to allow the product(s) to signal. Currently, there is virtually no knowledge of how much cleavage of any caspase substrate there should be for a biological or pathological outcome. Ultimately, in the transgenic models in which a caspase cleavage site has been replaced by a noncleavable one, usually an Asp–Ala mutation, there is the formal, albeit low-level, likelihood that a function of the protein other than proteolysis has been modified by the mutation. The list of caspase substrates that have been somewhat validated as important in driving part of the apoptotic phenotype includes proteins with diverse functions in cell structure, cell signaling, and cell motility (for review, see Crawford and Wells 2011).
A major objective of defining the inherent substrate preferences of caspases, using chemical scanning methods described earlier, has been to use this information to predict natural substrates—hence, the concept of predicting caspase natural substrates from consensus sequences (Timmer and Salvesen 2007). However, it turns out that this is not the only requirement for protein to become a caspase substrate. Protein substrates of caspase-3 show such discrepancies, a lot of them are cleaved at nonoptimal sites (Earnshaw et al. 1999; Nicholson 1999; Fischer et al. 2003; Fuentes-Prior and Salvesen 2004). This suggests that caspase exosite(s), regions involved in substrate binding that are remote from the active site, distinguish specific tertiary motifs in substrates (Fuentes-Prior and Salvesen 2004; Timmer and Salvesen 2007; Yin and Dong 2009). This additional condition sometimes annuls the requirements based on predictions from short synthetic peptides (Fuentes-Prior and Salvesen 2004; Timmer and Salvesen 2007). An interesting example of this propensity is seen in a biological diversity approach that sought to define the substrate preference of caspase-3 by phage display. An M13 bacteriophage gene III display fusion approach using fully randomized libraries of four and six residues was mostly in line with previously determined substrate specificity for caspase-3 and -8 using chemistry-based approach (Lien et al. 2004). Interestingly, some variations on the canonical motifs were discovered, such that peptide DLVD was cleaved by caspase-3 up to 170% faster than peptide DEVD. This method may generate much more conformational variability in peptide sequences, and also longer peptide sequences, possibly revealing the importance of structure and extended sequence in defining real caspase preferences that go beyond the residues lining the catalytic cleft, and this is discussed in more detail below. Currently there seems to be only one report of a defined caspase exosite, one in caspase-7 that distinguishes its preference over caspase-3 for the cleavage of PARP (Boucher et al. 2012). The existence of additional exosites is highly likely given that members of the inhibitor of apoptosis protein (IAP) family interact with regions distant from the caspase active-site cleft exosites (Srinivasula et al. 2001; Shiozaki et al. 2003; Scott et al. 2005; Timmer and Salvesen 2007).
The second issue to be considered is cleavage-site conformation of protein substrates. Unstructured and flexible loops are more readily accessible to caspases (Timmer and Salvesen 2007). As an example illustrating such a phenomenon, caspase-1 and its substrate pro-IL-1β may serve. The enzyme cleaves the substrate, pro-IL-1β, efficiently in a nonoptimal sequence regardless of whether the substrate is heat-denatured or folded (Sleath et al. 1990; Timmer and Salvesen 2007). Hence, it indicates that additional interactions during substrate cleavage might be unnecessary for some caspases, but important for others.
Several groups have deployed variations on a theme known as N-terminomics (and sometimes degradomics) by using focused proteomics technologies to interrogate natural caspase substrates (Van Damme et al. 2005; Enoksson et al. 2007; Dix et al. 2008; Mahrus et al. 2008; Timmer et al. 2009; Xu et al. 2009; Agard et al. 2010). Such technologies, reviewed in detail in Crawford and Wells (2011) and Impens et al. (2010), are capable of disclosing portions of the universe of substrates that have been cleaved by caspases in vivo, or at least in cellulo, and all have very high sensitivity and allow analysis of the caspase N-terminome in a very detailed way. Techniques used to identify natural substrates have been divided into two groups: forward (in vivo) and reverse (in vitro) (Crawford and Wells 2011). The first method allows definition of substrates that are cleaved in a specific biological process, whereas the second determination of substrates of a particular caspase—with the caveat that the natural context is lost. During investigations of an important cleavage, it is valid to apply these two kinds of techniques, because there may occur some discrepancies between cleaving substrates by caspase in vivo and in vivo. Moreover, in some cases, it may be obscure which caspase cleaves a specific substrate that is known to be cleaved during a certain process (Crawford and Wells 2011). Overall, there is still little confidence yet for the biological importance of cleavage events disclosed by these emerging technologies, although as data sets and validation procedures improve, we expect substantial progress in understanding the relevance of specific protein cleavages. Readers interested in the results of this line of research are pointed to the following database resources that capture most of the known cleavages of natural caspase (and other protease) substrates as well as predictive resources: wellslab.ucsf.edu/degrabase/; cutdb.burnham.org; clipserve.clip.ubc.ca/topfind; merops.sanger.ac.uk; bioinf.gen.tcd.ie/CASBAH/; sunflower.kuicr.kyoto-u.ac.jp/∼sjn/Cascleave/links.html.
SYNTHETIC CASPASE INHIBITORS
Biologists, chemists, and pharmaceutical scientists probably disagree about what it takes to be a good and selective enzyme inhibitor. The words “potent” and “specific” are overused in the literature, and this is probably because a chemist may consider a compound to be potent even though it doesn’t do much in vivo. Likewise, a biologist may consider a compound to be specific because it blocks a biological pathway of interest, even though it is broadly reactive with what a pharmaceutical chemist may consider “off targets.” The ideal inhibitor would have high potency (tight binding: Ki < 10 nm; or rapid reaction: kobs/I > 100,000 m−1 sec−1) and show selectivity for its target over all possible off-targets with a factor >1000 (and, of course, be bioavailable and pharmacologically active in vivo). Such a compound does not yet exist for any caspases. Why is this? The most likely reason is that the catalytic center and associated substrate binding cleft in proteases are designed to bind peptides, and thus any peptide or peptidomimetic is likely to bind more-or-less to every caspase. The catalytic center is just too good a thermodynamic sink for inhibitors that are competitive with substrate—so-called orthosteric inhibitors (Mattos et al. 2006; Drag and Salvesen 2010). However, less-than-ideal inhibitors have been highly informative in helping to define biological pathways and as lead compounds for anti-caspase therapy. Moreover, a rapidly growing interest in “allosteric” inhibitors—those that target highly specific sites away from the active site—is likely to achieve higher selectivity and specificity, although at the expense of potency (Drag and Salvesen 2010).
Orthosteric Inhibitors
The starting point for conventional, orthosteric, inhibitor design is to profile the inhibitor using substrate combinatorial library screening, caspase crystal structure analysis, and/or inhibitory structure–activity relationship studies. Detailed analyses of crystal structures of caspases bound to various peptide inhibitors have been used widely to help define whether there are any structural attributes dedicated to a particular caspase that can be useful for development-specific caspase inhibitors. Although the simple answer has been yes, in principle and on a coarse scale, the more important finding in comparing structures of inhibited and free caspases is that the structural plasticity and mobility of the active site have restrained the impact of structure-based probe design. The active sites and regions of caspases that direct inherent subsite occupancy are just too mobile for current design methods to deliver specificity based only on structural information; this is discussed further below.
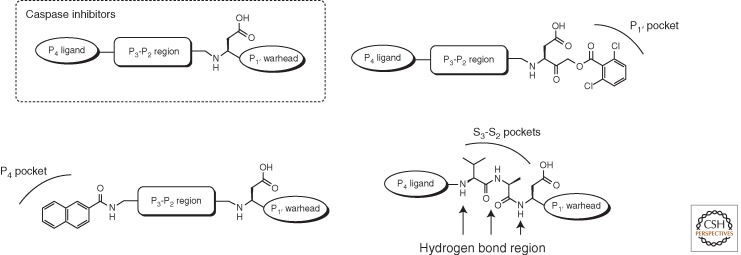
To address this problem, researchers have developed more tailored peptide and peptidomimetic backbones for caspase binding. An analysis of the literature reveals that caspase inhibitor structures can be divided into three regions: the amino-terminal P4 position, the dipeptide P3–P2 region, and a P1′ electrophilic group, sometimes called a “warhead” or “cysteine trap” (Fig. 5). In typical approaches, a series of inhibitors is synthesized in which a particular region is varied while the rest of the scaffold is held the same.
Figure 5.
General outline of caspase inhibitor architecture.
Warheads
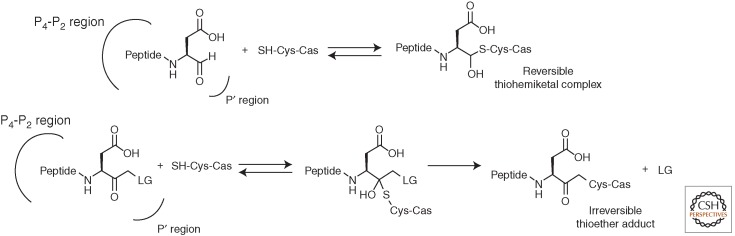
Various types of synthetic inhibitors have been described in the literature, including peptides, peptidomimetics, and nonpeptidic compounds. The guiding principle for many of these studies has been to generate therapeutic leads to modulate apoptotic and inflammatory pathways in disease. An orthogonal, sometimes overlapping, goal has been to generate probes that can be used to dissect the role of individual caspases, or caspases as a group, in biochemical events and networks that define cellular phenotypes and pathologies. Most synthetic caspase inhibitors are designed to use an electrophilic group (warhead) to modify the active-site cysteine residue covalently, leading to enzyme inactivation (Powers et al. 2002), and can be assigned as reversible (if the enzyme is inactivated through formation of reversible thiohemiketal) or irreversible (if the enzyme is permanently inactivated via formation of the thioether complex) (Fig. 6) (Talanian et al. 2000; Graczyk 2002). Some compounds display a bimodal pattern of inhibition, when a thiohemiketal is slowly transformed into a thioether adduct (Brady 1998; Brady et al. 1999).
Figure 6.
Reversible and irreversible caspase inhibition modes.
The main feature for efficient proteolysis is that caspases require an aspartate residue at P1 and recognize at least four amino acid residues to the left of the cleavage site (Thornberry et al. 1992; Pop and Salvesen 2009). As in the case of substrates, truncation of a tetrapeptide to a tripeptide or dipeptide leads to a dramatic decrease in inhibitor peptide-based activity (Graybill et al. 1997; James et al. 2004). Thus, inhibitor design for caspases has generally adhered to this consensus, with introduction of varied unnatural amino acid derivatives and variation of the chemistry of the warhead.
At the P1′ position, several different electrophiles have been tested among which aldehydes and ketones with their derivatives (halomethylketones, aryloxymethylketones, or acyloxymethylketones), warheads of high utility for design of inhibitors selective to cysteine proteases in general (Powers et al. 2002), have been heavily investigated. Interestingly, some electrophiles that are reactive with other families of cysteine proteases are very poor for caspases. Vinyl sulfone warheads, Michael acceptors that are highly reactive with cathepsins, and deubiquitinating enzymes (members of the protease clan CA) react very poorly with caspases (Newton et al. 2010). A different class of warhead, aza-epoxide and aza-Michael platforms, show excellent caspase inhibition and are suitable for being extended in the P1′ direction (Asgian et al. 2002; Ekici et al. 2004, 2006; James et al. 2004). There is no relevant reason that aza-expoxides or aza-Michaels acceptors could not be useful as therapeutic leads, although this appears not to have been followed up on. Exploration of the S1′ pocket of caspase-3 using aza-peptide epoxide inhibitors (Ganesan et al. 2006a) uncovered the basis for inhibition specificity on the right to the scissile bond. The S1′ pocket is large and can accommodate bulky, hydrophobic warheads, explaining in part the high reactivity of aryloxymethylketones or acyloxymethylketones with caspases.
Therapeutic Leads
Although the substrate specificity of a particular caspase cannot be used to discriminate it within the family, for reasons of overlap that also pertain to the substrates explained above, there are certain preferences that can be exploited. For instance, the finding that the caspase-1 inhibitor Ac-YVAD-CHO prevents the release of interleukin 1-β from monocytes was part of the initial evidence that caspase-1 is responsible for interleukin 1-β maturation, upon specific proteolysis (Thornberry et al. 1992). Unfortunately, most of these inhibitors cannot play therapeutic roles because of their toxic metabolites. A good example is Cbz-VAD-FMK, a broad-spectrum caspase inhibitor. Cbz-VAD-FMK (a.k.a. ZVAD) is an excellent probe for enzyme kinetic analysis and ablation of apoptosis and inflammation paradigms owing to its rapid inhibition of most caspases in vitro (Table 3), but is useless as a therapeutic lead because of production of highly toxic fluoroacetate (Van Noorden 2001).
Table 3.
Kinetic parameters of the widely used and heavily cited Cbz-VAD-FMK (often called ZVAD)
| kobs/I (m−1 sec−1) |
t/2 (sec) for I = 50 µm |
t/2 (sec) for I = 1 µm |
|
|---|---|---|---|
| Caspase-1 | 280,000 | 0.05 | 2.48 |
| Caspase-2 | 290 | 47.79 | 2389.66 |
| Caspase-3 | 16,000 | 0.87 | 43.31 |
| Caspase-4 | 5500 | 2.52 | 126.00 |
| Caspase-5 | 130,000 | 0.11 | 5.33 |
| Caspase-6 | 7100 | 1.95 | 97.61 |
| Caspase-7 | 18,000 | 0.77 | 38.50 |
| Caspase-8 | 280,000 | 0.05 | 2.48 |
| Caspase-9 | 180,000 | 0.08 | 3.85 |
The kobs/I values are adapted from Garcia-Calvo et al. (1998), and the half-time of inhibition (t/2) in seconds is calculated according to the relationship t/2 = ln 2/kobs, for the inhibitor concentration given at the head of the column (Bieth 1984).
Peptide to Peptidomimetic
A classic example of inhibitor development from peptide substrate specificity knowledge to a peptidomimetic that achieved clinical trial status is Pralnacasan (VX-740). The most common P3–P2 motif of early caspase-1 inhibitors is Val–Ala, but it was also reported that replacing the P2 Ala residue by other amino acids including secondary piperidine or proline does not affect inhibitory potency (Dolle et al. 1994). Such an example is VX-043198 with an unnatural tert-leucine in the P3 position and a secondary proline in P2 (Fig. 7). This inhibitor is an active metabolite of VX-765, an orally absorbed pro-drug. VX-765 is a rare example of a peptide-based caspase inhibitor possessing a good pharmacokinetic profile and displaying utility for treatment of inflammatory diseases (Wannamaker et al. 2007). To address potency issues of VX-765, a new, more flexible pyridazinodiazepine scaffold was described (Dolle et al. 1997). This moiety reduced the peptide character while maintaining a high affinity against caspase 1. The use of a pyridazinodiazepine group for mimicking the Val–Ala sequence resulted in the development of Pralnacasan (VX-740) (Fig. 7), which was tested in late clinical trials as a drug candidate for rheumatoid arthritis (unfortunately failing because of liver toxicity issues) (Dolle et al. 1997; Siegmund and Zeitz 2003; Linton 2005). The drug possesses a 2-naphthyl hydrophobic moiety in the P4 position, a pyridazinodiazepine-based bicyclic core mimicking the Val–Ala region (P3–P2 residues) and a lactone moiety at the carboxyl terminus, masking an aspartate residue at P1 (Fig. 7).
Figure 7.
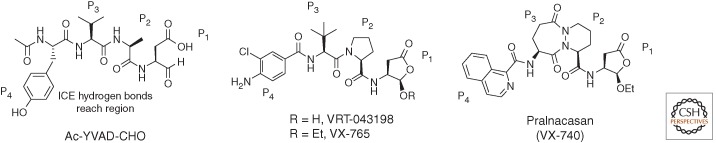
Structures of prototypic peptide-based (Ac-YVAD-CHO) and two peptidomimetic caspase-1 inhibitors.
Although the most potent caspase inhibitors occupy the S1–S4 subsites, it appears that the chemical nature of the P1′ leaving group can be a key determinant in the design of inhibitors. Simple dipeptide-based compounds can display nanomolar potency toward caspase-1 (Galatsis et al. 2010). The dipeptide-based IDN-6556, containing a 2,3,5,6-tetrafluorophenoxymethyl ketone warhead, was the first irreversible pan-caspase inhibitor to enter Phase II clinical trials targeting liver diseases. Taken together, it appears that inhibitors with appropriate P1′ leaving groups can act as broad spectrum irreversible inactivators targeting inflammatory, initiator, and apoptotic caspases. Such inhibitors, showing little selectivity, may be appropriate therapeutics in acute pathological situations, and that irreversibility per se may not be a barrier to development of drugs—depending on the therapeutic indication.
To enhance selectivity by exploring additional chemical space, screens using nonproteinogenic (unnatural) amino acids have been used to synthesize caspase peptide inhibitors by exploring libraries equipped with acyloxymethylketones (AOMKs) (Berger et al. 2006b) and 2,3,5,6-tetrafluorophenoxymethyl ketone warheads (Leyva et al. 2010). Although these screens have shown enhanced selectivity compared with natural amino acid libraries, there is still a way to go with this approach to achieve more optimal specificity.
Caspase Activity-Based Probes
Activity-based proteomics (ABP) is a relatively new field on the border of chemistry and biology. The main goal of ABP is to design small-molecule chemical tools that will be able to tag and monitor the active status of proteins within a complex proteome, providing links to their functions. The most commonly used probes possess an appropriate peptide or peptidomimetic sequence equipped with an electrophilic warhead that is used to modify the enzyme’s active site covalently (Serim et al. 2012). These sequences can be derived from substrate or inhibitor library screening or by iterative chemical synthesis approaches. Characteristic of activity-based probes is an amino-terminal tag, which reveals whether a probe binds to the active site or not. The most popular tags for this purpose are radioisotopes, fluorescent (or luminescent) groups (all three groups give direct readouts), and biotin (which is detected in a semidirect manner) (Fig. 8). All ABPs have this composition and are generally divided by the type of reporter. This concept was pioneered for proteases by James C. Powers and used at first to target serine proteases (Kam et al. 1993). Now this discipline has significantly evolved, confirmed by the number of recent review papers concentrating on distinct ABP approaches (Evans and Cravatt 2006; Fonovic and Bogyo 2008; Heal et al. 2008, 2011; auf dem Keller and Schilling 2010; Edgington et al. 2011; Serim et al. 2012).
Figure 8.
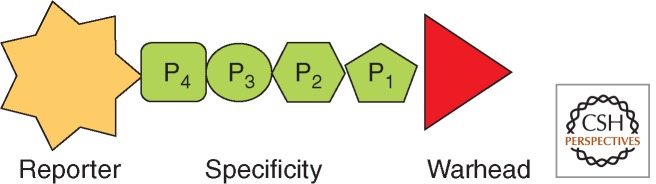
A generalized activity-based probe. The probe is designed to bind in the same manner as a synthetic substrate, but the red warhead binds irreversibly to the caspase catalytic residues.
Biotin Labels
In the early stage of the discovery of activity-based probes (ABPs) for caspases, commonly used inhibitors with appropriate peptide sequences were biotinylated and tested toward their protein targets (Thornberry 1994; Nicholson et al. 1995). Biotin is the tag of choice for ABPs because it displays diffusion-limited binding to streptavidin, allowing for high stringency and rapid probing (Sadaghiani et al. 2007). Because these probes label only active caspases, the principle is extremely useful for distinguishing active enzyme from the bulk of inactive precursors in cells and tissues. The use of biotin for affinity labeling has some limitations, the main problem being that biotin is not suitable for in vivo and in situ studies because of its poor cell permeability and nondirect visualization protocol and the presence of endogenous biotinylated polypeptides (Sadaghiani et al. 2007).
Although not suitable for use for intact cell imaging, biotin ABPs have been extremely useful in visualizing caspase activation in vitro following cell lysis and ligand blotting, usually with streptavidin conjugates. Thus biotin-VAD-CMK, biotin-YVAD-AOMK, Cbz-EK(biotin)D-FMK, and biotin-DEVD-CMK have been used to detect which caspases are being activated in response to various apoptosis-inducing triggers (Fearnhead et al. 1997; Cardone et al. 1998; Martins et al. 1998), heat shock (Tu et al. 2006), or nitrosylation (Li et al. 1997; Mannick et al. 2001). Although much of the earlier work was performed with FMK warheads, today AOMK warheads are preferred owing to their greater selectivity. Indeed, FMK warheads have lost popularity because of the substantial cross-reactivity with other families of cysteine proteases (Rozman-Pungercar et al. 2003). The sequence elements of early probes were based on prototypic caspase substrates and inhibitors. Unfortunately, these probes lack the specificity and produce high background labeling during crude proteome assays. Because of this, more specific caspase activity-based probes have been produced. For example, work from the Bogyo laboratory demonstrated that the commercially available biotin-VAD-FMK probe is widely reactive with caspases, but also produces high background when assayed with a caspase-3 mutant (C285A). This is not good news if the objective is to detect active enzyme. The tripeptide biotin-EVD-AOMK labels caspases well, with far less background (Kato et al. 2005). But replacing elements in the P3 and P2 positions with nonproteogenic (unnatural) amino acids begins to achieve a degree of selectivity for individual caspases. Although the absolute probe specificity has not been obtained, this work is an example of how selective labeling of specific caspases could be developed (Berger et al. 2006a,b).
Radioisotopes
Radioactive isotopes, especially 125I, have several advantages in ABPs: They can be easily introduced into an inhibitor structure, their relatively small size does not change the overall biochemical properties of a parent inhibitor, they can be easily detected by simple autoradiography methods, and they are sometimes used for whole-body imaging. The dipeptidyl general caspase inhibitor 125I-M808 (iodotyrosyl-VD-FMK) was used to show radiolabeling of caspases in tissues from animals following apoptotic challenges (Methot et al. 2004). A set of isatin-based analogs that are able to inhibit caspases, labeled with 18F or 11C, has been used in microPET imaging for detecting apoptosis in animal models, showing that caspase ABPs can act as radiotracers in living organisms (Zhou et al. 2006, 2009; Nguyen et al. 2009). A disadvantage of the isatin-based probes must be synthesized and used rapidly following synthesis. To overcome this limitation, a probe allowing introduction of a 64Cu atom via chelation via a 1, 4, 7, 10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) group has been shown to be a good tracer of caspases in a mouse model (Lee et al. 2009). The probe is bifunctional, because it also possesses a fluorescence-quencher pair that produces fluorescence upon proteolysis. Because the probe is really a substrate, not an inhibitor, the radio signal is always “on” and used to trace the compound, whereas the fluorescence generation reports caspase activity. Perhaps a combination of DOTA with a cell-penetrating caspase inhibitor would make an excellent, more widely available, probe for PET imaging.
Fluorescent (Luminescent) Dyes
The great advantage of fluorescent tags is their high water solubility and good cell-penetration properties. Likewise, some of them are pH insensitive, stable to solid-phase peptide synthesis, and give fast and high readouts (Sadaghiani et al. 2007). One of the first approaches in the use of fluorescently labeled probes for caspases, coined FLICA (for fluorochrome-labeled inhibitors of caspases), used carboxyfluorescein (FAM) as the tag in FAM-VAD-FMK (Bedner et al. 2000; Smolewski et al. 2001; Grabarek et al. 2002). FLICA offers a promising assay for studying caspase biology, but lack of specificity and background effects of other proteases with the FMK group limit its utility. To date, various types of FLICAs have been described in the literature. The commercially available FLICAs are usually tagged with FAM or fluorescein isothiocyanate (FITC), green dyes, or sulforhodamine B (SR), a red dye. Likewise, currently used FLICAs are designed for detection of individual caspases or a group of caspases separately (Smolewski et al. 2002; Teodori et al. 2002; Valero et al. 2005; Luthra et al. 2006; Jadhav et al. 2007; Kuzelova et al. 2007).
A developing fluorescent dye for caspase inhibitor labeling is Cy5, which belongs to the cyanine family. This fluorophore is also referred as a near-infrared fluorescent tag (NIRF) because of its spectral characteristic (maximum of absorbance at 649 nm, maximum of emission at 670 nm). Cy5 has been widely used to tag caspase inhibitors with the AOMK warhead, thereby improving selectivity. Caspase probes labeled with Cy5 can be used for in vivo apoptosis imaging by a noninvasive manner (Edgington et al. 2009, 2012). These initial studies with the use of Cy5 show that caspase activity can be detected in many biological assays, from cell-free apoptotic lysates, to tissues, to whole organisms. Moreover, potent and cell-permeable caspase fluorescent activity-based probes possess diagnostic and therapeutic potential. Enhancements in selectivity are likely to use nonproteinogenic amino acid strategies, and some success in this direction has been seen with a use to explore the role of pyroptosis in response to Salmonella infection in macrophages of primary mouse bone marrow, revealing the importance of apoptotic and inflammatory caspases (Puri et al. 2012).
Allosteric Inhibitors
Unlike orthosteric inhibitors, in which the starting point is to mimic substrate-binding modes, allosteric inhibitors are more difficult to design rationally. Screening with protein-based recombinant DARpin (designed ankyrin repeats) libraries revealed a highly selective and potent inhibitor of caspase-2. The caspase-2-specific DARpin targets a region that connected the active site and influenced the catalytic conformation of the protease (Schweizer et al. 2003). Although it would be difficult to render a small molecule from the DARpin, this showed that allosteric inhibitors can deliver exquisite specificity. Sometimes, experiments of nature often give reasonable starting points, and to this end, the dimer interface provides a point of leverage. Thus, the BIR3 domain of the endogenous caspase-9 inhibitor XIAP acts by preventing the activating dimerization of the enzyme (Shiozaki et al. 2003), and to this end, peptides mimicking this interaction have been described as allosteric caspase inhibitors (Huber et al. 2012). The dimerization interface of caspases-3 and -7 contains pockets that can be used to block the active conformation, by an allosteric mechanism (Hardy et al. 2004). Zinc has long been known as a caspase inhibitor (Perry et al. 1997; Stennicke and Salvesen 1997), and it has been revealed that this metal targets an allosteric site on caspase-6 (Velazquez-Delgado and Hardy 2012). Design and optimization of allosteric caspase inhibitors is in its infancy, but the promises in terms of selectivity are huge (Drag and Salvesen 2010).
CONCLUSION
The explosion of synthetic caspase substrates and inhibitors over the last few years has revealed the promise of defining the fundamental properties of caspases as enzymes, their role in promoting apoptosis and inflammation, and provided several excellent scaffolds for therapeutic leads. Innovative focused proteomics methods have disclosed a huge universe of potential natural protein caspase substrates in vivo. But these advances are not without caveats. Currently there seem to be no caspase inhibitors in advanced clinical trials, and this may be because of overlapping effects—due to the lack of appropriately selective inhibitors—or it may simply be because of the lack of confidence in targeting caspases for therapeutic ends. More selective inhibitors and a better understanding of the final outcomes of caspase inhibition in human disease are needed. The vast array of caspase natural protein substrates so far identified needs to be triaged into those that directly affect the outcome of apoptosis or inflammation from the innocent bystander substrates, and this will require substantial protein engineering and reconstitution experiments. We look forward to the emerging technologies that will provide resolution to these matters.
ACKNOWLEDGMENTS
This work is supported by the Foundation for Polish Science and the National Science Centre grant 2011/03/B/ST5/01048 in Poland and National Institutes of Health grant R01 GM09040.
Footnotes
Editors: Eric H. Baehrecke, Douglas R. Green, Sally Kornbluth, and Guy S. Salvesen
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
- Agard NJ, Maltby D, Wells JA 2010. Inflammatory stimuli regulate caspase substrate profiles. Mol Cell Proteomics 9: 880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK 2008. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell 30: 11–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J 1996. Human ICE/CED-3 protease nomenclature. Cell 87: 171. [DOI] [PubMed] [Google Scholar]
- Asgian JL, James KE, Li ZZ, Carter W, Barrett AJ, Mikolajczyk J, Salvesen GS, Powers JC 2002. Aza-peptide epoxides: A new class of inhibitors selective for clan CD cysteine proteases. J Med Chem 45: 4958–4960 [DOI] [PubMed] [Google Scholar]
- auf dem Keller U, Schilling O 2010. Proteomic techniques and activity-based probes for the system-wide study of proteolysis. Biochimie 92: 1705–1714 [DOI] [PubMed] [Google Scholar]
- Bedner E, Smolewski P, Amstad P, Darzynkiewicz Z 2000. Activation of caspases measured in situ by binding of fluorochrome-labeled inhibitors of caspases (FLICA): Correlation with DNA fragmentation. Exp Cell Res 259: 308–313 [DOI] [PubMed] [Google Scholar]
- Benkova B, Lozanov V, Ivanov IP, Mitev V 2009. Evaluation of recombinant caspase specificity by competitive substrates. Anal Biochem 394: 68–74 [DOI] [PubMed] [Google Scholar]
- Berger AB, Sexton KB, Bogyo M 2006a. Commonly used caspase inhibitors designed based on substrate specificity profiles lack selectivity. Cell Res 16: 961–963 [DOI] [PubMed] [Google Scholar]
- Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, Bogyo M 2006b. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell 23: 509–521 [DOI] [PubMed] [Google Scholar]
- Bieth JG 1984. In vivo significance of kinetic constants of protein proteinase inhibitors. Biochem Med 32: 387–397 [DOI] [PubMed] [Google Scholar]
- Blanchard H, Kodandapani L, Mittl PR, Marco SD, Krebs JF, Wu JC, Tomaselli KJ, Grutter MG 1999. The three-dimensional structure of caspase-8: An initiator enzyme in apoptosis. Structure 7: 1125–1133 [DOI] [PubMed] [Google Scholar]
- Boucher D, Blais V, Denault JB 2012. Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc Natl Acad Sci 109: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulware KT, Daugherty PS 2006. Protease specificity determination by using cellular libraries of peptide substrates (CLiPS). Proc Natl Acad Sci 103: 7583–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady KD 1998. Bimodal inhibition of caspase-1 by aryloxymethyl and acyloxymethyl ketones. Biochemistry 37: 8508–8515 [DOI] [PubMed] [Google Scholar]
- Brady KD, Giegel DA, Grinnell C, Lunney E, Talanian RV, Wong W, Walker N 1999. A catalytic mechanism for caspase-1 and for bimodal inhibition of caspase-1 by activated aspartic ketones. Bioorg Med Chem 7: 621–631 [DOI] [PubMed] [Google Scholar]
- Bullok K, Piwnica-Worms D 2005. Synthesis and characterization of a small, membrane-permeant, caspase-activatable far-red fluorescent peptide for imaging apoptosis. J Med Chem 48: 5404–5407 [DOI] [PubMed] [Google Scholar]
- Cai SX, Zhang HZ, Guastella J, Drewe J, Yang W, Weber E 2001. Design and synthesis of rhodamine 110 derivative and caspase-3 substrate for enzyme and cell-based fluorescent assay. Bioorg Med Chem Lett 11: 39–42 [DOI] [PubMed] [Google Scholar]
- Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC 1998. Regulation of cell death protease caspase-9 by phosphorylation. Science 282: 1318–1321 [DOI] [PubMed] [Google Scholar]
- Cen H, Mao F, Aronchik I, Fuentes RJ, Firestone GL 2008. DEVD-NucView488: A novel class of enzyme substrates for real-time detection of caspase-3 activity in live cells. FASEB J 22: 2243–2252 [DOI] [PubMed] [Google Scholar]
- Chen JM, Rawlings ND, Stevens RA, Barrett AJ 1998. Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases. FEBS Lett 441: 361–365 [DOI] [PubMed] [Google Scholar]
- Crawford ED, Wells JA 2011. Caspase substrates and cellular remodeling. Annu Rev Biochem 80: 1055–1087 [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC 1997. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388: 300–304 [DOI] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF 2008. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolle RE, Singh J, Rinker J, Hoyer D, Prasad CV, Graybill TL, Salvino JM, Helaszek CT, Miller RE, Ator MA 1994. Aspartyl α-((1-phenyl-3-(trifluoromethyl)-pyrazol-5-yl) oxy)methyl ketones as interleukin-1β converting enzyme inhibitors. Significance of the P1 and P3 amido nitrogens for enzyme-peptide inhibitor binding. J Med Chem 37: 3863–3866 [DOI] [PubMed] [Google Scholar]
- Dolle RE, Prasad CV, Prouty CP, Salvino JM, Awad MM, Schmidt SJ, Hoyer D, Ross TM, Graybill TL, Speier GJ, et al. 1997. Pyridazinodiazepines as a high-affinity, P2–P3 peptidomimetic class of interleukin-1β-converting enzyme inhibitor. J Med Chem 40: 1941–1946 [DOI] [PubMed] [Google Scholar]
- Drag M, Salvesen GS 2010. Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 9: 690–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH 1999. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 68: 383–424 [DOI] [PubMed] [Google Scholar]
- Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M 2009. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med 15: 967–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington LE, Verdoes M, Bogyo M 2011. Functional imaging of proteases: Recent advances in the design and application of substrate-based and activity-based probes. Curr Opin Chem Biol 15: 798–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington LE, van Raam BJ, Verdoes M, Wierschem C, Salvesen GS, Bogyo M 2012. An optimized activity-based probe for the study of caspase-6 activation. Chem Biol 19: 340–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici OD, Gotz MG, James KE, Li ZZ, Rukamp BJ, Asgian JL, Caffrey CR, Hansell E, Dvorak J, McKerrow JH, et al. 2004. Aza-peptide Michael acceptors: A new class of inhibitors specific for caspases and other clan CD cysteine proteases. J Med Chem 47: 1889–1892 [DOI] [PubMed] [Google Scholar]
- Ekici OD, Li ZZ, Campbell AJ, James KE, Asgian JL, Mikolajczyk J, Salvesen GS, Ganesan R, Jelakovic S, Grutter MG, et al. 2006. Design, synthesis, and evaluation of aza-peptide Michael acceptors as selective and potent inhibitors of caspases-2, -3, -6, -7, -8, -9, and -10. J Med Chem 49: 5728–5749 [DOI] [PubMed] [Google Scholar]
- Enoksson M, Li J, Ivancic MM, Timmer JC, Wildfang E, Eroshkin A, Salvesen GS, Tao WA 2007. Identification of proteolytic cleavage sites by quantitative proteomics. J Proteome Res 6: 2850–2858 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Cravatt BF 2006. Mechanism-based profiling of enzyme families. Chem Rev 106: 3279–3301 [DOI] [PubMed] [Google Scholar]
- Fischer U, Janicke RU, Schulze-Osthoff K 2003. Many cuts to ruin: A comprehensive update of caspase substrates. Cell Death Differ 10: 76–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonovic M, Bogyo M 2008. Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev Proteomics 5: 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS 2004. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 384: 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatsis P, Caprathe B, Downing D, Gilmore J, Harter W, Hays S, Kostlan C, Linn K, Lunney E, Para K, et al. 2010. Inhibition of interleukin-1β converting enzyme (ICE or caspase 1) by aspartyl acyloxyalkyl ketones and aspartyl amidooxyalkyl ketones. Bioorg Med Chem Lett 20: 5089–5094 [DOI] [PubMed] [Google Scholar]
- Ganesan R, Jelakovic S, Campbell AJ, Li ZZ, Asgian JL, Powers JC, Grutter MG 2006a. Exploring the S4 and S1 prime subsite specificities in caspase-3 with aza-peptide epoxide inhibitors. Biochemistry 45: 9059–9067 [DOI] [PubMed] [Google Scholar]
- Ganesan R, Mittl PR, Jelakovic S, Grutter MG 2006b. Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis. J Mol Biol 359: 1378–1388 [DOI] [PubMed] [Google Scholar]
- Garcia-Calvo M, Peterson EP, Leiting B, Ruel R, Nicholson DW, Thornberry NA 1998. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem 273: 32608–32613 [DOI] [PubMed] [Google Scholar]
- Garcia-Calvo M, Peterson EP, Rasper DM, Vaillancourt JP, Zamboni R, Nicholson DW, Thornberry NA 1999. Purification and catalytic properties of human caspase family members. Cell Death Differ 6: 362–369 [DOI] [PubMed] [Google Scholar]
- Grabarek J, Amstad P, Darzynkiewicz Z 2002. Use of fluorescently labeled caspase inhibitors as affinity labels to detect activated caspases. Hum Cell 15: 1–12 [DOI] [PubMed] [Google Scholar]
- Graczyk PP 2002. Caspase inhibitors as anti-inflammatory and antiapoptotic agents. Prog Med Chem 39: 1–72 [DOI] [PubMed] [Google Scholar]
- Graybill TL, Prouty CP, Speier G, Hoyer D, Dolle RE, Helaszek CT, Ator MA, Uhl J, Strasters J 1997. α-((Tetronoyl)oxy)- and α-((Tetramoyl)oxy)methyl ketone inhibitors of the interleukin-1B converting enzyme (ICE). Bioorg Med Chem Lett 7: 41–46 [Google Scholar]
- Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA 2004. Discovery of an allosteric site in the caspases. Proc Natl Acad Sci 101: 12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS 2000. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc Natl Acad Sci 97: 7754–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA 2000. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J Biol Chem 275: 27084–27093 [DOI] [PubMed] [Google Scholar]
- Heal WP, Wickramasinghe SR, Tate EW 2008. Activity based chemical proteomics: Profiling proteases as drug targets. Curr Drug Discov Technol 5: 200–212 [DOI] [PubMed] [Google Scholar]
- Heal WP, Dang TH, Tate EW 2011. Activity-based probes: Discovering new biology and new drug targets. Chem Soc Rev 40: 246–257 [DOI] [PubMed] [Google Scholar]
- Hickson J, Ackler S, Klaubert D, Bouska J, Ellis P, Foster K, Oleksijew A, Rodriguez L, Schlessinger S, Wang B, et al. 2010. Noninvasive molecular imaging of apoptosis in vivo using a modified firefly luciferase substrate, Z-DEVD-aminoluciferin. Cell Death Differ 17: 1003–1010 [DOI] [PubMed] [Google Scholar]
- Huber KL, Ghosh S, Hardy JA 2012. Inhibition of caspase-9 by stabilized peptides targeting the dimerization interface. Biopolymers 98: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impens F, Colaert N, Helsens K, Plasman K, Van Damme P, Vandekerckhove J, Gevaert K 2010. MS-driven protease substrate degradomics. Proteomics 10: 1284–1296 [DOI] [PubMed] [Google Scholar]
- Jadhav U, Ezhilarasan R, Vaughn SF, Berhow MA, Mohanam S 2007. Iberin induces cell cycle arrest and apoptosis in human neuroblastoma cells. Int J Mol Med 19: 353–361 [PMC free article] [PubMed] [Google Scholar]
- James KE, Asgian JL, Li ZZ, Ekici OD, Rubin JR, Mikolajczyk J, Salvesen GS, Powers JC 2004. Design, synthesis, and evaluation of aza-peptide epoxides as selective and potent inhibitors of caspases-1, -3, -6, and -8. J Med Chem 47: 1553–1574 [DOI] [PubMed] [Google Scholar]
- Kam CM, Abuelyaman AS, Li Z, Hudig D, Powers JC 1993. Biotinylated isocoumarins, new inhibitors and reagents for detection, localization, and isolation of serine proteases. Bioconjug Chem 4: 560–567 [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M 2005. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol 1: 33–38 [DOI] [PubMed] [Google Scholar]
- Kindermann M, Roschitzki-Voser H, Caglic D, Repnik U, Miniejew C, Mittl PR, Kosec G, Grutter MG, Turk B, Wendt KU 2010. Selective and sensitive monitoring of caspase-1 activity by a novel bioluminescent activity-based probe. Chem Biol 17: 999–1007 [DOI] [PubMed] [Google Scholar]
- Kushida Y, Hanaoka K, Komatsu T, Terai T, Ueno T, Yoshida K, Uchiyama M, Nagano T 2012. Red fluorescent scaffold for highly sensitive protease activity probes. Bioorg Med Chem Lett 22: 3908–3911 [DOI] [PubMed] [Google Scholar]
- Kuzelova K, Grebenova D, Hrkal Z 2007. Labeling of apoptotic JURL-MK1 cells by fluorescent caspase-3 inhibitor FAM-DEVD-fmk occurs mainly at site(s) different from caspase-3 active site. Cytometry A 71: 605–611 [DOI] [PubMed] [Google Scholar]
- Lazebnik YA 1997. Oncogene-dependent apoptosis in extracts from drug-resistant cells. Genes Dev 11: 1266–1276 [DOI] [PubMed] [Google Scholar]
- Lee H, Akers WJ, Cheney PP, Edwards WB, Liang K, Culver JP, Achilefu S 2009. Complementary optical and nuclear imaging of caspase-3 activity using combined activatable and radio-labeled multimodality molecular probe. J Biomed Opt 14: 040507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva MJ, Degiacomo F, Kaltenbach LS, Holcomb J, Zhang N, Gafni J, Park H, Lo DC, Salvesen GS, Ellerby LM, et al. 2010. Identification and evaluation of small molecule pan-caspase inhibitors in Huntington’s disease models. Chem Biol 17: 1189–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Billiar TR, Talanian RV, Kim YM 1997. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun 240: 419–424 [DOI] [PubMed] [Google Scholar]
- Lien S, Pastor R, Sutherlin D, Lowman HB 2004. A substrate-phage approach for investigating caspase specificity. Protein J 23: 413–425 [DOI] [PubMed] [Google Scholar]
- Linton SD 2005. Caspase inhibitors: A pharmaceutical industry perspective. Curr Top Med Chem 5: 1697–1717 [DOI] [PubMed] [Google Scholar]
- Liu J, Bhalgat M, Zhang C, Diwu Z, Hoyland B, Klaubert DH 1999. Fluorescent molecular probes V: A sensitive caspase-3 substrate for fluorometric assays. Bioorg Med Chem Lett 9: 3231–3236 [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Overall CM 2002. Protease degradomics: A new challenge for proteomics. Nat Rev Mol Cell Biol 3: 509–519 [DOI] [PubMed] [Google Scholar]
- Lozanov V, Ivanov IP, Benkova B, Mitev V 2009. Peptide substrate for caspase-3 with 2-aminoacridone as reporting group. Amino Acids 36: 581–586 [DOI] [PubMed] [Google Scholar]
- Luthra S, Fardin B, Dong J, Hertzog D, Kamjoo S, Gebremariam S, Butani V, Narayanan R, Mungcal JK, Kuppermann BD, et al. 2006. Activation of caspase-8 and caspase-12 pathways by 7-ketocholesterol in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci 47: 5569–5575 [DOI] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA 2008. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134: 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly DJ, Leonetti F, Backes BJ, Dauber DS, Harris JL, Craik CS, Ellman JA 2002. Expedient solid-phase synthesis of fluorogenic protease substrates using the 7-amino-4-carbamoylmethylcoumarin (ACC) fluorophore. J Org Chem 67: 910–915 [DOI] [PubMed] [Google Scholar]
- Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B 2001. S-Nitrosylation of mitochondrial caspases. J Cell Biol 154: 1111–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Green DR 1995. Protease activation during apoptosis: Death by a thousand cuts? Cell 82: 349–352 [DOI] [PubMed] [Google Scholar]
- Martins LM, Kottke TJ, Kaufmann SH, Earnshaw WC 1998. Phosphorylated forms of activated caspases are present in cytosol from HL-60 cells during etoposide-induced apoptosis. Blood 92: 3042–3049 [PubMed] [Google Scholar]
- Mattos C, Bellamacina CR, Peisach E, Pereira A, Vitkup D, Petsko GA, Ringe D 2006. Multiple solvent crystal structures: Probing binding sites, plasticity and hydration. J Mol Biol 357: 1471–1482 [DOI] [PubMed] [Google Scholar]
- Maxwell D, Chang Q, Zhang X, Barnett EM, Piwnica-Worms D 2009. An improved cell-penetrating, caspase-activatable, near-infrared fluorescent peptide for apoptosis imaging. Bioconjug Chem 20: 702–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay GP, Salvesen GS, Green DR 2008. Overlapping cleavage motif selectivity of caspases: Implications for analysis of apoptotic pathways. Cell Death Differ 15: 322–331 [DOI] [PubMed] [Google Scholar]
- Methot N, Vaillancourt JP, Huang J, Colucci J, Han Y, Menard S, Zamboni R, Toulmond S, Nicholson DW, Roy S 2004. A caspase active site probe reveals high fractional inhibition needed to block DNA fragmentation. J Biol Chem 279: 27905–27914 [DOI] [PubMed] [Google Scholar]
- Mittl PR, Di Marco S, Krebs JF, Bai X, Karanewsky DS, Priestle JP, Tomaselli KJ, Grutter MG 1997. Structure of recombinant human CPP32 in complex with the tetrapeptide acetyl-Asp-Val-Ala-Asp fluoromethyl ketone. J Biol Chem 272: 6539–6547 [DOI] [PubMed] [Google Scholar]
- Newton AS, Gloria PM, Goncalves LM, dos Santos DJ, Moreira R, Guedes RC, Santos MM 2010. Synthesis and evaluation of vinyl sulfones as caspase-3 inhibitors. A structure-activity study. Eur J Med Chem 45: 3858–3863 [DOI] [PubMed] [Google Scholar]
- Nguyen QD, Smith G, Glaser M, Perumal M, Arstad E, Aboagye EO 2009. Positron emission tomography imaging of drug-induced tumor apoptosis with a caspase-3/7 specific [18F]-labeled isatin sulfonamide. Proc Natl Acad Sci 106: 16375–16380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson DW 1999. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ 6: 1028–1042 [DOI] [PubMed] [Google Scholar]
- Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. 1995. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376: 37–43 [DOI] [PubMed] [Google Scholar]
- O’Brien MA, Daily WJ, Hesselberth PE, Moravec RA, Scurria MA, Klaubert DH, Bulleit RF, Wood KV 2005. Homogeneous, bioluminescent protease assays: Caspase-3 as a model. J Biomol Screen 10: 137–148 [DOI] [PubMed] [Google Scholar]
- Odake S, Kam CM, Narasimhan L, Poe M, Blake JT, Krahenbuhl O, Tschopp J, Powers JC 1991. Human and murine cytotoxic T lymphocyte serine proteases: Subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry 30: 2217–2227 [DOI] [PubMed] [Google Scholar]
- Pereira NA, Song Z 2008. Some commonly used caspase substrates and inhibitors lack the specificity required to monitor individual caspase activity. Biochem Biophys Res Commun 377: 873–877 [DOI] [PubMed] [Google Scholar]
- Perry DK, Smyth MJ, Stennicke HR, Salvesen GS, Duriez P, Poirier GG, Hannun YA 1997. Zinc is a potent inhibitor of the apoptotic protease, caspase-3. A novel target for zinc in the inhibition of apoptosis. J Biol Chem 272: 18530–18533 [DOI] [PubMed] [Google Scholar]
- Petrassi HM, Williams JA, Li J, Tumanut C, Ek J, Nakai T, Masick B, Backes BJ, Harris JL 2005. A strategy to profile prime and non-prime proteolytic substrate specificity. Bioorg Med Chem Lett 15: 3162–3166 [DOI] [PubMed] [Google Scholar]
- Pop C, Salvesen GS 2009. Human caspases: Activation, specificity, and regulation. J Biol Chem 284: 21777–21781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poreba M, Drag M 2010. Current strategies for probing substrate specificity of proteases. Curr Med Chem 17: 3968–3995 [DOI] [PubMed] [Google Scholar]
- Powers JC, Asgian JL, Ekici OD, James KE 2002. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev 102: 4639–4750 [DOI] [PubMed] [Google Scholar]
- Puri AW, Broz P, Shen A, Monack DM, Bogyo M 2012. Caspase-1 activity is required to bypass macrophage apoptosis upon Salmonella infection. Nat Chem Biol 8: 745–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rano TA, Timkey T, Peterson EP, Rotonda J, Nicholson DW, Becker JW, Chapman KT, Thornberry NA 1997. A combinatorial approach for determining protease specificities: Application to interleukin-1β converting enzyme (ICE). Chem Biol 4: 149–155 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ 2008. MEROPS: The peptidase database. Nucleic Acids Res 36: D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm M, Dussmann H, Janicke RU, Tavare JM, Kogel D, Prehn JH 2002. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3. J Biol Chem 277: 24506–24514 [DOI] [PubMed] [Google Scholar]
- Rozman-Pungercar J, Kopitar-Jerala N, Bogyo M, Turk D, Vasiljeva O, Stefe I, Vandenabeele P, Bromme D, Puizdar V, Fonovic M, et al. 2003. Inhibition of papain-like cysteine proteases and legumain by caspase-specific inhibitors: When reaction mechanism is more important than specificity. Cell Death Differ 10: 881–888 [DOI] [PubMed] [Google Scholar]
- Sadaghiani AM, Verhelst SH, Bogyo M 2007. Tagging and detection strategies for activity-based proteomics. Curr Opin Chem Biol 11: 20–28 [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Dixit VM 1997. Caspases: Intracellular signaling by proteolysis. Cell 91: 443–446 [DOI] [PubMed] [Google Scholar]
- Schilling O, Overall CM 2008. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol 26: 685–694 [DOI] [PubMed] [Google Scholar]
- Schweizer A, Briand C, Grutter MG 2003. Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway. J Biol Chem 278: 42441–42447 [DOI] [PubMed] [Google Scholar]
- Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS 2005. XIAP inhibits caspase-3 and -7 using two binding sites: Evolutionarily conserved mechanism of IAPs. EMBO J 24: 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serim S, Haedke U, Verhelst SH 2012. Activity-based probes for the study of proteases: Recent advances and developments. ChemMedChem 7: 1146–1159 [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y 2003. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell 11: 519–527 [DOI] [PubMed] [Google Scholar]
- Siegmund B, Zeitz M 2003. Pralnacasan (vertex pharmaceuticals). IDrugs 6: 154–158 [PubMed] [Google Scholar]
- Sleath PR, Hendrickson RC, Kronheim SR, March CJ, Black RA 1990. Substrate specificity of the protease that processes human interleukin-1β. J Biol Chem 265: 14526–14528 [PubMed] [Google Scholar]
- Smolewski P, Bedner E, Du L, Hsieh TC, Wu JM, Phelps DJ, Darzynkiewicz Z 2001. Detection of caspases activation by fluorochrome-labeled inhibitors: Multiparameter analysis by laser scanning cytometry. Cytometry 44: 73–82 [DOI] [PubMed] [Google Scholar]
- Smolewski P, Grabarek J, Lee BW, Johnson GL, Darzynkiewicz Z 2002. Kinetics of HL-60 cell entry to apoptosis during treatment with TNF-α or camptothecin assayed by the stathmo-apoptosis method. Cytometry 47: 143–149 [DOI] [PubMed] [Google Scholar]
- Snipas SJ, Drag M, Stennicke HR, Salvesen GS 2008. Activation mechanism and substrate specificity of the Drosophila initiator caspase DRONC. Cell Death Differ 15: 938–945 [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, et al. 2001. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410: 112–116 [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS 1997. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem 272: 25719–25723 [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Renatus M, Meldal M, Salvesen GS 2000. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J 350 (Pt 2): 563–568 [PMC free article] [PubMed] [Google Scholar]
- Sun J, Bottomley SP, Kumar S, Bird PI 1997. Recombinant caspase-3 expressed in Pichia pastoris is fully activated and kinetically indistinguishable from the native enzyme. Biochem Biophys Res Commun 238: 920–924 [DOI] [PubMed] [Google Scholar]
- Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW 1997. Substrate specificities of caspase family proteases. J Biol Chem 272: 9677–9682 [DOI] [PubMed] [Google Scholar]
- Talanian RV, Brady KD, Cryns VL 2000. Caspases as targets for anti-inflammatory and anti-apoptotic drug discovery. J Med Chem 43: 3351–3371 [DOI] [PubMed] [Google Scholar]
- Tang Y, Wells JA, Arkin MR 2011. Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis. J Biol Chem 286: 34147–34154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodori L, Grabarek J, Smolewski P, Ghibelli L, Bergamaschi A, De Nicola M, Darzynkiewicz Z 2002. Exposure of cells to static magnetic field accelerates loss of integrity of plasma membrane during apoptosis. Cytometry 49: 113–118 [DOI] [PubMed] [Google Scholar]
- Thornberry NA 1994. Interleukin-1β converting enzyme. Methods Enzymol 244: 615–631 [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Molineaux SM 1995. Interleukin-1β converting enzyme: A novel cysteine protease required for IL-1β production and implicated in programmed cell death. Protein Sci 4: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. 1992. A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature 356: 768–774 [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. 1997. A combinatorial approach defines specificities of members of the caspase family and Granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem 272: 17907–17911 [DOI] [PubMed] [Google Scholar]
- Timmer JC, Salvesen GS 2007. Caspase substrates. Cell Death Differ 14: 66–72 [DOI] [PubMed] [Google Scholar]
- Timmer JC, Zhu W, Pop C, Regan T, Snipas SJ, Eroshkin AM, Riedl SJ, Salvesen GS 2009. Structural and kinetic determinants of protease substrates. Nat Struct Mol Biol 16: 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, McStay GP, Boucher LM, Mak T, Beere HM, Green DR 2006. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat Cell Biol 8: 72–77 [DOI] [PubMed] [Google Scholar]
- Valero A, Merino F, Wolbers F, Luttge R, Vermes I, Andersson H, van den Berg A 2005. Apoptotic cell death dynamics of HL60 cells studied using a microfluidic cell trap device. Lab Chip 5: 49–55 [DOI] [PubMed] [Google Scholar]
- Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K 2005. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods 2: 771–777 [DOI] [PubMed] [Google Scholar]
- Van Noorden CJ 2001. The history of Z-VAD-FMK, a tool for understanding the significance of caspase inhibition. Acta Histochem 103: 241–251 [DOI] [PubMed] [Google Scholar]
- Velazquez-Delgado EM, Hardy JA 2012. Zinc-mediated allosteric inhibition of caspase-6. J Biol Chem 287: 36000–36011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Peter Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inzé D, Van Breusegem F 2004. Type-II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem 279: 45329–45336 [DOI] [PubMed] [Google Scholar]
- Walters J, Pop C, Scott FL, Drag M, Swartz P, Mattos C, Salvesen GS, Clark AC 2009. A constitutively active and uninhibitable caspase-3 zymogen efficiently induces apoptosis. Biochem J 424: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannamaker W, Davies R, Namchuk M, Pollard J, Ford P, Ku G, Decker C, Charifson P, Weber P, Germann UA, et al. 2007. (S)-1-((S)-2-{[1-(4-amino-3-chloro-phenyl)-methanoyl]-amino}-3,3-dimethyl-butanoy l)-pyrrolidine-2-carboxylic acid ((2R,3S)-2-ethoxy-5-oxo-tetrahydro-furan-3-yl)-amide (VX-765), an orally available selective interleukin (IL)–converting enzyme/caspase-1 inhibitor, exhibits potent anti-inflammatory activities by inhibiting the release of IL-1β and IL-18. J Pharmacol Exp Ther 321: 509–516 [DOI] [PubMed] [Google Scholar]
- Wei Y, Fox T, Chambers SP, Sintchak J, Coll JT, Golec JM, Swenson L, Wilson KP, Charifson PS 2000. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol 7: 423–432 [DOI] [PubMed] [Google Scholar]
- Winssinger N, Damoiseaux R, Tully DC, Geierstanger BH, Burdick K, Harris JL 2004. PNA-encoded protease substrate microarrays. Chem Biol 11: 1351–1360 [DOI] [PubMed] [Google Scholar]
- Wu X, Simone J, Hewgill D, Siegel R, Lipsky PE, He L 2006. Measurement of two caspase activities simultaneously in living cells by a novel dual FRET fluorescent indicator probe. Cytometry A 69: 477–486 [DOI] [PubMed] [Google Scholar]
- Xu G, Shin SB, Jaffrey SR 2009. Global profiling of protease cleavage sites by chemoselective labeling of protein N-termini. Proc Natl Acad Sci 106: 19310–19315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin XM, Dong Z 2009. Essentials of apoptosis: A guide for basic and clinical research, 2nd ed. Humana Press, Totowa, NJ [Google Scholar]
- Zhang HZ, Kasibhatla S, Guastella J, Tseng B, Drewe J, Cai SX 2003. N-Ac-DEVD-N′-(Polyfluorobenzoyl)-R110: Novel cell-permeable fluorogenic caspase substrates for the detection of caspase activity and apoptosis. Bioconjug Chem 14: 458–463 [DOI] [PubMed] [Google Scholar]
- Zhou D, Chu W, Rothfuss J, Zeng C, Xu J, Jones L, Welch MJ, Mach RH 2006. Synthesis, radiolabeling, and in vivo evaluation of an 18F-labeled isatin analog for imaging caspase-3 activation in apoptosis. Bioorg Med Chem Lett 16: 5041–5046 [DOI] [PubMed] [Google Scholar]
- Zhou D, Chu W, Chen DL, Wang Q, Reichert DE, Rothfuss J, D’Avignon A, Welch MJ, Mach RH 2009. [18F]- and [11C]-labeled N-benzyl-isatin sulfonamide analogues as PET tracers for apoptosis: Synthesis, radiolabeling mechanism, and in vivo imaging study of apoptosis in Fas-treated mice using [11C]WC-98. Org Biomol Chem 7: 1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]