Abstract
Genome integrity is challenged by DNA damage from both endogenous and environmental sources. This damage must be repaired to allow both RNA and DNA polymerases to accurately read and duplicate the information in the genome. Multiple repair enzymes scan the DNA for problems, remove the offending damage, and restore the DNA duplex. These repair mechanisms are regulated by DNA damage response kinases including DNA-PKcs, ATM, and ATR that are activated at DNA lesions. These kinases improve the efficiency of DNA repair by phosphorylating repair proteins to modify their activities, by initiating a complex series of changes in the local chromatin structure near the damage site, and by altering the overall cellular environment to make it more conducive to repair. In this review, we focus on these three levels of regulation to illustrate how the DNA damage kinases promote efficient repair to maintain genome integrity and prevent disease.
During the DNA damage response, kinases (1) modify the activities of repair proteins, (2) induce changes to chromatin structure near the damage site, and (3) make the cellular environment conducive to repair.
The DNA in each of our cells accumulates thousands of lesions every day. This damaged DNA must be removed for the DNA code to be read properly. Fortunately, cells contain multiple DNA repair mechanisms including: base excision repair (BER) that removes damaged bases, mismatch repair (MMR) that recognizes base incorporation errors and base damage, nucleotide excision repair (NER) that removes bulky DNA adducts, and cross-link repair (ICL) that removes interstrand cross-links. In addition, breaks in the DNA backbone are repaired via double-strand break (DSB) repair pathways including homologous recombination (HR) and nonhomologous end joining (NHEJ). Some of these mechanisms can operate independently to repair simple lesions. However, the repair of more complex lesions involving multiple DNA processing steps is regulated by the DNA damage response (DDR). For the most difficult to repair lesions, the DDR can be essential for successful repair.
The DDR consists of multiple pathways, but for the purposes of this review we will focus on the DDR kinase signaling cascades controlled by the phosphatidylinositol 3-kinase-related kinases (PIKK). These kinases include DNA-dependent protein kinase (DNA-PKcs), ataxia telangiectasia-mutated (ATM), and ATM and Rad3-related (ATR). DNA-PKcs and ATM are primarily involved in DSB repair, whereas ATR responds to a wide range of DNA lesions, especially those associated with DNA replication (Cimprich and Cortez 2008). ATR’s versatility makes it essential for the viability of replicating cells in mice and humans (Brown and Baltimore 2000; de Klein et al. 2000; Cortez et al. 2001). In the case of ATM, inherited biallelic mutations cause ataxia-telangiectasia—a disorder characterized by neurodegeneration, immunodeficiency, and cancer (Shiloh 2003; Lavin 2008). ATM mutations are also frequently found in several types of tumors (Negrini et al. 2010).
The DDR kinases share several common regulatory mechanisms of activation (Lovejoy and Cortez 2009). All three DDR kinases sense damage through protein–protein interactions that serve to recruit the kinases to damage sites. Once localized, posttranslational modifications and other protein–protein interactions fully activate the kinases to initate a cascade of phosphorylation events. The best-studied substrate of DNA-PKcs is actually DNA-PKcs itself, and autophosphorylation is an important step in direct religation of the DSB via nonhomologous end joining (NHEJ) (Weterings and Chen 2007; Dobbs et al. 2010). ATM and ATR have both unique and shared substrates that participate in DNA repair, checkpoint signaling, and determining cell fate decisions such as apoptosis and sensescence.
THREE LEVELS OF REPAIR REGULATION BY THE DDR KINASES
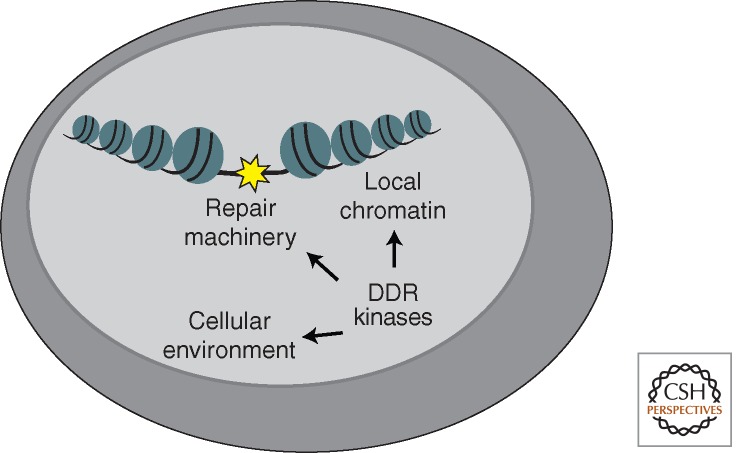
DDR kinases control DNA repair at three levels (Fig. 1). First, they regulate DNA repair enzymes directly through posttranslational modifications that alter their activity. These modifications appear to be especially important in the repair of complex lesions such as ICLs and repair associated with stalled replication forks. Second, the DDR kinases modify the chromatin near the DNA lesion to create a permissive local environment for repair. This chromatin response also provides a scaffolding function for the recruitment of additional DDR factors regulating both repair and signaling. Finally, the DDR kinases act at a more global level of the nucleus or even the entire cell to provide a cellular environment conducive to repair. This global response includes changes in transcription, the cell cycle, chromosome mobility, and deoxynucleotide (dNTP) levels. Controlling these processes may be most important for repair when damage is persistent.
Figure 1.
DDR kinases promote efficient DNA repair by directly regulating the DNA repair machinery, changing the local chromatin environment near the DNA lesion, and altering the cellular environment.
This review will highlight examples of each level of regulation. For the direct regulation of repair functions, we will discuss how DDR kinases regulate ICL repair and more general replication fork-associated repair. In discussing the local chromatin environment, we highlight the important role of chromatin modifications surrounding a DSB. Finally, at the global level, we discuss how the DDR alters nuclear architecture and maintains proper cellular dNTP pools to promote repair.
DDR KINASES DIRECTLY REGULATE THE REPLICATION-ASSOCIATED DNA REPAIR MACHINERY
DNA lesions pose an especially important problem when they interfere with DNA polymerases. Errors during DNA replication as well as mistakes in DNA repair cause mutations and chromosomal aberrations that are a source of genetic instability driving tumorigenesis. Additionally, many rare childhood diseases are the result of defects in replication-associated DNA repair. These include Seckel syndrome caused by mutations in ATR and other disorders caused by mutations in ATR substrates like BLM, WRN, and SMARCAL1 (Ciccia and Elledge 2010). Thus, the DNA damage response is particularly critical to ensure complete and accurate duplication of the genome.
ICL Repair during DNA Replication
Interstrand cross-links are perhaps the most difficult lesions to repair, requiring specialized repair mechanisms governed by genes mutated in patients with Fanconi anemia (FA), as well as components of nucleotide excision and DSB repair (Kim and D’Andrea 2012). In the context of DNA replication, interstrand cross-links are potent fork stalling lesions that activate ATR. Perhaps for these reasons, the ATR kinase has an especially critical function in initiating ICL repair (Fig. 2).
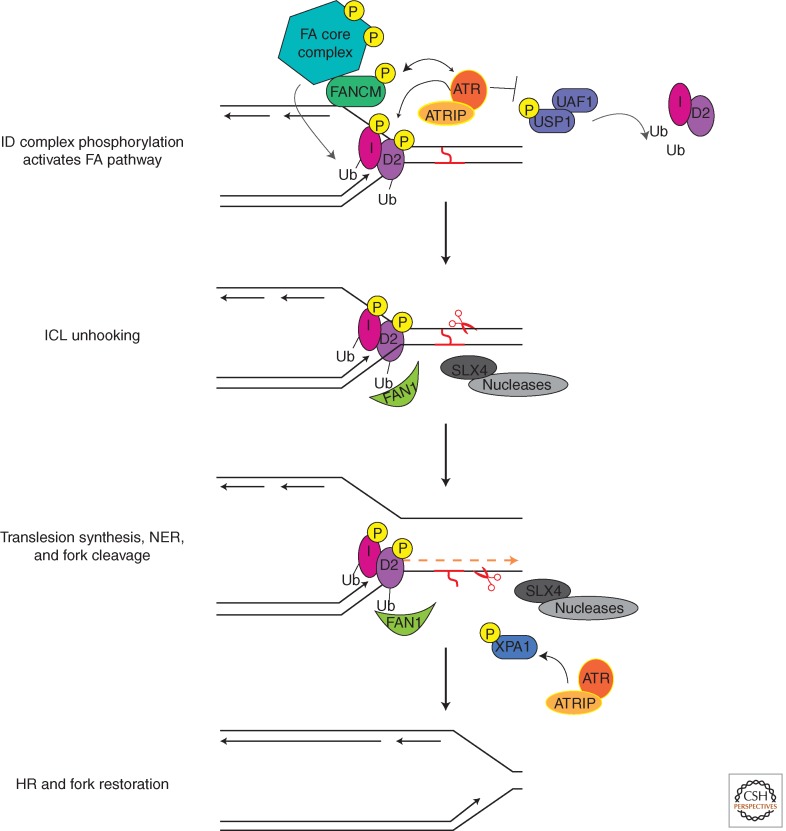
Figure 2.
A simplified model of ICL repair indicating steps regulated by ATR phosphorylation.
When the ICL stalls a replication fork, the DNA structure signals the recruitment of several Fanconi proteins beginning with the FANCM helicase (Meetei et al. 2005; Raschle et al. 2008; Knipscheer et al. 2009). FANCM may remodel the damaged fork to help recruit the FA core complex, a multisubunit ubiquitin ligase. An essential activity of the core complex is monoubiquitination of FANCD2 and FANCI within the FANCI-FANCD2 (ID) complex (Garcia-Higuera et al. 2001). Repair then initiates with synchronized incision on both sides of the cross-link. Incision may be mediated by the flap endonuclease FAN1 whose ubiquitin-binding motif recognizes mono-ub FANCD2 and is essential for ICL repair (Kratz et al. 2010; Liu et al. 2010; Smogorzewska et al. 2010). Additional nucleases such as those associated with SLX4 may also participate in ICL repair given that SLX4 mutations cause FA (Kim et al. 2011). Fork cleavage results in “unhooking” of the cross-link allowing error-prone polymerases to extend past the lesion and NER to remove the cross-linked base. The unhooking reaction also generates a DSB intermediate that is processed by HR to restore the replication fork (Long et al. 2011).
ATR controls the earliest events in the FA pathway and is essential for successful repair. Thus, ATR-deficiency yields high sensitivity to DNA cross-linking agents. ATR phosphorylates several FA proteins including FANCD2, FANCI, FANCA, FANCG, and FANCM (Andreassen et al. 2004; Ishiai et al. 2008; Wilson et al. 2008; Collins et al. 2009; Sobeck et al. 2009). The phosphorylation of FANCI is a particularly critical event for FA pathway activation, as it is needed for monoubiquitination and localization of FANCD2 to sites of damage. FANCI is phosphorylated on several conserved ATR and ATM consensus sites (Matsuoka et al. 2007), and mutants that cannot be phosphorylated prevent FANCD2 mono-ub and cause hypersensitivity to cross-linking reagents (Ishiai et al. 2008). Expression of FANCI mutants that mimic phosphorylation induce FANCD2 monoubiquitination even in the absence of exogenous DNA- damaging agents. These findings suggest that FANCI phosphorylation is a necessary and perhaps sufficient step for FANCD2 monoubiquitination and FA pathway activation. The mechanism by which phosphorylation induces ubiquitylation remains unknown. However, it should be noted that FANCI has WD40 repeats, which might act analogous to F-box proteins to recruit phosphorylated substrates for ubiquitination.
Analysis of the crystal structure of the FANC ID complex has revealed that the ubiquitination sites are buried in the ID interface (Joo et al. 2011). It is possible that ATR phosphorylation of ID in cis may inform ID of the presence of dsDNA and ssDNA junctions. A simple model would be that once phosphorylated at the cross-link, the ID complex alters its conformation allowing core complex recognition.
ATR may also regulate FANCD2 ubiquitylation by targeting the FANCD2 deubiquitination complex USP1-UAF1. Consistent with this notion, USP1 was identified as a putative ATM/ATR substrate (Matsuoka et al. 2007), and the interaction of USP1/UAF1 with FANCI is regulated by DNA damage (Yang et al. 2011). Furthermore, in response to DNA damage, USP1 undergoes inactivating autoproteolysis, further promoting FANC ID ubiquitination.
The activities of other FA proteins including FANCA and FANCG are also under the control of the ATR kinase. FANCA is a direct ATR substrate, and mutation of the phosphorylation site creates a protein that cannot fully complement FANCA-deficient cells (Collins et al. 2009). FANCG is phosphorylated on multiple sites and at least one (serine 7) is ATR-dependent (Wilson et al. 2008). Phosphorylation of FANCG regulates the interactions of BRCA2 with components of the core complex and FANCD2. FANCG S7 mutants fail to rescue the cross-link sensitivity of FANCG-deficient cells (Qiao et al. 2004).
In addition to controlling early events in cross-link repair, the ATR pathway may also regulate later steps. For example, ATR regulates the NER-dependent unhooking reaction pathway by regulating the localization of XPA (Wu et al. 2007; Shell et al. 2009). Also, ATR regulates the HR step by promoting the recruitment of the key RAD51 recombinase (Sorensen et al. 2005).
Thus, ATR regulates nearly every step of the ICL repair process. Why is this necessary? Perhaps the answer lies in the complexity of removing an ICL. ICL repair requires the coordinated activities of multiple repair steps often at a time of maximum vulnerability for the genome (when the replication fork reaches the cross-link). Perhaps ATR signaling provides a mechanism of ordering the repair steps to prevent undesirable DNA intermediates, which might yield aberrant repair products. In this context, it might be expected that the more difficult a DNA lesion is to repair, the more important the DDR pathways become for success.
DDR Kinase-Dependent Regulation of Replication Fork Repair Pathways
DDR regulation of ICL repair during DNA replication is a specialized version of a more general DDR response that coordinates repair of stalled forks. Base damage, dNTP depletion, and even difficult to replicate sequences that form secondary structures or RNA–DNA hybrids can cause fork damage. A stalled fork itself may not be a particularly devastating event to a cell because DNA replication will usually be completed from an adjacent origin of replication. In such cases, the DDR stabilizes the damaged fork to prevent aberrant DNA processing. In other cases, such as in replication of fragile sites that contain few replication origins, fork stabilization may be insufficient and DDR kinase-dependent restart of the stalled fork becomes essential (Casper et al. 2002).
The fork-stabilization activity of ATR is functionally defined either in terms of the ability to restart replication once a blockage is removed or by the changes in DNA or protein composition at the fork. Yeast mutants deficient in the ATR pathway lose the replicative polymerases from the fork (Cobb et al. 2003, 2005; Lucca et al. 2004) and accumulate abnormal DNA structures including long stretches of ssDNA and reversed fork structures resembling Holliday junctions (Lopes et al. 2001; Sogo et al. 2002). At least in yeast, the Exo1 nuclease is involved in generating the excess ssDNA at the stalled fork when the ATR pathway is inactivated (Cotta-Ramusino et al. 2005). Loss of ATR function in Xenopus extracts also causes loss of Pol epsilon and collapse of the fork into a DSB (Trenz et al. 2006).
Thus, one way ATR may stabilize a fork is by preventing dissociation of replisome proteins and thereby inhibiting aberrant enzymatic processing of the DNA. However, a recent paper by the Labib group has challenged this model (De Piccoli et al. 2012). This group monitored replisome stability in budding yeast lacking the Mec1ATR or Rad53Chk2 checkpoint kinases by immunoprecipitating a subunit of the replicative helicase and immunoblotting for other replisome proteins. In contrast to expectations, they did not observe disassembly of the replisome, and chromatin immunoprecipitation assays suggested that the replisome remained near origins in cells treated with high doses of hydroxyurea to stall forks. A subset of early origins lacked replisome proteins, but the authors concluded that this was as a result of replisome movement away from the earliest origins in the absence of DDR kinase activity instead of replisome disassembly. Thus, in this case, the ATR pathway may be important for restraining fork movement. If fork movement is not accompanied by productive leading and lagging strand synthesis, it could help generate the ssDNA gaps observed by electron microscopy in Mec1ATR-deficient yeast.
Exactly how ATR prevents replisome dissociation, movement, and aberrant fork processing is one of the least understood parts of the DDR. One DDR target is the downstream kinase CHK1, which is activated by ATR phosphorylation and needed to prevent fork collapse and regulate origin firing (Cimprich and Cortez 2008). Note that the mammalian and yeast functions of CHK1 and CHK2 have been reversed during evolution so that human CHK1 is the functional equivalent of yeast Rad53 with respect to replication fork regulation. ATR also directly phosphorylates replisome components including several Cdc45-MCM-GINS (CMG) helicase subunits (Cortez et al. 2004; Yoo et al. 2004; Matsuoka et al. 2007; Shi et al. 2007; Trenz et al. 2008; De Piccoli et al. 2012). Phosphorylation of CMG may regulate helicase activity to prevent excessive unwinding and is important to promote rescue of stalled forks from adjacent origins.
In addition, other replication fork proteins including RPA, CLASPIN, and members of the replication fork-pausing complex like TIMELESS, TIPIN, and AND1 are ATR substrates (Matsuoka et al. 2007). Deficiencies in these proteins cause hypersensitivity to replication stress agents (Chou and Elledge 2006; Errico et al. 2007; Unsal-Kacmaz et al. 2007; Yoshizawa-Sugata and Masai 2007, 2009; Leman et al. 2010). They act in part through promoting ATR-dependent CHK1 activation but may have additional roles in regulating the repair of damaged forks.
The DDR also targets several repair enzymes that remodel damaged forks including WRN, FANCM, and SMARCAL1. The WRN and FANCM proteins are helicases capable of unwinding a variety of complex DNA structures. SMARCAL1 is an SNF2 family ATPase that is activated by complex DNA structures and uses the energy of ATP hydrolysis to reanneal DNA strands (Yusufzai and Kadonaga 2008). All three enzymes are recruited to damaged forks and can catalyze fork regression generating a Holliday junction on model replication substrates (Machwe et al. 2006; Gari et al. 2008a; Betous et al. 2012; Ciccia et al. 2012). They can also branch migrate the Holliday junction, which could restore the normal fork structure (Gari et al. 2008b; Machwe et al. 2011; Betous et al. 2012). All three are targets of ATR phosphorylation (Yannone et al. 2001; Karmakar et al. 2002; Pichierri et al. 2003; Meetei et al. 2005; Bansbach et al. 2009; Sobeck et al. 2009; Ammazzalorso et al. 2010), and deficiencies in WRN and SMARCAL1 activity lead to MUS81-dependent fork cleavage and DSB formation (Franchitto et al. 2008; Betous et al. 2012).
ATR phosphorylation of WRN and FANCM promotes their recruitment to stalled forks (Sobeck et al. 2009; Ammazzalorso et al. 2010), and cells expressing a nonphosphorylatable mutant WRN show increased fork breakage (Ammazzalorso et al. 2010). SMARCAL1 phosphorylation by DDR kinases does not regulate its localization but does regulate its enzymatic activity (D Cortez, unpubl.). The exact substrates of these fork remodeling enzymes at stalled forks and how their activities promote fork restart in cells is not yet known. Additionally, many other helicases and DNA translocases including BLM have roles at damaged forks and are regulated by ATR phosphorylation (Davalos et al. 2004; Li et al. 2004; Sengupta et al. 2004; Rao et al. 2005; Tripathi et al. 2008).
Clearly, a great deal remains to be learned about how ATR promotes replication fork stability, replication-associated DNA repair, and fork restart. These are likely the most important functions of ATR in maintaining genome stability and cell viability based on results from separation of function mutants in both yeast and human ATR (Paciotti et al. 2001; Cobb et al. 2005; Nam et al. 2011). Yet, they are also arguably the least understood. The development of new reversible ATR inhibitors (Charrier et al. 2011; Reaper et al. 2011; Toledo et al. 2011), as well as new techniques to study DNA replication such as iPOND should accelerate the mechanistic studies (Sirbu et al. 2011, 2012). Such studies will be equally critical in defining the pathways that lead to the elevated levels of replication stress observed in cancer cells (Halazonetis et al. 2008). Combined with defects in other genome-maintenance activities, this stress creates an increased dependency on ATR for successful cell division. Thus, the ATR pathway is a promising target for new cancer drug development. Defining how ATR inhibition alters replication-associated DNA repair will be important for understanding the mechanism of action of these drugs.
DDR REGULATES LOCAL CHROMATIN STRUCTURE TO PROMOTE REPAIR
DNA lesions occur in various chromosomal contexts including compacted and opened chromatin, which influences both the activation of the DDR and DNA repair efficiency. For example, in highly condensed chromatin, repairing the damaged structure is more difficult presumably because repair proteins are physically occluded from accessing the damaged structure. Independently of DDR kinases, an ATP-dependent mechanism induces rapid chromatin relaxation around a DSB, and is required for recruitment of break-sensing proteins (Kruhlak et al. 2006). However, several DDR kinase-dependent local chromatin changes also promote a local environment conducive for repair. These activities include creation of a chromatin platform for recruitment of repair and signaling factors, regulating repair factor accessibility to the DNA, and inhibition of nearby transcription to prevent potential interference with DNA repair (Fig. 3). Here we discuss the DDR-dependent chromatin response as it relates to DSB repair.
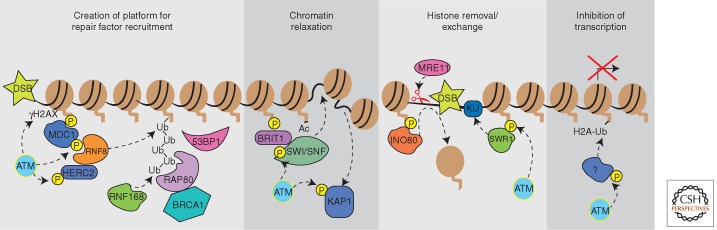
Figure 3.
DDR kinases regulate the chromatin near a double-strand break to provide a scaffold for the recruitment of DNA repair proteins, promote repair protein access through nucleosome remodeling, and inhibit local transcription.
γH2AX as a Platform for DSB Repair
One of the earliest consequences of ATM activation at a DSB is phosphorylation of the histone variant H2AX on an evolutionarily conserved serine (S139) producing γH2AX (Fernandez-Capetillo et al. 2004; Stucki and Jackson 2006; Dickey et al. 2009). A complex of MRN, MDC1, and γH2AX recruits additional ATM to flanking regions of chromatin and facilitates propagation of γH2AX to a large chromatin domain.
γH2AX-MDC1 is a platform for the recruitment of many additional chromatin modifying, DDR signaling, and DNA repair proteins. This scaffold recruits the RING ubiquitin ligases RNF8 and RNF168 to trigger a ubiquitylation cascade surrounding the DSB (Al-Hakim et al. 2010). This recruitment is mediated by ATM-dependent phosphorylation sites on MDC1, which are recognized by the FHA domain of RNF8. Along with the E2 enzyme UBC13, RNF8 catalyzes the formation of Lys63-linked polyubiquitin chains at DSBs (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007). Subsequently, RNF168, the protein encoded by the RIDDLIN syndrome gene recognizes and amplifies these ubiquitin chains (Doil et al. 2009; Stewart et al. 2009), whereas another ring finger protein RNF169 antagonizes the ubiquitin cascade (Chen et al. 2012; Poulsen et al. 2012). Another ATM substrate, HERC2, also regulates this process. HERC2 contains an ATM phosphorylation site that binds the RNF8 FHA domain and helps assemble the functional RNF8-UBC13 enzyme (Bekker-Jensen et al. 2010).
Ubiquitylation at the DSB regulates the recruitment of the DSB repair proteins BRCA1 and 53BP1 (Al-Hakim et al. 2010). BRCA1 is itself a ubiquitin ligase and is regulated by ATM and ATR-dependent phosphorylation (Cortez et al. 1999; Tibbetts et al. 2000). BRCA1 is recruited via an interaction with a complex of proteins containing the K63-linked ubiquitin binding protein Rap80 (Kim et al. 2007; Sobhian et al. 2007; Wang et al. 2007; Yan et al. 2007). Three distinct BRCA1 repair complexes (BRCA1-A, BRCA1-B, and BRCA1-C) are recruited, which contain different accessory proteins to regulate checkpoint activation or HR repair (Greenberg et al. 2006). 53BP1 accumulation near the DSB is also dependent on these ubiquitylation events although the mechanism is likely indirect. The overall effect of BRCA1 and 53BP1 recruitment downstream of histone phosphorylation and ubiquitylation is likely regulation of repair choice between NHEJ and HR.
In addition to recruiting repair factors to a DSB, DDR-dependent H2AX phosphorylation also induces changes to chromatin structure by recruiting ATP-dependent chromatin remodeling complexes including SWI/SNF, SWR1, and INO80. The SWI/SNF chromatin remodeling activity is targeted to DSBs through interactions with acetylated H3 (Lee et al. 2010) and BRIT1/MCPH1, a protein that binds γH2AX after damage (Wood et al. 2007; Peng et al. 2009). ATM and ATR phosphorylate a SWI/SNF subunit leading to an increased association with BRIT1 and DSBs (Peng et al. 2009). SWI/SNF presumably relaxes chromatin near the break to improve access of DNA repair enzymes to the damaged DNA.
The INO80 and SWR1 complexes are recruited to damage sites through direct interaction with γH2AX. At least in yeast, these complexes promote repair through two distinct mechanisms. INO80 catalyzes histone removal that facilitates Mre11 binding and DNA end resection to promote HR repair, whereas SWR1 promotes KU binding and NHEJ (van Attikum et al. 2007; van Attikum and Gasser 2009). INO80-dependent remodeling may also be important to promote the strand invasion step of HR through displacement of histones at the homologous donor sequences (Tsukuda et al. 2009).
H2AX-Independent but DDR Kinase-Dependent Regulation of Local Chromatin
Besides γH2AX-dependent regulation of repair, ATM controls other chromatin modifications to allow access for repair factors. H2B is monoubiquitylated near DSBs (Moyal et al. 2011). H2B-Ub is catalyzed by an RNF20-RNF40 heterodimer (the human ortholog of yeast Bre1), and this modification is typically associated with actively transcribed genes (Zhu et al. 2005). The levels of H2B-Ub increase near a DSB owing to recruitment of the RNF20-RNF40 proteins through a mechanism that may involve their interaction with ATM and NBS1 (Moyal et al. 2011). Both RNF20 and RNF40 are ATM substrates, and increased H2B-Ub surrounding the break is dependent on RNF20 phosphorylation. Both NHEJ and HR repair are impaired in cells when the damage-induced H2B-Ub is prevented (Moyal et al. 2011). The HR defect was traced to a defect in DNA end resection and could be rescued by experimentally inducing chromatin relaxation. Reduced NHEJ is associated with less XRCC4 and KU80 at the break in the absence of H2B-Ub.
In addition to modulating H2B-Ub, a second mechanism by which ATM relaxes chromatin to promote repair is through phosphorylation of KAP1 (Ziv et al. 2006). KAP1 is a transcriptional corepressor that works with histone methyltransferase and histone deacetylase complexes to promote chromatin compaction. ATM-dependent KAP1 phosphorylation disrupts an interaction between KAP1 and the CHD3 nucleosome remodeler thereby promoting chromatin relaxation (Goodarzi et al. 2011). As a result, ATM is particularly important for repair of DSBs that occur in heterochromatin (Goodarzi et al. 2008a; Noon et al. 2010).
In addition to the examples of local chromatin changes described here, there are changes in other histone modifications regulated by DDR kinases such as an ATM-dependent increase in H2A-Ub that inhibits transcription near DSBs (Shanbhag et al. 2010). There are also changes in the binding of chromatin proteins and the abundance of histone variants. Understanding how the DDR kinases regulate the local chromatin environment to promote repair of other types of DNA lesions, such as those encountered by elongating replication forks, will also be important. Some of the mechanisms may be similar. For example, γH2AX spreads away from stalled forks similarly to the spreading observed at DSBs (Sirbu et al. 2011). However, other mechanisms may be unique, adding to the complexity of the chromatin response to DNA damage.
DDR KINASES FACILITATE REPAIR BY CREATING AN OPTIMAL CELLULAR ENVIRONMENT
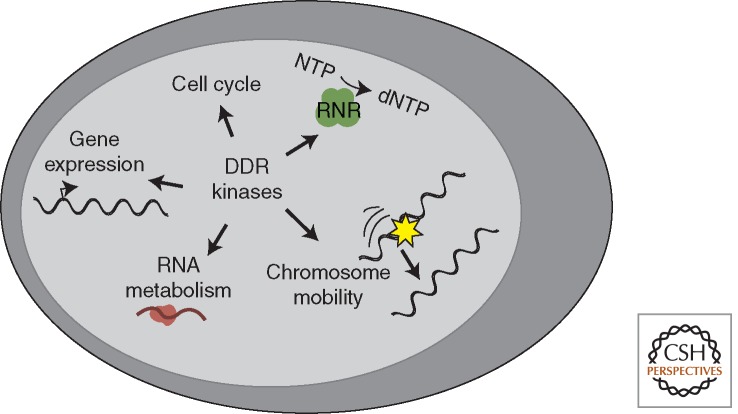
In addition to promoting DNA repair through direct regulation of repair proteins and changes in the chromatin near the DNA damage site, the DDR also facilitates repair through more global changes in the cellular environment (Fig. 4). The most obvious example of this mechanism is the checkpoint activity of the DDR kinases, which halts the cell cycle providing time to repair the DNA damage before DNA replication or mitosis. Checkpoint-dependent changes in cyclin-dependent kinase (CDK) activities also influence DNA repair more directly because many repair proteins are CDK substrates. A second example is the numerous DDR kinase-dependent changes in gene expression that are largely mediated through regulation of p53. In addition to inducing cell cycle arrest and apoptosis, these transcriptional changes can alter the levels of DNA repair proteins, as well as the nucleotides and histones needed for completing repair synthesis and restoring chromatin.
Figure 4.
DDR kinases regulate several aspects of nuclear and cellular physiology to provide an environment conducive for successful DNA repair.
Furthermore, results from functional genomic screens suggest a much broader regulation of cellular physiology by the DDR. For example, proteomic screens for ATM and ATR substrates and genetic screens for new DDR factors based on the level of ATM/ATR activity in undamaged cells identified proteins involved in a wide variety of cellular functions including intracellular protein trafficking, cellular immunity, and RNA metabolism (Matsuoka et al. 2007; Lovejoy et al. 2009; Paulsen et al. 2009; Bansbach and Cortez 2011). In many cases, the connection between these processes and the DDR kinases is likely to promote a cellular environment conducive to DNA repair.
Nuclear Organization and Chromosome Movements Facilitate DNA Repair
One of the important DDR kinase-dependent changes important for repair is regulation of nuclear organization. The nucleus is a highly organized organelle with compartments devoted to specific functions. A long-standing question is whether DNA repair occurs equally well anywhere within the nucleus or whether there are specific repair centers (Misteli and Soutoglou 2009). Recent studies on DSB repair in yeast suggest that repair centers exist and indicate that DDR-dependent changes in chromosome mobility promote HR repair.
Observations of DSBs marked with fluorescent proteins revealed that unrepairable DSBs move to the nuclear periphery and cells with two DSBs merge them into a single repair focus (Nagai et al. 2008; Oza et al. 2009). More recently, the Rothstein and Gasser groups have shown increased chromosomal mobility within the yeast nucleus because of a DSB (Dion et al. 2012; Mine-Hattab and Rothstein 2012). The increased movement depends on the Mec1ATR kinase, resection of the DNA end, and the RAD51 recombinase. Intriguingly, the Rothstein study also showed that the dynamics of unbroken, nonhomologous chromosomes is also increased in the presence of a DSB, suggesting that DDR kinases regulate global nuclear architecture (Mine-Hattab and Rothstein 2012).
The end-result of the increased chromosome mobility is an increase in repair efficiency. Likely this results from an increase in the ability of the RAD51-coated DNA end to find a homologous sequence. Flexibility of the RAD51-coated DNA fiber is important for an efficient homology search (Forget and Kowalczykowski 2012). It is also possible that the movement to or away from a specific nuclear location promotes repair. For example, movement out of a region containing heterochromatin or the nucleolus might increase repair efficiency.
Whether similar changes in chromosome dynamics occur in higher eukaryotes is less clear. Several studies indicate that most DNA ends are largely immobile in mammalian cells (Nelms et al. 1998; Kruhlak et al. 2006; Soutoglou et al. 2007; Jakob et al. 2009). However, deprotected telomere ends have increased mobility compared with protected telomeres (Dimitrova et al. 2008). This increased mobility depends on both ATM and 53BP1 and these ends are repaired through NHEJ. ATM and 53BP1 also control antigen receptor diversification, and chromosome movement may be needed especially in the context of long-range joining during class switch recombination (Nussenzweig and Nussenzweig 2010). DSBs induced by α particles are also mobile (Aten et al. 2004). Furthermore, breaks in heterochromatin in Drosophila cells cause an expansion of the heterochromatin domain followed by movement of the repair focus outside of the heterochromatin (Chiolo et al. 2011). These changes in heterochromatin are dependent on the DDR kinases and seem to be important after the resection step but before the RAD51-dependent homology search for HR repair (Chiolo et al. 2011). Thus, at least in some circumstances the increased mobility of broken chromosomes within the nucleus does occur in metazoan cells.
The mechanism by which the DDR promotes increased chromosome mobility is not known. One clue might be found in the recent observation that DNA attachments to the nuclear pore are regulated by the DDR (Bermejo et al. 2011). In this yeast study, the authors found that DDR kinase modification of nucleoporins releases the interaction between tethered chromosomes and the pore. Another possible mechanism could involve phosphorylation of KAP1, which is observed throughout the nucleus. KAP1 binds the heterochromatin protein HP1 and as mentioned earlier, KAP1 phosphorylation is important for the repair of breaks in heterochromatin (Goodarzi et al. 2008b). Finally, DSB recruitment of chromatin remodeling factors such as INO80 and histone variants such as H2A.Z may be important to promote the increase in mobility (Kalocsay et al. 2009; Neumann et al. 2012). Discovering the mechanisms by which the DDR kinases regulate chromosome dynamics will provide important information about nuclear architecture and how chromosomal domains are maintained. In addition, these studies have significant implications for the mechanisms driving chromosomal translocations and rearrangements that cause cancer.
Control of Cellular Nucleotide Levels for DNA Repair
Perhaps the best-documented example of how the DDR kinases create a cellular environment conducive for repair is through the regulation of nucleotide metabolism. In yeast, the intracellular concentration of dNTPs increases in response to DNA damage, whereas in mammalian cells increased production may be more localized (Chabes et al. 2003; Hakansson et al. 2006b). Higher concentrations of dNTPs cause an increase in mutation frequency (Chabes et al. 2003). Not surprisingly, maintaining an optimal balance of cellular dNTPs is a process strictly controlled at multiple levels by the DDR kinases.
The rate-limiting step in dNTP production is catalyzed by ribonucleotide reductase (RNR) (Nordlund and Reichard 2006). RNR contains two subunits, R1 and R2, encoded by multiple genes in most organisms. DDR kinases regulate RNR at almost every conceivable level. The transcriptional regulation of RNR subunits was one of the first documented functions of the DDR (Elledge et al. 1993). In human cells, a DDR kinase- and p53-dependent pathway induces expression of the catalytic RNR subunit p53R2 after prolonged exposure to DNA damage (Tanaka et al. 2000).
In addition to RNR gene expression, the DDR kinases directly regulate the stability of RNR subunits. For example, ATM phosphorylation of p53R2 increases its stability (Chang et al. 2008). Furthermore, ATR signaling inhibits Cyclin F-dependent R2 degradation, which may be a rapid way of increasing functional RNR enzyme levels (D’Angiolella et al. 2012).
The ATR pathway also controls the localization of the RNR subunits. In yeast, one of the RNR subunits is exported to the cytoplasm after damage to form an active RNR enzyme (Yao et al. 2003). In mammalian cells, RNR subunits may actually be recruited directly to sites of DNA damage to ensure dNTP production right where it is most needed (Niida et al. 2010).
Finally, in budding and fission yeast, small protein inhibitors of RNR including Dif1, Sml1, and Spd1 are regulated by DDR kinases. Dif1 and Spd1 control the localization of RNR subunits by regulating nuclear import (Liu et al. 2003; Lee et al. 2008) whereas Sml1 and Spd1 are direct inhibitors of RNR activity (Zhao et al. 1998; Hakansson et al. 2006a). The proteolysis of all three of these proteins is under control of the DDR pathway (Zhao et al. 2001; Liu et al. 2003; Lee et al. 2008; Wu and Huang 2008).
Thus, the DDR kinases control the timely and appropriate production of dNTPs for DNA repair through transcriptional, posttranscriptional, and localization mechanisms targeting RNR. The importance of this pathway to create an optimal cellular environment for repair and replication is illustrated by the observation that, in budding yeast, the lethality associated with deleting Mec1ATR can be rescued by increasing RNR activity (Desany et al. 1998; Zhao et al. 1998). Whether ATR regulation of RNR function is equally important in human cells is unknown.
CONCLUDING REMARKS
The basic DNA repair machinery is often sufficient to reconstitute simple repair reactions in vitro on naked DNA substrates. However, efficient repair often requires regulation by the DNA damage response. The DDR kinases directly modify repair proteins, change chromatin structure around the DNA lesion, and regulate nuclear and cellular environments. Failures at any of these levels cause genome instability and disease. Not surprisingly, the list of DDR kinase substrates is long and our understanding of their regulation is incomplete. Fortunately, new tools for discovery in multiple systems promise to rapidly move us toward an intimate understanding of mechanism. This knowledge may help in the design of cancer therapeutic opportunities based on manipulation of the DNA damage response, epigenetic therapies, and combinations with existing radiation and chemotherapies that work primarily by damaging DNA.
ACKNOWLEDGMENTS
Research in the Cortez laboratory on the DNA damage response and DNA repair is supported by NIH grants R01CA102729 and R01CA 136933. B.M.S. is funded by a Department of Defense Breast Cancer Research Program predoctoral fellowship (W81XWH-10-1-0226), and we thank Swim Across America for their support.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Al-Hakim A, Escribano-Diaz C, Landry MC, O’Donnell L, Panier S, Szilard RK, Durocher D 2010. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair 9: 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P 2010. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J 29: 3156–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, D’Andrea AD, Taniguchi T 2004. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev 18: 1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303: 92–95 [DOI] [PubMed] [Google Scholar]
- Bansbach CE, Cortez D 2011. Defining genome maintenance pathways using functional genomic approaches. Crit Rev Biochem Mol Biol 46: 327–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D 2009. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev 23: 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N 2010. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12: 80–86 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B, et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev 26: 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14: 397–402 [PMC free article] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW 2002. ATR regulates fragile site stability. Cell 111: 779–789 [DOI] [PubMed] [Google Scholar]
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391–401 [DOI] [PubMed] [Google Scholar]
- Chang L, Zhou B, Hu S, Guo R, Liu X, Jones SN, Yen Y 2008. ATM-mediated serine 72 phosphorylation stabilizes ribonucleotide reductase small subunit p53R2 protein against MDM2 to DNA damage. Proc Natl Acad Sci 105: 18519–18524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier JD, Durrant SJ, Golec JM, Kay DP, Knegtel RM, Maccormick S, Mortimore M, O'Donnell ME, Pinder JL, Reaper PM, et al. 2011. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem 54: 2320–2330 [DOI] [PubMed] [Google Scholar]
- Chen J, Feng W, Jiang J, Deng Y, Huen MS 2012. Ring finger protein RNF169 antagonises the ubiquitin-dependent signaling cascade at sites of DNA damage. J Biol Chem 287: 27715–27722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ 2006. Tipin and timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci 103: 18143–18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ 2010. The DNA damage response: Making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. 2012. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell 47: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM 2005. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NB, Wilson JB, Bush T, Thomashevski A, Roberts KJ, Jones NJ, Kupfer GM 2009. ATR-dependent phosphorylation of FANCA on serine 1449 after DNA damage is important for FA pathway function. Blood 113: 2181–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ 2001. ATR and ATRIP: Partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Cortez D, Glick G, Elledge SJ 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci 101: 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M 2005. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M 2012. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 149: 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos AR, Kaminker P, Hansen RK, Campisi J 2004. ATR and ATM-dependent movement of BLM helicase during replication stress ensures optimal ATM activation and 53BP1 focus formation. Cell Cycle 3: 1579–1586 [DOI] [PubMed] [Google Scholar]
- de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH 2000. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol 10: 479–482 [DOI] [PubMed] [Google Scholar]
- De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K 2012. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell 45: 696–704 [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev 12: 2956–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey JS, Redon CE, Nakamura AJ, Baird BJ, Sedelnikova OA, Bonner WM 2009. H2AX: Functional roles and potential applications. Chromosoma 118: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol 14: 502–509 [DOI] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, Lees-Miller SP 2010. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair 9: 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA 1993. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 15: 333–339 [DOI] [PubMed] [Google Scholar]
- Errico A, Costanzo V, Hunt T 2007. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc Natl Acad Sci 104: 14929–14934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A 2004. H2AX: The histone guardian of the genome. DNA Repair (Amst) 3: 959–967 [DOI] [PubMed] [Google Scholar]
- Forget AL, Kowalczykowski SC 2012. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature 482: 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P 2008. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol 183: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7: 249–262 [DOI] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A 2008a. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci 105: 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A 2008b. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29: 141–148 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA 2008a. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA 2008b. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA 2011. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol 18: 831–839 [DOI] [PubMed] [Google Scholar]
- Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM 2006. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev 20: 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson P, Dahl L, Chilkova O, Domkin V, Thelander L 2006a. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribonucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J Biol Chem 281: 1778–1783 [DOI] [PubMed] [Google Scholar]
- Hakansson P, Hofer A, Thelander L 2006b. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem 281: 7834–7841 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J 2008. An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. 2008. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol 15: 1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Taucher-Scholz G 2009. Positional stability of damaged chromatin domains along radiation tracks in mammalian cells. Radiat Res 171: 405–418 [DOI] [PubMed] [Google Scholar]
- Joo W, Xu G, Persky NS, Smogorzewska A, Rudge DG, Buzovetsky O, Elledge SJ, Pavletich NP 2011. Structure of the FANCI-FANCD2 complex: Insights into the Fanconi anemia DNA repair pathway. Science 333: 312–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33: 335–343 [DOI] [PubMed] [Google Scholar]
- Karmakar P, Piotrowski J, Brosh RM Jr, Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA 2002. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J Biol Chem 277: 18291–18302 [DOI] [PubMed] [Google Scholar]
- Kim H, D’Andrea AD 2012. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 26: 1393–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A 2011. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 43: 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142: 77–88 [DOI] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol 172: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF 2008. Ataxia-telangiectasia: From a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 9: 759–769 [DOI] [PubMed] [Google Scholar]
- Lee YD, Wang J, Stubbe J, Elledge SJ 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell 32: 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J 2010. A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J 29: 1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman AR, Noguchi C, Lee CY, Noguchi E 2010. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci 123: 660–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kim SM, Lee J, Dunphy WG 2004. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J Cell Biol 165: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 17: 1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J 2010. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329: 693–696 [DOI] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC 2011. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Cortez D 2009. Common mechanisms of PIKK regulation. DNA Repair (Amst) 8: 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Xu X, Bansbach CE, Glick GG, Zhao R, Ye F, Sirbu BM, Titus LC, Shyr Y, Cortez D 2009. Functional genomic screens identify CINP as a genome maintenance protein. Proc Natl Acad Sci 106: 19304–19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucca C, Vanoli F, Cotta-Ramusino C, Pellicioli A, Liberi G, Haber J, Foiani M 2004. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23: 1206–1213 [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Orren DK 2006. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45: 13939–13946 [DOI] [PubMed] [Google Scholar]
- Machwe A, Karale R, Xu X, Liu Y, Orren DK 2011. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) Holliday junctions to functional replication forks. Biochemistry 50: 6774–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 37: 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine-Hattab J, Rothstein R 2012. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol 14: 510–517 [DOI] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E 2009. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, et al. 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell 41: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam EA, Zhao R, Cortez D 2011. Analysis of mutations that dissociate G2 and essential S phase functions of human ataxia telangiectasia-mutated and Rad3-related (ATR) protein kinase. J Biol Chem 286: 37320–37327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD 2010. Genomic instability—An evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228 [DOI] [PubMed] [Google Scholar]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280: 590–592 [DOI] [PubMed] [Google Scholar]
- Neumann FR, Dion V, Gehlen LR, Tsai-Pflugfelder M, Schmid R, Taddei A, Gasser SM 2012. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev 26: 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Katsuno Y, Sengoku M, Shimada M, Yukawa M, Ikura M, Ikura T, Kohno K, Shima H, Suzuki H, et al. 2010. Essential role of Tip60-dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev 24: 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA 2010. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12: 177–184 [DOI] [PubMed] [Google Scholar]
- Nordlund P, Reichard P 2006. Ribonucleotide reductases. Annu Rev Biochem 75: 681–706 [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC 2010. Origin of chromosomal translocations in lymphoid cancer. Cell 141: 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP 2001. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol 21: 3913–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. 2009. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, Hu R, Li K, Lin SY 2009. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol 11: 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F, Franchitto A 2003. Werner’s syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene 22: 1491–1500 [DOI] [PubMed] [Google Scholar]
- Poulsen M, Lukas C, Lukas J, Bekker-Jensen S, Mailand N 2012. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J Cell Biol 197: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Mi J, Wilson JB, Zhi G, Bucheimer NR, Jones NJ, Kupfer GM 2004. Phosphorylation of fanconi anemia (FA) complementation group G protein, FANCG, at serine 7 is important for function of the FA pathway. J Biol Chem 279: 46035–46045 [DOI] [PubMed] [Google Scholar]
- Rao VA, Fan AM, Meng L, Doe CF, North PS, Hickson ID, Pommier Y 2005. Phosphorylation of BLM, dissociation from topoisomerase IIIα, and colocalization with γ-H2AX after topoisomerase I-induced replication damage. Mol Cell Biol 25: 8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134: 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR 2011. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 7: 428–430 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, et al. 2004. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J Cell Biol 166: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell SM, Li Z, Shkriabai N, Kvaratskhelia M, Brosey C, Serrano MA, Chazin WJ, Musich PR, Zou Y 2009. Checkpoint kinase ATR promotes nucleotide excision repair of UV-induced DNA damage via physical interaction with xeroderma pigmentosum group A. J Biol Chem 284: 24213–24222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Dodson GE, Mukhopadhyay PS, Shanware NP, Trinh AT, Tibbetts RS 2007. Identification of carboxyl-terminal MCM3 phosphorylation sites using polyreactive phosphospecific antibodies. J Biol Chem 282: 9236–9243 [DOI] [PubMed] [Google Scholar]
- Shiloh Y 2003. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer 3: 155–168 [DOI] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25: 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Cortez D 2012. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc 7: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, et al. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39: 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobeck A, Stone S, Landais I, de Graaf B, Hoatlin ME 2009. The Fanconi anemia protein FANCM is controlled by FANCD2 and the ATR/ATM pathways. J Biol Chem 284: 25560–25568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7: 195–201 [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T 2007. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9: 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP 2006. γH2AX and MDC1: Anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 5: 534–543 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y 2000. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404: 42–49 [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT 2000. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev 14: 2989–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O 2011. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 18: 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V 2006. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenz K, Errico A, Costanzo V 2008. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J 27: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Kaur S, Sengupta S 2008. Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis 29: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Lo YC, Krishna S, Sterk R, Osley MA, Nickoloff JA 2009. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair (Amst) 8: 360–369 [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK 2007. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol 27: 3131–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19: 207–217 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 26: 4113–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Chen DJ 2007. DNA-dependent protein kinase in nonhomologous end joining: A lock with multiple keys? J Cell Biol 179: 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JB, Yamamoto K, Marriott AS, Hussain S, Sung P, Hoatlin ME, Mathew CG, Takata M, Thompson LH, Kupfer GM, et al. 2008. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene 27: 3641–3652 [DOI] [PubMed] [Google Scholar]
- Wood JL, Singh N, Mer G, Chen J 2007. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J Biol Chem 282: 35416–35423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Huang M 2008. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol 28: 7156–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Shell SM, Liu Y, Zou Y 2007. ATR-dependent checkpoint modulates XPA nuclear import in response to UV irradiation. Oncogene 26: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM 2007. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res 67: 6647–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Moldovan GL, Vinciguerra P, Murai J, Takeda S, D’Andrea AD 2011. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev 25: 1847–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ 2001. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem 276: 38242–38248 [DOI] [PubMed] [Google Scholar]
- Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci 100: 6628–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Shevchenko A, Dunphy WG 2004. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J Biol Chem 279: 53353–53364 [DOI] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N, Masai H 2007. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem 282: 2729–2740 [DOI] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N, Masai H 2009. Roles of human AND-1 in chromosome transactions in S phase. J Biol Chem 284: 20718–20728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT 2008. HARP is an ATP-driven annealing helicase. Science 322: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20: 3544–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D 2005. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol Cell 20: 601–611 [DOI] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8: 870–876 [DOI] [PubMed] [Google Scholar]