Abstract
Position-effect variegation (PEV) results when a gene normally in euchromatin is juxtaposed with heterochromatin by rearrangement or transposition. When heterochromatin packaging spreads across the heterochromatin/euchromatin border, it causes transcriptional silencing in a stochastic pattern. PEV is intensely studied in Drosophila using the white gene. Screens for dominant mutations that suppress or enhance white variegation have identified many conserved epigenetic factors, including the histone H3 lysine 9 methyltransferase SU(VAR)3-9. Heterochromatin protein HP1a binds H3K9me2/3 and interacts with SU(VAR)3-9, creating a core memory system. Genetic, molecular, and biochemical analysis of PEV in Drosophila has contributed many key findings concerning establishment and maintenance of heterochromatin with concomitant gene silencing.
Position-effect variegation (PEV) in Drosophila has been used to study heterochromatin formation. To date, approximately 150 genes have been connected with the onset and/or maintenance of heterochromatic gene silencing in PEV.
OVERVIEW
Genes that are abnormally juxtaposed with heterochromatin, either by rearrangement or transposition, show a variegating phenotype. This is a result of the gene being silenced in some of the cells in which it is normally active. Because the change is caused by a change in the position of the gene in the genome, rather than a change in the gene itself, this phenomenon is termed “position-effect variegation” (PEV). The silencing that occurs in PEV can be attributed to the packaging of the reporter gene in a heterochromatic form, indicating that endogenous heterochromatin formation, once initiated, can spread to encompass nearby genes. Genetic, cytological, and biochemical analyses are all possible in Drosophila melanogaster. In this article we will show how these different approaches have converged to identify many contributors to this system, leading to characterization of both structural proteins and modifying enzymes that play key roles in establishing and maintaining heterochromatin.
Heterochromatin formation depends critically on methylation of histone H3 at lysine 9 (H3K9me2/3), with concomitant association of heterochromatin protein 1 (HP1a) and other interacting proteins, including H3K9 methyltransferases (HKMTs); the multiple interactions of these proteins are required for the spreading and maintenance of heterochromatin. Targeting of heterochromatin formation to particular regions of the genome appears to involve multiple mechanisms, from satellite DNA-specific binding proteins to utilization of the RNA interference (RNAi) machinery. Although heterochromatic regions (pericentric regions, telomeres, the Y chromosome, and the small fourth chromosome) share a common biochemistry, each is distinct, and each is complex in different ways. Heterochromatin in Drosophila is gene poor, but it is not devoid of genes, and counterintuitively, those genes that reside in heterochromatin are often dependent on this environment for full expression. A complete understanding of heterochromatin formation and maintenance (including targeting and spreading) will need to include an explanation for the varying responses of different genes to this chromatin environment.
1. GENES ABNORMALLY JUXTAPOSED WITH HETEROCHROMATIN SHOW A VARIEGATING PHENOTYPE
Large segments of the eukaryotic genome are packaged in a permanently inactive form of chromatin termed constitutive heterochromatin. This chromatin fraction was originally identified as that portion of the genome that remains condensed and deeply staining (heteropycnotic) in interphase; such material is generally associated with the telomeres and pericentric regions of the chromosomes. Heterochromatic regions tend to be late replicating and show little or no meiotic recombination. These domains are dominated by repetitious DNA sequences (∼30%–80%), both tandem repeats of short motifs (known as “satellite” DNA), and remnants of transposable elements (TEs), including DNA transposons and retroviruses. Although gene poor, these domains are not devoid of genes, and intriguingly, those genes that are present frequently are dependent on that environment for optimal expression. About one third of the Drosophila genome is considered heterochromatic, including the entire Y chromosome, most of the small fourth chromosome, the pericentric region that covers 40% of the X chromosome, and pericentric regions that cover 20% of the large autosomes (Smith et al. 2007). During the last few decades we have learned a great deal about the biochemistry of heterochromatin, and much of that understanding derives from our studies with Drosophila (see Schotta et al. 2003; Schulze and Wallrath 2007; Girton and Johansen 2008; Eissenberg and Reuter 2009 for prior reviews).
One of the first mutations identified in D. melanogaster was white, a mutation that results in a fly with a white eye, rather than the characteristic red pigmentation. Using X rays as a mutagen, Muller (1930) observed an unusual phenotype, in which the eye was variegating, with some patches of red and some patches of white facets (Fig. 1A). This phenotype suggested that the white gene itself was not damaged—after all, some facets remained red, and flies with entirely red eyes could be recovered as revertants, again using X rays as the mutagen. However, the white gene had clearly been silenced in some of the cells in which it is normally expressed. Subsequent examination of the polytene chromosomes indicated that such phenotypes are the consequence of an inversion or rearrangement with one breakpoint within the pericentric heterochromatin, and one breakpoint adjacent to the white gene (see Fig. 1A). Because the variegating phenotype is caused by a change in the position of the gene within the chromosome, this phenomenon is referred to as PEV. In Drosophila, virtually every gene that has been examined in an appropriate rearrangement has been shown to variegate, and rearrangements involving the pericentric heterochromatin of any chromosome can lead to PEV (Girton and Johansen, 2008). PEV has subsequently been observed in a variety of organisms, including yeasts (see Allshire and Ekwall 2014), flies, and mammals (see Blewitt and Whitelaw 2013; Brockdorff and Turner 2014), but has been used as a tool to study heterochromatin formation primarily in Drosophila.
Figure 1.
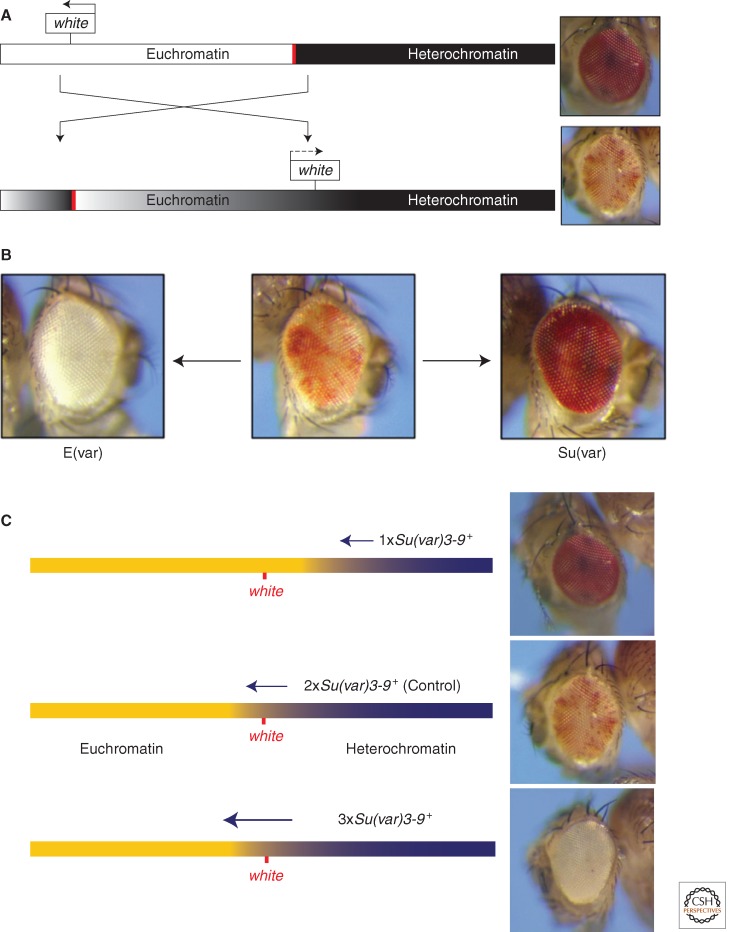
A schematic illustration of white variegation in the X chromosome inversion In(1)wm4. (A) Rearrangement attributable to an X-ray-induced inversion places the white locus, normally located in the distal euchromatin (white bar) of the X chromosome (see top line), ∼25 kb from a breakpoint in the pericentric heterochromatin (black bar; bottom rearranged line). Spreading of heterochromatin packaging into the euchromatic domain results in silencing (causing a white eye in this case); loss of silencing in some cells during differentiation results in a variegating phenotype (bottom line, right). (B) Given a variegating phenotype, screens for second site mutations can recover suppressors (Su(var)s) and enhancers (E(var)s) as described in the text. (C) Some Su(var) loci (e.g., Su(var)3-9, shown here) show an antipodal dosage-dependent effect, and are consequently thought to be structural proteins of heterochromatin. The presence of only one copy of the modifier gene results in less heterochromatin formation, and more expression from the reporter gene (suppression of PEV, top fly eye); conversely, the presence of three copies of such a modifier gene will drive more extensive heterochromatin formation, resulting in an enhancement of reporter gene silencing (enhancement of PEV, bottom fly eye).
PEV indicates that such rearrangements allow packaging of the newly positioned gene into a heterochromatic configuration, and suggests that this is the consequence of heterochromatin “spreading” along the chromosome from the adjacent constitutive heterochromatin region. Apparently, the rearrangement has removed a normally existing barrier or buffer zone. The consequence is an altered packaging with concomitant silencing of genes normally packaged in a euchromatic form. Visual inspection of the polytene chromosomes of larvae carrying such a rearrangement shows that the region carrying the reporter gene is packaged in a dense block of heterochromatin only in the cells in which the gene is inactive (Zhimulev et al. 1986). Patterns of variegated expression, observed as a consequence of rearrangement of white, vary in the number of pigmented cells, the size of the pigmented patches, and the level of pigment in the two different cell types observed, one with a high level of expression, and one with a low level or no expression (Fig. 1A). In a system using an inducible lac-Z gene as a reporter, investigators observed that silencing occurs in early embryogenesis, just after heterochromatin is first observed cytologically, and is epigenetically inherited in both somatic and germline lineages; the mosaic phenotype is determined during differentiation by variegated relaxation of silencing in third instar larvae (Lu et al. 1996). However, not all variegating genes remain silent until after differentiation, and the balance of factors leading to the on/off decision no doubt differs for different genes. (See Ashburner et al. 2005, Chapter 28 for a more detailed discussion.)
A fly line showing a PEV phenotype can be used to screen for dominant second site mutations that are either suppressors or enhancers of PEV. These second site mutations can be induced by chemical mutagens that cause point mutations or small insertions/deletions, but do not impact the chromosome rearrangement responsible for the PEV phenotype. A suppressor (denoted “suppressor of variegation,” Su(var)) results in a loss of silencing, whereas enhancers (denoted “Enhancer of variegation,” E(var)) result in an increase in silencing (Fig. 1B). About 150 loci have been identified from such screens, and of these ∼30 modifiers of PEV have been studied in detail. Where the gene has been cloned and the product characterized, one generally finds a chromosomal protein or a modifier of a chromosomal protein (see Table 1). A small subset of these loci cause both a haplo-abnormal and an opposite triplo-abnormal phenotype (i.e., if one copy of the gene results in suppression of PEV, three copies result in enhancement of PEV [Fig. 1C]). This suggests that the protein products of these genes play a structural role in heterochromatin, and that the spread of heterochromatic packaging can be driven by the dosage of these proteins in a stochastic manner (Locke et al. 1988). However, “spreading” is a complex process, not a simple linear continuum, and is most likely dependent on the organization of the region being silenced in addition to the adjacent heterochromatin mass (see Section 5).
Table 1.
Genetically defined Su(var) and E(var) genes and their molecular function
| Su(var)/E(var) gene | Cytological position | Molecular function, protein distribution, and phenotypic effects |
|---|---|---|
| Suv4-20 (Su(var)) | X; 1B13-14 | Histone lysine methyltransferase (HKMT), histone H4K20 trimethylation |
| Su(z)5 (Su(var)) | 2L; 21B2 | S-adenosylmethionine synthetase |
| chm (chameau) (Su(var)) | 2L; 27F3-4 | Myst domain histone acetylase; suppresses PEV but enhances Polycomb-group mutations |
| Su(var)2-5 (HP1a) | 2L; 28F2-3 | Heterochromatin Protein 1 (HP1a), binding of di- and trimethyl H3K9; binding of SU(VAR)3-9 |
| Su(var)2-HP2 | 2R; 51B6 | Heterochromatin-associated protein, binds HP1a |
| Su(var)2-10 | 2R; 45A8-9 | PIAS protein, negative regulators of JAK/STAT pathway |
| Su(var)3-64B (HDAC1 = RPD3) | 3L; 64B12 | Histone deacetylase HDAC1, deacetylation of H3K9 |
| SuUR (Su(var)) | 3L; 68A4 | Suppresses heterochromatin underreplication; heterochromatin-associated protein |
| Su(var)3-1 (JIL1) | 3L; 68A5-6 | Antimorphic JIL1 mutations, carboxy-terminal protein truncations, blocking of heterochromatin spreading |
| Su(var)3-3 | 3L; 77A3 | dLSD1, the H3K4me3 demethylase |
| Dom (Domina) (Su(var)) | 3R; 86B1-2 | Fork head winged-helix protein; heterochromatin-associated |
| Su(var)3-6 | 3R; 87B9-10 | PP1 protein serine/threonine phosphatase |
| Su(var)3-7 | 3R; 87E3 | Zinc-finger protein, heterochromatin-associated; interacts with HP1a and SU(VAR)3-9 |
| Su(var)3-9 | 3R; 89E6-8 | HKMT, histone H3-K9 methylation, heterochromatin-associated, interaction with HP1a |
| mod (modulo) (Su(var)) | 3R; 100E3 | DNA and RNA binding protein, phosphorylated Mod binds rRNA |
| E(var)3-64E/Ubp64Evar1 | 3L; 64E5-6 | Putative ubiquitin specific protease (Ubp46) |
| Trl (trithorax-like) (E(var)) | 3L; 70F4 | GAGA factor, binding of repetitive DNA sequences, abundant transcription factor |
| Mod(mdg4)/E(var)3-93D | 3R; 93D7 | Transcription regulator, more than 20 protein isoforms produced by trans-splicing |
| E(var)3-93E | 3R; 93E9-F1 | E2F transcription factor, haplo-enhancer, and triplo-suppressor |
The results observed on rearrangement of chromosomes suggest that a euchromatic gene inserted into a heterochromatic domain by transposition will also show a variegating phenotype. This has been found to be the case. The P element, a DNA transposon found in many strains of Drosophila in the wild, can be engineered for this purpose. A natural P element has distinctive inverted repeat sequences at each end, and codes for just one enzyme, the P-specific DNA transposase. Reporter constructs lacking the DNA transposase but containing other genes of interest can be inserted into the Drosophila genome in the presence of active transposase by coinjection into Drosophila embryos. A P-based TE, such as that shown (Fig. 2A) carrying an hsp70-driven copy of white, can be used in a fly with no endogenous copy of white to identify domains of heterochromatin. When the P element is inserted into euchromatin, the fly has a red eye. When this P is mobilized (by crossing in the gene encoding the transposase), approximately 1% of the lines recovered show a variegating eye phenotype. In situ hybridization shows that in these cases, the P element has jumped into the pericentric heterochromatin, the telomeres, the Y chromosome, or the small fourth chromosome (Wallrath and Elgin, 1995). This identification of heterochromatic domains is in agreement with earlier cytological studies.
Figure 2.
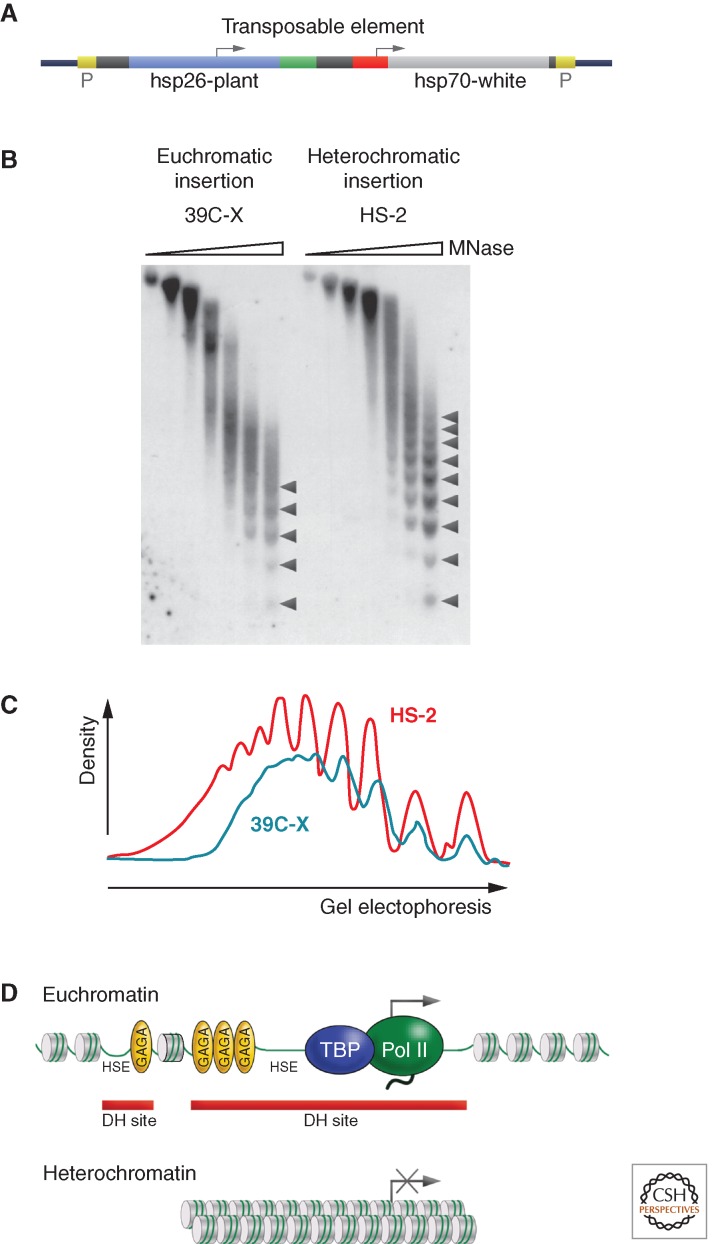
Heterochromatin is packaged into a regular nucleosome array. A TE such as that shown (A), carrying a marked copy of a heat shock gene for study and an hsp70-driven copy of white as a visual marker, can be used to study the same gene in different chromatin domains. (B) Nuclei from Drosophila lines carrying the transgene in a euchromatic domain (39C-X; red eye) and a heterochromatic domain (HS-2; variegating eye) were digested with increasing amounts of micrococcal nuclease (MNase), the DNA was purified and size-separated on an agarose gel, and the resulting Southern blot was hybridized with a probe unique to the transgene. Linker sites cleaved by MNase are marked with arrowheads. (C) Densitometer scans from the last lane of each sample are compared (top to bottom is left to right). An array of nine to 10 nucleosomes can be detected in heterochromatin (red line), compared with five to six in euchromatin (blue line), indicating more uniform spacing in the former case. (D) A diagrammatic representation of the results. DH site, deoxyribonuclease (DNase)-hypersensitive site; HSE, heat shock element. (B,C, Adapted from Sun et al. 2001, © American Society for Microbiology.)
The use of such P elements has allowed comparison of the packaging of the same reporter gene in heterochromatic and euchromatic environments. Heterochromatin is relatively resistant to cleavage by nucleases, whether nonspecific (e.g., DNase I) or specific (restriction enzymes), and is less accessible to other exogenous probes, such as dam methyltransferase. Analysis of the hsp26 transgene (marked with a fragment of unique plant DNA; Fig. 2A) in both euchromatin and pericentric heterochromatin using micrococcal nuclease (MNase) reveals a shift to a more ordered nucleosome array, indicating that spacing of the nucleosomes in heterochromatin is more regular (Fig. 2B,C). The MNase cleavage fragments are well defined, suggesting a smaller MNase target than usual in the linker region. The ordered nucleosome array extends across the 5′ regulatory region of the gene, a shift that no doubt contributes to the observed loss of 5′ hypersensitive sites (HS sites) (Sun et al. 2001). Indeed, although the mechanism of silencing is as yet incompletely understood, there is abundant evidence of transcriptional repression in strongly variegating genes, which includes the loss of binding of TFIID and other transcription factors (Cryderman et al. 1999a). Heterochromatin structure apparently minimizes contact or slows down the processes of nuclear complexes that facilitate transcription, replication, recombination, etc. Heterochromatin packaging has also been found to be critical in maintaining genome integrity; Su(var) mutations can result in disorganized nucleoli, a substantial increase in extrachromosomal circular repeated DNAs, and other forms of DNA damage, potentially related to errors in replication (Peng and Karpen 2009). This packaging also appears to be important in minimizing productive transcription of TEs, helping to keep them immobilized, so protecting genome integrity (e.g., Wang and Elgin 2011).
2. SCREENS FOR SUPPRESSORS AND ENHANCERS OF PEV HAVE IDENTIFIED CHROMOSOMAL PROTEINS AND MODIFIERS OF CHROMOSOMAL PROTEINS
PEV can be modified by a variety of factors. The temperature of development and the amount of heterochromatin within the genome were the first factors shown to affect the extent of variegation. As a rule, an increase in developmental temperature (from 25°C–29°C) results in suppression of variegation (loss of silencing), whereas lower temperatures (e.g., 18°C) cause enhancement of variegation (increase in silencing). Other changes in culture conditions that accelerate or slow the rate of development can have similar effects. Strong suppression is found in flies carrying an additional Y chromosome (XXY females and XYY males), whereas strong enhancement is shown in males without a Y chromosome (X0). In general, duplication of heterochromatic material suppresses whereas deletions of heterochromatic material enhance variegation. These observations suggest the titration of a fixed amount of key protein(s) required for heterochromatin packaging. One consequence is that Y chromosome polymorphisms that alter the amount of heterochromatic DNA in the genome can impact the expression of thousands of genes, presumably because of redistribution of key chromosomal proteins present in limited amounts. Interestingly, these Y polymorphisms have been shown to have a disproportionate influence on the expression of genes producing chromatin-associated protein products (Lemos et al. 2010).
The first mutations acting as a suppressor or enhancer of PEV were identified by Schultz (1950) and Spofford (1967). At present, approximately 150 such genes have been identified as causally connected with the onset and/or maintenance of the heterochromatic gene silencing seen in PEV. In most cases the modifying effect is dominant, and Su(var)/+ or E(var)/+ heterozygotes show a suppressed or enhanced PEV phenotype (Fig. 1B); not surprisingly, these mutations are often homozygous lethals. Efficient isolation and thorough genetic analysis of Su(var) and E(var) mutations has depended on the availability of an experimentally suitable PEV rearrangement. Of the many PEV rearrangements described (FlyBase 2012), one of the most useful lines for such experimental work is In(1)wm4 (Muller 1930). This rearrangement variegates for white, a phenotype easily recognizable in the eye of adult flies, as shown in Figure 1. Penetrance of white variegation in appropriate wm4 stocks is 100%, so every fly in the starting stock shows an eye with a white variegated phenotype, although the degree of variegation can differ considerably from one individual to the next. Inactivation of the white gene does not affect viability or fertility, allowing unlimited work with flies homozygous for wm4. Consequently, white has also been used as a reporter in P element constructs (such as that shown in Fig. 2A) used to examine the sensitivity of different heterochromatic domains to different modifiers.
In the wm4 rearrangement, the inversion results in juxtaposition of the white gene with heterochromatic material of the X chromosome, located at the distal border of the nucleolus organizer (Cooper 1959). This region contains tandem arrays of R1 type mobile elements; the heterochromatic breakpoint of In(1)wm4 has been suggested to fall within an R1 repeat unit (Tartof et al. 1984). Phenotypic w+ revertants of wm4 have been isolated after X-ray or EMS (ethane methyl sulfonate, a chemical mutagen) treatment. Analysis of a series of more than 50 of the w+ revertant chromosomes indicates that all show reinversion or translocation of the white gene to a euchromatic neighborhood, suggesting that the heterochromatic material immediately flanking the breakpoint causes the inactivation of the white gene in wm4. Most of the revertants show white variegation again if strong E(var) mutations are introduced, suggesting that some heterochromatic sequences remain associated with the white gene after relocation (Reuter et al. 1985), not surprising given that the breakpoint in the flanking DNA is randomly introduced. These studies implicate repetitious DNA (here R1, a retrotransposon) as a target for heterochromatin formation. In the fourth chromosome, 1360, a remnant of a DNA transposon, has been implicated as a target (see Section 7). Available data suggest that many, but not all, TEs can be targets for heterochromatin formation (e.g., Riddle et al. 2008; Wang and Elgin 2011).
Most of the known PEV modifier mutations have been isolated using In(1)wm4 or another reporter in a sensitized genetic background. For the isolation of dominant suppressor mutations, the test stock contains a dominant enhancer of PEV; consequently, the test stock has an almost entirely white eye, whereas the desired mutations (Su(var)s) result in a variegated or red eye. The converse is true for an E(var) screen; using a Su(var) mutation to generate a red eye in a variegating line, one screens for mutations that result in a variegated or white eye. More than 1 million flies have been inspected in different screens using this approach, and more than 140 Su(var) and 230 E(var) mutations have been isolated (Schotta et al. 2003). Mutations have been induced by EMS, by X-ray treatment, or by remobilization of P elements. Another set of Su(var) mutations has been isolated in a direct screen (no sensitization of the type described above) with wm4 (Sinclair et al. 1983). A screen using a Df(1;f) chromosome, which shows strong variegation for the yellow gene, a body color marker, resulted in the isolation of 70 PEV modifier mutations (Donaldson et al. 2002). In addition, screens for dominant modifiers of transposon reporter gene expression have identified several mutations with a Su(var) effect (Birchler et al. 1994). With modern technology, it is now possible to screen systematically for the impact of overproduction of proteins as well, and this strategy has identified many enhancers as well as suppressors of PEV (Schneiderman et al. 2010).
Altogether, these screens have identified approximately 500 dominant Su(var) and E(var) mutations. As noted above, based on the genetic analysis performed to date, the total number of Su(var) and E(var) genes can be estimated to be around 150. In naming these genes, the Su(var) and E(var) symbols are usually combined with numbers, indicating the chromosome where the mutation is located, the gene number, and the number of the allele. Thus Su(var)3-917 symbolizes allele 17 of the ninth Su(var) gene identified on the third chromosome. At present, only ca. 30 of the corresponding genes have been carefully mapped and alleles identified (Table 1). Dosage dependent effects (Fig. 1C) have been inferred for 15–20 of the 150 identified loci, either by using overlapping deficiencies and duplications (e.g., Schotta et al. 2003), or by using transgenes (e.g., Eissenberg et al. 1992); this suggests a structural role for these gene products in heterochromatin formation. As we have come to appreciate the modification of PEV as a means for identifying genes that code for chromosomal proteins, reverse screens have been increasingly used to test mutations in candidate genes for Su(var) or E(var) activity (e.g., Pal-Bhadra et al. 2004). Of course, a genetic test by itself does not tell one whether the effect of the mutation is direct or indirect; further characterization is always needed.
Analysis of the identified genes to date suggests that although a discrete set of proteins is required for heterochromatin formation and concomitant gene silencing (loci showing Su(var) mutations), the proteins involved in gene activation are used more broadly. For example, a subset of critical regulatory genes found in the euchromatic arms is maintained in a silent state by the Polycomb (Pc) group genes, and up-regulated by the trithorax group (trxG) genes (see Grossniklaus and Paro 2014; Kingston and Tamkun 2014). In direct tests, relatively few mutations in Pc group genes result in suppression of PEV (e.g., Sinclair et al. 1998). In contrast, many mutations in the trxG genes are enhancers of PEV (Dorn et al. 1993; Farkas et al. 1994). This indicates that the Pc and heterochromatin silencing mechanisms are distinct, although gene activation processes frequently share common components.
Three loci, Su(var)2-5, Su(var)3-7, and Su(var)3-9, can be used to describe some of the different approaches taken to investigate putative suppressors of PEV. Su(var)2-5 was cloned by screening a copy DNA expression library with a monoclonal antibody that recognizes heterochromatin (Fig. 3A) (James and Elgin 1986). The encoded heterochromatin-associated protein was consequently designated HP1, heterochromatin protein 1 (now called HP1a). In situ hybridization analysis, using the isolated cloned DNA, identified a gene in region 28–29 of the polytene chromosomes, where Su(var)2-5 had been previously mapped by Sinclair et al. (1983). DNA sequence analysis of the mutant alleles confirmed that the Su(var)2-5 locus at chromosome position 28F1-2 encodes HP1a (Eissenberg et al. 1990, 1992). HP1a contains two conserved domains, an amino-terminal chromo and a carboxy-terminal chromo-shadow domain (see Patel 2014 for further discussion of the chromodomain), and interacts with many other chromosomal proteins.
Figure 3.
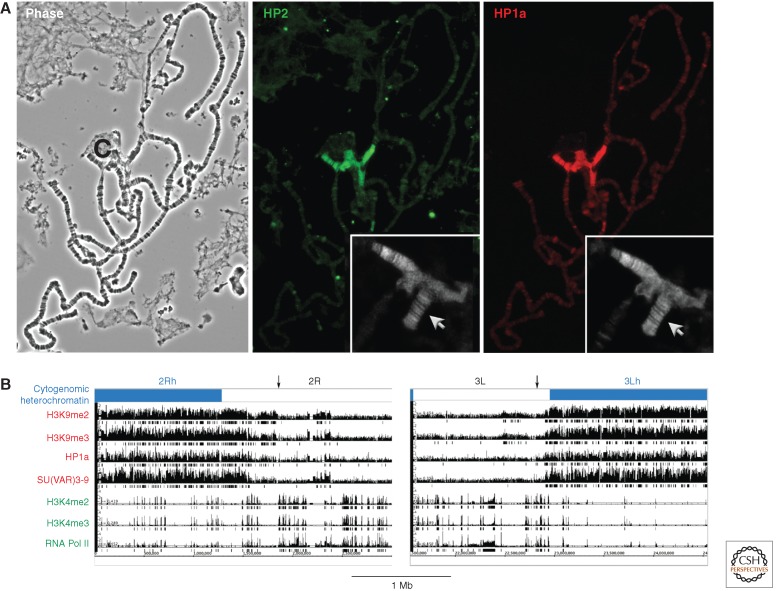
The distribution of chromosomal proteins and histone modifications defines different chromatin domains. (A) Immunofluorescent staining of the polytene chromosomes identifies proteins predominantly associated with heterochromatin. The polytene chromosomes, prepared by fixation and squashing of the larval salivary gland (shown by phase contrast microscopy, left; C = chromocenter) are “stained” by incubating first with antibodies specific for a given chromosomal protein, and then with a secondary antibody coupled to a fluorescent tag. HP1a (right) and HP2 (center) have similar distribution patterns, showing prominent association with the pericentric heterochromatin (found in the condensed chromocenter), small fourth chromosome (inset, arrow), and a small set of sites in the long euchromatin arms. (Adapted from Shaffer et al. 2002.) Note that the efficacy of any antibody can be affected by the choice of fixation protocol (see Stephens et al. 2003). (B) Chromatin marks define the epigenomic border between heterochromatin and euchromatin (indicated with an arrow). The border can be delineated based on chromatin immunoprecipitation (ChIP)-array data using antibodies to proteins known to be associated with heterochromatin (HP1a) or euchromatin (RNA polymerase II [RNA Pol II]), and to key histone modifications. Enrichment values are shown for the centromere-proximal 3 Mb of chromosome arms 2R and 3L (in BG3 cells). Boxes underneath the bar graphs indicate significant enrichment (p < 0.001). The cytologically defined heterochromatin is shown by the blue bar. The border, indicated here by the black arrow, is fairly well defined by the congruence of silencing marks. (B, Adapted from Riddle et al. 2011, © Cold Spring Harbor Laboratory Press.)
Su(var)3-7 was first cytogenetically mapped to region 87E1-4 in the third chromosome using a series of overlapping deletions and duplications. It was further assigned to within a 7.8 kb DNA fragment based on the triplo-enhancer effect it had on a variegating reporter (Reuter et al. 1990). Su(var)3-7 encodes a protein with seven regularly spaced zinc fingers, domains that have been shown to function in DNA binding (Cleard and Spierer 2001).
Su(var)3-9 was cloned by P element transposon tagging (Tschiersch et al. 1994). The Su(var)3-9 gene in Drosophila forms a bicistronic unit with the gene encoding eIF2γ, which can complicate genetic analysis. The SU(VAR)3-9 protein, like SU(VAR)2-5, contains a chromodomain in its amino-terminal region, but has a SET domain (identified first in the proteins SU(VAR)3-9, ENHANCER OF ZESTE [E(Z)], and TRITHORAX) at its carboxyl terminus. The SET domain allows this protein to function as a histone lysine methyltransferase (HKMT), specifically methylating histone H3 at lysine 9 (H3K9).
Immunocytological analyses using specific antibodies or transgene-expressed fusion proteins have shown that all three proteins—HP1a (encoded by Su(var)2-5), SU(VAR)3-7, AND SU(VAR)3-9—are preferentially associated with pericentric heterochromatin (see Fig. 3A for an example; James et al. 1989; Cleard et al. 1997; Schotta et al. 2002). Strong colocalization is particularly evident for HP1a and SU(VAR)3-9. Association of these proteins with each other has also been shown by coimmunoprecipitation (Delattre et al. 2000; Schotta et al. 2002). Thus, these proteins potentially form a core heterochromatin complex. Surprisingly, HP1a is also found at a number of euchromatic sites, and is involved in the positive regulation of a small set of euchromatic genes (Cryderman et al. 2005; Piacentini et al. 2009). Mutations in other genes encoding histone variants, chromatin-modifying enzymes, chromatin binders, or nucleosome remodeling factors often result in dominant PEV modifier effects (cf. Fodor et al. 2010). However, in the majority of these cases, causal analysis of the mutant effects on gene silencing in PEV is still missing, so it is possible that the effects are indirect. Despite intensive work on PEV modifiers, we still do not have a clear picture of the macromolecular assemblies in pericentric heterochromatin.
P element insertions carrying the w+ reporter gene into other heterochromatic domains—telomeric regions, the Y chromosome, or the fourth chromosome—also show white variegation (e.g., Wallrath and Elgin 1995; Phalke et al. 2009). Genetic analysis of such reporters has revealed that although the different heterochromatic domains share some common features, they also can rely on different chromatin factors. For example, heterochromatin-like packaging is observed at TAS (telomere associated satellite) sequences, which are clusters of repetitious DNA elements just proximal to the HeT-A and TART retroviral elements that make up Drosophila telomeres (Cryderman et al. 1999b). Surprisingly, HP1a mutations do not show an effect on such reporters, although HP1a is important for telomere integrity (Fanti et al. 1998). Characteristics of this domain are sufficiently distinct that variegation here is referred to as telomere position effect (TPE). In another example, several groups have observed that silencing of reporters on the fourth chromosome is often sensitive to mutations in the gene for dSETDB1, and not to mutations in Su(var)3-9, although both encode an H3K9 HKMT (Seum et al. 2007; Tzeng et al. 2007; Brower-Toland et al. 2009). Investigations using a range of modifier loci to look at PEV reporters in different heterochromatin domains have shown that a unique complement of proteins is required for each domain to maintain silencing in somatic cells (Donaldson et al. 2002; Phalke et al. 2009).
3. DISTRIBUTION AND ASSOCIATION PATTERNS OF CHROMOSOMAL PROTEINS
One of the advantages of working with Drosophila is the ability to examine the polytene chromosomes, which provide a visual road map of the genome. Polyteny occurs during the larval stage when the chromosomes in many terminally differentiated cells are replicated, but do not go through mitosis; rather the chromatin strands remain paired, in perfect synapsis, with all copies aligned. The most extreme case is found in the salivary glands, where the euchromatic arms of the chromosomes have undergone 10 rounds of replication, generating ∼1000 copies. Replication is not uniform, however; many repetitious sequences are underreplicated, and satellite DNA sequences are not replicated at all. All of the chromosome arms fuse in a common chromocenter (see Fig. 3A) (reviewed in Ashburner et al. 2005).
Polytene chromosomes provide an opportunity to determine the distribution pattern of chromosomal proteins by immunofluorescent staining, achieving much higher resolution than can be obtained using metaphase spreads (Silver and Elgin 1976). The approach has been used to discover heterochromatin-associated proteins (e.g., HP1a; James and Elgin 1986) and to determine whether other candidates, identified genetically (by Su(var) phenotype) or by interaction with known heterochromatin proteins, show such localization (Fig. 3A) (e.g., HP2, identified by interaction with HP1a; Shaffer et al. 2006). Whereas genome-wide ChIP techniques described below have higher resolution, polytene chromosome staining remains a quick and inexpensive way (given specific antibodies in hand) to show distribution patterns. Approximately 10 heterochromatin-specific proteins have been identified using this approach; if mutations in the genes encoding these proteins are available, one often observes dominant suppression of PEV (e.g., Greil et al. 2007; reviewed in Ashburner et al. 2005, Chapter 28). These proteins are thus candidate structural components of heterochromatin.
With the availability of oligonucleotide arrays and high-throughput sequencing, it has become possible to map both chromosomal proteins and histone modifications across the sequenced genome by ChIP. In the most common approach, chromatin is cross-linked using formaldehyde, sonicated to obtain 500- to 1000-bp fragments, and the desired fragments pulled down using antibodies fixed to beads. The recovered fraction of genomic DNA is characterized either by qPCR (quantitative polymerase chain reaction, used if only a few loci are to be queried), hybridization to a genomic array (ChIP-chip), or deep sequencing (ChIP-seq). The validity of the results is critically dependent on the specificity of the antibodies used. As the commercial producers do not always provide sufficient quality control, the experimenters must do the controls themselves (Egelhofer et al. 2011). The technique depends on the ability to map the recovered DNA fragments to the assembled genome. In the case of D. melanogaster, only ∼25% of the heterochromatic regions have been sequenced and assembled (not including satellite DNA, for example; Hoskins et al. 2007; Smith et al. 2007), and this limitation must be kept in mind in interpreting the results.
The modENCODE project, sponsored by the National Institutes of Health (USA), has reported genome-wide chromatin profiling in Drosophila for 25 histone modifications and many more chromosomal proteins and transcription factors, looking at several cell culture lines as well as several developmental stages. These data, as well as maps of transcription patterns, DNase I HS sites, and other chromatin features can be accessed through FlyBase (2012) or modMine (2011). Looking at the base of the chromosome arms in BG3 cells, for example, one observes a transition from a domain with abundant HP1a and SU(VAR)3-9 (pericentric heterochromatin) to a region where these proteins are only sporadically observed (euchromatin) (Riddle et al. 2011) (Fig. 3B). These results are in good agreement with the cytogenetically defined border between euchromatin and heterochromatin, but provide much higher resolution. The border clearly reflects the shift in genome organization around this point, with heterochromatic domains showing a decrease in gene density and increase in repeat density. However, the border can shift by hundreds of kb in S2 cells, suggesting that it is not fixed by a particular DNA sequence, but rather reflects the balance of chromosomal proteins or other factors specific to the cell type.
4. HISTONE MODIFICATION PLAYS A KEY ROLE IN HETEROCHROMATIN SILENCING
Analysis of SU(VAR)3-9 identified the key function of H3K9 methylation in heterochromatic gene silencing (Tschiersch et al. 1994). The protein contains a SET domain that enzymatically functions to methylate histone H3K9. That this protein is a histone methyltransferase (HKMT) targeting H3K9 was first shown by characterization of the human SUV39H1 homolog (Rea et al. 2000). In Drosophila, SU(VAR)3-9 is a major, but not the only, H3K9 HKMT (Schotta et al. 2002; Ebert et al. 2004). SU(VAR)3-9 contributes to di- and trimethylation of H3K9 (H3K9me2 and me3) in the bulk of the pericentromeric heterochromatin, but not in the majority of the fourth chromosome, telomeres, or euchromatic sites. The bulk of the dimethylation of these latter regions is independent of SU(VAR)3-9, as is monomethylation of H3K9 in pericentromeric heterochromatin (Ebert et al. 2004). dSETDB1 (“eggless”) plays a major role in H3K9 methylation on the fourth chromosome (Seum et al. 2007; Tzeng et al. 2007; Brower-Toland et al. 2009; Riddle et al. 2012); G9a and potentially other HKMTs could also contribute, but the specifics are still unknown. The importance of H3K9 dimethylation in heterochromatic gene silencing is shown by the strong dosage-dependent effect of SU(VAR)3-9 on the PEV phenotype, as well as by the finding that suppression of gene silencing by Su(var)3-9 mutations correlates with HKMT activity. The enzymatically hyperactive Su(var)3-9ptn mutation is a strong enhancer of PEV and causes elevated H3K9 di- and trimethylation (H3K9me2 and H3K9me3) at the chromocenter, as well as generating prominent H3K9me2 and me3 signals at many euchromatic sites (ectopic heterochromatin) (Ebert et al. 2004). S-adenosylmethionine functions as the methyl donor for all of these methylation reactions; consequently, mutations in the gene encoding S-adenosylmethionine synthase, Su(z)5, are dominant suppressors of PEV (Larsson et al. 1996).
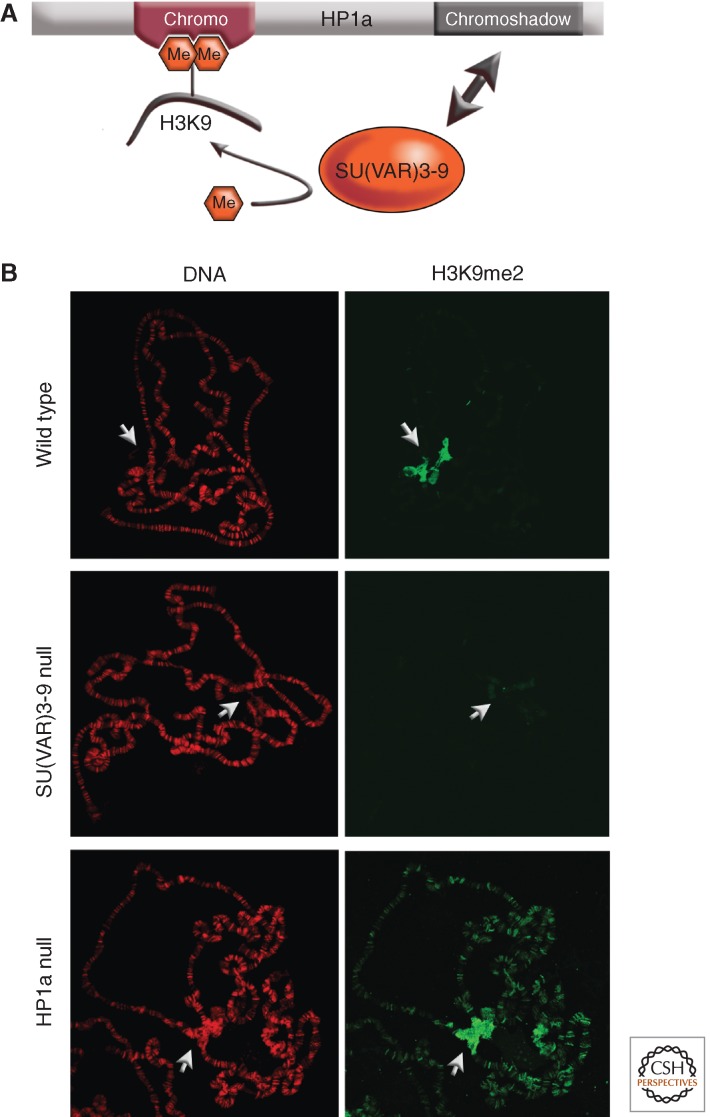
Studies using mutations in SU(VAR) genes have begun to reveal the sequence of molecular reactions required to establish heterochromatic domains. SU(VAR)3-9 binding at heterochromatic sequences depends on both its chromo and its SET domains (see Patel 2014 for details of protein structure; Schotta et al. 2002). How SU(VAR)3-9 binding is controlled is not yet understood. The act of methylating H3K9 by SU(VAR)3-9 establishes binding sites for HP1a. The HP1a chromo domain specifically binds H3K9me2 and H3K9me3 (Jacobs et al. 2001). That SU(VAR)3-9 also binds HP1a has been shown by yeast two-hybrid tests and by immunoprecipitation (Schotta et al. 2002). In fact, the region of SU(VAR)3-9 amino-terminal to its chromodomain interacts with the chromoshadow domain of HP1a, and this interaction stabilizes HP1a binding to H3K9me2/3 (Fig. 4A) (Eskeland et al. 2007). This region of SU(VAR)3-9 also interacts with the carboxy-terminal domain of SU(VAR)3-7. The SU(VAR)3-7 protein interacts at three different sites with the chromoshadow domain of HP1a (Delattre et al. 2000). This pattern of interactions suggests that the three proteins—HP1a, SU(VAR)3-7, and SU(VAR)3-9—physically associate in multimeric heterochromatin protein complexes.
Figure 4.
Interaction of SU(VAR)3-9 and HP1a in setting the distribution pattern of H3K9 methylation. (A) HP1a interacts with H3K9me2/3 through its chromodomain, and with SU(VAR)3-9 through its chromoshadow domain. By recognizing both the histone modification and the enzyme responsible for that modification, HP1a provides a mechanism for heterochromatin spreading and epigenetic inheritance. (B) SU(VAR)3-9 is responsible for much of the dimethylation of H3K9 (H3K9me2); loss of the enzyme results in loss of this modification in the pericentric heterochromatin, as shown by loss of antibody staining of the polytene chromosomes (compare middle panel with top panel). Loss of HP1a results in a loss of targeting of SU(VAR)3-9; high levels of H3K9me are consequently now seen throughout the chromosome arms (bottom panel).
Association of SU(VAR)3-9 and HP1a with pericentric heterochromatin is interdependent (Schotta et al. 2002). SU(VAR)3-9 causes H3K9 di- and trimethylation, which are specifically recognized by the chromodomain of HP1a (Jacobs et al. 2011). Consequently, in Su(var)3-9 null larvae, HP1a binding to pericentric heterochromatin is impaired. As H3K9 dimethylation does not depend exclusively on SU(VAR)3-9 in the inner chromocenter, the fourth chromosome, telomeres and euchromatic sites, HP1a continues to be found at all of these sites in the mutant lines. Thus although SU(VAR)3-9 associates with these sites in wild-type cells, it appears to be relatively inactive.
Conversely, if HP1a is not present (having been depleted by mutations), SU(VAR)3-9 is no longer associated primarily with the pericentric heterochromatin, but is found along the euchromatic chromosome arms. It is now seen at almost all bands, where it causes ectopic mono- and dimethylation of H3K9 (H3K9me1 and H3K9me2) (Fig. 4B). Thus HP1a is essential for the restricted binding of SU(VAR)3-9 to pericentric heterochromatin. These data suggest a sequence of reactions starting with SU(VAR)3-9 association with heterochromatic domains and consequent generation of H3K9me2/3. This mark is recognized by the chromodomain of HP1a; binding of SU(VAR)3-9 to the HP1a chromoshadow domain ensures its association with heterochromatin (Fig. 4A). Interestingly, a chimeric HP1a-Pc protein has been generated in which the chromodomain of HP1a is replaced with the chromodomain of the Pc protein (Platero et al. 1996). The chromodomain of Pc binds strongly to H3K27me3 (Fischle et al. 2003), and the HP1a-Pc chimeric protein binds these sites in the euchromatic arms. In the presence of such a chimeric HP1a-Pc protein, the SU(VAR)3-9 protein is also found at Pc binding sites, demonstrating its strong association with the chromoshadow domain of HP1a (Schotta et al. 2002).
In SU(VAR)3-9 null cells another heterochromatin-specific methylation mark, H4K20 trimethylation (H4K20me3), is strongly reduced. The interdependence between H3K9 dimethylation and H4K20 trimethylation in heterochromatin has been shown to reflect an interaction between the SU(VAR)3-9, HP1a, and SUV4-20 proteins. SUV4-20 is a histone lysine methyl transferase (HKMT) that controls H4K20 methylation in heterochromatin. This heterochromatin-specific methylation mark is also strongly impaired in HP1a null cells, suggesting association of SU(VAR)3-9, HP1a, and SUV4-20 in a mutually dependent protein complex, although such a complex has not yet been isolated from flies. Mutations in the Suv4-20 gene cause suppression of PEV-induced gene silencing, indicating that the H4K20me3 mark is required for this process (Schotta et al. 2004).
Taken together, the evidence argues that the HP1a protein has a central function in pericentric heterochromatin formation and associated gene silencing; it binds H3K9me2 and H3K9me3, and interacts directly with SU(VAR)3-9 (one of the H3K9 HKMTs) as well as several other key chromosomal proteins. The resulting complexes probably include several additional heterochromatin-specific proteins. Variations on this theme apply to other heterochromatic domains, such as the fourth chromosome (Riddle et al. 2012). However, given the number of identified Su(var) loci, the model is certain to become more complex!
In mammals and plants, histone H3K9 methylation and DNA methylation represent interrelated marks of repressed chromatin (Martienssen and Colot 2001; Bird 2002). Whether or not DNA methylation occurs at all in Drosophila has been a point of contention for many years. Recent reports showing low levels of DNA methylation in the early embryo have renewed this discussion (reviewed in Krauss and Reuter 2011). In Drosophila the only recognizable DNA methyltransferase present is Dnmt2. Mutations in this gene have a significant impact on retrotransposon silencing in somatic cells (Phalke et al. 2009). However, many inbred laboratory strains show only a very low level of Dnmt2 expression; variation of this sort could explain conflicting results concerning DNA methylation in Drosophila (O Nickel, C Nickel, and G Reuter, unpubl.).
5. CHROMOSOMAL PROTEINS FORM MUTUALLY DEPENDENT COMPLEXES TO MAINTAIN AND SPREAD HETEROCHROMATIC STRUCTURE
PEV reflects a change in gene expression, specifically a loss in expression of a reporter gene in some of the cells in which it is normally active, as a consequence of a genetic rearrangement or transposition. Several different models, not all mutually exclusive, have been suggested to explain PEV. One possibility originally considered was the random loss of the gene, perhaps as a consequence of late replication (Karpen and Spradling 1990). Quantitative Southern blot analysis has shown that this explanation is not generally applicable; variegating genes are generally fully replicated in diploid tissue (Wallrath et al. 1996). A second test can be performed using a variegating reporter transgene (white) flanked by FRT sites; by inducing FLP recombinase, the gene is excised from the chromosome, forming an independent closed circle. A reporter gene subject to variegation (PEV) can be relieved of that silencing by excision from the chromosome in cells where it would otherwise be inactive (Ahmad and Golic, 1996). This shows not only the continuing presence of the gene, but that the heterochromatic state can be reversed once the reporter is removed from the heterochromatic environment.
Other models have focused on the association of the variegating gene with a heterochromatic compartment in the nucleus (see Section 6), and/or on the spreading of heterochromatic structure from the newly adjacent heterochromatin. The spreading model, which is based on extensive genetic and cytological data, explains gene silencing as a consequence of heterochromatin packaging spreading across the breakpoint into normally euchromatic domains. In normal chromosomes, euchromatic and heterochromatic regions appear to be differentiated by the higher density of repetitious sequences in heterochromatin, and are potentially insulated from each other by specific sequences or buffer zones. Because these “insulating sequences” (never well-defined in Drosophila) are not present at the euchromatic-heterochromatic junction in PEV rearrangements (see Fig. 1A), heterochromatinization of euchromatic sequences is variably induced. This heterochromatinization is cytologically visible in the polytene chromosomes as a shift from a banded to an amorphous structure at the base of the chromosome arms (Fig. 5A) (Hartmann-Goldstein 1967); the extent of this change can be modified by Su(var) and E(var) mutations (Reuter et al. 1982).
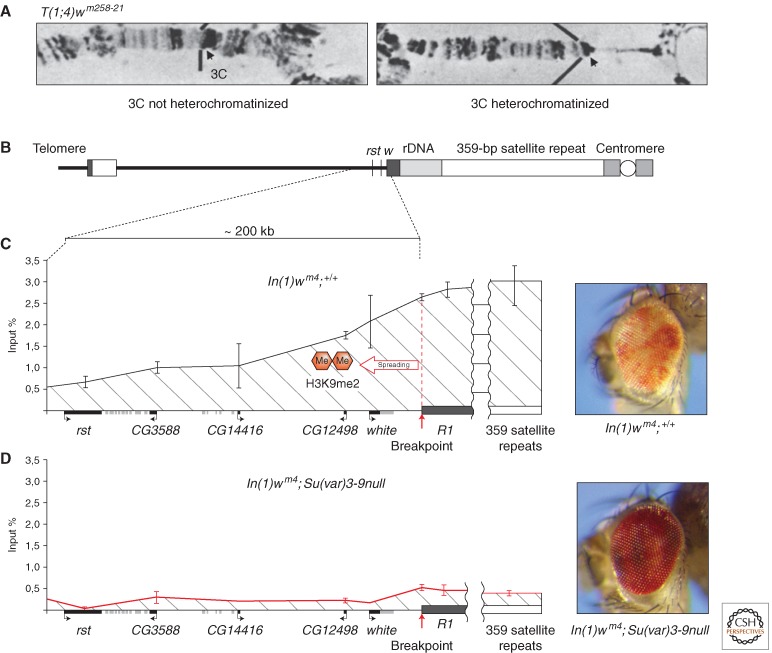
Figure 5.
Spreading of histone H3K9me2 and cytological heterochromatinization at the white locus in PEV rearrangements. (A) In translocation T(1;4)wm258-21 (with a breakpoint in the heterochromatin of chromosome 4), heterochromatinization becomes cytologically visible in the polytene larval salivary gland chromosomes as a loss of banding, the apparent consequence of condensation and underreplication (right portion of the right panel). (Reprinted from Reuter et al. 1982, with kind permission from Springer Science+Business Media.) (B) In the In(1)wm4 chromosome, spreading of the heterochromatic chromatin state over ∼200 kb of the adjacent euchromatic region is initiated by the segment of heterochromatin located distal to the rDNA cluster (dark gray box). (C) ChIP with an antibody specific for H3K9me2 detects spreading of this heterochromatic histone mark along the euchromatic region between the roughest (rst) gene and the breakpoint of wm4. (D) Flies homozygous for a null mutation of the Su(var)3-9 gene lose H3K9me2 in pericentric heterochromatin as well as in the white gene region, restoring wild-type activity of the white gene in In(1)wm4 flies. (Adapted from Rudolph et al. 2007.)
Inactivation of euchromatic genes over a distance along the chromosome can be genetically shown (Demerec and Slizynska 1937). The affected regions become associated with HP1a (Belyaeva et al. 1993; Vogel et al. 2009) and show dimethylation of histone H3 at lysine 9 (H3K9me2) (Ebert et al. 2004; Rudolph et al. 2007). The spreading model postulates a competition between packaging into euchromatin versus packaging into heterochromatin; the recovery of dosage-dependent modifiers, as discussed above, supports such a model (Locke et al. 1988; Henikoff 1996). However, spreading does not seem to be a simple matter of mass action, in which the regions closest to the heterochromatic mass show the greatest silencing. One can identify cells in which a marker closer to the pericentric heterochromatin in a rearrangement is active and one further away is silenced (Talbert and Henikoff 2000). Inspection using ChIP, followed by qPCR, shows that there is a gradient of the H3K9me2 heterochromatin mark declining with distance from the pericentriomeric heterochromatin (Fig. 5C) (Rudolph et al. 2007). Ironically, the regulatory region of the white gene appears to be particularly vulnerable to the accumulation of silencing marks, shown in particular as an accumulation of HP1a in a DamID mapping study (method described below; Vogel et al. 2007). This regulatory region is present in reporter transgenes marked with mini-white, but not in those such as shown in Fig. 2, which use an hsp70 promoter to drive white expression. Such differences should be kept in mind in evaluating studies using transgene reporters.
The heterochromatin spreading effect clearly depends on a series of molecular reactions within the euchromatic regions. Several histone modifications are now known to be mutually exclusive in defining alternative chromatin states. Acetylation of H3K9 (H3K9ac), di- and trimethylation of H3K4 (H3K4me2/3), and phosphorylation of H3S10 (H3S10ph) are typical marks of active euchromatin, whereas H3K9me2/3 and H4K20me3 are specific marks of silenced regions. Heterochromatinization of euchromatic regions therefore requires specific deacetylation, demethylation, and dephosphorylation reactions within euchromatin, as illustrated in Figure 6A. This transition depends initially on H3K9 deacetylation by HDAC1. Mutations in the rpd3 gene, encoding the histone H3K9-specific deacetylase HDAC1, are strong suppressors of PEV (Mottus et al. 2000), antagonizing the effect of SU(VAR)3-9 in gene silencing (Czermin et al. 2001). HDAC1 has been shown to be associated in vivo with the SU(VAR)3-9/HP1a complex; the two enzymes work cooperatively to methylate previously acetylated histones.
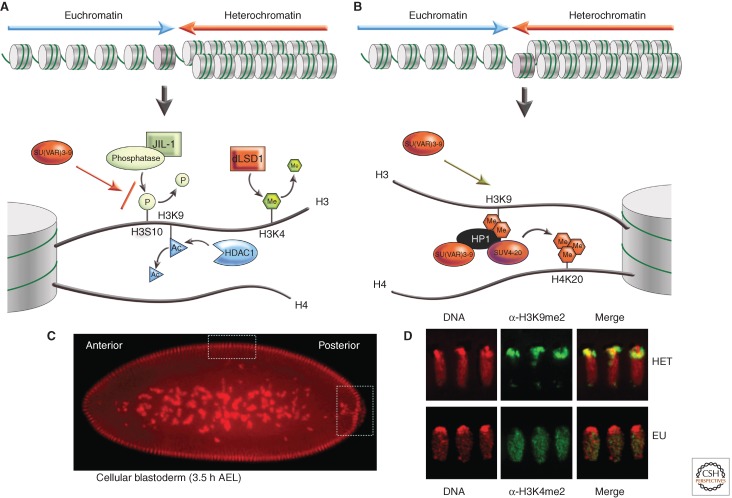
Figure 6.
The transition from a euchromatic state to a heterochromatic state requires a series of changes in histone modification. (A) Active genes are marked by H3K4me2/3; if present, this mark must be removed by LSD1. H3K9 is normally acetylated in euchromatin; this mark must be removed by a histone deacetylase, HDAC1. Phosphorylation of H3S10 can interfere with methylation of H3K9; dephosphorylation appears to involve a phosphatase targeted by interaction with the carboxyl terminus of the JIL1 kinase. These transitions set the stage for acquisition of the modifications associated with silencing, shown in B, including methylation of H3K9 by SU(VAR)3-9, binding of HP1a, and subsequent methylation of H4K20 by SUV4-20, an enzyme potentially recruited by HP1a. (C) Differentiation of euchromatin and heterochromatin is initiated in early embryogenesis around cell cycle 10 and is completed when cellular blastoderm (top box) and primordial germline cells (right-hand box) are formed. (D) Blastoderm nuclei show an apicobasal polarity (Rabl conformation). Heterochromatin (H3K9me2 staining) is established at the apical site, whereas euchromatin (H3K4me2 staining) is organized toward the basal site. (Immunofluorescent images provided by Sandy Mietzsch.)
Spreading of heterochromatin into euchromatin is completely blocked in Su(var)3-1 mutations (Ebert et al. 2004). Su(var)3-1 mutations are frame shift mutations within the gene encoding JIL1 kinase that result in expression of a truncated JIL1 protein, lacking the carboxyl terminus region. The JIL1 protein contains two kinase domains, and catalyzes H3S10 phosphorylation in euchromatin. The JIL1Su(var)3-1 mutations do not affect H3S10 phosphorylation, but probably impair dephosphorylation of H3S10, effectively inhibiting methylation of H3K9. This suggests involvement of a phosphatase. Whether the PP1 enzyme (that has been identified with Su(var)3-6 mutations [Baksa et al. 1993]) is directly involved in this reaction is not yet known.
Demethylation of H3K4 appears to be another prerequisite for heterochromatinization of euchromatic regions (Fig. 6A). Work in mammals has shown that the LSD1 amine oxidase functions as an H3K4 demethylase (Shi et al. 2005). Mutations in the Drosophila LSD1 homolog antagonize spreading of heterochromatin into euchromatic regions in all PEV rearrangements tested. In SU(VAR)3-3 null cells, lacking LSD1, the acquisition of H3K9 methylation in the euchromatin flanking a break point is eliminated, although constitutively heterochromatic regions are not affected (Rudolph et al. 2007). In the syncytial blastoderm, dLSD1 is concentrated in the nuclei at the boundary between heterochromatic and euchromatic domains (Fig. 6D). These findings show that the coordinated function of several enzymes is required to remove euchromatin-specific histone modification marks before the transition to heterochromatin packaging can take place (see Fig. 6A). It seems likely that the required enzymes will be found to form complexes with SU(VAR)3-9/HP1a, as has already been shown for HDAC1.
The above patterns of association among histone marks and chromosomal proteins suggest that the chromatin landscape could be described in terms of these patterns of coassociation, and indeed this has proved to be the case. Two different approaches have been taken for mapping genome-wide distribution patterns of chromosomal proteins: ChIP (described above) and DamID. In the latter case, DNA adenine methyltransferase (Dam, a bacterial specific protein) fused to a chromatin protein of interest deposits a stable adenine-methylation “footprint” in vivo at those sites where the chromatin protein associates. The modified DNA fragments are recovered and the pattern of association assessed using oligonucleotide microarrays. A study using 53 broadly selected chromosomal proteins in Drosophila Kc cells identified five major chromatin types: heterochromatin (enrichment of HP1a and H3K9me2), Pc silenced domains (enrichment in PC and H3K27me3), additional inactive regions, and two types of active domains, both associated with high levels of RNA polymerase but distinguished by molecular organization and H3K36 methylation levels. Principle component analysis shows that this classification can largely be achieved (85.5% agreement) using five proteins: histone H1, HP1a, PC, MRG15, and BRM (the latter two associated with nucleosome remodeling) (Filion et al. 2010).
The modENCODE study used the enrichment patterns of 18 different histone modifications to generate a model based on 30 alternative states, in contrast to the five categories by Filion et al. (2010) described above. Distribution patterns were determined by ChIP-chip experiments (with assessment of the captured DNA on oligonucleotide arrays); this approach, although requiring fixation, provides higher resolution. Much of the overall complexity is captured in nine prevalent combinatorial patterns (states), illustrated in Figure 7A (Kharchenko et al. 2011). The analysis shows the presence of correlated features, including those associated with the transcription start site (state 1), the body of the transcribed gene (state 2), and regulatory regions (states 3 and 4). Distinctive states associated with large domains include that found on the male X chromosome (presumably related to dosage compensation—see Kuroda and Lucchesi 2014) (state 5), that associated with the Pc silencing complex (state 6; see Grossniklaus and Paro 2014), and two associated with heterochromatin marks, one common to pericentric heterochromatin (state 7) and the other (with lower concentrations of H3K9me2/3) found in the euchromatic arms (state 8). (The pattern of state 8 domains is cell-type specific, suggesting “facultative” heterochromatin [Kharchenko et al. 2011].) Those domains with no distinctive features are grouped together in state 9 (see Fig. 7A).
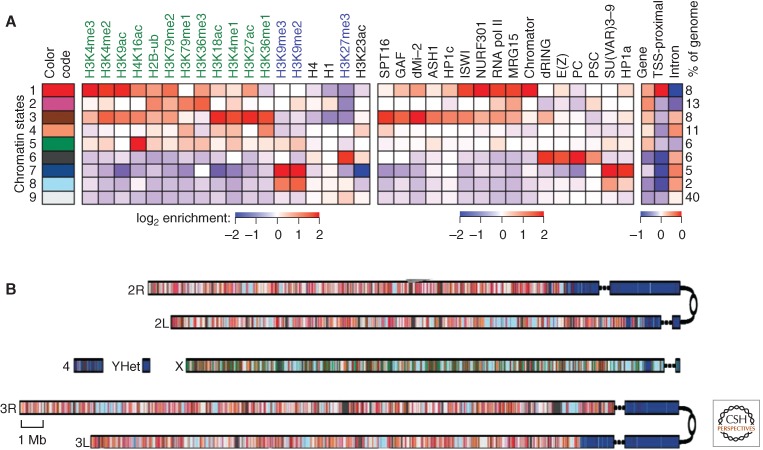
Figure 7.
Chromatin annotation of the D. melanogaster genome. (A) A nine-state model of prevalent chromatin states was generated using data from S2 cells. Each chromatin state (row) is defined by a combinatorial pattern of enrichment (red) or depletion (blue) for specific histone modification marks. First (left) panel, color code for mapping; second panel, histone modification marks used (active marks labeled in green, repressive in blue, general in black); third panel, enrichment or depletion of chromosomal proteins found in that state; fourth (right) panel, fold over-/underrepresentation of genic and transcription start site (TSS)-proximal (±1 kb) regions relative to the entire tiled genome. (B) A genome-wide karyotype view of the domains defined by the nine-state model in S2 cells. Centromeres are shown as open circles; dashed lines span gaps in the genome assembly. Note the association of pericentric heterochromatin and the fourth chromosome distal arm with state 7 and the association of state 5 with the male X chromosome. (Adapted, by permission from Macmillan Publishers Ltd: NATURE, from Kharchenko et al. 2011, © 2011.)
Although the generation of this nine-state model is based solely on the map of histone modification marks, one observes distinctive patterns of enrichment and depletion for chromosomal proteins. For example, HP1a and SU(VAR)3-9 are greatly enriched in state 7, and moderately enriched in state 8, states that are depleted for proteins associated with gene expression. Mapping these states back across the genome provides both an overview of chromatin organization (Fig. 7B) and a detailed characterization of individual genes. The latter can be viewed on FlyBase (2012) by choosing the GBrowse option. The nine-state model allows us to see general patterns at the level of the chromosome or large domains, whereas going to more complex models (such as the 30-state model) can resolve more detail at the gene level.
Results from the two approaches, classifying by histone marks or by chromosomal proteins, are clearly in agreement in identifying alternative silencing domains (H3K9me2/HP1a vs. H3K27me3/Pc). However, the classification of active genes differs, with that based on histone modifications identifying different gene regions (1-transcription start site, 2-body of the transcribed region, 3/4-regulatory regions), whereas that based on chromosomal proteins identifies two groups of genes differing in remodeling strategies (Filion et al. 2010; Kharchenko et al. 2011). Analysis using key components identified by each study should provide a powerful approach in the future.
6. NOT ALL HETEROCHROMATIN IS IDENTICAL: SPATIAL ORGANIZATION MATTERS
In D. melanogaster, constitutive heterochromatin is arranged in large blocks that flank the centromeres, smaller blocks associated with the telomeres, the whole of the Y chromosome, and most of the small fourth chromosome. The centromeric regions are made up of large (0.2–1 Mb) blocks of satellite DNA interspersed with “islands” of complex sequences, primarily TEs (Le et al. 1995). Although gene-poor, these regions are not devoid of genes; a minimum of 230 protein-coding genes (conserved in other Drosophilids), as well as 32 pseudogenes and 13 noncoding RNAs reside in the pericentromeric heterochromatin (Smith et al. 2007). The telomeres of Drosophila do not have the typical G-rich repeats seen in other eukaryotes, but are composed of copies of HeT-A and TART retrotransposons. TAS, blocks of 102–103 nucleotide repeats, are found just proximal, and white transgene reporters inserted in these regions display the TPE variegating phenotype. Although the Y chromosome does carry the genes for a number of male fertility factors, the bulk of the chromosome is made up of satellite DNA, and it remains condensed in cells other than the male germline. Thus, although all of these domains are characterized by a high density of repeats, the type of repeats (and interspersion of different types) varies. The consequences have been seen in studies that look at the impact of different blocks of pericentric heterochromatin on expression from a reporter—one observes that the severity of the phenotype does not depend simply on the amount of heterochromatin in cis, but varies depending on the local heterochromatin environment (Howe et al. 1995). Heterochromatin-associated proteins that might play a role in specific subdomains include the AT-hook protein D1, preferentially associated with the 1.688 g/cm3 satellite III (Aulner et al. 2002), and DDP1, a multi-KH-domain protein homologous to vigilin that binds the pyrimidine-rich C strand of the dodeca satellite (Cortes and Azorin 2000).
The small fourth chromosome is perhaps the most complex heterochromatic domain. It is on the order of 4.3 Mb in size, with ∼3 Mb made up of satellite DNA. The distal 1.2 Mb can be considered euchromatic in that it is polytenized in the salivary gland (see Fig. 3A), but it appears heterochromatic by virtue of its late replication, its complete lack of meiotic exchange, and its association with HP1a, HP2, and H3K9me2/3 (Figs. 3A, 4B, and 7B). This region has a six- to sevenfold higher density of transposon fragments than found in the euchromatic arms, similar to regions at the junction of pericentric heterochromatin and euchromatin on the other chromosomes (Kaminker et al. 2002). Nonetheless, approximately 80 genes are present, a density similar to that of the long chromosome arms. An investigation of the fourth chromosome using the white reporter P element discussed above (Fig. 2) found primarily heterochromatic domains (resulting in a variegating phenotype), with a few permissive domains (resulting in a red eye phenotype) interspersed (Sun et al. 2004). Analysis using modENCODE data indicates that the permissive domains are under Pc regulation (Riddle et al. 2012).
The differences in DNA sequence organization described above are reflected in differences in the chromatin biochemistry and/or the enzymes used to achieve it. Examination of the impacts of mutations in 70 different modifiers on different variegating genes (including wm4, bwD, or P element reporters in pericentric heterochromatin or in a TAS array) showed that there is substantial overlap in the targets of modifiers, but there is also surprising complexity. This set of tests divided the modifiers into seven different groups in terms of their ability to impact silencing in a given compartment (Donaldson et al. 2002). The only modifier in this group to impact silencing in the TAS array was a new allele of Su(var)3-9. TAS silencing is also sensitive to alleles of Psc and Su(z)2, two Pc group genes (Cryderman et al. 1999b). Using a similar approach, Phalke et al. (2009) identified modifiers that distinguished pericentric heterochromatin, the fourth chromosome, retrotransposons, and TAS sequences from each other.
Heterochromatic masses are seen mostly at the nuclear periphery and around the nucleolus. In Drosophila embryos, this tendency is even more pronounced. Early development in Drosophila is syncytial until nuclear division cycle 14, when cell walls form between the nuclei, creating the typical blastula, a ball of cells. Heterochromatic masses are first seen in early embryogenesis, as the nuclei move to the periphery of the egg. The heterochromatic material (centromeres, chromosome four) is concentrated at one side of the nucleus, oriented to the exterior surface of the egg (Foe and Alberts, 1985) (see Fig. 6C,D). Such spatial subdivision of the nucleus persists during development, leading to the concept of heterochromatin “compartments” within the nucleus (for more discussion of nuclear organization, see Dekker and Misteli 2014). These compartments might maintain a high concentration of factors required for heterochromatin formation (such as HP1a and the H3K9 HKMTs), while being depleted in factors required for euchromatin assembly and gene expression (such as HATs and RNA Pol II). Indeed, proximity to heterochromatic masses, both in position along the chromosome and in three dimensions, has been shown to be a factor in PEV.
Chromosomal proximity to the mass of pericentric heterochromatin has been shown to have an impact on variegation both for euchromatic genes (of which white is an example), and for heterochromatic genes, the best studied examples being light and rolled. Genes that normally reside in heterochromatic domains can be observed to variegate when a rearrangement places them in juxtaposition with euchromatin; generally, they show the opposite dependencies, requiring normal levels of HP1a for full expression, and showing an enhancement of variegation when HP1a is depleted. Variegation of light depends not only on its juxtaposition to euchromatin, but also on the position of the breakpoint, relative to the distance along the chromosome arm from heterochromatin (Wakimoto and Hearn 1990). Similar results have been reported for rolled. Investigations of bwD, a euchromatic gene induced to variegate by insertion of repetitious DNA, have shown that a shift in proximity of the locus to the pericentric heterochromatin can result in enhancement of silencing (if closer), or suppression of silencing (if further away) (Henikoff et al. 1995). Similarly, a reciprocal translocation that moves a fourth chromosome arm carrying a white reporter to the distal end of chromosome arm 2L or 2R results in a dramatic loss of silencing. The loss of silencing is correlated with a change in the position of the fourth chromosome fragment (now at the tip of the second chromosome arm) in the nucleus; it now frequently occupies sites in the salivary gland nucleus distant from the chromocenter (Cryderman et al. 1999b). These results suggest that proximity to a heterochromatic mass is necessary for effective silencing.
A recent study using high-resolution microscopy examined both gene activity (using antibodies specific for the product) and nuclear location of a reporter (using FISH, fluorescence in situ hybridization) in the same cell during the normal time frame of expression. A white variegating inversion, bwD, and a variegating lacZ transgene were studied in differentiating eye discs or adult eyes. This investigation found a strong inverse correlation between the position of the reporter gene in the cell relative to pericentric heterochromatin and the level of expression, supporting the idea that a heterochromatic “compartment” exists, and that positioning within this compartment is correlated with gene silencing (Harmon and Sedat 2005). However, the correlation is not absolute. This is not surprising, given the stochastic nature of PEV. Further, studies with an hsp70-white reporter indicate the presence of both permissive and silencing domains interspersed on the small fourth chromosome (which is always close to the mass of pericentromeric heterochromatin in wild-type cells), indicating that local determinants also contribute to the decision to package the chromatin in one form or the other (Sun et al. 2004).
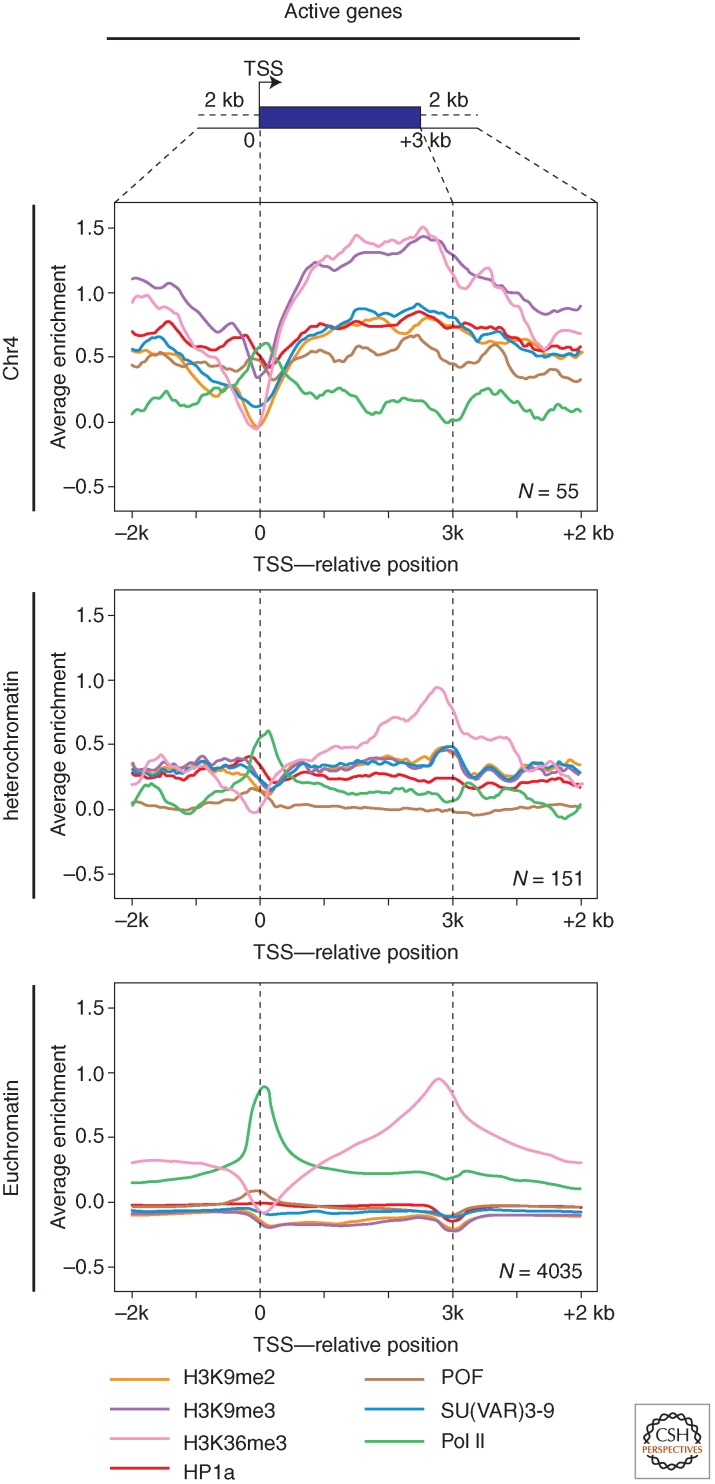
Genes normally residing in heterochromatin (light and rolled) function best in that domain, and show a loss of expression on depletion of HP1a. This is the opposite of what we see in PEV, in which our reporter genes (which normally function in euchromatin) show HP1a-dependent silencing. How do the genes in pericentric heterochromatin, or residing on the fourth chromosome, function in this presumably “hostile” environment? Examination of light using ChIP showed that although the region is generally enriched for H3K9me2, that mark is specifically depleted at the 5′ end of the gene (Yasuhara and Wakimoto 2008). The modENCODE project has allowed us to systematically examine the chromatin packaging for most pericentric and fourth chromosome genes to extend this analysis. There is indeed a conspicuous loss of silencing marks at the TSS of active genes in these domains (Fig. 8), although the usual heterochromatic marks (including H3K9me2) are still present upstream and across the body of the gene. As expected, the TSSs are occupied by RNA Pol II and are flanked downstream by nucleosomes with H3K4me2/3. Thus, these genes have “state 1” chromatin at their 5′ ends (around the TSSs), but state 7 chromatin over the body of the gene (Fig. 8). The presence of H3K9me3 and HP1a across the body of an active gene seems contradictory, but these marks are actually enriched there in preference to other sites (including intergenic spacer) on the fourth chromosome. The majority of the fourth chromosome genes show a loss of expression on HP1a depletion, showing a dependence on this chromatin structure (Riddle et al. 2012). How then is the gene expressed? POF (painting of fourth; Larsson et al. 2001), a protein found uniquely on the fourth chromosome, binds to nascent RNA and may play a role in transcript elongation (Johansson et al. 2007a; Johansson et al. 2012). And HP1a itself has been implicated in transcript elongation at some euchromatic sites (Piacentini et al. 2012). Although HP1a appears to bind to clusters of repeats on chromosome 4, as elsewhere in heterochromatin (utilizing H3K9me2/3 generated by SU(VAR)3-9), its association with fourth chromosome genes is dependent on POF (Johansson et al. 2007b; Riddle et al. 2012). This interaction may play a key role in facilitating transcription of fourth chromosome genes.
Figure 8.
Packaging of active genes in chromosome 4, pericentric heterochromatin, and euchromatin. The plots show log2 enrichment (y-axis) for RNA Pol II (green), H3K36me3 (pink), H3K9me2 (yellow), H3K9me3 (purple), SU(VAR)3-9 (blue), POF (brown), and HP1a (red) for a scaled metagene and 2 kb flanking region created by averaging data for all active genes in a given compartment. Fourth chromosome (top) and pericentric (middle) genes show a similar depletion in silencing marks at the TSS; these marks reappear over the body of the gene only in the case of the fourth chromosome. As expected, euchromatic genes do not show association with any of the silencing marks (bottom). (Adapted from Riddle et al. 2012.)
7. HOW IS HETEROCHROMATIN FORMATION TARGETED IN DROSOPHILA?
Although we have learned a great deal about the biochemistry of heterochromatin structure, this leaves open the question of how heterochromatin formation is targeted to the desired regions of the genome in its normal configuration. The analysis of the fourth chromosome suggests that the presence of local elements in the DNA can signal the formation or stabilize the presence of heterochromatin. Genetic screens for a switch in phenotype (from red to variegating or vice versa) have shown that local deletions or duplications of 5–80 kb of DNA flanking a fourth chromosome transposon reporter can lead to the loss or acquisition of variegation, pointing to short-range cis-acting determinants for silencing in this domain. This silencing is dependent on HP1a, and correlates with a change in chromatin structure from an accessible (euchromatic) to a closed (heterochromatic) state based on changes in the nucleosome array, as shown by nuclease accessibility assays (Sun et al. 2001). Mapping data in one region of the fourth chromosome implicate the 1360 transposon (and other TEs) as a target for heterochromatin formation, and suggest that once heterochromatin formation is initiated at dispersed repetitive elements, it can spread along the fourth chromosome for ∼10 kb, or until it encounters competition from a euchromatic determinant (Sun et al. 2004; Riddle et al. 2008). Short-range cis-acting determinants related to copy number are also implied by the observation that tandem or inverted repeats of reporter P elements result in heterochromatin formation and gene silencing (Dorer and Henikoff, 1994).
Such cis-acting elements in the DNA might function by sequence-specific binding of a protein capable of triggering heterochromatin formation. Proteins that bind specifically to some of the satellite DNAs have been identified, including D1, which is associated with the 1.672- and 1.688-g/cm3 AT-rich satellite repeats (Aulner et al. 2002). Their importance has been inferred from the impact of satellite-specific DNA-binding drugs. For example, the P9 polyamide binds the X-chromosome 1.688-g/cm3 satellite III, displacing the associated D1 and HP1a; this causes suppression of PEV in wm4h, indicating a mechanistic link (Blattes et al. 2006). Other specific DNA binding proteins of this type could well be responsible for heterochromatin formation at satellite DNAs in the pericentric heterochromatin.
The findings in yeast and plants (see Allshire and Ekwall 2014; Martienssen and Moazed 2014; Pikaard and Mittelsten Scheid 2014) suggest an additional model for dealing with TEs and their remnants by heterochromatization—one based on the RNAi system, able to recognize a diverse set of elements. Work from many laboratories has shown that the RNAi system is present in Drosophila and plays several important roles via posttranscriptional gene silencing. For example, there is a Dicer-1 dependent miRNA system impacting developmental regulation by messenger RNA degradation or translational inhibition, and a Dicer-2 dependent small interfering RNA (siRNA) system active in viral defense (reviewed in Kavi et al. 2008; Huisinga and Elgin 2009).
A Dicer-independent piRNA pathway exists, generating small piRNAs (24–30 nt) based on the cleavage activity of Piwi, Argonaute 3 (Ago3), and Aubergine (Aub), acting either alone or in a “ping-pong” mechanism to achieve a robust signal. rasiRNAs (repeat associated small interfering RNAs) generated by this pathway have been identified from 40% of the known TEs (including 1360) and other repeated sequences (Aravin et al. 2003). To test the idea of targeted silencing, a P element carrying a 1360 copy immediately adjacent to an hsp70-driven white reporter was inserted at many different sites in the genome. The presence of a single copy of this TE was insufficient to induce silencing at most sites in the eukaryotic arms, but a variegating phenotype was observed when the P element inserted into a repeat-rich region at the base of chromosome arm 2L. Thus, formation of stable heterochromatin appears to be dependent on the nuclear location as well as a specific target; perhaps the spatial requirements are tied to the need for an abundant pool of heterochromatin proteins. The degree of silencing was dependent on the presence of the 1360 copy. This targeted silencing is dependent both on HP1a and SU(VAR)3-9, and on components of the rasiRNA pathway (Haynes et al. 2006). A larger screen identified many more 1360-sensitive sites, some in euchromatic domains lying close to heterochromatic masses (base of chromosome 2L). In this type of insertion site, 1360 can drive heterochromatin formation (HP1a accumulation) at a normally euchromatic position. Use of a “landing pad” construct that allows one to replace the 1360 element with an altered copy led to the conclusion that the piRNA hotspots, but not the repetitious end sequences or putative TSSs, are critical for this 1360-dependent silencing (Sentmanat and Elgin, 2012). This suggests a recognition event dependent on the piRNA system.
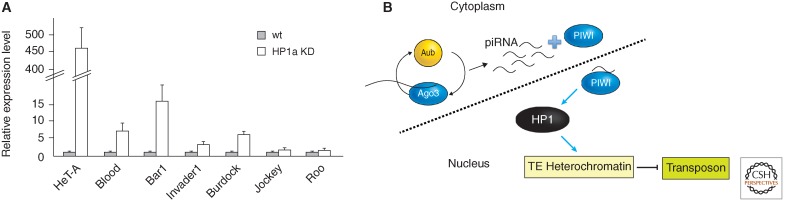
rasiRNAs (including piRNAs) are abundant in the female germline, where they clearly play a role in silencing TEs (Senti and Brennecke, 2010). Whether this role encompasses transcriptional silencing as well as posttranscriptional silencing is the question of interest here. In a genetic test, Pal-Bhadra et al. (2004) found that mutations in piwi (a member of the PAZ domain family) and homeless (a DEAD box helicase) suppress the PEV associated with tandem arrays of the white gene. Mutations in piwi, aubergine, and homeless (aka spn-e) suppress silencing of the white transgene P[hsp70-w] in pericentric heterochromatin or the fourth chromosome. The amount of rasiRNAs produced for a wide range of retroelements is significantly reduced in ovaries of flies carrying a mutation in spn-E, with concomitant depletion of HP1a at these TEs (Klenov et al. 2007). In the fly female germline, among the argonaute family of proteins (that bind rasiRNA), only Piwi is found to be a predominantly nuclear protein, and it has been reported to interact with HP1a (Brower-Toland et al. 2007). Specific depletion of HP1a in the female germline results in overexpression of some (but not all) TEs assayed, indicating a role for heterochromatin in silencing these elements (Fig. 9A). Germline depletion of Piwi also leads to a loss of silencing for this group of TEs, with concomitant loss of HP1a and H3K9me2 association. Piwi appears to function downstream of Aub here, suggesting that it is utilizing the products of the ping-pong rasiRNA system (Wang and Elgin 2011). A mutation that removes the nuclear localization signal of Piwi similarly leads to TE overexpression, with a shift in chromatin structure, demonstrating that Piwi is required to be in the nucleus for these functions (Klenov et al. 2011). These results support a model in which piRNAs, generated in the female germline and transported to the nucleus by Piwi, can promote HP1a deposition at target TEs (Fig. 9B). However, although HeT-A, Blood, Bari, and many other TEs are reported to be significantly affected, other TEs are not, including Jockey and Roo. It is apparent that there are multiple, redundant ways to silence TEs; it is not yet clear what determines the sensitivity of a given TE to a given mechanism.
Figure 9.
The piRNA system may drive silencing of some TEs through heterochromatin formation. (A) Depletion of HP1a (shown here), or of Piwi, results in overexpression of a subset of the TEs in the female germline. (B) This suggests that Piwi can promote transcriptional gene silencing, directly or indirectly, by targeting HP1a deposition at these TEs, presumably by a piRNA recognition event. (Adapted from Wang and Elgin 2011.)
Although a model using germline Piwi with associated rasiRNAs to direct silencing by heterochromatin formation at TEs is attractive, the evidence is as yet indirect. Any mutation that disrupts TE silencing (whether at the transcriptional or the posttranscriptional level) will result in TE mobilization and trigger a DNA damage response, which could lead to further destabilization of the genome (Khurana et al. 2011). The above experiments have focused on the germline, where the need for TE silencing is greatest, and Piwi is most abundant; similar effects are not seen in adult somatic cells (Klenov et al. 2007). However, the nurse cells deposit both RNA and protein into the oocyte, and could supply the needed materials for such Piwi-dependent targeted heterochromatin formation at blastoderm. Alternatively, an siRNA pathway has been suggested to operate in somatic cells based on the observation that expression of viral suppressor proteins results in suppression of PEV, with associated changes in chromatin structure (Fagegaltier et al. 2009).
8. PEV, HETEROCHROMATIN FORMATION, AND GENE SILENCING IN DIFFERENT ORGANISMS
The phenomenon of position-effect variegation was initially detected in Drosophila, simply because this was one of the first organisms for which X irradiation was used to induce mutations. X irradiation is much more likely than other commonly used mutagens to induce chromosomal rearrangements that can result in PEV. Similar mutations have been isolated from the mouse, in which variegating coat color indicates PEV (see Blewitt and Whitelaw 2013). For example, the insertion of an autosomal region carrying a fur color gene onto the X chromosome results in variable silencing of the allele (Russel and Bangham 1961; Cattanach 1961). Variegation, however, is only observed in females carrying this insertion combined with a homozygous mutation in the original coat color gene. This is because the translocated wild-type allele becomes inactivated as a consequence of X inactivation by heterochromatinization (see Brockdorff and Turner 2014). In plants the only unequivocal case of PEV that has been described was reported in Oenothera blandina (Catcheside 1939). In these cases, as is the case in Drosophila, PEV silencing of euchromatic genes is connected with placement of those genes into a new heterochromatic neighborhood.
Transcriptional gene silencing has also been observed for repeated sequences (repeat induced gene silencing), particularly in plants (Baulcombe and Dean 2014). Analysis of the affected sequences has revealed the appearance of similar epigenetic marks (histone modifications and DNA methylation) as found in heterochromatin and in regions silenced by PEV. For example, if DNA fragments containing tandemly arranged luciferase genes are introduced into Arabidopsis, one observes either complete silencing or variegated luciferase expression. In the variegated line, the Arabidopsis SU(VAR)3-9 homolog SUVH2 shows a dosage dependent effect on silencing, suggesting that heterochromatin formation is responsible for the gene silencing observed and that the underlying molecular mechanisms are similar to those seen in other higher eukaryotic organisms (Naumann et al. 2005).
A central feature of pericentric heterochromatic gene silencing in Drosophila is the interaction of HP1a with H3K9me2/me3 and SU(VAR)3-9, a histone H3K9 HKMT. These three components of heterochromatin are highly conserved and found in the majority of eukaryotic taxa. HP1a is conserved from the fission yeast Schizosaccharomyces pombe to man, and is consistently associated with pericentric heterochromatin. The human HP1a genes can even be used to rescue the deficiency in Drosophila (Ma et al. 2001). However, a protein that binds to H3K9me2/me3, like HP1a, has not been identified in plants as such, and a clear homolog is lacking in some metazoans, such as Caenorhabditis elegans. SU(VAR)3-9 is even more widely represented, having been identified in fission yeast (clr4p), Neurospora (DIM5), Arabidopsis and mammals (SUV39h). All of these SU(VAR)3-9 homologs catalyze H3K9 methylation and function in heterochromatin formation. Also, as is the case for HP1a, a human SUV39H1 transgene can completely compensate for the loss of the endogenous Drosophila protein in mutant lines (Schotta et al. 2002). In higher plants (rice, Arabidopsis, and maize), there are in fact several SU(VAR)3-9 homologous proteins (SUVH) (Baumbusch et al. 2001). The high number of HKMTs may reflect the plasticity of plant development, or the need to respond to environmental factors (discussed further in Pikaard and Mittelsten Scheid 2014).
Several other genes identified by Drosophila Su(var) mutations encode proteins with conserved functions. For instance, many of the key enzymes controlling histone modification are evolutionarily conserved, supporting the idea of a histone code (Jenuwein and Allis 2001). However, examination of the heterochromatin-specific histone modification marks observed in Drosophila, mammals, and plants (Arabidopsis) also identifies some genus-specific features (cf. Ebert et al. 2006), arguing that the histone code is not completely universal, but rather exists in different dialects.
9. SUMMING UP: THERE IS MUCH THAT WE DO NOT KNOW ABOUT HETEROCHROMATIN
Although PEV has provided us with an extraordinary opportunity to study heterochromatin formation and gene silencing, the development of the phenotype itself remains puzzling. Studies with an hsp70-driven lacZ reporter (that can be scored at any point in development and in most tissues) show that silencing begins in early embryogenesis, and is most extensive at cellular blastoderm, after which islands of inducible cells emerge (Lu et al. 1996). Events that occur during embryogenesis can impact the adult phenotype (Hartmann-Goldstein 1967). This persistence is of course implied by the concept of an epigenetic state: once formed at a specific site, heterochromatin will be maintained at that site through many rounds of mitosis by replication of the chromatin assembly. However, this “memory” has not yet been directly tested in Drosophila.
Many questions remain. Why do we observe a variegating pattern of silencing? What tips the balance, leading to a switch from the silent to the active state? When can that happen? What sets the patterns that indicate clonal inheritance, but are not easily related to the developmental process? Can faulty heterochromatin be restored? PEV is generally analyzed as a problem of maintaining the reporter gene “on” or “off,” but in many instances (particularly when using P element based reporters) one observes red facets on a yellow or pale orange background, suggesting that gene expression has been reduced uniformly, but that that down-regulation has been lost in some cells. Careful analysis of such lines might lead to identification of chromatin states with an intermediate impact on gene expression. Although the data support a crude model for loss or maintenance of silencing based on mass action, the final model will be complex, involving numerous interacting proteins (see, e.g., the proposal by Henikoff, 1996). One is tempted to consider the nucleosome as a summation device, collecting modifications and displaying the results within a chromosomal domain in terms of both particular protein binding patterns, and facility for remodeling. The chromatin state might then reflect the results of competition for achieving different modifications. Such a model could be useful in sorting out the effects noted above. It is also compatible with observations demonstrating that the frequency of silencing of a GAL4-dependent reporter is sensitive to GAL4 levels (Ahmad and Henikoff 2001).
As might be anticipated for a system dependent on approximately 150 modifier loci, PEV phenotypes frequently differ in penetrance and expression of variegation in different inbred laboratory strains. For example, several genetically stable lines of In(1)wm4 have been identified that differ significantly in expression of white variegation. In some cases the difference is linked to the heterochromatic breakpoint, suggesting that spontaneous alterations at the inactivating heterochromatic sequences are involved (Reuter et al. 1985). Variations in copy number of repeated sequences among strains could also have a significant effect on the degree of variegation caused by the titration of a fixed amount of key proteins required for heterochromatic packaging. Such changes may be more frequent in lines carrying Su(var) mutations, where heterochromatin structure is less stable (Peng and Karpen 2009).
It is standard technique to use inbred Drosophila lines with strong variegation when screening for or assessing putative Su(var) loci. However, fly genetics is not performed with homozygous lines and genetic background must always be carefully controlled. Different laboratories will make different choices of which reporter loci etc. to test, resulting in the presence of different chromosomes. It seems likely that the resulting differences in genetic background are the basic cause of some recent controversies in the field. The observed redundancies in the silencing system coupled with the absence of a recent transposon challenge in laboratory stocks appear to have allowed more variation in silencing system components than we might anticipate. This provides us with a very rich set of starting materials for investigating differences in penetrance and expressivity, a critical issue in understanding the health impact of such mutations in humans. We note that many basic features of the heterochromatin silencing system, first shown in Drosophila, have proven to be generally applicable.
The RNAi system provides a plausible mechanism for targeting heterochromatin formation to silence TEs in the germline, presumably by targeting a complex including HP1a, an H3K9 HKMT, or both. RNAi systems have repeatedly been found to play a role in genome surveillance and modification (see Chalker et al. 2013). However, many questions remain for Drosophila. What is the source of the dsRNA? Must it be produced in cis (as implied by the results in S. pombe), or can it operate in trans (as suggested by results in plants) (i.e., can the production of dsRNA from one 1360 site result in targeting of all 1360 sites)? Must the target site be transcribed? Are all repetitious elements potential targets? The latter seems unlikely from the fourth chromosome analysis described above. If a subset of repetitious elements plays a key role, what determines that choice? The results obtained on the fourth chromosome indicate that the density and distribution of critical repetitious elements will impact expression of the genes in the vicinity. Thus, sequencing of an entire genome, not just the protein coding regions, is likely to be necessary when studying a novel organism.
How is spreading of heterochromatin accomplished, and what are the normal barriers to spreading? Note that there is no evidence for transitive RNAi, originally discovered in plants, in Drosophila (i.e., the spread of silencing to target genes that lie upstream of an introduced sequence that generates dsRNA [Celotto and Gravely 2002]). This is congruent with the lack of evidence for any RNA-dependent RNA polymerase in this system. An assembly system based on the interactions of HP1a, H3K9me2/3, and an HKMT might well account for the spread of heterochromatin for approximately 10 kb, as observed on the fourth chromosome; this type of spreading could be limited by a site of histone acetylation. But what about the spreading that occurs in rearrangements, which has been found to extend for hundreds of kb, as shown in Figure 5C? This form of spreading is not contiguous, but again appears to depend critically on chromatin proteins, notably JIL-1, in a role that does not depend on its kinase activity.
These and other questions remain unanswered. There is much to do to understand the heterochromatin system in Drosophila.
ACKNOWLEDGMENTS
We thank Gabriella Farkas for creating several of the original drawings used here, and the members of our research groups for a critical review of this article. Our work is supported by Deutsche Forschungsgemeinschaft (DFG) (G.R.) and by grants from the National Institutes of Health (S.C.R.E.).
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ahmad K, Golic KG 1996. Somatic reversion of chromosomal position effects in Drosophila melanogaster. Genetics 144: 657–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S 2001. Modulation of a transcription factor counteracts heterochromatin gene silencing in Drosophila. Cell 104: 839–847 [DOI] [PubMed] [Google Scholar]
- *.Allshire R, Ekwall K 2014. Epigenetics in Saccharomyces pombe. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T 2003. The small RNA profile during Drosophila melanogaster development. Dev Cell 5: 337–350 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS 2005. Chromosomes and position effect variegation. In Drosophila: A laboratory handbook, 2nd ed., Chaps. 4, 28 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Aulner N, Monod C, Mandicourt G, Jullien D, Cuvier O, Sall A, Janssen S, Laemmli UK, Kas E 2002. The AT-hook protein D1 is essential for Drosophila melanogaster development and is implicated in position-effect variegation. Mol Cell Biol 22: 1218–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksa K, Morawietz H, Dombradi V, Axton M, Taubert H, Szabo G, Török I, Gyurkovics H, Szöör B, Gloover D, et al. 1993. Mutations in the phosphatase 1 gene at 87B can differentially affect suppression of position-effect variegation and mitosis in Drosophila melanogaster. Genetics 135: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Baulcombe D, Dean C 2014. Epigenetic regulation in plant responses to the environment. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen R 2001. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionary conserved classes. Nucleic Acid Res 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva ES, Demakova OV, Umbetova GH, Zhimulev IF 1993. Cytogenetic and molecular aspects of position-effect variegation in Drosophila melanogaster. V. Heterochromatin-associated protein HP1 appears in euchromatic chromosomal regions that are inactivated as a result of position-effect variegation. Chromosoma 102: 583–590 [DOI] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Rabinow L, Linsk R, Nguyen-Huyuh AT 1994. Weakener of white (Wow), a gene that modifies the expression of white eye color locus and that suppresses position effect variegation in Drosophila melanogaster. Genetics 137: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Blattes R, Monod C, Susbielle G, Cuvier O, Wu J-H, Hsieh T-S, Laemmli UK, Kas E 2006. Displacement of D1, HP1 and topoisomerase II from satellite heterochromatin by a specific polyamide. EMBO J 25: 2397–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Blewitt M, Whitelaw E 2013. The use of mouse models to study epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Brockdorff N, Turner B 2014. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SCR, Lin H 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21: 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Riddle NC, Jiang H, Huisinga KL, Elgin SCR 2009. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 181: 1303–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catcheside DG 1939. A position effect in Oenothera. J Genet 38: 345–352 [Google Scholar]
- Cattanach BM 1961. A chemically-induced variegated-type position effect in the mouse. Z Vererbungsl 92: 165–182 [DOI] [PubMed] [Google Scholar]
- Celotto AM, Graveley BR 2002. Exon-specific RNAi: A tool for dissecting the functional relevance of alternative splicing. RNA 8: 718–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chalker DL, Meyer E, Mochizuki K 2013. Epigenetics of ciliates. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F, Spierer P 2001. Position-effect variegation in Drosophila: The modifier Su(var)3-7 is a modular DNA-binding protein. EMBO Rep 21: 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard F, Delattre M, Spierer P 1997. SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J 16: 5280–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KW 1959. Cytogenetic analysis of major heterochromatic elements (especially Xh and Y) in Drosophila melanogaster and the theory of “heterochromatin”. Chromosoma 10: 535–588 [DOI] [PubMed] [Google Scholar]
- Cortes A, Azorin F 2000. DDP1, a heterochromatin-associated multi-KH-domain protein of Drosophila melanogaster, interacts specifically with centromeric satellite DNA sequences. Mol Cell Biol 20: 3860–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman DE, Tang H, Bell C, Gilmour DS, Wallrath LL 1999a. Heterochromatic silencing of Drosophila heat shock genes acts at the level of promoter potentiation. Nucleic Acids Res 27: 3364–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman DE, Morris EJ, Biessmann H, Elgin SCR, Wallrath LL 1999b. Silencing at Drosophila telomeres: Nuclear organization and chromatin structure play critical roles. EMBO J 18: 3724–3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman DE, Grade SK, Li Y, Fanti L, Pimpinelli S, Wallrath LL 2005. Role of HP1 in euchromatic gene expression. Dev Dyn 232: 767–774 [DOI] [PubMed] [Google Scholar]
- Czermin B, Schotta G, Hülsmann BB, Brehm A, Becker PB, Reuter G, Imhof A 2001. Physical and functional interaction of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep 2: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dekker J, Misteli T 2014. Long-range chromatin interactions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Spierer A, Tonka CH, Spierer P 2000. The genomic silencing of position-effect variegation in Drosophila melanogaster: Interaction between the heterochromatin-associated proteins Su(var)3-7 and HP1. J Cell Sci 113: 4253–4261 [DOI] [PubMed] [Google Scholar]
- Demerec M, Slizynska H 1937. Mottled white 258-18 of Drosophila melanogaster. Genetics 22: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Lui A, Karpen GH 2002. Modifiers of terminal deficiency-associated position effect variegation in Drosophila. Genetics 160: 995–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S 1994. Expansion of transgene repeats causes heterochromatin formation and gene silencing in Drosophila. Cell 77: 993–1002 [DOI] [PubMed] [Google Scholar]
- Dorn R, Krauss V, Reuter G, Saumweber H 1993. The enhancer of position-effect variegation E(var)93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proc Natl Acad Sci 90: 11376–11380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G 2004. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev 18: 2973–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, Lein S, Schotta G, Reuter G 2006. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res 14: 377–392 [DOI] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, et al. 2011. An assessment of histone-modification antibody quality. Nat Struct Mol Biol 18: 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Reuter G 2009. Cellular mechanism for targeting heterochromatin formation in Drosophila. Int Rev Cell Mol Biol 273: 1–47 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SCR 1990. A mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position effect variegation in Drosophila melanogaster. Proc Natl Acad Sci 87: 9923–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Morris GD, Reuter G, Hartnett T 1992. The hetero-chromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Eberharter A, Imhof A 2007. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol Cell Biol 27: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D, Bouge A-L, Berry B, Poisot E, Sismeiro O, Coppee J-Y, Theodore L, Voinnet O, Antoniewski C 2009. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci 106: 21258–21263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L, Pimpinelli S 2004. Immunostaining of squash preparations of chromosomes of larval brains. Methods Mol Biol 247: 353–361 [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 2: 527–538 [DOI] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Krach F 1994. The trithorax-like gene encodes the Drosophila GAGA factor. Nature 371: 806–808 [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase 2012. The Drosophila database. Available from the World Wide Web at the URLs http://morgan/harvard.edu and http://www.ebi.ac.uk/flybase/.
- Fodor DB, Shukeir N, Reuter G, Jenuwein T 2010. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol 26: 471–501 [DOI] [PubMed] [Google Scholar]
- Foe VE, Alberts BM 1985. Reversible chromosome condensation induced in Drosophila embryos by anoxia: Visualization of interphase nuclear organization. J Cell Biol 100: 1623–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girton JR, Johansen KM 2008. Chromatin structure and the regulation of gene expression: The lessons of PEV in Drosophila. Adv Genet 61: 1–43 [DOI] [PubMed] [Google Scholar]
- Greil F, de Wit E, Bussemaker HJ, van Steensel B 2007. HP1 controls genomic targeting of four novel heterochromatin proteins in Drosophila. EMBO J 26: 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R 2014. Transcriptional silencing by Polycomb group proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon B, Sedat J 2005. Cell-by-cell dissection of gene expression and chromosomal interactions reveals consequences of nuclear reorganization. PLoS Biol 3: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Goldstein IJ 1967. On the relationship between heterochromatization and variegation in Drosophila, with special reference to temperature sensitive periods. Genet Res 10: 143–159 [DOI] [PubMed] [Google Scholar]
- Haynes KA, Caudy AA, Collins L, Elgin SCR 2006. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr Biol 16: 2222–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S 1996. Dosage-dependent modification of position-effect variegation in Drosophila. Bioessays 18: 401–409 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Jackson JM, Talbert PB 1995. Distance and pairing effects on the brownDominant heterochromatic element in Drosophila. Genetics 140: 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, Wan KH, Park S, Mendez-Lago M, Rossi F, et al. 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M, Dimitri P, Berloco M, Wakimoto BT 1995. Cis-effects of heterochromatin on heterochromatic and euchromatic gene activity in Drosophila melanogaster. Genetics 140: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Elgin SCR 2009. Small RNA-directed heterochromatin formation in the context of development: What flies might learn from fission yeast. Biochim Biophys Acta 1789: 3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Eissenberg JC, Allis CD, Khorasanizadeh S 2001. Specificity of the HP1 chromo domain fort he methylated N-terminus of histone H3. EMBO J 20: 5232–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Elgin SCR 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol 6: 3862–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SCR 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol 50: 170–180 [PubMed] [Google Scholar]
- Jenuwein T, Allis CD 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Johansson AM, Stenberg P, Pettersson F, Larsson J 2007a. POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet 3: e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson AM, Stenberg P, Bernhardsson C, Larsson J 2007b. Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J 26: 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson AM, Stenberg P, Allgardsson A, Larsson J 2012. POF regulates the expression of genes on the fourth chromosome in Drosophila melanogaster by binding to nascent RNA. Mol Cell Biol 32: 2121–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. 2002. The transposable elements of the Drosophila melanogaster genome: A genomics perspective. Genome Biol 3: RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC 1990. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell 63: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi HH, Fernandez H, Xie W, Birchler JA 2008. Genetics and biochemistry of RNAi in Drosophila. Curr Top Microbiol Immunol 320: 37–75 [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Wenig Z, Theurkauf WE 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147: 1551–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kingston R, Tamkun J 2014. Transcriptional regulation by Trithorax group proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA 2007. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35: 5430–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA 2011. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci 108: 18760–18765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss V, Reuter G 2011. DNA methylation in Drosophila—A critical evaluation. Prog Mol Biol Transl Sci 101: 177–191 [DOI] [PubMed] [Google Scholar]
- *.Kuroda M, Lucchesi J 2014. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebesorger A, Popkin D, Pillus L, Jenuwein T 1997. Mammalian homologs of Enhancer of zeste mediate position-effect variegation in Drosophila and restore telomeric silencing in S. cerevisiae. EMBO J 16: 3219–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Zhang J, Rasmuson-Lestander A 1996. Mutations in the Drosophila melanogaster S-adenosylmethionine synthase suppress position-effect variegation. Genetics 143: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Chen JD, Rasheva V, Rasmuson-Lestander A, Pirrotta V 2001. Painting of fourth, a chromosome-specific protein in Drosophila. Proc Natl Acad Sci 98: 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MH, Duricka D, Karpen GH 1995. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics 141: 283–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Branco A, Hartl D 2010. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci 107: 15826–15831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J, Kotarski MA, Tartof KD 1988. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics 120: 181–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BY, Bishop CP, Eissenberg JC 1996. Developmental timing and tissue specificity of heterochromatin-mediated silencing. EMBO J 15: 1323–1332 [PMC free article] [PubMed] [Google Scholar]
- Ma J, Hwang KK, Worman HJ, Courvalin JC, Eissenberg JC 2001. Expression and functional analysis of three isoforms of human heterochromatin-associated protein HP1 in Drosophila. Chromosoma 109: 536–544 [DOI] [PubMed] [Google Scholar]
- Martienssen RA, Colot V 2001. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293: 1070–1074 modMine. 2011. Available on the World Wide Web at the URL http://intermine.modencode.org/ [DOI] [PubMed] [Google Scholar]
- *.Martienssen R, Moazed D 2014. RNAi and heterochromatin assembly. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottus R, Sobels RE, Grigliatti TA 2000. Mutational analysis of a histone deacetylase in Drosophila melanogaster: Missence mutations suppress gene silencing associated with position effect variegation. Genetics 154: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ 1930. Types of visible variations induced by X-rays in Drosophila. J Genet 22: 299–334 [Google Scholar]
- Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, Hause G, Aurich AC, Dorn R, Jenuwein T, et al. 2005. Pivotal role of AtSUVH2 in control of heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J 24: 1418–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SCR 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672 [DOI] [PubMed] [Google Scholar]
- *.Patel D 2014. A structural perspective on readout of epigenetic H and DM marks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Karpen G 2009. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet 5: e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalke S, Nickel O, Walluscheck D, Hortig F, Onorati MC, Reuter G 2009. Epigenetic control of retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nature Genet 41: 696–702 [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Negri R, Del Vescovo V, Fatica A, Altieri F, Pimpinelli S 2009. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet 5: e1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pikaard C, Mittelsten Scheid O 2014. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero JS, Sharp EJ, Adler PN, Eissenberg JC 1996. In vivo assay for protein-protein interaction using Drosophila chromosomes. Chromosoma 104: 393–404 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun Z-W, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Reuter G, Werner W, Hofmann HJ 1982. Mutants affecting position-effect heterochromatinization in Drosophila melanogaster. Chromosoma 85: 539–551 [DOI] [PubMed] [Google Scholar]
- Reuter G, Wolff I, Friede B 1985. Functional properties of the heterochromatic sequences inducing wm4 position-effect variegation in Drosophila melanogaster. Chromosoma 93: 132–139 [Google Scholar]
- Reuter G, Giarre N, Farah J, Gausz J, Spierer A, Spierer P 1990. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature 344: 219–223 [DOI] [PubMed] [Google Scholar]
- Riddle NC, Leung W, Haynes KA, Granok H, Wuller J, Elgin SCR 2008. An investigation of heterochromatin domains on the fourth chromosome of Drosophila melanogaster. Genetics 178: 1177–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, Gorchakov AA, Kennedy C, Linder-Basso D, Jaffe JD, et al. 2010. Plasticity in patterns of histone modifications and chromosomal proteins in the Drosophila heterochromatin. Genome Res 21: 147–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, Gorchakov AA, Jaffe JD, Kennedy C, Linder-Basso D, et al. 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res 21: 147–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Jung Y, Gu T, Alekseyenko AA, Asker D, Gui H, Kharchenko PV, Minoda A, Plachetka A, Schwartz YB, et al. 2012. Enrichment of HP1a on Drosophila chromosome 4 genes creates an alternate chromatin structure critical for regulation in this heterochromatic domain. PLoS Genet 8: e1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt Q, Jenuwein T, et al. 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell 13: 103–115 [DOI] [PubMed] [Google Scholar]
- Russel LB, Bangham JW 1961. Variegated type position effects in the mouse. Genetics 46: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiderman JI, Goldstein S, Ahmad K 2010. Perturbation analysis of heterochromatin-mediated gene silencing and somatic inheritance. PLoS Genet 6: e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Reuter G 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J 21: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Dorn R, Reuter G 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol 14: 67–75 [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J 1950. Interrelations of factors affecting heterochromatin-induced variegation in Drosophila. Genetics 35: 134 [Google Scholar]
- Schulze SR, Wallrath LL 2007. Gene regulation by chromatin structure: Paradigms established in Drosophila melanogaster. Annu Rev Entomol 52: 171–192 [DOI] [PubMed] [Google Scholar]
- Senti KA, Brennecke J 2010. The piRNA pathway: A fly’s perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentmanat MF, Elgin SCR 2012. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc Natl Acad Sci 109: 14104–14109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Pauli D, Delattre M, Jaquet Y, Spierer A, Spierer P 2002. Isolation of Su(var)3-7 mutations by homologous recombination in Drosophila melanogaster. Genetics 161: 1125–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Reo E, Peng H, Rauscher FJ, Spierer P, Bontron S 2007. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet 3: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer CD, Stephens GE, Thompson BA, Funches L, Bernat JA, Craig CA, Elgin SCR 2002. Heterochromatin protein 2 (HP2), a partner of HP1 in Drosophila heterochromatin. Proc Natl Acad Sci 99: 14322–14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer C, Cenci G, Thompson B, Stephens GE, Slawson E, Adu-Wusu K, Gatti M, Elgin SCR 2006. The large isoform of Drosophila melanogaster heterochromatin protein 2 plays a critical role in gene silencing and chromosome structure. Genetics 174: 1189–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y 2005. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Silver LM, Elgin SCR 1976. A method for determination of the in situ distribution of chromosomal proteins. Proc Natl Acad Sci 73: 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DAR, Mottus RC, Grigliatti TA 1983. Genes which suppress position effect variegation in Drosophila melanogaster are clustered. Mol Gen Genet 191: 326–333 [Google Scholar]
- Sinclair DAR, Clegg NJ, Antonchuk J, Milner TA, Stankunas K, Ruse C, Grigliatti TA, Kassis J, Brock HW 1998. Enhancer of Polycomb is a suppressor of position-effect variegation in Drosophila melanogaster. Genetics 148: 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Shu S, Mungall CJ, Karpen GH 2007. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science 316: 1586–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spofford JB 1967. Single-locus modification of position-effect variegation in Drosophila melanogaster. I. White variegation. Genetics 57: 751–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens GE, Craig CA, Li Y, Wallrath LL, Elgin SCR 2003. Immunofluorescent staining of polytene chromosomes: Exploiting genetic tools. Methods Enzymol 376: 372–393 [DOI] [PubMed] [Google Scholar]
- Sun F-L, Cuaycong MH, Elgin SCR 2001. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentromeric heterochromatin. Mol Cell Biol 21: 2867–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F-L, Haynes K, Simpson CL, Lee SD, Collins L, Wuller J, Eissenberg JC, Elgin SCR 2004. cis-Acting determinants of heterochromatic formation on Drosophila melanogaster chromosome four. Mol Cell Biol 24: 8210–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S 2000. A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics 154: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof KD, Hobbs C, Jones M 1984. A structural basis for variegating position effects. Cell 37: 869–878 [DOI] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G 1994. The protein encoded by the Drosophila position effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J 13: 3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng TY, Lee CH, Chan LW, Shen CK 2007. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methylatransferase dSETDB1. Proc Natl Acad Sci 104: 12691–12696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Peric-Hupkes D, van Steensel B 2007. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc 2: 1467–1478 [DOI] [PubMed] [Google Scholar]
- Vogel MJ, Pagie L, Talhout W, Nieuwland M, Kerkhoven RM, van Steensel B 2009. High-resolution mapping of heterochromatin redistribution in a Drosophila position-effect variegation model. Epigenetics Chromatin 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto BT, Hearn MG 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SCR 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9: 1263–1277 [DOI] [PubMed] [Google Scholar]
- Wallrath LL, Gunter VP, Rosman LE, Elgin SCR 1996. DNA representation of variegating heterochromatic P element inserts in diploid and polytene tissue of Drosophila melanogaster. Chromosoma 104: 519–527 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM 2001. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105: 433–443 [DOI] [PubMed] [Google Scholar]
- Wang SH, Elgin SCR 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci 108: 21164–21169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT 2008. Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones. PLoS Genet 4: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev IF, Belyaeva ES, Fomina OV, Protopopov MO, Bolshkov VN 1986. Cytogenetic and molecular aspects of position effect variegation in Drosophila melanogaster. Chromosoma 94: 492–504 [Google Scholar]