Abstract
Patterns of replication within eukaryotic genomes correlate with gene expression, chromatin structure, and genome evolution. Recent advances in genome-scale mapping of replication kinetics have allowed these correlations to be explored in many species, cell types, and growth conditions, and these large data sets have allowed quantitative and computational analyses. One striking new correlation to emerge from these analyses is between replication timing and the three-dimensional structure of chromosomes. This correlation, which is significantly stronger than with any single histone modification or chromosome-binding protein, suggests that replication timing is controlled at the level of chromosomal domains. This conclusion dovetails with parallel work on the heterogeneity of origin firing and the competition between origins for limiting activators to suggest a model in which the stochastic probability of individual origin firing is modulated by chromosomal domain structure to produce patterns of replication. Whether these patterns have inherent biological functions or simply reflect higher-order genome structure is an open question.
Replication timing is influenced at two levels: (1) local sequence and chromatin composition, and (2) the 3D structure of chromosomes. In organisms with large genomes, the latter seems to be more important.
Replication of eukaryotic chromosomes takes place in segments that generally replicate in a predictable temporal order. Because the rate of elongation of replication forks varies little throughout S phase, this “replication timing program” is largely mediated by the time of initiation of replication within the corresponding segments. However, it is the temporal order of replication, not the sites of initiation, that is conserved among species (Aladjem et al. 2002; Farkash-Amar et al. 2008; Liachko et al. 2010; Ryba et al. 2010; Yaffe et al. 2010; Di Rienzi et al. 2012; Muller and Nieduszynski 2012; Xu et al. 2012), suggesting that replication timing is regulated independently of mechanisms specifying origins. Despite this evolutionary conservation, the biological significance of replication timing has remained elusive. In multicellular but not unicellular organisms, early replication is correlated with transcriptional activity and is developmentally regulated (Hiratani et al. 2009), but causal links have not been established. Recent findings establish the importance of large-scale chromatin folding in the regulation of replication timing in both yeasts and mammals and provide new promise for our understanding of both the significance and mechanism of the replication program. These results suggest a unifying model of how structural compartmentalization of the genome in the eukaryotic nucleus can influence overall functional output.
In this chapter, we will begin by summarizing the important contributions of genome-scale methods to the field. Next we will examine the relationships between replication timing and the three-dimensional (3D) organization of chromosomes in the nucleus. We will follow this with a discussion of mechanistic insights and conclude with speculation on the potential biological significance of a replication timing program. Along the way, we will refer the reader to many outstanding recent reviews for more detailed discussion of various aspects of this complex topic.
CONTRIBUTIONS FROM GENOME-SCALE METHODS
Although pioneering genome-scale studies of replication timing were performed more than 10 years ago (Donaldson and Schildkraut 2006), they have more recently been applied to many different cell types and experimental conditions, providing a comprehensive view of the temporal program and a robust tool for experimentation. In fact, replication timing profiles are such a stable characteristic of particular cell types that they can be used for cell type identification (Pope et al. 2011; Ryba et al. 2011). Details of these methods have been described in several recent reviews (Farkash-Amar and Simon 2010; Raghuraman and Brewer 2010; Ryba et al. 2011). Here we will focus on the salient findings from these studies. Table 1 compiles a list of published genome-wide replication timing data sets for various species at the time of this writing.
Table 1.
Genome-wide replication profiles
| Species | Cell type | Cell line | References |
|---|---|---|---|
| Homo sapiens | hESC | H1 | Desprat et al. 2009; Ryba et al. 2011 |
| H7 | Ryba et al. 2010 | ||
| H9 | Ryba et al. 2010 | ||
| BG01 | Ryba et al. 2010 | ||
| BG02 | Hansen et al. 2010; Ryba et al. 2010 | ||
| hiPSC | iPSC4 | Ryba et al. 2010 | |
| iPSC5 | Ryba et al. 2010 | ||
| Definitive endoderm | BG02-derived | Ryba et al. 2011 | |
| Mesendoderm | BG02-derived | Ryba et al. 2011 | |
| Mesoderm | BG02-derived | Ryba et al. 2011 | |
| Smooth muscle | BG02-derived | Ryba et al. 2011 | |
| Neural precursors | BG01-derived | Ryba et al. 2010 | |
| Myoblast | 7 individuals | Pope et al. 2011 | |
| Lymphoblastoid | CO2O2 (male) | Ryba et al. 2010 | |
| GM06990 (female) | Hansen et al. 2010; Ryba et al. 2011 | ||
| TL010 | Hansen et al. 2010 | ||
| HO287 (male) | Hansen et al. 2010 | ||
| MOLT-4 | Yaffe et al. 2010 | ||
| Fibroblast | IMR90 | Pope et al. 2011 | |
| BJ | Hansen et al. 2010 | ||
| FFT | Yaffe et al. 2010 | ||
| HeLa | Chen et al. 2010 | ||
| Erythroid | K562 | Hansen et al. 2010 | |
| CD4+-derived basophilic erythroblasts | Desprat et al. 2009 | ||
| Mus musculus | mESC | D3 | Hiratani et al. 2008 |
| 46C | Hiratani et al. 2008 | ||
| TT2 | Hiratani et al. 2008 | ||
| TT2 flox G9a ESC: mock and OHT | Yokochi et al. 2009 | ||
| miPSC | iPS | Hiratani et al. 2008 | |
| iPSC1D4 | Hiratani et al. 2010 | ||
| iPSC2D4 | Hiratani et al. 2010 | ||
| pmiPSC | piPSC1A2 | Hiratani et al. 2010 | |
| piPSC1B3 | Hiratani et al. 2010 | ||
| piPSC1V3 | Hiratani et al. 2010 | ||
| Early primitive ectoderm | D3-derived as embryoid bodies | Hiratani et al. 2010 | |
| Early primitive ectoderm | D3-derived as monolayer | Hiratani et al. 2010 | |
| Definitive ectoderm | D3-derived as embryoid bodies | Hiratani et al. 2010 | |
| Neural precursors | D3-derived as embryoid bodies | Hiratani et al. 2008 | |
| 46C-derived as monolayer | Hiratani et al. 2008 | ||
| TT2-derived as monolayer | Hiratani et al. 2008 | ||
| TT2 flox G9a-derived: mock and OHT | Yokochi et al. 2009 | ||
| Endoderm | GscSox17-EB5-derived | Hiratani et al. 2010 | |
| Mesoderm | GscSox17-EB5-derived | Hiratani et al. 2010 | |
| mEpiSC | EpiSC5 | Hiratani et al. 2010 | |
| EpiSC7 (female) | Hiratani et al. 2010 | ||
| Lymphoblastoid | L1210 | Farkash-Amar et al. 2008; Hiratani et al. 2010 | |
| Fibroblast | C127 mammary | Lu et al. 2010 | |
| MEF male and female | Hiratani et al. 2010; Yaffe et al. 2010 | ||
| Myoblast | J185a | Hiratani et al. 2010 | |
| Drosophila melanogaster | Wing imaginal disc | Cl8 | Schwaiger et al. 2009 |
| Embryonic | Kc | Schwaiger et al. 2009; Eaton et al. 2011 | |
| Kc HP1 knockdown | Schwaiger et al. 2010 | ||
| S2 | Eaton et al. 2011 | ||
| Neuronal | Bg3 | Eaton et al. 2011 | |
| Chicken | Lymphoid | DT40 | Hassan-Zadeh et al. 2012 |
| Arabidopsis thalania | Col-0 (chromosome 4 only) | Lee et al. 2010 | |
| Saccharomyces cerevisiae | wt | Raghuraman et al. 2001 | |
| wt | Yabuki et al. 2002 | ||
| wt | Feng et al. 2006 | ||
| rad35Δ | Feng et al. 2006 | ||
| wt | Alvino et al. 2007 | ||
| wt + HU | Alvino et al. 2007 | ||
| wt | McCune et al. 2008 | ||
| clb5Δ | McCune et al. 2008 | ||
| clb5Δ clb6Δ | McCune et al. 2008 | ||
| wt | Knott et al. 2009 | ||
| rpd3Δ | Knott et al. 2009 | ||
| eaf3Δ (Rpd3S) | Knott et al. 2009 | ||
| set2Δ (Rpd3S) | Knott et al. 2009 | ||
| rco1Δ (Rpd3S) | Knott et al. 2009 | ||
| dep1Δ (Rpd3L) | Knott et al. 2009 | ||
| cti6Δ (Rpd3L) | Knott et al. 2009 | ||
| rpd3Δ set2Δ | Knott et al. 2009 | ||
| rpd3Δ cti6Δ | Knott et al. 2009 | ||
| rpd3Δ eaf3Δ | Knott et al. 2009 | ||
| cti6Δ eaf3Δ | Knott et al. 2009 | ||
| dep1Δ eaf3Δ | Knott et al. 2009 | ||
| rco1Δ eaf3Δ | Knott et al. 2009 | ||
| eaf3Δ set2Δ | Knott et al. 2009 | ||
| wt | Crabbe et al. 2010 | ||
| rad9 | Crabbe et al. 2010 | ||
| rev3 rad30 | Crabbe et al. 2010 | ||
| eco1 | Crabbe et al. 2010 | ||
| ctf4 | Crabbe et al. 2010 | ||
| ddc1 | Crabbe et al. 2010 | ||
| rad24 | Crabbe et al. 2010 | ||
| pol2-12 | Crabbe et al. 2010 | ||
| elg1 | Crabbe et al. 2010 | ||
| tof1 | Crabbe et al. 2010 | ||
| mrc1AQ | Crabbe et al. 2010 | ||
| mrc1AQ rad9 | Crabbe et al. 2010 | ||
| ctf8 | Crabbe et al. 2010 | ||
| ctf18 | Crabbe et al. 2010 | ||
| mrc1 | Crabbe et al. 2010 | ||
| ddc1 | Crabbe et al. 2010 | ||
| ctf18 rad9 | Crabbe et al. 2010 | ||
| mec1-100 | Crabbe et al. 2010 | ||
| mec1-100 | Crabbe et al. 2010 | ||
| mec1-100 | Crabbe et al. 2010 | ||
| mec1-100 | Crabbe et al. 2010 | ||
| rad53 | Crabbe et al. 2010 | ||
| wt | Lian et al. 2011 | ||
| yku70Δ | Lian et al. 2011 | ||
| wt | Koren et al. 2010 | ||
| sic1Δ | Koren et al. 2010 | ||
| dia2Δ | Koren et al. 2010 | ||
| clb5Δ | Koren et al. 2010 | ||
| mrc1Δ | Koren et al. 2010 | ||
| rrm3Δ | Koren et al. 2010 | ||
| dpb3Δ | Koren et al. 2010 | ||
| dpb4Δ | Koren et al. 2010 | ||
| rnr1Δ | Koren et al. 2010 | ||
| rad27Δ | Koren et al. 2010 | ||
| ura7Δ | Koren et al. 2010 | ||
| tda3Δ | Koren et al. 2010 | ||
| met7Δ | Koren et al. 2010 | ||
| gln3Δ | Koren et al. 2010 | ||
| arg2Δ | Koren et al. 2010 | ||
| wt | Tanaka et al. 2011 | ||
| Overexpression of Sld3, Sld7, and Cdc45 | Tanaka et al. 2011 | ||
| Overexpression of Dbf4 | Tanaka et al. 2011 | ||
| Overexpression of Cdc7 and Dbf4 | Tanaka et al. 2011 | ||
| wt | Knott et al. 2012 | ||
| fkh1Δ | Knott et al. 2012 | ||
| fkh2Δ | Knott et al. 2012 | ||
| fkh1Δ fkh2Δ | Knott et al. 2012 | ||
| Schizosaccharomyces pombe |
wt |
Heichinger et al. 2006 |
|
| wt | Feng et al. 2006 | ||
| wt | Hayashi et al. 2007 | ||
| wt | Eshaghi et al. 2007 | ||
| wt | Mickle et al. 2007b | ||
| wt | Mickle et al. 2007a | ||
| wt | Kumar and Huberman 2009 | ||
| wt | Wu and Nurse 2009 | ||
| wt | Hayano et al. 2011 | ||
| wt | Hayano et al. 2011 | ||
| wt | Xu et al. 2012 |
hESC, human embryonic stem cells; hiPSC, human-induced pluripotent stem cells; mESC, mouse embryonic stem cells; OHT, 4-hydoxytamoxifen; miPSC, mouse-induced pluripotent stem cells; pmiPSC, partially reprogrammed mouse-induced pluripotent stem cells; mEpiSC, mouse epiblast-derived stem cells; wt, wild type; HU, hydroxyurea.
Interpreting Genome-Wide Replication Timing Profiles
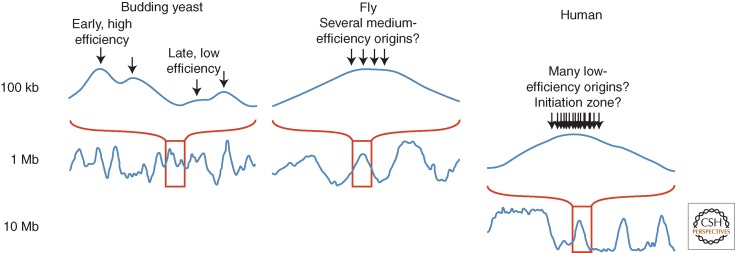
Figure 1 shows exemplary profiles of replication timing in human, Drosophila, and yeast cells, and Figure 2 shows a schematic to help with the interpretation of these profiles. In the large genomes of multicellular organisms, the profiles consist of broad, computationally defined constant timing regions (CTRs), or “replication domains,” punctuated by slopes of progressive change in timing, commonly referred to as timing transition regions (TTRs). The appearance of constant timing across large replication domains may in part be due to the resolution of existing methodology, but the prevailing working model (Fig. 2) is that the similar timing on the population level is due to the averaging of heterogeneous firing on the individual cell level from clusters of multiple origins or initiation zones (regions within which different cells initiate at different sites) that fire with similar timing. The evidence for this model is indirect, inferred from a combination of genome-scale data that show similar average replication times at 100-kb resolution and individual DNA fiber origin-mapping data that reveal evidence for clusters of initiation at sites that are heterogeneous between cells (Lebofsky et al. 2006; Cayrou et al. 2011; Letessier et al. 2011; Besnard et al. 2012).
Figure 1.
Scales of replication timing in species with different-sized genomes. Smoothed replication profiles of segments of the human (Ryba et al. 2010), fly (Schwaiger et al. 2009), and budding yeast genomes (Alvino et al. 2007; Yang et al. 2010). Although the profiles look qualitatively similar, they show features on very different scales. In yeasts, the peaks in the replication profiles represent individual origins and the average replication times of origins are determined by a combination of their average firing times and the frequency with which they are passively replicated by forks originating at neighboring origins. In mammals, what appear to be sharp peaks of early replication flatten at higher resolution to broad, near-megabase-sized domains, which contain many unresolved individual replicons. This lack of resolution can be accounted for by spatial or temporal heterogeneity in origin firing within each domain. Fly genomes are an order of magnitude smaller than human, and their domains of coordinate replication are similarly smaller but still probably contain multiple unresolved replicons. The slope of the curves moving away from early-replicating regions is often interpreted as being proportional to the rate of replication in that region. However, even in budding yeasts, where origins can be very efficient, this correlation is not strong; the slope is determined more by the ratio of fork directions than by the rate of those forks (Sekedat et al. 2010; Retkute et al. 2011).
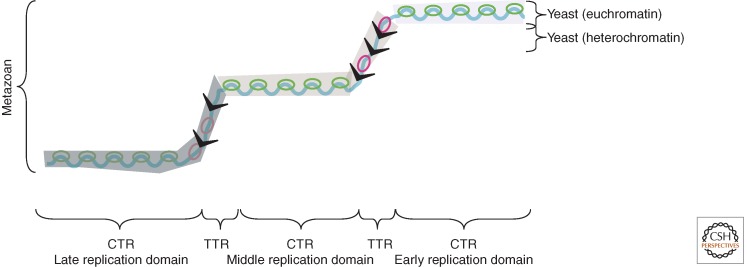
Figure 2.
Interpreting replication profiles in different species. A hypothetical segment of a replication timing profile contains regions of constant timing (CTRs), or “replication domains,” that replicate at different times during S phase, and regions of temporal transition (TTRs). However, these regions can be interpreted differently depending on genome size and computational parameters. In metazoan genomes, replication domains can be operationally defined regions where replication timing differs by 10%–20% of the length of S phase. The similarity of replication timing within such domains is proposed to be due in part to the heterogeneous, population-averaged firing of adjacent origins (green bubbles) with similar firing times, but in the case of large CTRs can be due to the nearly simultaneous firing of adjacent but independently regulated replication domains. The actual number of initiation sites within each domain that can potentially be used in a population of cells is believed to be in the dozens (origin clusters) to hundreds (initiation zones). TTRs are regions of suppressed origin activity (indicated as a gray “slime”), which may be replicated either by a single fork (black arrows) moving unidirectionally through time (y-axis) or—if slow-moving forks stimulate firing of inefficient or “dormant” origins—by sequentially activated origins (red bubbles). In contrast, the entire genome in less complex organisms such as yeasts can be thought of as a single replication domain, with the majority of regulation controlled by more origin-proximal mechanisms. The exception is the late-replicating heterochromatin, such as telomeres, which form the equivalents of TTRs, being passively replicated by forks originating in neighboring euchromatic regions.
TTRs are the regions between CTRs that have a gradient of replication timing from their early-replicating sides to their late-replicating sides (Fig. 2). Their replication timing is consistent with either unidirectional forks emanating from the early CTRs toward the late CTRs (Farkash-Amar et al. 2008; Hiratani et al. 2008; Desprat et al. 2009; Ryba et al. 2010) or sequentially firing origins (Guilbaud et al. 2011). At least part of one TTR has been shown to replicate predominantly as a single long unidirectional fork in different human and mouse cell lines (Norio et al. 2005; Schultz et al. 2010), and sequences that can function as origins at their normal locations rarely fire when inserted into this TTR (Guan et al. 2009). Even strong transcription or tethering of a potent histone acetyltransferase near an origin within a TTR did not alter its firing rate, whereas tethering similar molecules near origins that do not reside within a TTR strongly affects origin firing (Goren et al. 2008; Hassan-Zadeh et al. 2012). On the other hand, the rate of replication through TTRs genome-wide has been estimated to vary by two- to threefold more than global rate of fork elongation (Takebayashi et al. 2005; Frum et al. 2009; Ryba et al. 2010; Guilbaud et al. 2011), suggesting that sequential firing of origins occurs within at least some TTRs (Guilbaud et al. 2011). Furthermore, origin activity has been detected within TTRs (Cadoret et al. 2008; Cayrou et al. 2011). The frequency of initiation within TTRs may be influenced by factors that slow replication fork movement, which may induce the firing of dormant origins within the TTRs. TTRs terminate by merging with forks coming from an opposing TTR or a later-replicating CTR (Fig. 2). Merging TTRs create a U-shaped valley in the replication profiles resulting from the population average of forks terminating at different sites, without the need to invoke late-firing origins. Some of these late-replicating regions coincide with common fragile sites and fail to activate origins even under conditions of replicational stress (Le Tallec et al. 2011; Letessier et al. 2011; Debatisse et al. 2012). Large late replication domains, however, likely contain late-firing origins that fire heterogeneously but with similar average timing, much as early CTRs do (Frum et al. 2009; Guilbaud et al. 2011; Le Tallec et al. 2011; Letessier et al. 2011; Debatisse et al. 2012). Ultimately, accurate interpretation of these temporal profiles will require more extensive studies of origin site selection that combine ensemble and DNA fiber origin-mapping methods (Cayrou et al. 2011).
In multicellular organisms with smaller genomes, such as Drosophila and Arabidopsis (Lee et al. 2010), replication timing profiles are qualitatively similar but at a different scale. Whereas replication domains in mammals range from several hundred kilobases to many megabases (Hiratani et al. 2008; Ryba et al. 2010), domains in these organisms range from 75 to 250 kb (MacAlpine et al. 2004; Schwaiger et al. 2009), which is small enough to be initiated from one or a few closely spaced origins (Fig. 1). Unfortunately, there are no studies of fork rates in Drosophila or Arabidopsis that can inform the interpretation of replication timing profiles in these species.
In the budding and fission yeasts, in which the genomes are 100-fold smaller than in mammals, the replication profiles look qualitatively similar to those of metazoans (Fig. 1). However, because origins are well-defined loci in yeasts, it is possible to resolve individual origins. Therefore the peaks in the yeast profiles correspond to actual origins, not just early-replicating regions. This resolution allows information about the efficiency and timing of individual origin firing to be extracted from yeast replication timing profiles. Although at the population level origins show distinct average firing times, at the single-cell level origin firing is heterogeneous, in that a different subset of origins is activated in a different timing pattern in each cell (Patel et al. 2006; Czajkowsky et al. 2008). Therefore, forks from neighboring origins can passively replicate even the earliest origins at some frequency, and caution must be used in interpreting the profiles. For instance, variations in the slope of replication profiles is generally due to the ratio of fork directions, not due to variations in individual fork speed (Sekedat et al. 2010; Retkute et al. 2011). Likewise, the probability of an origin firing is not directly proportional to its efficiency (i.e., the fraction of cells in which it fires); efficiency is determined by a combination of an origin’s firing probability and its proximity to other origins with higher or lower probabilities of firing earlier and generating a replication fork that passively replicates the origin (de Moura et al. 2010). As in metazoans, there are early- and late-replicating segments of the yeast genome (Heichinger et al. 2006; McCune et al. 2008); however, these segments are not due to the clustering of early or late origins per se (Yang et al. 2010). Instead, the early-replicating segments have both early and late origins, but the late origins are usually passively replicated by forks from their early neighbors. In another similarity to metazoans, the latest-replicating sequences in the budding and fission yeast genomes are heterochromatin. However, because heterochromatin constitutes such a small portion of yeast genomes, these regions do not form late-replicating CTRs, as in metazoans, but rather form TTRs, which are often passively replicated by forks originating from neighboring euchromatic segments.
The availability of high-resolution, genome-wide replication timing profiles has permitted the development of mathematical models of replication kinetics. These models have been used to interpret population-averaged replication timing profiles, taking into account single-molecule DNA fiber data, and have generated testable hypotheses (Lygeros et al. 2008; de Moura et al. 2010; Hyrien and Goldar 2010; Luo et al. 2010; Yang et al. 2010). Many of these models have concluded that replication timing profiles can be accounted for by a combination of the probability with which origins in a region fire and their proximity to other origins with higher or lower probabilities, which determines the frequency of inactivation of neighboring origins by passive replication. In such models, factors that establish the probability of origin firing determine the replication timing program.
Replication Timing and DNA Sequence Composition
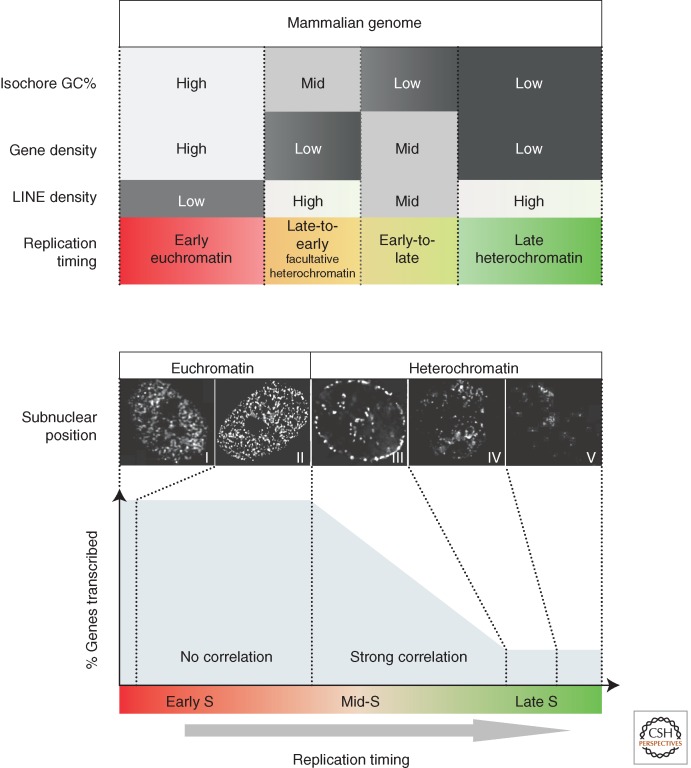
In multicellular organisms there are statistically significant links between replication timing and features of primary genomic sequence. Gene density correlates with early replication, while the density of repetitive elements associated with heterochromatin correlates with late replication. In vertebrates a general correlation exists between early replication and gene-rich, LINE-poor, GC-rich regions of the genome (Fig. 3, top), often segmented into regions called isochores (Schmegner et al. 2007; Costantini et al. 2009). However, these DNA sequence features are clearly not sufficient to dictate replication timing. Changes in replication timing during development (see below) can strongly influence the genome-wide alignment of replication timing to isochores (Hiratani et al. 2010; Ryba et al. 2010). Moreover, homologous loci can replicate at different times in the same cells (Karnani et al. 2007; Farkash-Amar et al. 2008; Hansen et al. 2010). Isochores that exhibit the most extreme of the above-mentioned sequence compositions tend to replicate at the same time in all tissues, whereas regions with intermediate or mixed sequence features are more prone to change replication timing during development (Fig. 3, top), suggesting that sequence composition has some indirect influence on replication timing. There is also evidence that replication timing can influence sequence composition. At about 1000 sites in the human genome, there are significant transitions in nucleotide composition that coincide with peaks of earliest regional replication timing (Touchon et al. 2005; Huvet et al. 2007); a similar phenomenon is seen in yeast genomes (Agier and Fischer 2012; Marsolier-Kergoat and Goldar 2012). These transitions are believed to arise from different mutation frequencies experienced by leading versus lagging strands, leaving an evolutionary imprint of replication fork polarity.
Figure 3.
Relationship between isochore properties and replication timing regulation, subnuclear positioning, and transcription. (Top) Isochores with unusual sequence properties are subject to replication timing regulation. The mammalian genome is partitioned into isochores with different GC content, LINE composition, and gene density, which are generally correlated. Isochores that are high in GC and gene density but low in LINE density are replicated early in S phase, whereas the alternate extremes are replicated late. Isochores with intermediate or mixed sequence features are frequently subject to replication timing regulation during differentiation (speculatively labeled “Facultative heterochromatin”) (Hiratani et al. 2008; Ryba et al. 2010). (Bottom) Changes in replication timing that traverse the middle of S phase accompany changes in subnuclear position and transcriptional potential. Replication early in S phase (patterns I and II) takes place within the interior euchromatic compartment, whereas replication later in S phase (patterns III, IV, and V) takes place at the nuclear periphery (pattern III), at the nucleolar periphery (pattern III), and at internal blocks of heterochromatin (patterns IV and V) (Berezney et al. 2000; McNairn and Gilbert 2003). Note the dramatic transition from euchromatic to heterochromatic replication pattern during mid-S phase (from pattern II to III). In addition, a strong relationship between earlier replication timing and transcription is observed for genes that replicate during mid- to late stages of S phase (“strong correlation”; corresponds to pattern III) (Hiratani et al. 2008). Very few genes are replicated at the end of S phase. In contrast, genes that are replicated in the first third of S phase have equally high probability of being expressed (“no correlation”), and thus even large changes within this period may not accompany changes in subnuclear repositioning and/or transcriptional competence may be inconsequential for transcription. The figure illustrates how the durations of different spatial replication patterns are likely to relate to the probability of transcription based on published results (Dimitrova and Gilbert 1999; Hiratani et al. 2008). These relationships imply that genome-wide replication timing analyses provide a means to infer changes in subnuclear position and transcriptional potential (Hiratani et al. 2008). Together these observations strongly suggest that changes in replication timing that traverse the mid-S phase (i.e., pattern II to III or vice versa) accompany subnuclear repositioning and altered transcriptional potential, with the latter likely confined to certain classes of genes. We submit that although genome-wide analysis of subnuclear position changes by fluorescence in situ hybridization is impractical, these spatiotemporal patterns of replication provide a proxy of their 3D distribution. Genome-wide replication timing analyses provide a means to infer changes in subnuclear position and even transcriptional potential using these relationships (Hiratani et al. 2008). Photos are deconvolved images from Chinese hamster cells pulse-labeled with bromodeoxyuridine at different times during S phase (J Lu and DM Gilbert, unpubl.). (From Hiratani et al. 2009; adapted, with permission, from Elsevier.)
Relationships to Transcription: Divergence of Multicellular and Single-Celled Organisms
Genome-wide studies have revealed a strong positive correlation between early replication and transcriptional activity in multicellular organisms, including plants, particularly when transcriptional output is integrated across replication domains (MacAlpine et al. 2004; Hiratani et al. 2008; Lee et al. 2010). In contrast, no such correlation can be found in budding or fission yeasts, except for the fact that the few silent heterochromatic regions of yeasts, such as telomeres, are late-replicating (Raghuraman et al. 2001; Heichinger et al. 2006). One way of reconciling this discrepancy is to consider that in higher eukaryotes there is no correlation between transcription and replication timing throughout the entire first half of S phase; the observed correlation is accounted for by genes that replicate in the second half of S (Fig. 3, bottom). Moreover, in multicellular organisms, any given cell type replicates 75% of genes in the first half of S phase. In this sense the bulk of the yeast genome can be likened to early-replicating domains in multicellular organisms, with only the heterochromatic regions, which comprise a small fraction of the yeast genome, acting as later-replicating domains. Perhaps the “exploitation” of heterochromatin by multicellular organisms led to the expansion of the heterochromatic chromatin compartment and more extensive late-replicating domains (Gilbert 2002).
Developmental Control and Stable Epigenetic States
Another property of replication timing in multicellular organisms is developmental control. At least half the mammalian genome is subject to developmentally regulated changes in replication timing that occur in units of 400–800 kb and are generally coordinated with changes in transcriptional regulation of a certain class of promoters (Hiratani et al. 2008; Ryba et al. 2010). The smaller and more uniform size of these developmentally regulated replication domains suggests that larger domains of constant replication timing observed in any given cell type consist of multiple units that can be independently regulated during development. In fact, replication domains in pluripotent cells are smaller and their replication timing becomes aligned to consolidate them into larger domains during differentiation. Some differences in replication timing have also been observed between Drosophila melanogaster cell lines, and these also occur in smaller units than the replication domains found within a cell type (Schwaiger et al. 2009). As expected from the correlation to transcription, changes in replication timing generally coincide with changes in transcription. However, this coordination is promoter-specific in mammals; changes in transcription of genes with low- but not high-CpG-content promoters correlate with replication timing changes (Hiratani et al. 2008, 2010). Given that replication domains are 400–800 kb, many genes can change replication timing coordinately, with some showing coordinated transcriptional changes and others not, which has made dissection of this relationship difficult. However, when the analysis is focused on low-CpG-content promoters during the course of stem cell differentiation, transcription and replication switches are so closely coordinated that it is still difficult to answer the long-standing question of which change occurs first (Hiratani et al. 2010).
What is clear, however, is that switches in replication timing are correlated with epigenetically stable transitions in cell state and gene responsiveness. Dosage compensation in mammals occurs by the inactivation of one of the two female X chromosomes, accompanied by a switch to late replication. Interestingly, monotremes, marsupials, and eutherian mammals have widely different molecular mechanisms to silence transcription on the inactive X, but a switch to late replication is common to all of these clades (Hiratani and Gilbert 2010). Moreover, genome-wide studies uncovered a specific set of autosomal replication domains that switch from early to late replication coincident with X inactivation (Hiratani et al. 2010). These regions remain late-replicating in all committed tissues queried, such that their early replication constitutes a “pluripotency fingerprint” (Ryba et al. 2011). These timing changes coincide with a loss in the ability of cells to colonize blastocysts, make chimeras, or revert to the embryonic state. Importantly, they reflect a discrete epigenetic barrier to cellular reprogramming; both early replication and transcription of genes within these regions are blocked in somatic cells that fail to reprogram to induced pluripotent stem cells (iPSCs), and the expressed transcription factors Oct4, Sox2, Nanog, and Klf4 fail to bind to their promoters (Hiratani et al. 2010).
Relationships to Chromatin Proteins
Early studies reported a close correlation of replication timing with R and G chromosome banding patterns, which presumably arise from heterogeneous protein composition along the lengths of chromosomes, but genomic analyses revealed that this alignment breaks down at molecular resolution (Pope et al. 2010). However, such studies prompted the search for other known properties of chromatin that correlate with replication timing. As expected given the correlation to transcriptional activity, early replication correlates well with general nuclease accessibility (Gilbert et al. 2004; Bell et al. 2010), histone modification, and chromatin proteins associated with active transcription (Hiratani et al. 2008; Ryba et al. 2010). However, the correlation with repressive or heterochromatin histone marks was found to be highly cell-type-specific and quite poor in some cell types (Hiratani et al. 2008; Ryba et al. 2010), consistent with earlier cytogenetic findings (Wu et al. 2005). To date, none of these chromatin marks were confined to the boundaries of replication timing, although some were found enriched at the borders of early replication domains (Ryba et al. 2010). Surprisingly, despite a strong correlation of early replication to chromatin accessibility (Prendergast et al. 2007; Bell et al. 2010), accessibility does not change coordinately with replication timing; regions that change replication timing remain relatively inaccessible whether early- or late-replicating (Takebayashi et al. 2012). It is important to point out, however, that computational methods that integrate the density of proteins or protein modifications could miss important localized features of chromatin.
A more stringent test of the role of a protein or histone modification is to determine whether it is necessary to maintain replication timing. In the past such studies were limited to testing effects of gene knockouts on the replication timing of a few specific regions (Stevenson and Gottschling 1999; Vogelauer et al. 2002; Aparicio et al. 2004; Jorgensen et al. 2007). Genomic methods are now robust enough to routinely perform an unbiased search for regions of the genome affected by mutations. In mammals the timing program is surprisingly resilient to such mutations (Jorgensen et al. 2007; Yokochi et al. 2009). For example, a strong statistical and spatial correlation between late replication and H3K9me2 in mouse embryonic stem cells (ESCs) was tested using a conditional knockout of the histone methyltransferase G9a. This mutation caused a near complete ablation of this mark on late-replicating chromatin but had no effect on replication timing or developmentally regulated changes in replication timing during differentiation (Yokochi et al. 2009). It is possible that multiple redundant modifications maintain a robust replication timing program, and studies combining mutations with chemical inhibitors of chromatin-modifying enzymes are consistent with this conclusion (Casas-Delucchi et al. 2012). Moreover, replication timing is not a binary early or late phenomenon but rather a continuum throughout S phase; hence it is not surprising that many mutations in both mammals and yeasts usually have only minor effects on replication timing. However, some chromatin mutations do exhibit profound localized effects on replication timing (Li et al. 2005; Wu et al. 2006; Bergstrom et al. 2007; Hayashi et al. 2009; Schwaiger et al. 2010), and a few examples of gene disruptions causing broad misregulation of mammalian replication timing are emerging (Cornacchia et al. 2012; Yamazaki et al. 2012).
Results measuring replication timing in yeasts that have been disrupted for chromatin modification enzymes have revealed similar complex effects. Disruption of the histone acetyltransferase Rpd3L advances the firing time of approximately 100 origins (about 30% of detected origins) in Saccharomyces cerevisiae (Knott et al. 2009), and deletion of budding yeast Rif1 causes earlier replication of telomeres (Lian et al. 2011). Moreover, recent results in fission yeasts suggest that deletion of either of the telomeric proteins Taz1 (Tazumi et al. 2012) or Rif1 (Hayano et al. 2012) results in a dramatic shift in the time of initiation of a set of nontelomeric origins. A similar effect occurs when Rif1 is knocked down in mammalian cells (Cornacchia et al. 2012; Yamazaki et al. 2012), although the mammalian Rif1 protein has a different function than its yeast homolog (de Lange 2004). A profound effect on replication timing was recently reported after knockout of the forkhead transcription factors in budding yeasts (Knott et al. 2012).
3D CHROMATIN ORGANIZATION AND REPLICATION TIMING
A far-reaching conclusion from genome-wide studies is the uncanny alignment of replication timing to maps of 3D chromatin interactions. This observation has shifted paradigms in our understanding of a long-standing enigmatic relationship between replication timing and large-scale chromosome structure. Moreover, very recent results have shed light on the similarities and differences in this level of regulation between yeasts and metazoans. In this section, we briefly summarize the classic observations and propose a unified working model in which the structural organization of chromosomes can regulate replication timing in unicellular and multicellular eukaryotes.
Spatial Compartmentalization of Early and Late Replication
X-chromosome inactivation in mammals is accompanied by a chromosome-wide switch to late replication, dramatic compaction, and movement of the entire chromosome to the nuclear periphery (Hiratani and Gilbert 2010). Similarly, autosomal chromatin that replicates at different times during S phase is globally compartmentalized, with early replication taking place in the interior and chromatin at the periphery being replicated later in S phase (Figs. 3, bottom, 4B). Compartmentalization appears to result from the anchorage of chromatin to immobile structures such as the nuclear periphery (Gilbert and Gasser 2006; Steglich et al. 2012) or nucleolus (Nemeth and Langst 2011). In organisms with large nuclei, chromatin movement is constrained throughout the nucleus. In yeasts, the entire nucleus is similar in scale to the range of chromatin motion in mammals, but some regions, particularly those containing late-replicating origins, are more constrained and molecules that anchor chromatin to specific subnuclear locations have been elucidated (Gilbert and Gasser 2006). The significance of compartmentalization to the replication timing program remains elusive, and under some experimental conditions, tethering budding yeast replication origins to the nuclear periphery is neither necessary (Hiraga et al. 2006) nor sufficient (Lian et al. 2011) for late replication.
Determinants of Replication Timing Established during G1 Are Lost before G2 Phase
A compelling link between replication timing and the spatial compartmentalization of chromatin is the finding that replication timing is established coincident with the global repositioning and anchorage of chromatin as the nucleus is reassembled during early G1 phase (Dimitrova and Gilbert 1999). The molecular events occurring at this timing decision point (TDP) remain elusive. Recently it was shown that when a single complete round of replication is initiated during G2 phase, replication proceeds in a random spatial and temporal order despite maintenance of chromatin spatial organization (Lu et al. 2010), implying that determinants of replication timing are lost during S phase. Possible mechanisms include the dilution of preexisting chromatin components during replication or the disruption of normal chromatin interactions when two identical sister chromatids share the same nuclear space. Together these results suggest that the global spatial organization of chromatin is not sufficient to dictate timing during G2 phase, although it may contribute to the establishment of replication timing. This observation is consistent with a similar finding in budding yeasts that chromosomal context is important during early G1 to establish late replication but is not necessary thereafter (Raghuraman et al. 1997; Heun et al. 2001).
Changes in Subnuclear Position Linked to Replication Timing and Cell-Fate Transitions
The spatial patterns of DNA synthesis change dramatically and abruptly during the middle of S phase in all multicellular species that have been examined (Fig. 3, bottom). Likewise, there seems to be a similar change in subnuclear position when replication domains change replication timing during development. Fluorescence in situ hybridization detects the movement of domains toward or away from the periphery during stem cell differentiation when replication timing becomes later or earlier, respectively (Williams et al. 2006; Hiratani et al. 2008). As predicted from global spatial patterns, these changes only occur when replication timing changes through mid- to late S phase (Hiratani et al. 2010), which is also the same period of S phase during which a correlation exists between replication timing and transcription (Fig. 3, bottom). Hence, replication timing can change substantially during the first half of S phase without a detectable change in subnuclear position, but a switch through mid- to late S phase is highly predictive of a subnuclear position change, suggesting that replication timing may provide a convenient genome-wide readout of subnuclear position changes (Fig. 3, bottom). What is not clear, however, is which occurs first during the course of differentiation, and whether the subnuclear position change is confined to the region of replication timing change. Intuitively it seems that without major chromatin unfolding, subnuclear position changes would need to involve chromosomal regions larger than the size of a 400- to 800-kb replication domain, but this remains to be tested.
Replication Foci: Cytogenetic Units of Replication Timing Regulation?
A common feature of the spatial patterns of DNA synthesis is a punctate appearance of the labeled sites of DNA synthesis, or “replication foci.” In mammals each of these foci takes 45–60 min to complete replication (Berezney et al. 2000). Based on the number of foci observed over the course of S phase (hundreds at any one time), it is estimated that each contains 0.5–1 Mb of DNA (Berezney et al. 2000), close to the size of developmentally regulated replication domains. When pulse-labeled cells are chased through multiple cell cycles, labeled foci do not change size, shape, or intensity of label, indicating that the DNA that is synthesized together remains together as a structural unit (Sadoni et al. 2004). In many cases, adjacent foci are genetically contiguous and replicate sequentially (Sporbert et al. 2002; Maya-Mendoza et al. 2010).
Models for the structure of chromatin within replication foci have been rapidly evolving. A long-standing model is that replication foci represent “factories” of DNA organized in such a way that multiple replicons throughout hundreds of kilobases are simultaneously spooled through a single common replication complex (Fig. 4) (Gilbert and Gasser 2006), although evidence for this model has been inconclusive. Recently evidence has been presented for the presence of replication foci containing multiple replication forks in budding (Kitamura et al. 2006) and fission (Meister et al. 2007) yeasts, suggesting that they may share a similar structural organization. However, a study in cell-free Xenopus egg extracts has shown that individual molecules tethered at both ends can be efficiently replicated, showing that neither large immobilized complexes nor complexes simultaneously engaged with both bidirectional replication forks are necessary for in vitro DNA replication (Yardimci et al. 2010). Moreover, high-resolution light microscopy methods (Cseresnyes et al. 2009; Baddeley et al. 2010) applied to the study of replication foci reveal that each “focus” appears to resolve into many separate but clustered subfoci that are consistent with the amount of DNA in a single bidirectional replicon (Fig. 4D). In short, although replication foci likely represent the cytological manifestation of replication domains, their structure remains unresolved.
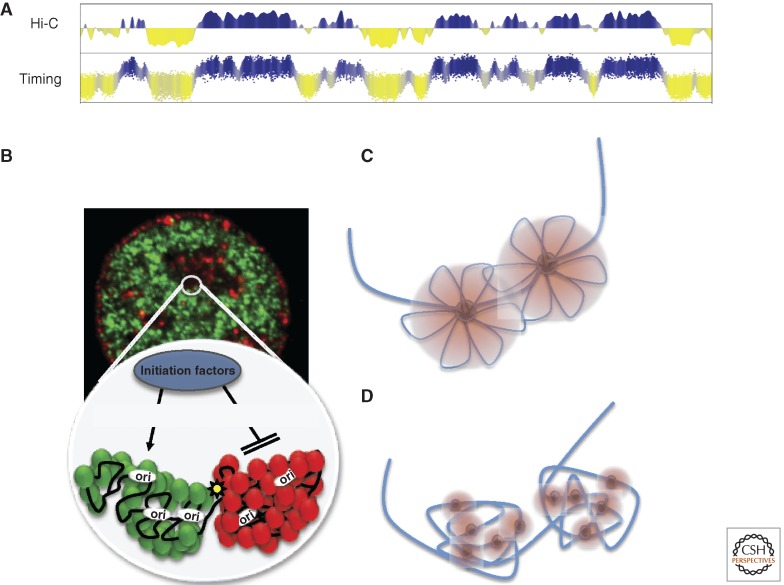
Figure 4.
Units of replication timing regulation correspond to units of large-scale chromatin organization. (A) Replication timing profiles align better to eigenvector displays of chromatin conformation capture profiles than to any other structural or functional property of chromosomes examined to date (Ryba et al. 2010). (B) Regions that replicate at different times are spatially segregated and may form self-interacting domains that may set thresholds for the accessibility of S-phase promoting factors. (C,D) Two interpretations of replication foci. (C) Concept of a replication factory where several replicons in a spatially contiguous chromosome region are replicated by a common, fixed replication protein complex. (D) Current super-resolution microscopy methods suggest that replication foci consist of clusters of smaller foci that were not resolved by prior light microscopy. Together the data suggest that foci are the result of several separate replication complexes, possibly replicating both bidirectional leading and lagging strands together (Heun et al. 2001; Kitamura et al. 2006; Meister et al. 2007), but rather than being a single fixed complex, they consist of a spatially clustered group of independent replication complexes that initiate replication nearly synchronously, possibly as a result of their common presence within a single self-interacting unit of large-scale chromatin organization. (A and B from Ryba et al. 2010; reprinted, with permission, © Cold Spring Harbor Laboratory Press.)
Importantly for our discussion, although the ultrastructure of replication foci is not resolved, evidence suggests that replication timing is regulated at the level of replication foci. As discussed above, coordinated changes in replication timing during differentiation are regulated at the level of 400- to 800-kb replication domains, which are in line with the sizes of foci defined cytogenetically. Moreover, studies in Xenopus egg extracts have shown that the timing program can be compressed or elongated, without modulating the temporal order of replication, by increasing or decreasing the number of simultaneously activated replication foci through modulation of cyclin-dependent kinase (CDK) activity, independent of the numbers of origins or rate of elongation (Thomson et al. 2010). Together these results have strengthened the view of the replication focus as a unit of replication regulation.
An Uncanny Link between Replication Timing and Chromatin Interaction Maps
Replication timing correlates better with genome-wide maps of 3D chromatin interactions than any other chromosomal property analyzed to date (Ryba et al. 2010); in fact, it is one of the strongest correlations in genomics (Fig. 4A). Chromatin interaction maps are made by cross-linking cellular chromatin, digesting it with a restriction enzyme, and then religating to covalently link all pieces of DNA that are close together in the nucleus. In Hi-C (Lieberman-Aiden et al. 2009), a genome-wide chromosome-interaction mapping technique, the junctions of all of these novel ligation events are sequenced to reveal all the interactions (near the restriction sites) in a population of cells. These studies have shown that chromatin is organized into two spatially distinct compartments such that chromatin in one compartment rarely interacts with chromatin in the alternate compartment, with distinct boundaries separating interactions between alternate compartments. Hi-C data sets not only correlate strongly with replication timing but align precisely at both large and small replication timing transition boundaries, suggesting that the chromatin between segments of self-interacting chromatin may be the TTRs. The fact that chromosome interaction correlates better with replication timing than either of them do with any single histone modification or chromatin-binding protein suggests that replication timing is connected to chromatin structure through a complex combination of histone modifications and chromatin-binding proteins, no one of which alone captures the overall large-scale structure. This observation also suggests that replication timing may be a convenient proxy for chromatin interaction maps.
These results provide molecular evidence supporting conclusions from cytogenetic data that early- and late-replicating DNAs reside in spatially separate compartments of the nucleus and that the DNA that is in close spatial proximity is replicated together as a structural and functional unit. Data from Hi-C can be fit to models in which chromosomes are organized into self-interacting domains that are remarkably similar in size to replication domains and are predicted to fold into structures resembling replication foci (Fig. 4B). It remains to be seen whether this correlation will also be found in yeasts, but chromatin interaction maps in budding yeasts show that early replication origins frequently interact (Duan et al. 2010). Moreover, a recent study shows that forkhead transcription factors bind near a subset of early replication origins in budding yeasts, interact to organize early origins in nuclear space, and are required for their early replication (Knott et al. 2012).
MECHANISMS REGULATING REPLICATION TIMING
The strong correlation between replication timing and chromatin structure suggests a mechanistic connection. However, timing appears to be controlled at the level of large replication domains, whereas the regulation of replication initiation is at the level of individual origins. How the chromatin structure of large domains is related to the firing times of individual origins within those domains remains one of the central questions in the field.
Establishment versus Execution
The regulation of replication timing can be thought of as having two mechanistically distinct processes: establishment and execution. Establishment, which occurs during late mitosis and G1, sets the timing program by committing origins to their characteristic firing times. Execution, which occurs during S, realizes the timing program by activating origins at those times. Manipulating either process can affect the timing of replication, but in mechanistically different ways. Manipulations that affect establishment tend to affect large-scale attributes, such as chromatin structure and subnuclear localization, whereas manipulations that affect execution affect the firing of individual origins without affecting chromatin structure and subnuclear localization.
Execution Reflects Competition for Rate-Limiting Factors
Recent work has shed light on the execution of replication timing: how and when individual origins are selected to fire. Underlying this work is the heterogeneous nature of origin firing. In budding and fission yeasts, in which origins are well-defined genetic loci and the replication timing pattern of each origin is known, the execution of the replication program during S phase involves the stochastic firing of individual origins, which nonetheless results in reproducible patterns of average firing times (Patel et al. 2006; Rhind 2006; Czajkowsky et al. 2008; Lygeros et al. 2008; de Moura et al. 2010; Yang et al. 2010). Likewise in metazoans, origin firing is heterogeneous, with individual origins often firing with very low efficiency (Anglana et al. 2003; Lebofsky et al. 2006; Labit et al. 2008; Cayrou et al. 2011). It is important to emphasize that stochastic firing of origins does not imply that all origins fire with equal probability; indeed, different origins can have very different probabilities of firing. Rather, the term “stochastic” simply implies that the firing of origins is governed by probability, such that an origin may fire in one cell but not another. Such behavior can be explained if each origin has a characteristic probability of firing (Rhind 2006; Rhind et al. 2010). Origins with high firing probabilities are more likely to fire early in S and therefore, on average, have earlier firing times. Likewise, origins with lower firing probabilities will, on average, have later replication times. Mathematical models based on stochastic origin firing account well for observed replication kinetics in yeasts, frog embryos, and mammalian cells (Goldar et al. 2008; Lygeros et al. 2008; de Moura et al. 2010; Yang et al. 2010; Gauthier et al. 2012), and recent high-resolution DNA fiber analysis of replication kinetics at the mouse Igh locus is consistent with stochastic origin firing (Demczuk et al. 2012).
The probability of origin firing is affected by competition for rate-liming factors. In budding and fission yeasts, initiation factors—such as Cdc45, Sld2, and Sld3—and the Dbf4-dependent kinase Cdc7(DDK) initiation kinase are limiting for origin firing; overexpression of these factors increases the probability of origin firing, and tethering them to specific loci can increase the firing of local origins (Patel et al. 2008; Wu and Nurse 2009; Mantiero et al. 2011; Tanaka et al. 2011). In addition, limiting the expression of S-phase cyclins can limit the firing of late origins (Donaldson et al. 1998; McCune et al. 2008; Katsuno et al. 2009). The idea that varying the level of limiting activators affects the execution but not the establishment of replication timing is supported by the observation that slowing replication, either by slowing fork rates or deleting replication factors, elongates the replication timing program but preserves the relative order of average replication times (Alvino et al. 2007; Koren et al. 2010). Stochastic firing of origins in mammalian cells suggests that they are also regulated by rate-limiting factors (Lebofsky et al. 2006; Cayrou et al. 2011; Letessier et al. 2011). Consistent with this possibility, varying the level of CDK affects replication timing in vertebrates (Krasinska et al. 2008; Katsuno et al. 2009; Thomson et al. 2010).
The activation of origins by limiting factors predicts that as S phase progresses and fewer unfired origins remain, the remaining origins will be able to compete more efficiently for the limiting activators and so their firing probability will increase (Herrick et al. 2000; Hyrien et al. 2003; Rhind 2006). Such an increase in firing probabilities as S phase progresses has been observed in yeasts and metazoans (Herrick et al. 2000; Patel et al. 2006; Eshaghi et al. 2007) and has been proposed to be a universal feature of eukaryotic replication (Goldar et al. 2009). The probability of origin firing is predicted to increase until it becomes limited by the diffusion rates of the activators, a prediction consistent with quantitative analysis (Gauthier and Bechhoefer 2009). The increasing probability of origin firing has been also been invoked as a guarantee that late-replicating regions of the genome will replicate efficiently and that replication will finish in a timely manner, despite stochastic origin firing (Yang and Bechhoefer 2008; Rhind et al. 2010).
Establishment Is Set at the Level of Chromosome Domains
If the execution of origin timing reflects the probability of origins competing for limiting initiation factors, the establishment of origin timing sets those probabilities. The difference in timing between different replication domains correlates with the chromatin structure of those domains and is not intrinsic to the origins themselves. As described above, targeting chromatin regulators near an origin or moving an origin to new chromatin contexts can affect origin timing (Ferguson and Fangman 1992; Friedman et al. 1996; Vogelauer et al. 2002; Zappulla et al. 2002; Goren et al. 2008). Furthermore, the mammalian TDP occurs before the point at which specific origins are selected (origin decision point; ODP), suggesting that timing is regulated independently of individual origins. This separation is consistent with the observation that timing patterns are more strongly conserved across species than origin locations (Ryba et al. 2010; Di Rienzi et al. 2012; Muller and Nieduszynski 2012; Xu et al. 2012). For instance, mice and humans have a different distribution of origins at the β-globin locus but similar developmentally regulated timing programs (Aladjem et al. 2002). Likewise, different growth conditions can change origin usage without affecting timing patterns (Courbet et al. 2008).
These observations have led to a model in which the establishment of replication timing involves regulation of the chromatin structure affecting large chromosomal regions. Chromatin context established during G1 could affect the ability of origins to compete for limiting factors during S. In fission yeasts, centromeric heterochromatin replicates early, in contrast to the late replication of most heterochromatin (Kim et al. 2003). However, early replication is dependent on the recruitment of the DDK replication kinase to heterochromatin by interaction with the fission yeast HP1 homolog (Hayashi et al. 2009). In the absence of this interaction, centromeres replicate late, in a heterochromatin-dependent manner, suggesting that heterochromatin generally enforces late replication by reducing access of origins to initiation factors such as DDK.
In addition, chromatin context during G1 could affect the licensing of origins in ways that affect the timing of origin firing in S. For instance, the time at which the origin-recognition complex (ORC) binds to origins in fission yeasts correlates with origin timing, with origins binding ORC earlier in G1 also firing earlier in S (Wu and Nurse 2009). Origin function correlates well with nucleosome-free regions (Eaton et al. 2010; MacAlpine et al. 2010; Lubelsky et al. 2011), so nucleosome density may play an important role in regulating ORC binding. How the regulation of ORC binding could affect the timing of origin firing is unclear. However, it has been proposed that firing time is regulated by the number of minichromosome maintenance (MCM) complexes loaded at an origin, with early origins having more MCMs loaded and thus being more likely to fire early (Yang et al. 2010). In such a scenario, the amount of time ORC spends bound to an origin in G1 could affect the number of MCMs loaded at an origin, which could in turn affect the average firing time of the origin. In mammals, MCMs are bound to both early- and late-replicating chromatin before the timing program is set (Dimitrova and Gilbert 1999), but MCMs continue to load during the course of G1 phase and it is not clear whether that continued loading is homogeneous (Kuipers et al. 2011).
There can also be significant differences in timing between origins within one replication domain. The lower resolution of mammalian timing profiles and origin locations has not allowed for systematic analysis of intradomain timing. However, in budding yeasts, substantial differences in origin timing are detected between euchromatic origins (Fig. 1) (Friedman et al. 1997; Raghuraman et al. 2001). Furthermore, deviations from the correlation between timing and chromatin contexts, and experimental manipulations that can modify specific origin firing (Lin et al. 2003; Goren et al. 2008; Hassan-Zadeh et al. 2012), make it clear that although regional chromatin context strongly biases replication timing, it does not impose absolute control. It seems likely that the timing of firing of individual origins is a combination of intrinsic probability of the origin firing modified by the origin’s chromosomal context.
Checkpoints Affect Replication Timing
In addition to the execution of the replication timing program established during G1, replication timing can also be affected by feedback regulation during S phase. The best-understood of these mechanisms involve the S-phase DNA damage checkpoint, which is activated by fork stalling and inhibits subsequent origin firing (Shirahige et al. 1998; Costanzo et al. 2003; Merrick et al. 2004). Such a mechanism could act to balance fork termination with origin firing to maintain a uniform fork density (Diaz-Martinez and Clarke 2003). The damage checkpoint acts to restrain origin firing by inhibiting the function of limiting activators, such as Cdc45, Sld2, Sld3, and DDK (Lopez-Mosqueda et al. 2010; Zegerman and Diffley 2010). In budding yeasts, a similar checkpoint-dependent mechanism acts to maintain the temporal order of origin firing when S phase is elongated by limiting nucleotides or mutant replication factors (Alvino et al. 2007; Koren et al. 2010). However, these intra-S-phase timing mechanisms function primarily to maintain the appropriate fork density by enforcing the relative order of origin firing, but are not involved in determining replication timing patterns per se.
WHAT IS THE BIOLOGICAL FUNCTION OF A TIMING PROGRAM?
Replication timing is universally observed in eukaryotes, suggesting an important and conserved function. Nonetheless, that function is largely mysterious. Moreover, it is possible that replication timing per se does not contribute to fitness but is an inevitable consequence of some other factor, such as the domain architecture of the genome. To understand the biological function of replication timing, we have to consider two questions. First, why is S phase longer than its minimal possible length? Second, what is the reason for the particular conserved order of replication? Although there is little direct evidence to address either question, several plausible hypotheses are consistent with existing data.
Possible Reasons for an Extended S Phase
S phase in metazoan somatic cells can be more than 10 times longer than embryonic S phase. In fly and frog embryos, replication takes less than 20 min (Graham 1966; Blumenthal et al. 1974). Yet in somatic cells of the same species, replication takes as long as 8 h (Blumenthal et al. 1974). A slower S phase may be more economical; by extending the length of S phase, cells reduce the number of concurrent replication forks and thus the number of fork proteins that need to be synthesized. Even in frog embryos, which are transcriptionally silent and in which cellular energy is focused on genome duplication, replication kinetics appear to be optimized to use the fewest number of forks required to complete replication in the allotted time (Yang and Bechhoefer 2008). However, the theoretical energy savings may not be biologically significant compared with the total energy budget of the cell.
Another possible advantage to pacing the rate of replication is to allow for late-firing origins to serve as backups. Licensing more origins than are normally required for S phase provides a robust solution to replication fork stalling (Blow et al. 2011). If two approaching forks both stall, the only way to replicate the intervening sequence is to activate new forks from an origin between them. And because origins can only be established during G1, such backup origins must be positioned throughout the genome and not fired early in S phase. Thus, dramatically reducing the number of licensed origins does not affect normal replication, but does make cells much more sensitive to replication stress (Woodward et al. 2006; Ge et al. 2007). In a system of redundant, inefficient origins, such backup origins need not be qualitatively different “dormant” origins, they can simply be whichever origins do not happen to fire in a particular cell.
Reducing the number of concurrent forks may also enhance genome stability by avoiding overtaxing DNA repair and fork stability mechanisms. In bacteria, increasing the number of replication forks can saturate the mismatch repair capacity and lead to increased mutation rates (Schaaper and Radman 1989). Also, concurrent fork collapses may increase the chance of illegitimate repair events involving nonhomologous replication structures, which could lead to translocations and other genome rearrangements. In addition, higher numbers of active forks increases the flux in nucleotide pool levels, which, because polymerase fidelity is sensitive to nucleotide concentration (Kumar et al. 2010), may lead increased misincorporation. Although there is little direct evidence to support the idea the extending S phase reduces genome instability, increasing the rate of origin firing moderately increases genome instability (Patel et al. 2008).
Finally, extending S phase may facilitate specific regulatory mechanisms that control gene expression or other aspects of chromatin metabolism (Gilbert 2002; Hiratani et al. 2009). If extending S phase does have a regulatory role, the order in which various loci are replicated becomes an important parameter. Therefore, the fact that patterns of replication timing are conserved has led to speculation that these patterns have regulatory significance.
Possible Reasons for Temporal Order of Replication
The most compelling model for a regulatory role for replication timing is that the time during S phase that a locus is replicated can influence its chromatin structure. Because chromatin is assembled at the replication fork, if different chromatin conformations are favored during different parts of S phase, the replication timing of a locus could influence its chromatin structure. This model is supported by the observation that the time at which a plasmid is injected into a cell during S phase influences its chromatin structure and its expression level (Zhang et al. 2002; Lande-Diner et al. 2009). An attractive feature of this model is its self-reinforcing nature. Chromatin context influences replication time, while replication at that time may reestablish chromatin context. This model also provides a mechanism by which replication timing and chromatin context could spread. If a localized alteration causes early replication of a small region in an otherwise late-replicating domain, the passive early replication of surrounding loci could alter chromatin context and reprogram the replication timing of the entire replication domain. Thus, this model suggests mechanisms for both the epigenetic inheritance of replication timing and its developmental reprogramming.
It is also possible that replication timing regulates other aspects of genome metabolism. For instance, mutation rates are lower in early-replicating regions (Stamatoyannopoulos et al. 2009; Chen et al. 2010; Lang and Murray 2011). If replication timing directly influences mutation rates, perhaps by concentrating repair capacity early in S phase or altering the balance of nucleotide pools during S, there might be evolutionary pressure to replicate gene-rich regions earlier in S phase. Likewise, recombination rates are higher in earlier-replicating regions, allowing the possibility that the time of replication during premeiotic S phase could regulate crossover frequency.
The inevitable effect of replication on gene dosage could also have regulatory consequences. Genes that are duplicated early will be present at twice the copy number of late-replicating genes for most of the duration of S phase, increasing transcriptional output. Early-replicating sequences are also the first to have sister-chromatid templates for homologous recombination. The ability to perform DNA repair by homologous recombination for a greater fraction of the cell cycle may contribute to the fact the early-replicating DNA has lower rates of mutation. These dosage effects would be most pronounced in cells in which S phase is a substantial fraction of the cell cycle, such as mammalian ESCs.
It is important to keep in mind that the strong correlation between replication timing and important aspects of genome metabolism may simply be an indirect consequence of how closely replication timing reflects the structure of chromatin. For example, mutation rates are also known to be lowest in open chromatin (Prendergast et al. 2007). It is possible then that a particular temporal sequence to replication may serve no specific biological function, but rather just be a reflection of the large-scale organization of the genome. This model explains the bulk of available data with no extraneous assumptions or mechanisms and thus should be considered the null hypothesis against which any more complicated model for functional significance should be weighed.
CONCLUDING REMARKS
In comparison with our sophisticated knowledge of most fundamental cellular processes, our understanding of replication timing—both its biological significance and mechanism—has remained primitive and experimental progress has mainly underscored its complexity. The timing program is established at a minimum of two levels, both of which act heterogeneously in different individual cells to establish a probability for any given region to replicate at a particular time. At one level local sequence and chromatin composition influence whether a prereplicative complex is formed, the number of MCM complexes loaded, or the local affinity for initiation proteins such as Cdc45, Sld2, Sld3, and DDK. Mutations in several gene products affecting this local level of regulation have been identified, which exhibit partial or localized effects on the timing program. At a second level the global timing program of large replication domains appears to be established by the large-scale folding of chromosomes, a more challenging problem to dissect. The execution of the timing program results from an integration of these two levels of influence, which may play more or less important roles in different species. For example, local regulation likely plays a more important role in the small genomes of single-celled organisms. In fact, we propose that the majority of the yeast genome can be thought of as the equivalent of early replication domains in mammals (Fig. 1), with the large-scale structure of early-replicating regions spatially clustered by a few central organizers such as the Fkh proteins and centromere-proximal regions (Knott et al. 2012), whereas late-replicating regions such as telomeres and mating-type loci are organized into spatially separate clusters. In progressively larger genomes, large-scale folding is a major organizing principle, diminishing the importance of localized effects to the overall timing program. Elucidating these complex influences will remain a challenge, but perhaps even more challenging and important will be assessing the degree to which cell-to-cell heterogeneity plays a role in the overall timing program. This challenge will require advancements in our ability to evaluate replication of individual molecules.
As new experimental systems to probe mechanism emerge, the elephant in the room remains our complete lack of appreciation of the biological significance of this program. We have summarized many of the hypotheses that have been proposed; however, any proposed biological function for replication timing must be tested against the simple possibility that the program results from a combination of limiting resources for replication, executed in a temporal sequence that reflects the assembly of chromatin for other cellular functions. Even if the simple possibility should be the case, replication timing has emerged as a robust means to assess the large-scale epigenetic state of specific cell types and identify regions of large-scale structural reorganization during differentiation or disease (Ryba et al. 2012).
ACKNOWLEDGMENTS
We thank Oscar Aparicio, John Bechhoefer, David MacAlpine, Conrad Nieduszynski, and Paolo Norio for valuable comments on the manuscript. Research in the Gilbert lab is supported by National Institutes of Health (NIH) grants GM083337 and GM085354. Research in the Rhind lab is supported by NIH grants GM069957 and GM098815 and an American Cancer Society Research Scholar Grant.
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Agier N, Fischer G 2012. The mutational profile of the yeast genome is shaped by replication. Mol Biol Evol 29: 905–913 [DOI] [PubMed] [Google Scholar]
- Aladjem MI, Rodewald LW, Lin CM, Bowman S, Cimbora DM, Brody LL, Epner EM, Groudine M, Wahl GM 2002. Replication initiation patterns in the β-globin loci of totipotent and differentiated murine cells: Evidence for multiple initiation regions. Mol Cell Biol 22: 442–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK 2007. Replication in hydroxyurea: It’s a matter of time. Mol Cell Biol 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M 2003. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 [DOI] [PubMed] [Google Scholar]
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley D, Chagin VO, Schermelleh L, Martin S, Pombo A, Carlton PM, Gahl A, Domaing P, Birk U, Leonhardt H, et al. 2010. Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res 38: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D 2010. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol 17: 894–900 [DOI] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Bergstrom R, Whitehead J, Kurukuti S, Ohlsson R 2007. CTCF regulates asynchronous replication of the imprinted H19/Igf2 domain. Cell Cycle 6: 450–454 [DOI] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM 2012. Unraveling cell type–specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19: 837–844 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA 2011. How dormant origins promote complete genome replication. Trends Biochem Sci 36: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol 38: 205–223 [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Delucchi CS, van Bemmel JG, Haase S, Herce HD, Nowak D, Meilinger D, Stear JH, Leonhardt H, Cardoso MC 2012. Histone hypoacetylation is required to maintain late replication timing of constitutive heterochromatin. Nucleic Acids Res 40: 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, et al. 2011. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res 21: 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Rappailles A, Duquenne L, Huvet M, Guilbaud G, Farinelli L, Audit B, d’Aubenton-Carafa Y, Arneodo A, Hyrien O, et al. 2010. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res 20: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacchia D, Dileep V, Quivy JP, Foti R, Tili F, Santarella-Mellwig R, Anthony C, Almouzni G, Gilbert DM, Buonomo SB 2012. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J 31: 3678–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini M, Cammarano R, Bernardi G 2009. The evolution of isochore patterns in vertebrate genomes. BMC Genomics 10: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell 11: 203–213 [DOI] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- Crabbe L, Thomas A, Pantesco V, De Vos J, Pasero P, Lengronne A 2010. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol 17: 1391–1397 [DOI] [PubMed] [Google Scholar]
- Cseresnyes Z, Schwarz U, Green CM 2009. Analysis of replication factories in human cells by super-resolution light microscopy. BMC Cell Biol 10: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky DM, Liu J, Hamlin JL, Shao Z 2008. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J Mol Biol 375: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O 2012. Common fragile sites: Mechanisms of instability revisited. Trends Genet 28: 22–32 [DOI] [PubMed] [Google Scholar]
- Demczuk A, Gauthier MG, Veras I, Kosiyatrakul S, Schildkraut CL, Busslinger M, Bechhoefer J, Norio P 2012. Regulation of DNA replication within the immunoglobulin heavy-chain locus during B cell commitment. PLoS Biol 10: e1001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moura AP, Retkute R, Hawkins M, Nieduszynski CA 2010. Mathematical modelling of whole chromosome replication. Nucleic Acids Res 38: 5623–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprat R, Thierry-Mieg D, Lailler N, Lajugie J, Schildkraut C, Thierry-Mieg J, Bouhassira EE 2009. Predictable dynamic program of timing of DNA replication in human cells. Genome Res 19: 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Martinez L, Clarke DJ 2003. Self-regulating model for control of replication origin firing in budding yeast. Cell Cycle 2: 576–578 [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell 4: 983–993 [DOI] [PubMed] [Google Scholar]
- Di Rienzi SC, Lindstrom KC, Mann T, Noble WS, Raghuraman MK, Brewer BJ 2012. Maintaining replication origins in the face of genomic change. Genome Res 22: 1940–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Schildkraut CL 2006. Temporal order of DNA replication. In DNA replication and human disease (ed. DePamphilis ML), pp. 197–216 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell 2: 173–182 [DOI] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS 2010. A three-dimensional model of the yeast genome. Nature 465: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, Macalpine DM 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi M, Karuturi RK, Li J, Chu Z, Liu ET, Liu J 2007. Global profiling of DNA replication timing and efficiency reveals that efficient replication/firing occurs late during S-phase in S. pombe. PLoS ONE 2: e722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash-Amar S, Simon I 2010. Genome-wide analysis of the replication program in mammals. Chromosome Res 18: 115–125 [DOI] [PubMed] [Google Scholar]
- Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I 2008. Global organization of replication time zones of the mouse genome. Genome Res 18: 1562–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ 2006. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol 8: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BM, Fangman WL 1992. A position effect on the time of replication origin activation in yeast. Cell 68: 333–339 [DOI] [PubMed] [Google Scholar]
- Friedman KL, Diller JD, Ferguson BM, Nyland SV, Brewer BJ, Fangman WL 1996. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev 10: 1595–1607 [DOI] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ, Fangman WL 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2: 667–678 [DOI] [PubMed] [Google Scholar]
- Frum RA, Khondker ZS, Kaufman DG 2009. Temporal differences in DNA replication during the S phase using single fiber analysis of normal human fibroblasts and glioblastoma T98G cells. Cell Cycle 8: 3133–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier MG, Bechhoefer J 2009. Control of DNA replication by anomalous reaction-diffusion kinetics. Phys Rev Lett 102: 158104. [DOI] [PubMed] [Google Scholar]
- Gauthier MG, Norio P, Bechhoefer J 2012. Modeling inhomogeneous DNA replication kinetics. PLoS ONE 7: e32053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2002. Replication timing and transcriptional control: Beyond cause and effect. Curr Opin Cell Biol 14: 377–383 [DOI] [PubMed] [Google Scholar]
- Gilbert DM, Gasser SM 2006. Nuclear structure and DNA replication. In DNA replication and human disease (ed. DePamphilis ML), pp. 175–196 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA 2004. Chromatin architecture of the human genome: Gene-rich domains are enriched in open chromatin fibers. Cell 118: 555–566 [DOI] [PubMed] [Google Scholar]
- Goldar A, Labit H, Marheineke K, Hyrien O 2008. A dynamic stochastic model for DNA replication initiation in early embryos. PLoS ONE 3: e2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldar A, Marsolier-Kergoat MC, Hyrien O 2009. Universal temporal profile of replication origin activation in eukaryotes. PLoS ONE 4: e5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H 2008. DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Genes Dev 22: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CF 1966. The regulation of DNA synthesis and mitosis in multinucleate frog eggs. J Cell Sci 1: 363–374 [DOI] [PubMed] [Google Scholar]
- Guan Z, Hughes CM, Kosiyatrakul S, Norio P, Sen R, Fiering S, Allis CD, Bouhassira EE, Schildkraut CL 2009. Decreased replication origin activity in temporal transition regions. J Cell Biol 187: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilbaud G, Rappailles A, Baker A, Chen CL, Arneodo A, Goldar A, d’Aubenton-Carafa Y, Thermes C, Audit B, Hyrien O 2011. Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput Biol 7: e1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA 2010. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan-Zadeh V, Chilaka S, Cadoret JC, Ma MK, Boggetto N, West AG, Prioleau MN 2012. USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. PLoS Biol 10: e1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Masai H 2011. Mrc1 marks early-firing origins and coordinates timing and efficiency of initiation in fission yeast. Mol Cell Biol 31: 2380–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H 2012. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H 2007. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Takahashi TS, Nakagawa T, Nakayama J, Masukata H 2009. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat Cell Biol 11: 357–362 [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P 2006. Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick J, Stanislawski P, Hyrien O, Bensimon A 2000. Replication fork density increases during DNA synthesis in X. laevis egg extracts. J Mol Biol 300: 1133–1142 [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM 2001. The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol 152: 385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S, Robertson ED, Donaldson AD 2006. The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. EMBO J 25: 1505–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Gilbert DM 2010. Autosomal lyonization of replication domains during early mammalian development. Adv Exp Med Biol 695: 41–58 [DOI] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi SI, Lu J, Gilbert DM 2009. Replication timing and transcriptional control: Beyond cause and effect—part II. Curr Opin Genet Dev 19: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvet M, Nicolay S, Touchon M, Audit B, d’Aubenton-Carafa Y, Arneodo A, Thermes C 2007. Human gene organization driven by the coordination of replication and transcription. Genome Res 17: 1278–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Goldar A 2010. Mathematical modelling of eukaryotic DNA replication. Chromosome Res 18: 147–161 [DOI] [PubMed] [Google Scholar]
- Hyrien O, Marheineke K, Goldar A 2003. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25: 116–125 [DOI] [PubMed] [Google Scholar]
- Jorgensen HF, Azuara V, Amoils S, Spivakov M, Terry A, Nesterova T, Cobb BS, Ramsahoye B, Merkenschlager M, Fisher AG 2007. The impact of chromatin modifiers on the timing of locus replication in mouse embryonic stem cells. Genome Biol 8: R169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A 2007. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res 17: 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M 2009. Cyclin A–Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci 106: 3184–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA 2003. Early-replicating heterochromatin. Genes Dev 17: 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU 2006. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell 125: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavare S, Aparicio OM 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM 2012. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Soifer I, Barkai N 2010. MRC1-dependent scaling of the budding yeast DNA replication timing program. Genome Res 20: 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinska L, Besnard E, Cot E, Dohet C, Mechali M, Lemaitre JM, Fisher D 2008. Cdk1 and Cdk2 activity levels determine the efficiency of replication origin firing in Xenopus. EMBO J 27: 758–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers MA, Stasevich TJ, Sasaki T, Wilson KA, Hazelwood KL, McNally JG, Davidson MW, Gilbert DM 2011. Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J Cell Biol 192: 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma D, Viberg J, Nilsson AK, Chabes A 2010. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res 38: 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Huberman JA 2009. Checkpoint-dependent regulation of origin firing and replication fork movement in response to DNA damage in fission yeast. Mol Cell Biol 29: 602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Viberg J, Nilsson AK, Chabes A 2010. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res 38: 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labit H, Perewoska I, Germe T, Hyrien O, Marheineke K 2008. DNA replication timing is deterministic at the level of chromosomal domains but stochastic at the level of replicons in Xenopus egg extracts. Nucleic Acids Res 36: 5623–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande-Diner L, Zhang J, Cedar H 2009. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell 34: 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW 2011. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol Evol 3: 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A 2006. DNA replication origin interference increases the spacing between initiation events in human cells. Mol Biol Cell 17: 5337–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Pascuzzi PE, Settlage SB, Shultz RW, Tanurdzic M, Rabinowicz PD, Menges M, Zheng P, Main D, Murray JA, et al. 2010. Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet 6: e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tallec B, Dutrillaux B, Lachages AM, Millot GA, Brison O, Debatisse M 2011. Molecular profiling of common fragile sites in human fibroblasts. Nat Struct Mol Biol 18: 1421–1423 [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123 [DOI] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I 2005. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 24: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Bhaskar A, Lee C, Chung SC, Tye BK, Keich U 2010. A comprehensive genome-wide map of autonomously replicating sequences in a naive genome. PLoS Genet 6: e1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HY, Robertson ED, Hiraga S, Alvino GM, Collingwood D, McCune HJ, Sridhar A, Brewer BJ, Raghuraman MK, Donaldson AD 2011. The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol Biol Cell 22: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CM, Fu H, Martinovsky M, Bouhassira E, Aladjem MI 2003. Dynamic alterations of replication timing in mammalian cells. Curr Biol 13: 1019–1028 [DOI] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maas NL, Jonsson ZO, Defazio-Eli LG, Wohlschlegel J, Toczyski DP 2010. Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li F, Murphy CS, Davidson MW, Gilbert DM 2010. G2 phase chromatin lacks determinants of replication timing. J Cell Biol 189: 967–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Sasaki T, Kuipers MA, Lucas I, Le Beau MM, Carignon S, Debatisse M, Prinz JA, Dennis JH, Gilbert DM 2011. Pre-replication complex proteins assemble at regions of low nucleosome occupancy within the Chinese hamster dihydrofolate reductase initiation zone. Nucleic Acids Res 39: 3141–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Li J, Eshaghi M, Liu J, Karuturi RK 2010. Genome-wide estimation of firing efficiencies of origins of DNA replication from time-course copy number variation data. BMC Bioinformatics 11: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygeros J, Koutroumpas K, Dimopoulos S, Legouras I, Kouretas P, Heichinger C, Nurse P, Lygerou Z 2008. Stochastic hybrid modeling of DNA replication across a complete genome. Proc Natl Acad Sci 105: 12295–12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier-Kergoat MC, Goldar A 2012. DNA replication induces compositional biases in yeast. Mol Biol Evol 29: 893–904 [DOI] [PubMed] [Google Scholar]
- Maya-Mendoza A, Olivares-Chauvet P, Shaw A, Jackson DA 2010. S phase progression in human cells is dictated by the genetic continuity of DNA foci. PLoS Genet 6: e1000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune HJ, Danielson LS, Alvino GM, Collingwood D, Delrow JJ, Fangman WL, Brewer BJ, Raghuraman MK 2008. The temporal program of chromosome replication: Genomewide replication in clb5Δ Saccharomyces cerevisiae. Genetics 180: 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNairn AJ, Gilbert DM 2003. Epigenomic replication: Linking epigenetics to DNA replication. Bioessays 25: 647–656 [DOI] [PubMed] [Google Scholar]
- Meister P, Taddei A, Ponti A, Baldacci G, Gasser SM 2007. Replication foci dynamics: Replication patterns are modulated by S-phase checkpoint kinases in fission yeast. EMBO J 26: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick CJ, Jackson D, Diffley JF 2004. Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem 279: 20067–20075 [DOI] [PubMed] [Google Scholar]
- Mickle KL, Oliva A, Huberman JA, Leatherwood J 2007a. Checkpoint effects and telomere amplification during DNA re-replication in fission yeast. BMC Mol Biol 8: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickle KL, Ramanathan S, Rosebrock A, Oliva A, Chaudari A, Yompakdee C, Scott D, Leatherwood J, Huberman JA 2007b. Checkpoint independence of most DNA replication origins in fission yeast. BMC Mol Biol 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CA, Nieduszynski CA 2012. Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res 22: 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Langst G 2011. Genome organization in and around the nucleolus. Trends Genet 27: 149–156 [DOI] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL 2005. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20: 575–587 [DOI] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N 2006. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell 17: 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Kommajosyula N, Rosebrock A, Bensimon A, Leatherwood J, Bechhoefer J, Rhind N 2008. The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol Biol Cell 19: 5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Hiratani I, Gilbert DM 2010. Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Res 18: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Tsumagari K, Battaglia D, Ryba T, Hiratani I, Ehrlich M, Gilbert DM 2011. DNA replication timing is maintained genome-wide in primary human myoblasts independent of D4Z4 contraction in FSH muscular dystrophy. PLoS ONE 6: e27413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast JG, Campbell H, Gilbert N, Dunlop MG, Bickmore WA, Semple CA 2007. Chromatin structure and evolution in the human genome. BMC Evol Biol 7: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Brewer BJ 2010. Molecular analysis of the replication program in unicellular model organisms. Chromosome Res 18: 19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL 2001. Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Retkute R, Nieduszynski CA, de Moura A 2011. Dynamics of DNA replication in yeast. Phys Rev Lett 107: 068103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N 2006. DNA replication timing: Random thoughts about origin firing. Nat Cell Biol 8: 1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Yang SC, Bechhoefer J 2010. Reconciling stochastic origin firing with defined replication timing. Chromosome Res 18: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM 2011. Genome-scale analysis of replication timing: From bench to bioinformatics. Nat Protoc 6: 870–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Chang BH, Shirley JW, Buckley Q, Pope BD, Devidas M, Druker BJ, Gilbert DM 2012. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res 22: 1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D 2004. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci 117: 5353–5365 [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Radman M 1989. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J 8: 3511–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmegner C, Hameister H, Vogel W, Assum G 2007. Isochores and replication time zones: A perfect match. Cytogenet Genome Res 116: 167–172 [DOI] [PubMed] [Google Scholar]
- Schultz SS, Desbordes SC, Du Z, Kosiyatrakul S, Lipchina I, Studer L, Schildkraut CL 2010. Single-molecule analysis reveals changes in the DNA replication program for the POU5F1 locus upon human embryonic stem cell differentiation. Mol Cell Biol 30: 4521–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schubeler D 2009. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev 23: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Kohler H, Oakeley EJ, Stadler MB, Schubeler D 2010. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res 20: 771–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekedat MD, Fenyo D, Rogers RS, Tackett AJ, Aitchison JD, Chait BT 2010. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol Syst Biol 6: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T 2004. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 18: 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol Cell 10: 1355–1365 [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR 2009. Human mutation rate associated with DNA replication timing. Nat Genet 41: 393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich B, Filion G, van Steensel B, Ekwall K 2012. The inner nuclear membrane proteins Man1 and Ima1 link to two different types of chromatin at the nuclear periphery in S. pombe. Nucleus 3: 77–87 [DOI] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE 1999. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev 13: 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S, Sugimura K, Saito T, Sato C, Fukushima Y, Taguchi H, Okumura K 2005. Regulation of replication at the R/G chromosomal band boundary and pericentromeric heterochromatin of mammalian cells. Exp Cell Res 304: 162–174 [DOI] [PubMed] [Google Scholar]
- Takebayashi S, Dileep V, Ryba T, Dennis JH, Gilbert DM 2012. Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc Natl Acad Sci 109: 12574–12579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H 2011. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol 21: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Tazumi A, Fukuura M, Nakato R, Kishimoto A, Takenaka T, Ogawa S, Song JH, Takahashi TS, Nakagawa T, Shirahige K, et al. 2012. Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev 26: 2050–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AM, Gillespie PJ, Blow JJ 2010. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J Cell Biol 188: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Nicolay S, Audit B, Brodie of Brodie EB, d’Aubenton-Carafa Y, Arneodo A, Thermes C 2005. Replication-associated strand asymmetries in mammalian genomes: Toward detection of replication origins. Proc Natl Acad Sci 102: 9836–9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M 2002. Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. 2006. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci 119: 132–140 [DOI] [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ 2006. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 173: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Nurse P 2009. Establishing the program of origin firing during S phase in fission yeast. Cell 136: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Terry AV, Singh PB, Gilbert DM 2005. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol Biol Cell 16: 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Singh PB, Gilbert DM 2006. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J Cell Biol 174: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yanagisawa Y, Tsankov AM, Hart C, Aoki K, Kommajosyula N, Steinmann KE, Bochicchio J, Russ C, Regev A, et al. 2012. Genome-wide identification and characterization of replication origins by deep sequencing. Genome Biol 13: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K 2002. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789 [DOI] [PubMed] [Google Scholar]
- Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I 2010. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Ishii A, Kanoh Y, Oda M, Nishito Y, Masai H 2012. Rif1 regulates the replication timing domains on the human genome. EMBO J 31: 3667–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SC, Bechhoefer J 2008. How Xenopus laevis embryos replicate reliably: Investigating the random-completion problem. Phys Rev E Stat Nonlin Soft Matter Phys 78: 041917. [DOI] [PubMed] [Google Scholar]
- Yang SC, Rhind N, Bechhoefer J 2010. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol Syst Biol 6: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC 2010. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell 40: 834–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM 2009. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci 106: 19363–19368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Sternglanz R, Leatherwood J 2002. Control of replication timing by a transcriptional silencer. Curr Biol 12: 869–875 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF 2010. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H 2002. Establishment of transcriptional competence in early and late S phase. Nature 420: 198–202 [DOI] [PubMed] [Google Scholar]