Abstract
Salmonella and Shigella bacteria require the type III secretion system (T3SS) to inject virulence proteins into their hosts and initiate infections. The tip proteins SipD and IpaD are critical components of the Salmonella and Shigella T3SS, respectively. Recently, SipD and IpaD have been shown to interact with bile salts, which are enriched in the intestines, and are hypothesized to act as environmental sensors for these enteric pathogens. Bile salts activate the Shigella T3SS but repress the Salmonella T3SS, and the mechanism of this differing response to bile salts is poorly understood. Further, how SipD binds to bile salts is currently unknown. Computer modeling predicted that IpaD binds the bile salt deoxycholate in a cleft formed by the N-terminal domain and the long central coiled coil of IpaD. Here, we used NMR methods to determine which SipD residues are affected by the interaction with the bile salts deoxycholate, chenodeoxycholate and taurodeoxcholate. The bile salts perturbed nearly the same set of SipD residues, however, the largest chemical shift perturbations occurred away from what was predicted for the bile salt binding site in IpaD. Our NMR results indicate that that bile salt interaction of SipD will be different from what was predicted for IpaD, suggesting a possible mechanism for the differing response of Salmonella and Shigella to bile salts.
The type III secretion system (T3SS) is essential in the pathogenesis of many Gram-negative bacteria (1) and it consists of a needle apparatus, which is a nanometer-scale bacterial injector found on the bacterial surface. A structural component of the T3SS needle apparatus is the tip protein (2, 3), which is known as SipD in Salmonella typhimurium, IpaD in Shigella flexneri (3), and BipD in Burkholderia pseudomallei. The tip proteins are critical in the assembly of the T3SS needle apparatus and in bacterial pathogenesis (4–6). Recently, IpaD was shown to bind the bile salts deoxycholate, taurodeoxycholate and chenodeoxycholate (7); and this specific binding was suggested to function as an environmental signal to activate the T3SS in Shigella flexneri (8). The same study also showed that SipD binds to deoxycholate (7), however, other studies have shown that bile salts suppressed the invasiveness of Salmonella (9, 10). The crystal structures of IpaD (11) and SipD (to be reported elsewhere) are highly similar (with Cα RMSD of 1.4 Å), thus it is not clear how bile salts would interact with IpaD and SipD to induce opposing effects – activation of the Shigella T3SS and inactivation the Salmonella T3SS.
Using computer docking simulation, Stensrud et al. (7) predicted that deoxycholate binds IpaD in a cleft formed by the central coiled-coil and the N-terminal domain of IpaD, which was then confirmed by fluorescence spectroscopy and mutagenesis (7). Here, we used NMR methods to identify which SipD residues are involved in the interaction with bile salts deoxycholate, taurodeoxycholate and chenodeoxycholate. Our NMR results indicated that bile salts bind SipD in a different site from what is predicted for IpaD, suggesting a basis for the differing mechanism of bile salt interaction observed for Shigella and Salmonella.
EXPERIMENTAL PROCEDURES
Protein expression and purification
The coding region for wild type Salmonella typhimurium SipD residues 39 to 343 was PCR amplified and subcloned into pET-21a with a His6-tagged GB1 domain and a tobacco etch virus (TEV) protease cleavage site appended at the N-terminus of SipD39–343 (after cleavage of the fusion tag, SipD39–343 retained a three-residue cloning artifact “GHM” at its N-terminus). All growth media contained 30 µg/mL kanamycin and 100 µg/mL carbenicillin. For protein expression, the SipD39–343 expression plasmid was transformed into E. coli BL21(DE3) DNAY and a starter culture was grown at 37°C overnight in 20 mL LB medium followed by centrifugation. Perdeuterated 15N,13C-labeled SipD39–343 was obtained resuspending the starter culture in 1 L M9 minimal media dissolved in 99% D2O (Cambridge Isotopes) and supplemented with 2 g/L of 13C-glucose (Isotec) and 1 g/L of 15N-ammonium chloride (Isotec). Isotopically 15N-labeled SipD39–343 was obtained by resuspending the starter culture in 1 L M9 minimal medium supplemented with 1 g/L 15N-ammonium chloride. Cells were grown at 37°C until A600 ~ 0.6 to 0.8, induced with 1 mM IPTG and cell growth was continued in a 15°C shaker incubator overnight. Cells were harvested by centrifugation, resuspended in 30 mL binding buffer (500 mM NaCl, 20 mM Tris-HCl, 500 mM NaCl, 5 mM imidazole, pH 8.0) and stored at −20°C. To purify SipD39–343, cells were thawed on ice and 40 µL of 1 mM phenylmethylsulfonyl fluoride was added before sonication. The cell lysate was centrifuged (18,200 ×g, 10 min, 4°C), and 1/10 volume of 1% polyethyleneimine (pH 8) was added to the supernatant to precipitate the nucleic acids, then centrifuged (18,200 ×g, 10 min, 4°C). The supernatant was loaded onto a 5 mL Ni2+-affinity column (Sigma), washed with 35 mL binding buffer, and eluted in 1 mL fractions with a total of 15 mL elution buffer (250 mM imidazole, 500 mM NaCl, 20 mM Tris, pH 8.0). Fractions containing the GB1-SipD39–343 fusion protein were combined (into a total volume of 20 mL) and dialyzed overnight at room temperature in 1 L buffer (50mM Tris, 0.5 mM EDTA, 1mM DTT, 20 mM NaCl pH 8) in the presence of 50 uL 0.07 mM recombinant TEV protease that was purified as described (12). The digest was loaded onto a 5 mL Ni2+-affinity column, which retained the GB1 tag and eluted SipD39–343. Purified SipD39–343 was dialyzed in buffer (10 mM sodium phosphate pH 6.5 and 10 mM NaCl), concentrated using Amicon Ultra 3K (Millipore) and protein concentration was determined by UV absorbance at 280 nm.
Amino acid specific labeling
In total, eight 15N-amino acid-specific labeled SipD39–343 were prepared using 15N-labeled Leu, Val, Ile, Ala, Phe, Tyr, Met and Lys. To label SipD39–343 with 15N-Leu, for example, E. coli BL21(DE3) DNAY harboring the SipD39–343 expression plasmid was grown in 1 L LB medium overnight at 37°C. The cells were harvested by centrifugation and resuspended in 500 mL M9 minimal medium supplemented with 125 mg/L of 15N-Leu (Isotec) and 600 mg/L of the rest of the 19 unlabeled amino acids. The cells were grown at 37°C for ~45 min until A600 ~0.8, induced with 1 mM IPTG and cell growth was continued at 37 °C for 4 hours before harvest. Protein was purified as described above.
Mutagenesis of tryptophan residues
To assign the side chain resonances of the four tryptophan residues in SipD (W135, W177, W234 and W290), each tryptophan was mutated into tyrosine by the Stratagene Quickchange method. Recombinant 15N-labeled SipD39–343 with point mutation W135Y, W177Y, W234Y or W290Y was expressed and purified as described above.
NMR spectroscopy
NMR data were acquired at 30°C on a Bruker Avance 800 MHz equipped with cryogenic triple-resonance probe, processed using NMRPipe (13) and analyzed using NMRView (14). Proteins were dissolved in NMR buffer (10 mM sodium phosphate pH 6.5, 10 mM NaCl, and 10% D2O). For backbone assignments, 0.5 mM perdeuterated 15N,13C-SipD39– 343 was used to acquire 2D 1H-15N TROSY-HSQC (15), 3D TROSY-HNCA (16), 3D TROSY-HNCACB (17), 3D TROSY-HNCO (17) and 3D TROSY-HN(CA)CO (17). In addition, 2D 1H-15N TROSY-HSQC (15) was acquired using 0.3 – 0.6 mM 15N-amino acid-labeled SipD39–343. Tryptophan side chains were assigned by acquiring 2D 1H-15N HSQC or 2D 1H-15N TROSY-HSQC (15) using 15N-labeled SipD39–343 W135Y, W177Y, W234Y and W290Y. For NMR chemical shift mapping, 2D 1H-15N TROSY-HSQC (15) spectra were acquired using 15N-labeled SipD39–343 that was titrated with varying amounts of deoxycholate (Amresco), chenodeoxycholate (Sigma), taurodeoxycholate (Sigma) or cholate hydrate (Sigma). NMR samples for chemical shift mapping were prepared as follows: 0.6 mM 15N-SipD39–343 was dialyzed overnight in 1 L of NMR buffer containing increasing concentrations of deoxycholate (0, 0.7, 1.4 or 2.1 mM) or chenodeoxycholate (0.0, 0.3, 0.6, 1.3 or 2.6 mM); and 0.7 mM 15N-SipD39– 343 in NMR buffer with taurodeoxycholate (0.0, 0.3, 0.7, 1.3 or 2.6 mM) or cholate hydrate (0.0, 0.3, 0.7, 1.3 or 2.6 mM).
RESULTS
Protein expression and purification
Full-length SipD yielded poor NMR spectra (Fig. S1) and was not amenable for NMR characterization. However the crystal structures of BipD (11, 18) and IpaD (11) showed that the N-terminal 29 – 38 residues were disordered and these proteins were predominantly alpha helical in secondary structure. Thus, the N-terminal 38 residues of SipD were truncated, forming the SipD39–343 construct used in this study. Recombinant SipD39–343 was overexpressed as a fusion protein with the GB1 domain, purified by Ni2+-affinity chromatography and digested with TEV protease to remove the fusion tag. The circular dichroism (CD) spectra of full-length SipD and SipD39–343 were both indicative of highly alpha helical proteins (Fig. S2). Further, the CD thermal denaturation curves for full-length SipD and SipD39–343 (Fig. S2) were similar in pattern and showed two transition temperatures at 59°C and 75°C. Thus, CD spectroscopy indicated that SipD39–343 retained the core structured domain of SipD. In contrast to full-length SipD, the truncated SipD39–343 yielded well-dispersed NMR spectra (Fig. 1 & Fig. S1C), which allowed further NMR characterization.
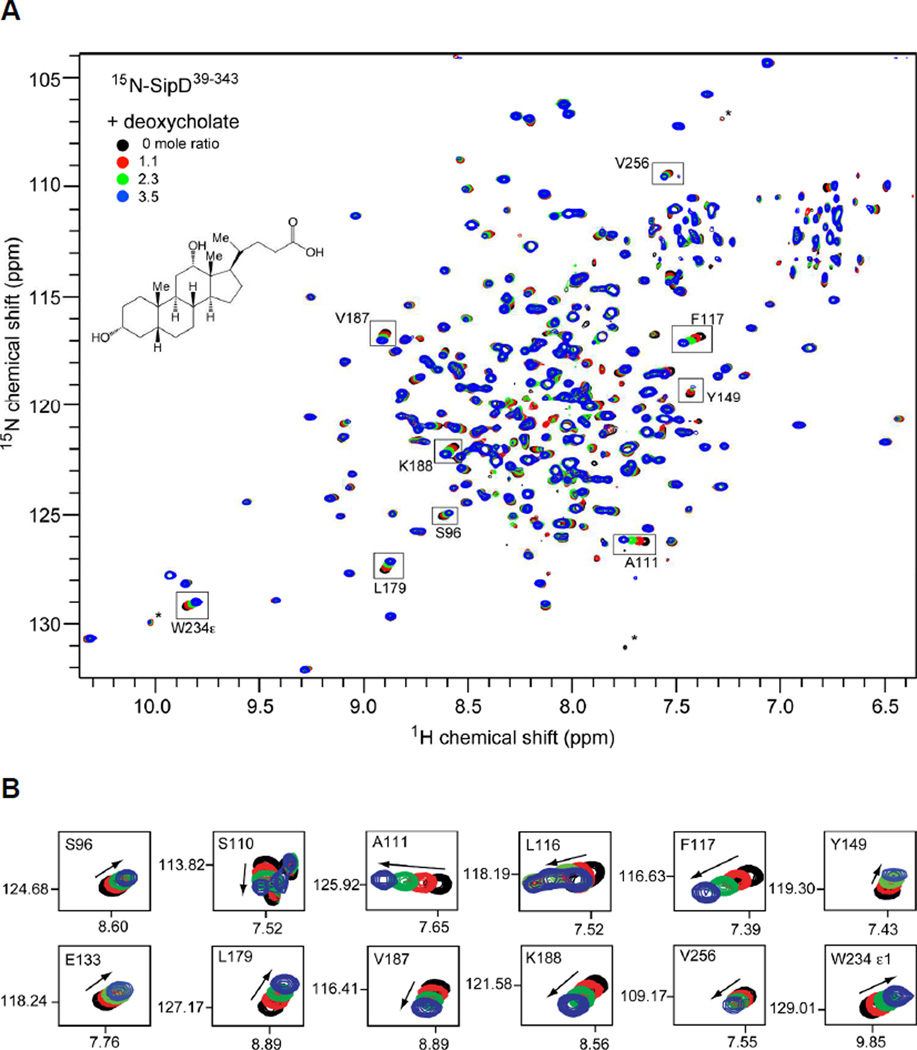
Fig. 1.
(A) Overlay of four 2D 1H-15N TROSY spectra of [2H,15N,13C]-labeled SipD39–343 with increasing amounts of deoxycholate. Some of the assignments are indicated as well as the noise peaks (*). (B) Expanded regions of the 2D 1H-15N TROSY spectra for specific residues that showed chemical shift changes with deoxycholate (arrows indicate the direction of the chemical shift change). Similar NMR titration data for SipD39–343 with taurodeoxycholate (Fig. S7), chenodeoxycholate (Fig. S8) and cholate hydrate (Fig. S9) are shown in the Supporting Information.
NMR assignment of SipD39–343
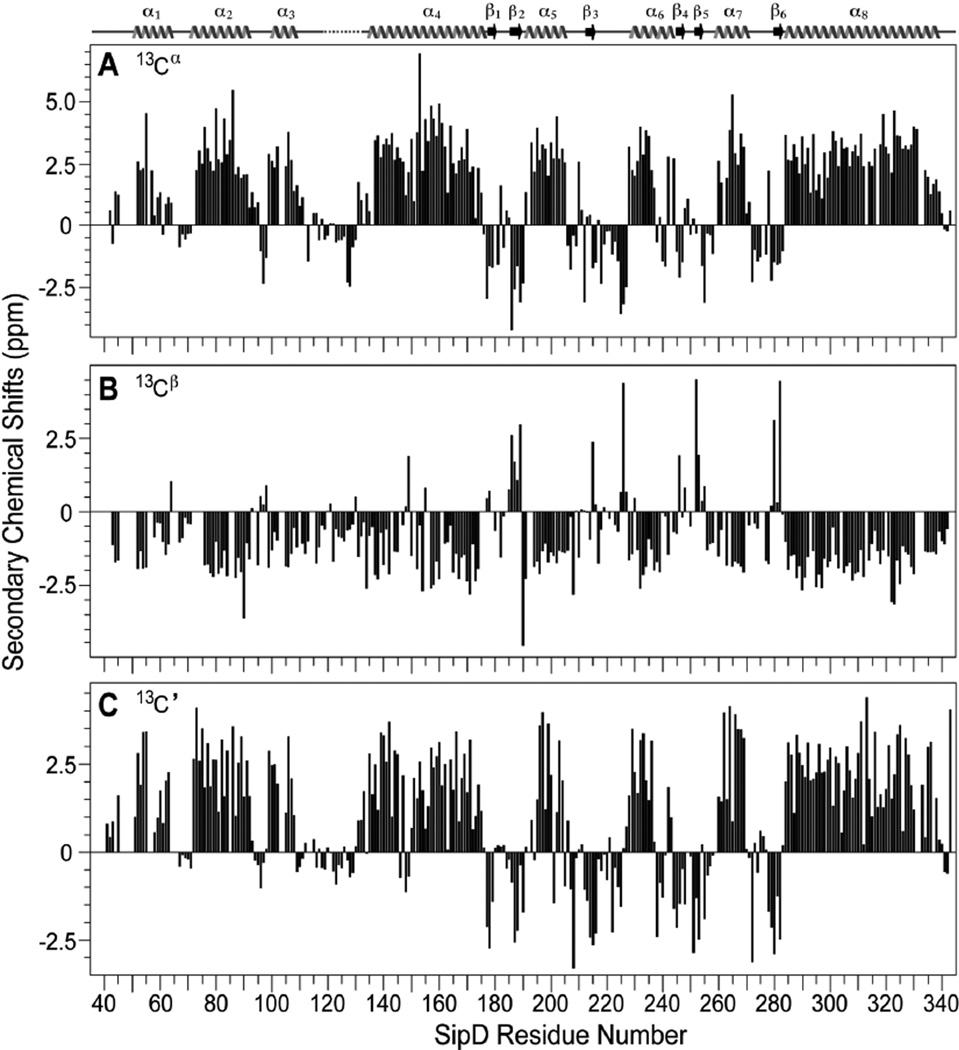
The backbone resonances of SipD39–343 (305 residues) were assigned using perdeuteration, TROSY NMR (Fig. S3) and 15N-amino acid-specific labeling (Fig S4). The backbone amides of 274 residues, or 93% of 295 non-proline residues in SipD39–343, were assigned. The remaining 7% of backbone amides that could not be assigned were due to peak overlap or peak broadening, and these residues were: Arg (41); Leu (48); Thr (65); Glu (103); His (40 and 56); Gln (57 and 262); Asn (72 and 104) and Ser (47, 49, 86, 143, 243, 250, 313, 317, 332 and 333). Nevertheless, nearly complete assignment of the backbone amides allowed the assignment of 93% of the Cα, Cβ and C’ resonances of SipD39–343. The secondary Cα, Cβ and C’ chemical shifts of SipD39–343 (Fig. 2) showed a predominantly alpha helical protein with several short beta strands, which correlated well with the secondary structures of BipD (11, 18) and IpaD (11). The chemical shifts suggested that the three dimensional structure of SipD in solution will be similar to that of the crystal structures of BipD (11, 18) and IpaD (11). Indeed, our 1.9 Å resolution crystal structure of SipD39–343 (to be reported elsewhere) showed similar secondary structures as indicated by the Cα, Cβ and C’ chemical shifts. Additionally, our SipD crystal structure showed a similar pattern of secondary structures with the IpaD crystal structure (Fig. S5) indicating that SipD and IpaD are structurally similar. However, in our SipD39–343 crystal structure, residues 118–133, which connect the N-terminal domain with the central coiled coil, lacked electron density and were disordered in the SipD39–343 crystal. To assign the tryptophan side chain, the four tryptophan residues (W135, W177, W234 and W290) were each mutated into a tyrosine residue. Proton-nitrogen correlation spectra showed that the tryptophan to tyrosine point mutants were folded, which allowed the assignment of the tryptophan residues in SipD (Fig. S6).
Fig. 2.
Secondary structures of SipD39–343 based on the (A) Cα, (B) Cβ and (C) C’ secondary NMR chemical shifts. The secondary structures of the SipD39–343 crystal are also shown and are denoted by arrow (beta strand), wavy line (helix); solid line (loop), and broken line (disordered loop). Residues 118–133 lacked electron density in the SipD39–343 crystal.
NMR titrations of SipD39–343 with bile salts
We used NMR chemical shift mapping to investigate the SipD interaction with bile salts deoxycholate, taurodeoxycholate, chenodeoxycholate and cholate hydrate. 15N-labeled SipD39–343 was dialyzed in buffer with increasing amounts of bile salts and 2D 1H-15N TROSY spectra were acquired. The protein-bile salt molar ratios used in the NMR titrations were limited by the critical micelle concentrations of deoxycholate (5 mM) (19), taurodeoxycholate (2.5 mM) (20) and chenodeoxycholate (7 mM) (19) (in contrast, the critical micelle concentration of cholate hydrate is 18 mM (19)). SipD39–343 titrated with deoxycholate (Fig. 1), taurodeoxycholate (Fig. S7) and chenodeoxycholate (Fig. S8) showed many residues with chemical shift perturbations with increasing amounts of bile salts; whereas titration with cholate hydrate (Fig. S9) showed only minor chemical shift changes for one residue (L179) and generally no chemical shift perturbation for the rest of SipD39–343. Thus, NMR titrations suggested specific interaction of SipD39–343 with the deoxycholate (Fig. 1), taurodeoxycholate (Fig. S7) and chenodeoxycholate (Fig. S8) but not with cholate hydrate (Fig. S9). Further, the interactions were in fast exchange NMR time scale as indicated by the progressive changes in the SipD39–343 peak positions with increasing amounts of deoxycholate (Fig. 1), taurodeoxycholate (Fig. S7) and chenodeoxcholate (Fig. S8).
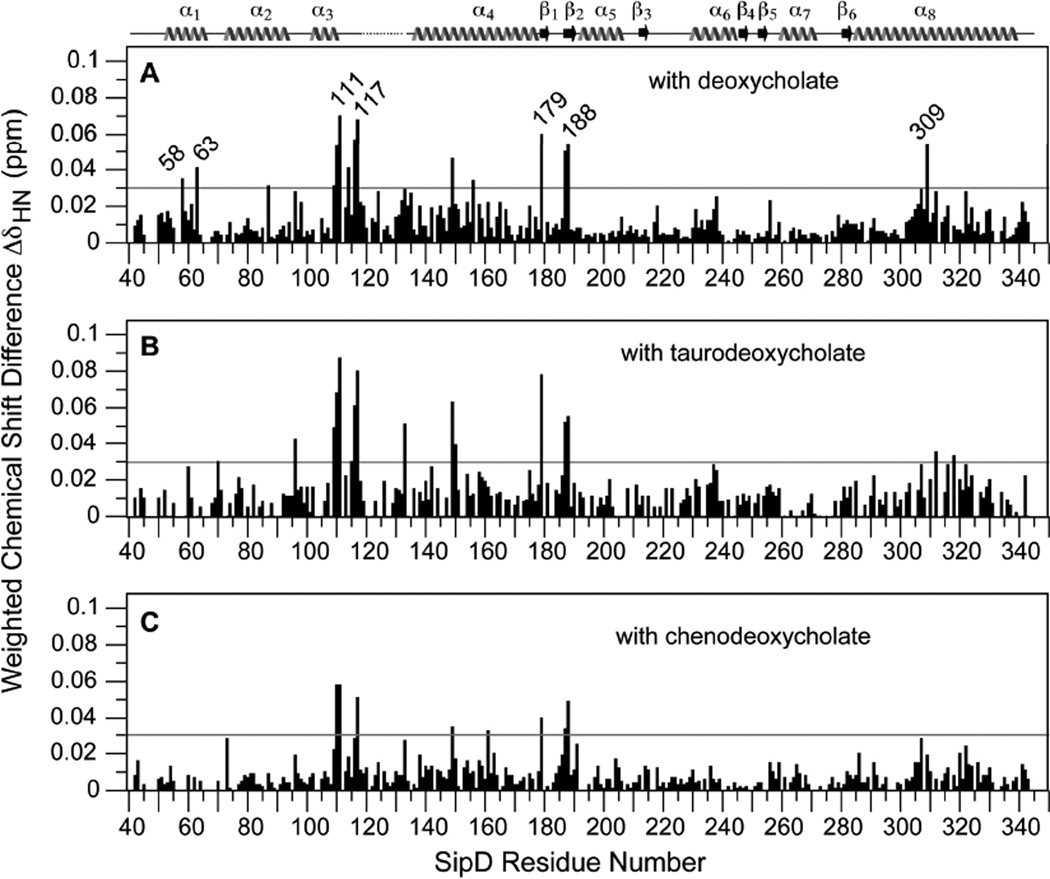
We identified all the backbone amides and the tryptophan side chain of SipD39–343 that were perturbed by deoxycholate, taurodeoxycholate and chenodeoxycholate. In the presence of deoxycholate, the SipD39–343 residues that showed the largest chemical shift perturbations (with ΔδHN > 0.05) were: S110, A111, L116, F117, Y149, L179, V187, K188 and T309 (Fig. 3A). Residues that showed significant chemical shift perturbations (with ΔδHN between 0.03 – 0.05) were: A58, Q63, L87, and F109 (Fig. 3A). Taurodeoxycholate strongly perturbed the same residues (with ΔδHN > 0.05) as deoxycholate in addition to E133 (Fig. 3B). For taurodeoxycholate, residues that showed significant chemical shift perturbations (with ΔδHN between 0.03 – 0.05) were: E70, S96, F109, A115, L150, Y312 and L318 (Fig. 3B). Compared to deoxycholate and taurodeoxycholate, chenodeoxycholate had an overall smaller effect on the chemical shifts of SipD (Fig. 3C). For chenodeoxycholate, the strongest perturbations (having ΔδHN > 0.05) occurred in residues S110, A111 and F117; with significant perturbations (having ΔδHN between 0.03–0.05) in residues Y149, T161, L179, V187 and K188. Chenodeoxycholate perturbed generally the same subset of residues perturbed by deoxycholate and taurodeoxycholate. Regarding the tryptophan side chain, both deoxycholate (Fig. 1B) and taurodeoxycholate (Fig. S7B) perturbed the W234 side chain whereas chenodeoxycholate (Fig. S8B) did not.
Fig. 3.
Weighted chemical shift difference (ΔδHN) of the 1H and 15N resonances of SipD39–343 when titrated with (A) deoxycholate, (B) taurodeoxycholate and (C) chenodeoxycholate (ΔδHN = [½ (δH2 + 1/25δN2)]1/2 (25)). The 1H and 15N chemical shifts were extracted from the 2D 1H-15N TROSY spectra of the free protein and the protein with the highest molar ratios of titrants at (A) 3.5, (B) 3.7 and (C) 4.3, respectively. For the deoxycholate titration (A), the average ΔδHN for all residues was 0.011 ppm with a standard deviation (σ) of 0.011 ppm. Thus, ΔδHN values above 3σ (or 0.03 ppm, indicated by a horizontal line) were deemed significant. Horizontal lines at ΔδHN 0.03 ppm in (B) and (C) were drawn for comparison with (A).
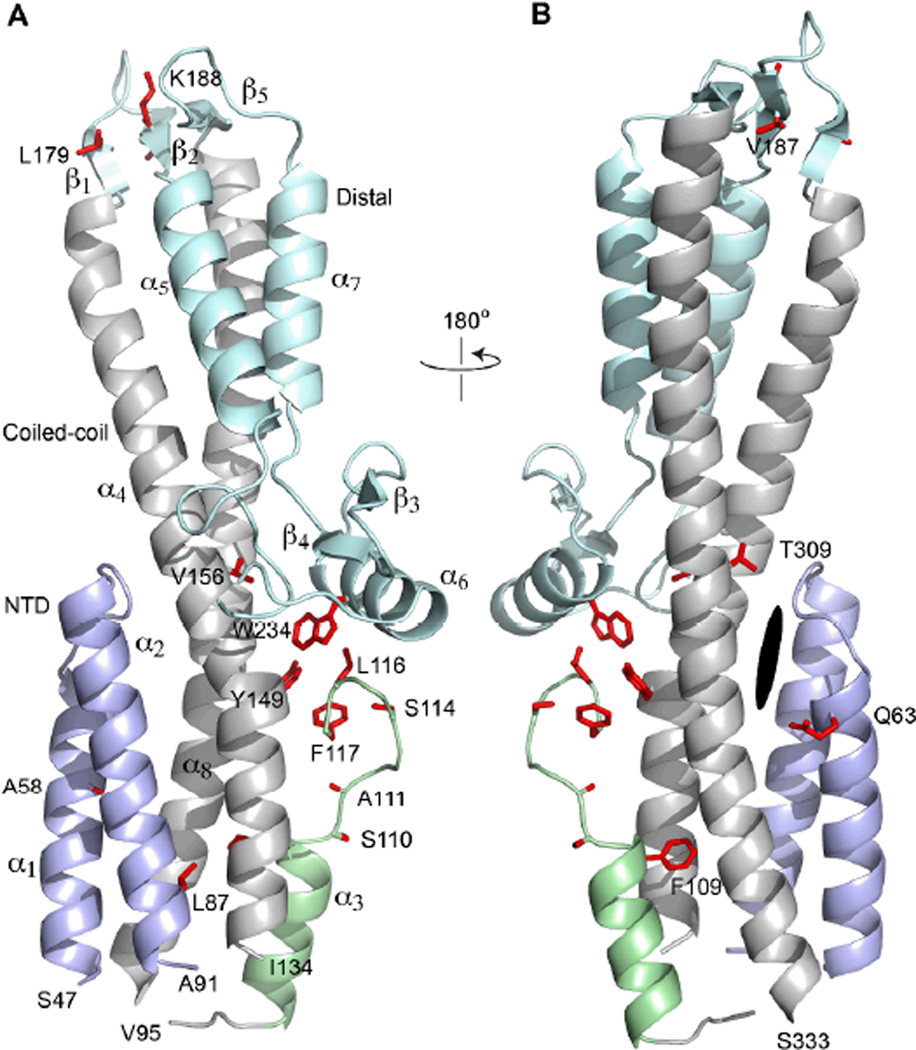
A majority of the residues that were strongly perturbed by deoxycholate, taurodeoxycholate and chenodeoxycholate were located on a 23-residue loop between residues 110–134, which connects the N-terminal helix α1, α2 and α3 with the long central coiled-coil of SipD (Fig. 4). Another subset of residues perturbed by biles salts (residues L179, V187 and K188) was located on beta strands β1 and β2 on the far end of the central coiled-coil and away from loop 110–134 (Fig. 4). The N-terminal helix α1 and α2 also showed chemical shift perturbations with deoxycholate (residues A58, Q63, and L87) and with taurodeoxycholate (residues E70 and S96), albeit with lesser ΔδHN values compared to loop 110–134 and the beta strands β1 and β2.
Fig. 4.
Ribbon representation of the crystal structure of SipD39–343 (to be reported elsewhere). The side chains of residues that showed significant chemical shift perturbation (ΔδHN > 0.03) with deoxycholate were colored red (legend: α, α-helix; β, beta strand; dark oval represents the equivalent region in IpaD that was predicted by computer docking to be deoxycholate binding site in IpaD (7)). The various regions of SipD were colored as follows: N-terminal helix α1 and α2 (light blue), helix α3 and loop 110–117 (light green), the central coiled coil formed by helix α4 and α8 (gray), and the rest of SipD (pale cyan).
DISCUSSION
The major significance of this work is the NMR identification of SipD residues that were perturbed by bile salts (Fig. 1, Fig. S7 & Fig. S8). This was made possible by our NMR assignment of the backbone resonances of SipD (Fig. 2). To date, no other tip protein has been assigned by NMR, and this is due to the challenge posed by NMR studies of the tip proteins. SipD, IpaD and BipD have 343, 332, and 310 residues, respectively. Further, these proteins were soluble only in submillimolar range, aggregated in NMR solution conditions at high concentrations and showed poor quality NMR spectra that were unsuitable for NMR characterization. The SipD N-terminal 38 residues, which were expected to be disordered based on structural homology with BipD (11, 18) and IpaD (11), decreased the quality of NMR data of full-length SipD (Fig. S1). A truncated construct, SipD39–343, retained the core structure of SipD (Fig. S2) and showed the best quality NMR spectra (Fig. S1). Perdeuteration, TROSY (Fig. S3) and amino acid-specific labeling (Fig. S4) were used to obtain nearly complete backbone assignments for SipD39–343. All the SipD39–343 backbone amide peaks that showed chemical shift perturbation with bile salts were assigned (Fig. 1, Fig. S7 & Fig. S8). In addition, the tryptophan side chain peaks of SipD (Fig. S6), one of which was perturbed by deoxycholate and taurodeoxycholate, were also assigned.
Another major finding of this work is that the bile salt binding site in SipD obtained from NMR chemical shift mapping is inconsistent with the published model of IpaD-deoxycholate interaction (7). Using the computer program AutoDock, Stensrud et al. (7) reported a model of IpaD-deoxycholate complex where deoxycholate binds in a pocket at the interface of the IpaD Nterminal region (equivalent to helix α1 and α2 of SipD, see Fig. S5) and the long central coiled coil (equivalent to helix α4 and α8 of SipD) (Fig. 4). If the model of the IpaD-deoxycholate interaction is applied to SipD, the N-terminal helix α1 and α2 of SipD (residues 47–91) and the middle part of the coiled coil (helix α4 and α8 facing helix α1 and α2) would be expected to show relatively the strongest chemical shift perturbations with deoxycholate. However, in our NMR titrations, the N-terminal helix α1 and α2 of SipD did not show large chemical shift perturbations with deoxycholate (Fig. 3). The helix α1 and α2 residues (A58, Q63 and L87) that were perturbed by deoxycholate showed relatively smaller chemical shift perturbations compared to residues that showed the largest chemical shift perturbations (Fig. 3). And in helix α8, only one residue (T309) showed chemical shift perturbation above the threshold of significance (at ΔδHN = 0.03). The relatively smaller chemical shift perturbations and the paucity of residues affected lead us to conclude that helix α1, α2 and α8 are not the primary binding site for deoxycholate in SipD. Instead, for deoxycholate, taurodeoxychoate and chenodeoxycholate, the strongest chemical shift perturbations occurred in loop 110–134 (Fig. 3 & Fig. 4), which connects the N-terminal region (helix α1 – α3) to the central coiled coil (helix α4 and α8). Our NMR results suggest that in SipD, loop 110–134 is most likely involved in interaction with bile salts. Residues L179, V187 and K188 located on beta strands β1 and β2 (Fig. 4) also showed consistently significant chemical shift perturbations (Fig. 3), however these residues are far away from loop 110–134 (Fig. 4). Thus, the chemical shift perturbation of L179, V187 and K188 is possibly due to non-specific interaction with bile salts. Based on our chemical shift mapping, we conclude that SipD bind bile salts in a different manner as predicted for IpaD (7) despite strong structural similarity between IpaD and SipD (with Cα RMSD of 1.4 Å) (Fig. S5).
We propose that the difference in the interaction between IpaD and SipD to bile salts reflects the differing responses of Shigella and Salmonella T3SS to bile salts. Bile salts activate the Shigella T3SS (7, 8), whereas they repress the Salmonella T3SS (9). In Salmonella, bile salts repress the transcription of T3SS genes and reduce the secretion of T3SS effectors (9, 10, 21). The precise target of bile salts vis-à-vis T3SS transcriptional regulation is currently unknown (9, 10). Bile salts do not interact directly with any of the T3SS transcriptional regulators (HilA and SirC) (9). Prouty and Gunn (9) hypothesized that bile salts enter the T3SS regulatory pathway through the two-component system SirA-BarA (which regulates HilA and SirC) or some unknown factor that affects the activity of SirA-BarA. On the other hand, others have shown recently that IpaD is present at the tip of the Shigella needle prior to contact with eukaryotic cells (3). In the presence of bile salts, bile salts interact with IpaD and trigger the release of IpaB (8), a protein involved in the assembly of the translocon (a membrane spanning structure that punctures a hole in the eukaryotic cell membrane to allow the passage of bacterial proteins). Similarly in Salmonella, Lara-Tejero & Galan (22) have shown that SipD is present on the bacterial surface before contact with eukaryotic cells, and upon contact with eukaryotic cells, the translocator proteins SipB and SipC are released. Thus, in Shigella and Salmonella, there is a period when IpaD and SipD are present at the tip of the needle (3, 8, 22) when they could act as environmental sensors by interacting directly with bile salts. Our results, together with the results of Stensrud et al. (7) suggest a model of tip protein-bile salt interaction where IpaD and SipD have different binding sites for bile salts. One site (the IpaD site), results to activation of the T3SS by allowing the assembly of the transcolon, and another site (the SipD site), results to inactivation of the T3SS.
Although the use of truncated SipD facilitated the NMR studies reported here, it also prevented the determination of the role of the SipD N-terminal 38 residues with respect to bile salt interaction. It is expected, however, that the main function of the N-terminal region is as a type III secretion signal (23, 24) rather than for bile-salt interaction. Another limitation of the current work is that the NMR method used here could not determine the Kd of the SipD-bile salt interaction, which is needed for functional studies of SipD-bile salt interaction. The reason for this is that the chemical shift changes did not reach saturation during the titration. Increasing the amount of bile salts was not possible because of the limitation imposed by the millimolar CMCs of bile salts and the requirement for submillimolar amounts of Sipd39–343 for NMR. Because of the poor quality of the NMR spectrum of the full-length SipD (Fig. S1B), it was also not possible to use the full-length SipD in binding studies by NMR methods. Isothermal calorimetry also failed to determine the Kd because the heat generated by (full-length and truncated) SipD upon titration with bile salts was lower than the heat generated by micelle formation of bile salts (19). Thus, a more sensitive method such as fluorescence spectroscopy, which was successfully used in studies of IpaD-bile salt interaction (7), would have to be used to determine the Kd of SipDbile salt interaction.
In summary, we have used NMR methods to investigate the interaction of SipD with bile salts and found that the interaction is different from what was predicted for IpaD-bile salt interaction. The differing response of SipD and IpaD to bile salts is most likely related to the opposing responses of Shigella and Salmonella to bile salts. Our results presented herein contribute to our understanding of the molecular interactions involved in type III secretion.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. Gianluigi Veglia (University of Minnesota) for assistance in specific labeling, to Chet Egan for CD spectroscopy and to Asokan Anbanandam for NMR spectroscopy.
NMR assignments have been deposited in the BioMagResBank, BMRB #16729.
Supported by NIH grants R01AI074856 and RR017708.
ABBREVIATIONS AND TEXTUAL FOOTNOTES
- CMC
critical micelle concentration
- HSQC
heteronuclear single quantum coherence spectroscopy
- Kd
dissociation constant
- NMR
nuclear magnetic resonance spectroscopy
- RMSD
root mean square deviation
- TEV
tobacco etch virus
- T3SS
type III secretion system
- T3SS
type III secretion system
- TROSY
transverse relaxation spectroscopy
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supporting information includes HSQC optimization of SipD constructs (Fig. S1); CD spectroscopy of SipD (Fig. S2); 3D NMR data used in the backbone assignments of SipD (Fig. S3); amino acid specific labeling of SipD (Fig. S4); alignment of secondary structures of IpaD and SipD (Fig. S5); assignment of tryptophan side chain by mutagenesis (Fig. S6); and titration of SipD with taurodeoxycholate (Fig. S7), chenodeoxycholate (Fig. S8) and cholate hydrate (Fig. S9). Supplemental information may be accessed free of charge online at http://pubs.acs.org.
REFERENCES
- 1.Cornelis GR. The type III secretion injectisome. Nat. Rev. Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 2.Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 3.Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect. Immun. 2006;74:4391–4400. doi: 10.1128/IAI.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect. Immun. 2005;73:1432–1440. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevens MP, Haque A, Atkins T, Hill J, Wood MW, Easton A, Nelson M, Underwood-Fowler C, Titball RW, Bancroft GJ, Galyov EE. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology. 2004;150:2669–2676. doi: 10.1099/mic.0.27146-0. [DOI] [PubMed] [Google Scholar]
- 6.Kaniga K, Trollinger D, Galan JE. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J. Biol. Chem. 2008;283:18646–18654. doi: 10.1074/jbc.M802799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect. Immun. 2007;75:2626–2629. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prouty AM, Gunn JS. Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infect. Immun. 2000;68:6763–6769. doi: 10.1128/iai.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prouty AM, Brodsky IE, Manos J, Belas R, Falkow S, Gunn JS. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 2004;41:177–185. doi: 10.1016/j.femsim.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. Selfchaperoning of the type III secretion system needle tip proteins IpaD and BipD. J. Biol. Chem. 2007;282:4035–4044. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisbrecht BV, Bouyain S, Pop M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expr. Purif. 2006;46:23–32. doi: 10.1016/j.pep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 14.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 15.Czisch M, Boelens R. Sensitivity enhancement in the TROSY experiment. J. Magn. Reson. 1998;134:158–160. doi: 10.1006/jmre.1998.1483. [DOI] [PubMed] [Google Scholar]
- 16.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J. Am. Chem. Soc. 1999;121:844–848. [Google Scholar]
- 18.Erskine PT, Knight MJ, Ruaux A, Mikolajek H, Sang NW, Withers J, Gill R, Wood SP, Wood M, Fox GC, Cooper JB. High Resolution Structure of BipD: An Invasion Protein Associated with the Type III Secretion System of Burkholderia pseudomallei. J. Mol. Biol. 2006;363:125–136. doi: 10.1016/j.jmb.2006.07.069. [DOI] [PubMed] [Google Scholar]
- 19.Simonovic BR, Momirovic M. Determination of critical micelle concentration of bile acid salts by micro-calorimetric titration. Mikrochimica Acta. 1997;127:101–104. [Google Scholar]
- 20.DeLong LJ, Nichols JW. Time-resolved fluorescence anisotropy of fluorescent-labeled lysophospholipid and taurodeoxycholate aggregates. Biophys J. 1996;70:1466–1471. doi: 10.1016/S0006-3495(96)79707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Velkinburgh JC, Gunn JS. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 1999;67:1614–1622. doi: 10.1128/iai.67.4.1614-1622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 2009;77:2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karavolos MH, Roe AJ, Wilson M, Henderson J, Lee JJ, Gally DL, Khan CM. Type III secretion of the Salmonella effector protein SopE is mediated via an N-terminal amino acid signal and not an mRNA sequence. J. Bacteriol. 2005;187:1559–1567. doi: 10.1128/JB.187.5.1559-1567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold R, Brandmaier S, Kleine F, Tischler P, Heinz E, Behrens S, Niinikoski A, Mewes HW, Horn M, Rattei T. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 2009;5:e1000376. doi: 10.1371/journal.ppat.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu JS, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 1996;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.