Abstract
Nonparametric Bayesian methods are employed to constitute a mixture of low-rank Gaussians, for data x ∈ ℝN that are of high dimension N but are constrained to reside in a low-dimensional subregion of ℝN. The number of mixture components and their rank are inferred automatically from the data. The resulting algorithm can be used for learning manifolds and for reconstructing signals from manifolds, based on compressive sensing (CS) projection measurements. The statistical CS inversion is performed analytically. We derive the required number of CS random measurements needed for successful reconstruction, based on easily-computed quantities, drawing on block-sparsity properties. The proposed methodology is validated on several synthetic and real datasets.
Index Terms: Beta process, compressive sensing, Dirichlet process, low-rank Gaussian, manifold learning, mixture of factor analyzers, nonparametric Bayes
I. Introduction
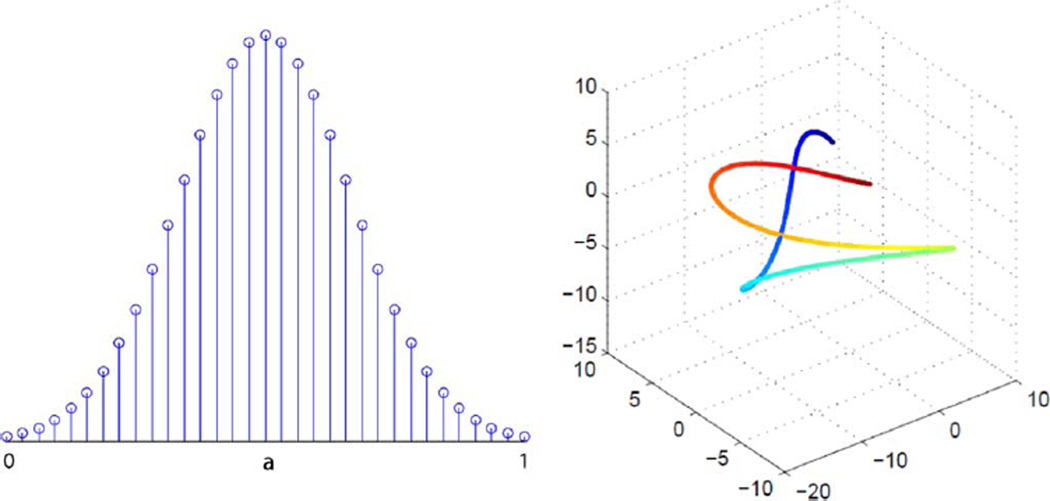
Compressive sensing (CS) theory [1] shows that if a signal may be sparsely rendered in some basis, it may be recovered perfectly based on a relatively small set of random-projection measurements. In practice most signals are compressible in an appropriate basis (not exactly sparsely represented), and in this case highly accurate CS reconstructions are realized based on such measurements. Recent work has extended the notion of sparsity-based CS to the more general framework of manifold-based CS [2]–[4]. In manifold-based CS, the signal is assumed to belong to a manifold, and low information content corresponds to low intrinsic dimension of the manifold. For a simple example (taken from [4]), consider signals generated by taking samples of a truncated and shifted Gaussian pulse, as shown in the left panel of Fig. 1. Although the ambient dimension (number of samples) of the signals is high, the only degree of freedom is the scalar shift a; therefore, the signals belong to a one-dimensional manifold. If we have a collection of signals, corresponding to different shifts, and project them onto, say, a random three-dimensional subspace, then the signals will, with high probability, form a twisting curve that does not self-intersect (right panel of Fig. 1). Thus, very few measurements are required to capture the characteristics of a signal on this manifold. We will return to this illustrative example in our experimental results. There are many more real examples of low-dimensional manifold signals, such as digit and face images (see, e.g., [5]).
Fig. 1.
Left: samples from a truncated and shifted Gaussian, with peak shift a. Right: projections of multiple such signals onto a random 3-D subspace, with the different points on the curve corresponding to different shifts a.
Although a theoretical analysis for CS on manifolds has been established in [2] and [3], very few algorithms exist for practical implementation. Moreover, existing performance guarantees depend on quantities that are not easily computable, such as the manifold condition number. In this paper, we propose a statistical framework for CS on manifolds, using a well-studied statistical model: a mixture of factor analyzers (MFA) [6], [7]. We model a manifold as a finite mixture of Gaussians, but we depart from conventional Gaussian mixture models (GMMs) by imposing a very particular structure—the covariances should be approximately low-rank, and the rank should equal the intrinsic dimension of the manifold. We employ nonparametric statistical methods [8], [9] to infer an appropriate number of mixture components for a given data set, as well as the associated rank of the Gaussians.
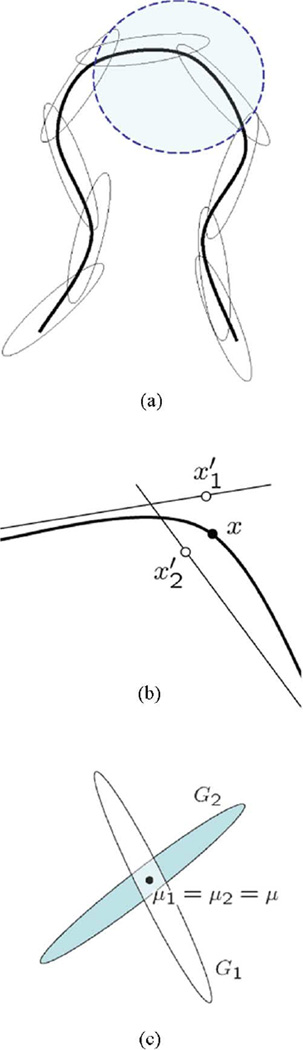
This model class is rich and can be used for modeling compact manifolds (see, for instance, [10]). To give some intuition why this is the case, note that if a manifold is compact (which among other things implies that it is bounded) then it admits a finite covering by topological disks whose dimensionality equals the intrinsic dimension. We can equate these topological disks to the principal hyperplanes of sufficiently flat ellipsoids corresponding to high-probability mass sets of our Gaussians. If there are a sufficiently high number of ellipsoids, then the hyperplanes are approximately tangent to the manifold and (by definition of manifold) we can establish locally valid one-to-one mappings between points on the manifold and points on the hyperplanes, with arbitrarily small distance between those points and their mappings. Fig. 2(a) and (b) illustrates this. Moreover, there are certain sets that are not manifolds but can still be well modeled by an MFA—a simple example consists of two Gaussians with the same mean but differently oriented principal hyperplanes, which do not constitute a manifold due to self intersection, as shown in Fig. 2(c). Thus, while the proposed model is appropriate for learning the statistics of manifolds, it is more generally applicable to data that reside in a low-dimensional region of a high-dimensional space.
Fig. 2.
Modeling a manifold with a mixture of Gaussians: (a) shows a covering of the manifold by high-probability mass ellipsoids; the shaded blue area is zoomed in (b), where we can see that point x on the manifold can be made arbitrarily close to projections if the mixture contains enough Gaussians; in (c), Gaussians G1 and G2 with common mean μ model a union of hyperplanes which is not a manifold due to self-intersection.
In this paper, we show how to nonparametrically learn the MFA based upon available training data, and how to reconstruct signals based on limited random-projection measurements (while motivated by manifold learning, we emphasize, as discussed above, the MFA may also be applied to other signals not necessarily restricted to a manifold). We obtain guarantees similar to those in CS for sparse signals, using sub-Gaussian random projections that are, with high probability, incoherent with a certain block-sparsity dictionary. Unlike typical CS results, the dictionary is not, in general, an orthonormal basis. Namely, we derive a restricted isometry property (RIP) for the composite measurement-dictionary ensemble, drawing on one of two assumptions: i) separability of the Gaussian means and ii) block-incoherence of the low-rank hyperplanes spanned by the principal directions of the covariances. Importantly, it is shown that only one of the conditions, i) or ii), needs to hold.
An important issue is that, as Fig. 2 suggests, a manifold can be covered by an infinite number of Gaussian mixtures, i.e., the entire model is not uniquely identifiable. However, the posterior log-probability of different mixture models with similar quality and parsimony should be similar—we believe learnability is a more interesting property than identifiability in the context of learning the MFA. Our method favors MFA models that have a small number of components and a small number of factors (enforced by the corresponding priors), while maintaining high reconstruction quality, as measured by posterior log-probability. Additionally, and very importantly, given the mixture parameters, our results guarantee that (with high probability) a signal drawn from that mixture is uniquely identifiable from random projections.
Our contributions are as follows.
We develop a hierarchical Bayesian algorithm that learns an MFA for the manifold based on training data. Unlike existing MFA inference algorithms [7], [11], [12], we adopt nonparametric techniques to simultaneously infer the number of clusters and the intrinsic subspace dimensionality.
We present a method for reconstructing out-of-sample data using compressed random measurements. By using the probability density function learned from the MFA as the prior distribution, the reconstruction can be found analytically by Bayes’ rule.
We derive bounds on the number of required random-projection measurements, in terms of easily-computable quantities, such as the rank of the covariances, number of clusters, separation of the Gaussians, coherence and subcoherence of the dictionary.
The remainder of the paper is organized as follows. In Section II we develop the nonparametric mixture of factor analyzers, and in Section III we discuss how this model may be employed for CS inversion. Section IV provides performance bounds for CS assuming that the signals of interest are drawn from a known low-rank Gaussian mixture model, with example results presented in Section V. Conclusions are provided in Section VI.
II. Nonparametric Mixture of Factor Analyzers
We assume the signals x ∈ ℝN under study are drawn from a Gaussian mixture model, and that the rank of the covariance of each mixture component is small relative to N. We represent this statistical model as a mixture of factor analyzers (MFA) [7], [13]–[15], with the MFA parameters learned from training data. As a special case this model may be applied to data drawn from a manifold or manifolds. In the discussion that follows, for conciseness, we will continually refer to data drawn from manifolds, since this is an important and motivating subproblem.
Many existing inference algorithms for learning an MFA [7], [11], [12] require one to a priori fix the subspace dimension and the number of mixture components (clusters). Unfortunately, these quantities are usually unknown in advance. We address this issue by placing Dirichlet process (DP) [8], [16] and Beta process (BP) [9], [17] priors on the MFA model, to infer the above mentioned quantities in a data-driven manner. As discussed further below, the DP is a nonparametric tool for mixture modeling, for inferring an appropriate number of mixture components [18]. The BP is a tool that allows one to uncover an appropriate number of factors [9], [17]. By integrating DP and BP into a single algorithm, we address both of the aforementioned problems associated with previous development of mixtures of factor analyzers.
A. Beta Process for Inferring Number of Factors
Assume access to n samples xi ∈ ℝN, with i = 1,…,n (all vectors are column vectors). The data are said to be drawn from a factor model if for all i
| (1) |
where A ∈ ℝN × J, μ ∈ ℝN and IN is the N × N dimensional identity matrix (with IJ similarly defined). The precision α ∈ ℝ+, and one typically places a gamma prior on this quantity (discussed further below).We initially assume we know the number of factors J, and at this point we do not consider a mixture model.
By first considering (1) we may motivate the model that follows. Specifically, we may re-express (1) as
| (2) |
If J ≤ N and the columns of A are linearly independent, we may express , with orthonormal υj ∈ ℝN and singular values ζj ∈ ℝ+, where ζ1 through ζJ are ordered in decreasing amplitude. If 1/α is small relative to ζJ, then xi is drawn approximately from a Gaussian of covariance rank J, and if J ≪ N these Gaussians corresponds to localized, low-dimensional “pancakes” in ℝN (see Fig. 2). As is well known [19], the vectors υj define the principal coordinates of the low-rank Gaussian, which is centered about mean position μ.
Before generalizing (1) to a mixture of factor analyzers, we wish to address the problem of inferring J, which defines the rank of the Gaussians, with this related to the dimensionality of the manifold. Toward this end, we modify (1) as
| (3) |
where K is an integer chosen to be large relative to the number of anticipated factors (e.g., we may set K = N), πk is the kth component of π, and ○ represents the point-wise (Hadamard) vector product. We use Bernoulli(·) Beta(·) and Mult(·) to refer respectively to Bernoulli, Beta and Multinomial densities parameterized by the arguments inside parentheses. This is a hierarchical model, in the sense that the priors for the parameters of the density of xi | wi, A, μ have their own priors (hyper-priors) with hyper-parameters.
As discussed in [9], when K → ∞ the number of non-zero components in z is drawn from Poisson(a/b). For finite K, of interest here, one may show that the number of non-zero components of z is drawn from Binomial(K, a/(a + b(K − 1))), and therefore one may set a and b to impose prior belief on the number of factors that will be important per mixture component. The expected number of non-zero components in z is aK/[a + b(K − 1)]. This construction has been related to a Beta Process, as discussed in [9]. Briefly, we draw z from a Bernoulli process parameterized by a Beta process over a measurable space Ω. Note that a Beta process, like a Dirichlet process, admits a representation as an infinite sum. We use a finite approximation to the Beta process (denoted, say, H(ω) for ω ∈ Ω) of the form , with π specified in the second line of (3) and where ω1,…,ωK is a partition of Ω. In our problem Ω represents the space of possible columns of the factor-loading matrix A, and the {ωk}k=1,K represent possible columns of A (factor loadings), drawn from a base measure H0. The design of these columns and H0 are discussed further below. The truncated Beta process procedure defines K candidate columns of A, with respective probability of usage {πk}k=1,K, and via the sparseness-inducing properties of this construction (the vector z is sparse), only a subset of the candidate columns are selected, those that best fit the data, as quantified via the likelihood.
When one performs Bayesian inference using a construction like (3), the posterior density function on the binary vector z defines the number of columns of A that contribute to the factor analysis model, and hence this yields the rank of the Gaussian. As discussed further below, Bayesian inference may be performed relatively simply via this construction.
B. Dirichlet Process for Mixture of Factor Analyzers
The hierarchical model in (3) may be used to infer the number of factors in a factor model, with all xi residing in a single associated subspace. However, to capture the nonlinear shape of a manifold, we are interested in a mixture of low-rank Gaussians (see Fig. 2). In general one does not know a priori the proper number of mixture components. We consequently employ the Dirichlet process (DP) [8], [16]. A draw G from a DP may be expressed as G ~ DP(ηG0), where η ∈ ℝ+ and G0 is a base probability measure. A constructive representation for such a draw may be written as [8]
| (4) |
where we note that, by construction, ; the expression is a point measure situated at . The observed data samples {xi}i=1, N may be drawn from a parametric model f(θi) with associated parameter θi, with θi ~ G. Since G is of the form in (4), for a relatively large number of samples N, many of the xi will share the same parameters , and therefore the {xi}i=1, N are drawn from a mixture model. In our problem f(·) is a Gaussian, and hence we yield a Gaussian mixture model. While there are an infinite number of components (in principle) within G, via posterior inference we infer an appropriate number of mixture components for the data {xi}i=1, N.
The DP favors a small number of mixture components, via the “stick” weights λt, which become small as t increases (only a relatively small set of mixture components are probable, with the number of components selected impacted by the data via the likelihood function). The base measure G0 is here a prior on the components of each mixture-component-dependent factor model. Specifically, the G0 is a prior on the factor loading A, and mean μ. The component of G0 associated with A is represented as discussed above in terms of a truncated Beta process construction (the number of used columns of A may be different for each of the mixture components). Our hierarchical model may be represented as
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
with , where we have truncated the DP sum to T terms (properties of this truncation are discussed in [20]). The notation is meant to mean that the K diagonal elements of Δt are drawn from , with k = 1, 2,…,K. The diagonal matrix Δt encodes the importance of each column in Ãt, playing the same role of singular values in SVD. The expression Mult(1; λ1,…,λT) represents drawing one sample from a multinomial distribution defined by (λ1,…,λT), and t(i) corresponds to the mixture component associated with the ith draw. The expression 𝒩t(i) (0, IK) is meant to indicate that the factor score associated with a given sample is explicitly linked to a particular mixture component (this is important for yielding a mixture-component-dependent posterior density function of w, and impacts the Gibbs-sampler update equations). The vector μ represents the mean computed based on all training data used to design the model, i.e., .
The expression means that each of the K columns of At are drawn independently from 𝒩(0, (1/N)IN) (this corresponds to the base measure H0 in the Beta process), implying that on average these columns will have unit norm (although any given draw will not exactly have unit norm). The covariance associated with mixture component t, as constituted via the prior, is , where Δ̃t = Δt diag(zt1,…,ztK). Hence, the number of non-zero components in the binary vector zt defines the approximate rank of Σt, assuming the smallest diagonal element of is large relative to . There are other ways one may draw At from a hierarchical generative model, but this construction appears to be the most stable among many we have examined.
Note that in (12)–(16), within the prior the T components {zt}t=1,T, {πt}t=1,T, {μt}t=1,T, {Ãt}t=1,T and {Δt}t=1,T are drawn once for all samples {xi}i=1,N, and these effectively correspond to the draws from the DP base measure G0; (10)–(11) are also drawn once, these yielding the T mixture weights in the truncated “stick-breaking” representation of the DP [20]. The ŵi is drawn separately for each of the N samples. Hence, all data drawn from a given mixture component t share the factor-loading matrix At ∈ ℝN × K and the same set of important columns (factor loadings) defined by zt, but each draw from a given mixture component has unique weights wi.
Concerning the way in which μt ∈ ℝN and At ∈ ℝN × K are drawn, the use of independent Gaussians allows for convenient inference. It is important to note that (14) and (15) simply constitute convenient priors, while the posterior Bayesian analysis will infer the correlations within these terms. The same is true for the factor score ŵi.
C. Inference and Hyper-Parameter Settings
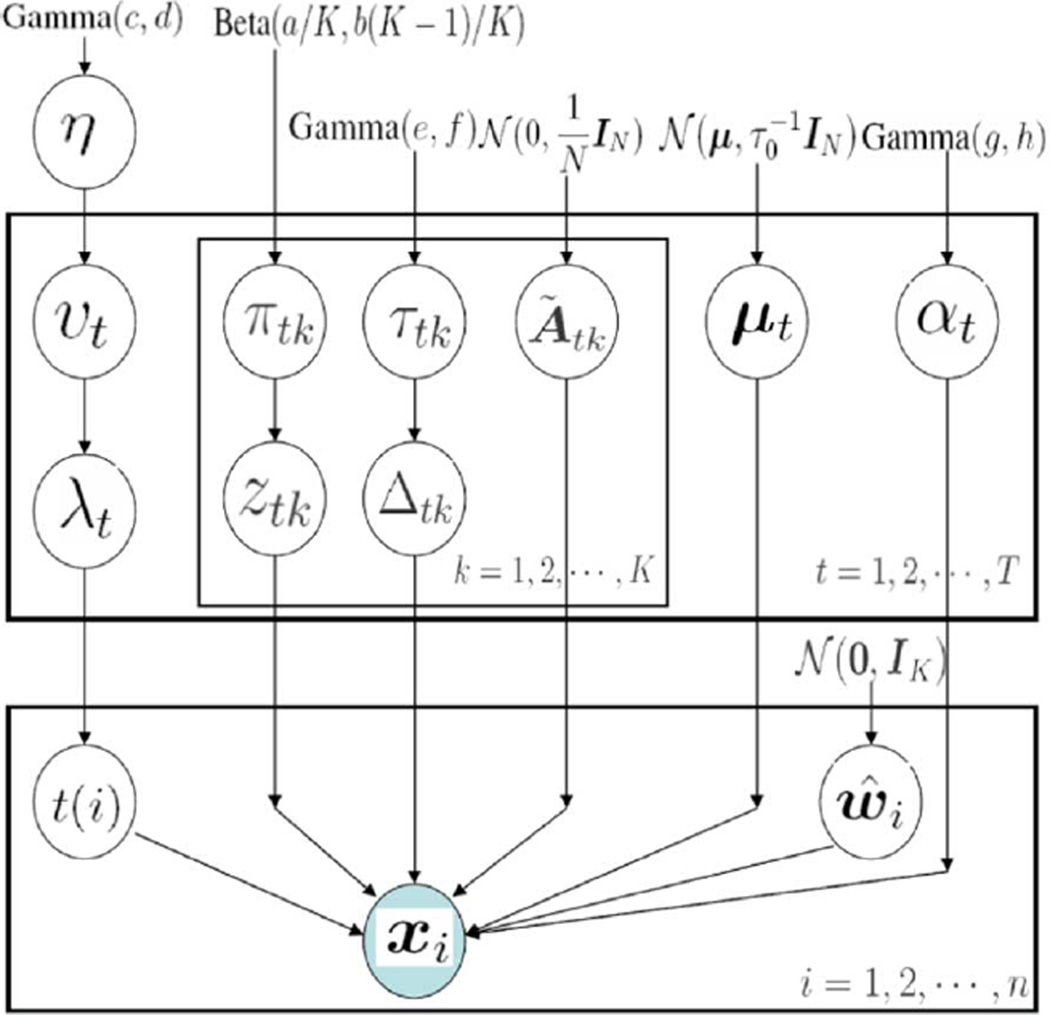
To complete the model, we assume η ~ Gamma(c, d), τtk ~ Gamma(e, f), αt ~ Gamma(g, h). Non-informative hyper-parameters are employed throughout, setting c = d = e = f = g = h = 10−6. For further illustration, we present the corresponding complete graphical model in Fig. 3. The precision τ0 = 10−6, implying that the means μt were drawn almost from a uniform prior. Further, we set a = b = 1 in all examples. While there may appear to be a large number of hyper-parameters, all of these settings are “standard” [9], [21] and there has been no parameter tuning for any of the examples. In all examples below, we set the truncations as K = T = 50.
Fig. 3.
The model parameters can be estimated using Gibbs sampling—we provide a detailed explanation in the Appendix. It is important to note that Gibbs sampling can sometimes suffer from slow mixing, becoming trapped in local modes. Other sampling techniques for DP have been developed, such as split-merge MCMC [22], that attempt to overcome this limitation. However, [23] is based on a Chinese Restaurant Process (CRP) DP construction, while here we employ a stick-breaking formulation. An advantage of the CRP is that it truly allows an infinite number of mixture components (in principle), while the stick-breaking construction discussed above employed a truncation. To remove this truncation, we also have considered a retrospective Gibbs sampler [23] for the stick-breaking construction, with this compared below to the truncated stick-breaking representation. The method in [23] also allows an infinite number of mixture components in principle, like [22]. The relative effectiveness of the truncated stick-breaking DP representation, and the associated Gibbs-sampler implementation, is discussed in detail in [24].
Efore, we decided to retain the truncated stick-breaking construction.
One may also perform variational Bayesian (VB) [11] analysis for this model. However, learning of the model need only be performed once, and therefore Gibbs sampling has been employed for this purpose. In all examples presented below, we employed 2000 burn-in iterations, and 1000 collection iterations, and we document the convergence behavior by showing log-probability plots. One may clearly use more collection iterations if desired, but we found this unnecessary for the CS application which is the focus of this paper.
D. Probability Density Estimation From the Above Nonparametric MFA Model
The above model can be interpreted as a Bayesian local PCA model, in which the signal manifold is approximated by a mixture of local subspaces. After model inference, the probability density function (pdf) of the signal can be estimated as follows:
| (16) |
| (17) |
This is the explicit form of the low-rank Gaussian mixture model density we estimate. If we use the prior distribution for ŵ, i.e., ξt = 0 and Λt = IK, then χt = μt and Ωt = Σt with Σt defined in Section II-B. However, this estimator may not be accurate since ŵ has its own posterior distribution in each mixture component. Thus, we use the following posterior mean and covariance estimate for ŵ from Gibbs sampling:
where is the mth collected Gibbs sample. We use the mean of the Gibbs samples to estimate Ãt. The estimated pdf in (16) will be used as the prior distribution for new testing signal in the compressive sensing application, which is illustrated in the following section.
III. Compressive Sensing Using a Low-Rank MFA
A. CS Inversion for Data Drawn From a Low-Rank GMM
Using the procedure discussed in the previous section, assume access to an MFA for a low-rank Gaussian mixture model of interest (e.g., for representation of a manifold). Let x ∈ ℝN be a vector drawn from this distribution. Rather than measuring x directly, we perform a projection measurement y = Φx + ν, where Φ ∈ ℝm × N is a measurement projection matrix, and we are interested in the case m ≪ N. Details on the design of Φ are provided in Section IV. The vector ν ∈ ℝN represents measurement noise. Our goal is to recover x from y, with this done effectively with m ≪ N measurements because x is known to be drawn from a low-dimensional MFA model. Bounds on requirements for m, based upon the properties of the MFA, are discussed in Section IV. Our objective here is to describe a general-purpose algorithm for recovering x from y.
Let p(x) represent the MFA learned via the procedure discussed in Section II. Note that the nonparametric learning procedure discussed there infers a full posterior density function on all parameters of the mixture model. Within the CS inversion we utilize the inferred mean of each mixture component, and an approximation to the covariance matrix based on averaging across all collection samples. Specifically, we have where χt represents the mean for mixture component t, and Ωt is the approximate inferred covariance matrix defined in (16) and (17). The λt are the mean mixture weights learned via the DP analysis, noting that in practice many of these will be very near zero (hence, we infer the proper number of meaningful mixture components, with T simply a large-valued truncation of the DP stick-breaking representation [20]).
The noise ν is assumed drawn from a zero-mean Gaussian with precision matrix (inverse covariance) R. The condition distribution for y given x may be evaluated analytically as
| (18) |
with
In the above computations, the following identity for normal distributions is used:
with the reader referred to [21] for a fuller description of a related derivation. In the results presented below we consider the case for which the components of R−1 tend to zero, therefore assuming noise-free measurements. If the measurements are noisy one may infer R within a hierarchical Bayesian analysis, but the inversion for x is no longer analytic (unless the noise covariance R−1 is known a priori).
It is interesting to note that the MFA mixture model may be relatively computationally expensive to learn, depending on the number of samples one has available for learning the properties of p(x) (it is desirable that the number of samples be as large as possible, to improve model quality). However, once p(x) is learned “offline,” the CS recovery y → x is analytic, in the sense that we have closed-form expressions for all the parameters of the posterior distribution p(x|y). Moreover, rather than simply yielding a single “point” estimate for x, we recover the full distribution p(x | y). When presenting results we plot the mean value of the inferred x.
B. Illustration Using Simple Manifold Data
To make the discussion more concrete, we return to the shifted Gaussian manifold example discussed in the Introduction. This simple example is considered to examine learning p(x), as well as inference of p(x | y) for CS inversion. The training set consists of n = 900 shifted Gaussian samples, each with dimension N = 128.
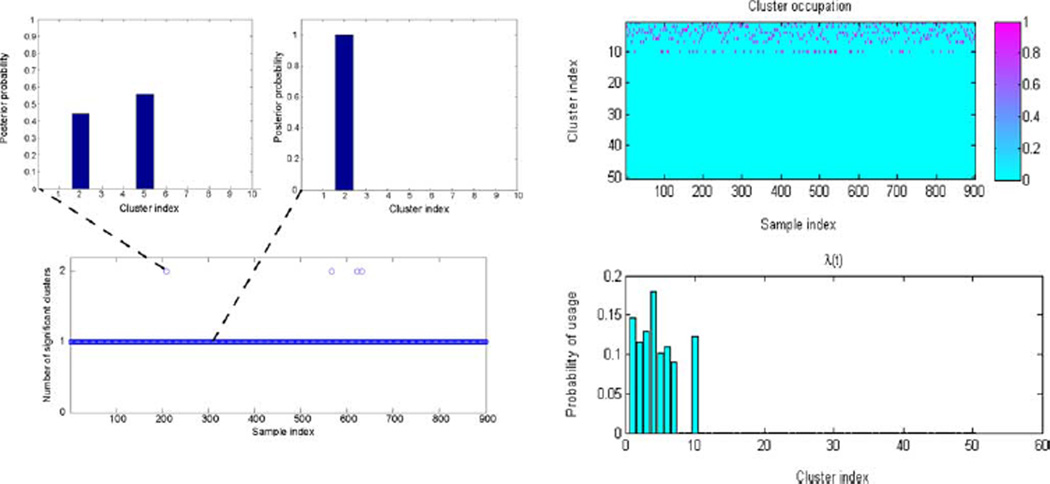
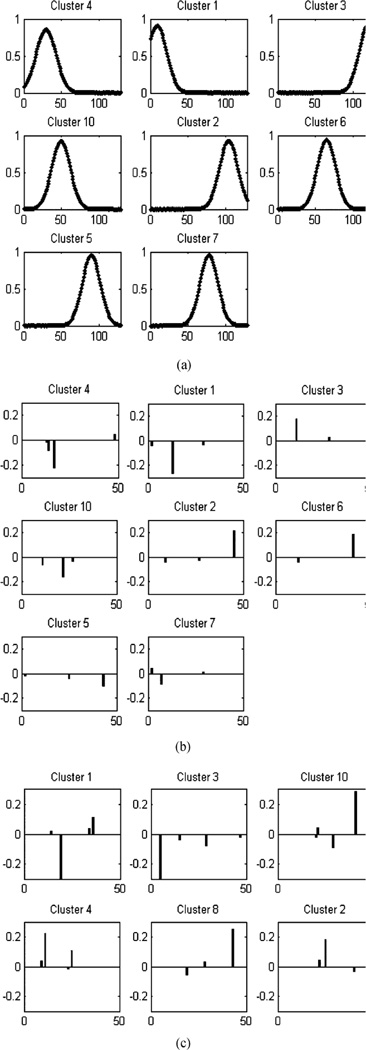
Fig. 4 examines the number of mixture components inferred, as well as the properties of any particular data point as viewed from the MFA. Specifically, a total of eight important mixture components are inferred, and the vast majority of data samples (896 of 900) are associated with only one mixture component (cluster). This implies that the signals possess block sparsity, in that a given signal x only employs factors from one of the mixture components. We exploit this property when deriving bounds for the number of required CS measurements. These results were obtained using a truncated stick-breaking DP representation.
Fig. 4.
Learned MFA mixture-component occupancy for the shifted Gaussian data. Bottom left: Number of mixture components (clusters) with significant (>0.1) posterior probability, for each training point. Note how only four out of 900 points on the manifold have more than one significant cluster. Top left: plots of the posterior p(ti | xi, –), for i = 208 and i = 300. In most cases, the posterior behaves as shown for i = 300. Only weights for mixture components 1–10 are shown, with the remaining components having negligible mixture weights for these samples. Top right: Cluster occupation probability for each training point. Bottom right: Learned weights of the clusters in the mixture (in total, only eight dominant mixture components are inferred).
In Fig. 5 are shown the properties of the individual factor models (for a given mixture component) of the MFA. We utilized both a truncated stick-breaking construction and retrospective sampling, as discussed in Section II-C. Note in Fig. 5(b) how the truncated stick-breaking construction results in each mixture component being dominated by a single factor, consistent with the one-dimensional character of the manifold. This is less evident in Fig. 5(c), corresponding to retrospective sampling, where less clusters are used but each cluster requires more factors. This behavior in undesirable, since it dilutes the low-rank structure of the Gaussians and, in our experiments it yields worse reconstruction performance, especially for real data. Hence, all subsequent results are presented using the truncated stick-breaking DP representation. We note that it may be possible to improve the retrospective Gibbs sampler performance by careful parameter tuning, but given the effectiveness of the truncated stick-breaking construction, such tuning was not exhaustively considered.
Fig. 5.
Properties of the individual (mixture-component-dependent) factor models for the MFA, considering the shifted Gaussian data. (a) Cluster centers (χt) using truncated stick-breaking; (b) Factor usage (Δt diag(zt)) of each cluster using truncated stick-breaking; (c) Factor usage of each cluster using retrospective sampling.
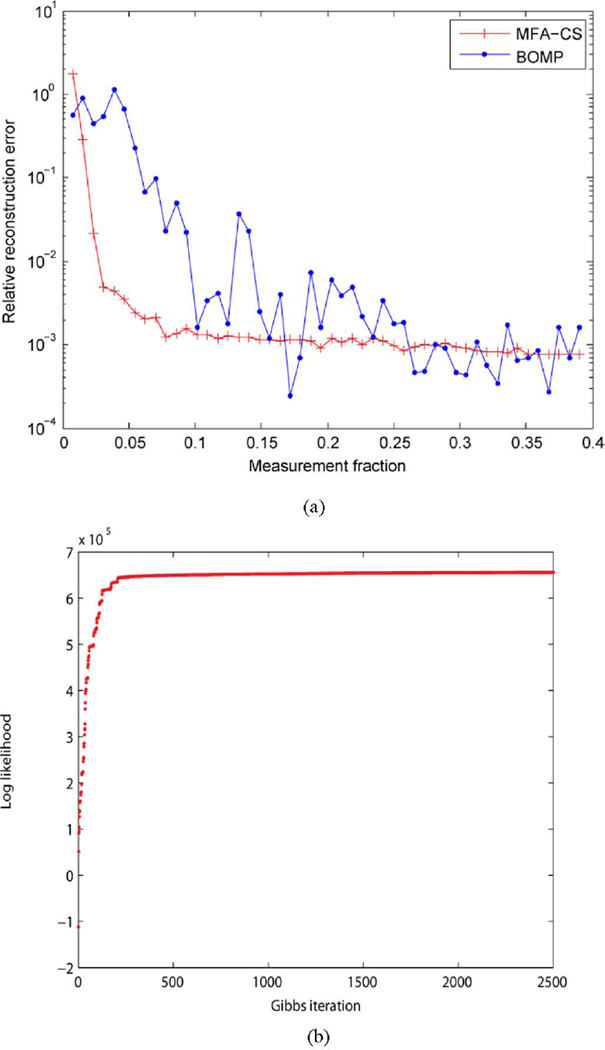
After the training process using the n = 900 samples, we have an MFA model for p(x), which we may now employ in CS inversion. To test the CS inversion, we generated 100 new (noise-free) test signals with different peak positions a, and manifested associated compressive projection measurements y (again, with the components of Φ drawn i.i.d. from 𝒩(0, 1)). The performance is shown in Fig. 6(a) and examples of the reconstruction are given in Fig. 7 for m = 5 CS measurements. In Fig. 6(a) (and in related results in Section V), the relative reconstruction error is defined as follows. If all n testing signals consist of the matrix X ∈ ℝN × n and the associated reconstructed signals are represented as X̂ ∈ ℝN × n, the relative reconstruction error is defined as (‖X − X̂‖F)/(‖X‖F). Using very few measurements (m ≪ N), we reconstruct x almost perfectly. Also shown is the performance of Block-OMP (BOMP) [25], a CS method for block-sparse signals which, as we explain in Section IV, utilizes some similar ideas to ours.
Fig. 6.
(a) Relative reconstruction error for the shifted Gaussian data, as a function of the number of measurements (fraction of the N = 128). We compare MFA-CS (our method) with BOMP. (b) Log-probability plot as a function of the number of Gibbs iterations (for learning the MFA model).
Fig. 7.
CS reconstruction result for the shifted Gaussian data. The left figure shows examples of the test signals, and the right figure shows the reconstructed signals with 3.9% measurements (recovery of x ∈ ℝ128 based on y ∈ ℝ5).
C. Gibbs Sampling and Label Switching
When performing Gibbs sampling to learn the MFA, we infer the number of factors per mixture component, as well as the number of mixtures. Further, we set the truncation levels for the number of factors and mixture components to large values (K = T = 50). In principle, the indexes on the factors and mixtures are exchangeable, and therefore within the Gibbs sampler the indexes (between the 50 possible values) of the factors and mixture components may be interchanged between consecutive Gibbs samples (in fact, this would be an indication of good mixing, since the labels are exchangeable). Plots like the right figures in Fig. 4 indicate that the labels of the factors and mixtures converge to a local mode, as the associated labels are stable after sufficient number of Gibbs burn-in iterations. We also show, in Fig. 6(b), a plot of the posterior log-probability as a function of the number of Gibbs iterations; while this does not assure convergence of the Gibbs sampler to the posterior of all model parameters, the convergence in Fig. 6(b) may explain why our learned MFA works well in practice for the objective of CS recovery. We note that if label switching becomes a problem for particular MFA examples, techniques are available to address this [26], [27], such that one may recover an analytic expression for the mixture model. In all examples considered here label switching was not found to be a problem.
IV. Bounds on the Number of Random Measurements
We derive sample-complexity bounds for CS reconstruction under the MFA model. In practice we assume available training data for images of interest, and using these data we learn the MFA, using the procedure discussed in the previous subsections. We now assume access to this learned MFA, and we wish to examine the required number of compressive measurements on new data, required to assure accurate recovery of the underlying signal (which is assumed drawn from the learned MFA).
Each observation xi is assumed drawn from an MFA as in (6), where in the analysis below we consider the case αt(i) → ∞; hence, we ignore additive measurement noise for simplicity. For notational simplicity, we additionally assume that each mixture component is composed of d factors. We define a matrix Ψ ∈ ℝN × (d+1)T, where consecutive blocks of d+1 contiguous columns correspond to the associated columns in the mixture-component-dependent factor matrices At, for t = 1,…,T(T mixture components). The first column in each block corresponds to the respective normalized (to unit Euclidean distance) mean vector, and the remaining d columns in each block are defined by the columns of A (which are assumed to be linearly independent). Any x satisfies x = Ψθ, where θ is assumed to have only d + 1 non-zero components, corresponding to the respective block. Note that this assumption comes from the way in which we have arranged Ψ to match the MFA model: each contiguous block of d + 1 columns corresponds to one mixture component; within each block the first column is the normalized mean of that component and the other d columns are the factors. This block structure in Ψ naturally imposes the same structure on θ, which we exploit. Thus, the dictionary Ψ contains a subset of the information present in the MFA parameters, namely the means and covariances of the mixture component s.
Recovering x from random projections amounts to recovering θ, which we assume to follow a particular sparsity pattern: θ is block-sparse, meaning that the non-zero coordinates of θ appear in predefined (d + 1)-sized blocks; in this case, it also happens that the first element is always equal to ‖μt‖ for the tth block.
The role of block-sparsity has recently been noted in the problem of reconstructing signals that live in a union of linear subspaces [28], [29]. The related notion of block-coherence, introduced in the same work, will be of use here, although in our setting we are interested in a union of affine spaces rather than linear subspaces (our hyperplanes will generally not include the origin). An additional difference is that our dictionaries can be, and usually are under-complete, i.e., (d + 1)T < N. Also of crucial importance is the role of separability, which has been explored by Dasgupta [19] in the context of learning mixtures of high dimensional Gaussians (not necessarily low-rank) from random projections. We build on these results and extend them to certain cases where separability does not hold.
We now define block-sparsity, block-coherence and separability more precisely, following [28] and [19], respectively.
a) Block-sparsity: [25] Let x ∈ ℝN be represented in a dictionary Ψ ∈ ℝN × (d+1)T, so that x = Ψθ, where θ ∈ ℝ(d+1)T is a parameter vector. We say that x is block L-sparse in Ψ, with blocks of size d + 1, if θ can be written as
| (19) |
with at most L of the θ[1]… θ[T] blocks having non-zero norm. We will assume L = 1, so only one block is active for each x.
b) Block-coherence: [25] Let denote the spectral radius of some matrix A, where λmax(A⊤ A) is the largest eigenvalue of the positive semidefinite matrix A⊤ A. Let us also express dictionary Ψ in block form as
| (20) |
and write M[l, r] = Ψ[l]TΨ[r], with M[l, r] ∈ ℝ(d+1) × (d+1). The block coherence of Ψ is defined as μB = maxl,r≠l(1)/(d+1)ρ(M[l, r]).
It should be stressed that the subdictionaries Ψ[1],…,Ψ[T] do not need to be orthonormal bases themselves. However, they can be orthogonalized without changing the block-sparsity of Ψ, as long as the columns of Ψ[t] are linearly independent, for all t. This can be ensured by the following proposition from [28].
Proposition 1: The representation x = Ψθ is unique iff Ψg ≠ 0 for every g ≠ 0 that is block 2L-sparse.
In other words, no 2L subdictionaries (or less) can be linearly dependent. In our setting, with L = 1, this condition excludes pairs of linearly dependent subdictionaries. It also excludes degenerate subdictionaries where, for instance, the mean lies in the span of the vectors of a given factor.
Additionally, we introduce the concept of subcoherence as in [25]. Subcoherence is defined as , for ψi, ψj ∈ Ψ[ℓ]. Clearly, if all subdictionaries Ψ[ℓ] are orthonormal, then ν = 0.
c) Separation: [19] Two Gaussians, 𝒩(μ1, Σ1) and 𝒩(μ2, Σ2) in ℝN, are c-separated if .
A. Separability Result
Our first result stems almost directly from [19]. Namely, we draw from [19, Lemma 2, p. 23] and from [19, Lemmas 18 and 19, p. 57].
Theorem 1: Let x come from Gaussian component t in an MFA where each component has d factors. Choose accuracy and confidence parameters ε, δ > 0. Assume that all the Gaussians are 1-separated, and that we observe a vector y ∈ ℝm of sub-Gaussian random projections with m ≥ max(d, (4)/(ε2) log(T2)/(2δ)). The original means and covariances will also be projected onto known m-dimensional vectors and m × m matrices . Assign to y the label t* such that . Then, Pr(t* ≠ t), i.e., the probability that x is misclassified into Gaussian j whose separation (in low dimensions, with probability >1 − δ) from t is , is at most
| (21) |
Consequently, for the T-component mixture, with probability at least
| (22) |
if we reconstruct x by mapping y back onto ℝN, more specifically to the d-dimensional hyperplane associated with Gaussian t*, and denote the reconstruction as x̂, then we have ‖x − x̂‖ ~ 𝒩(0, λd (Σt)), where λd (Σt) is the dth largest eigenvalue of Σt.
The proofs are essentially those in [19], but we have made a few changes, as follows.
Lemmas 18 and 19 assume that we observe x and have estimates for the mixture parameters. In contrast, we assume known mixture parameters but unknown x—we only observe y, therefore we must work with projections in ℝm, not with the full data in ℝN;
Lemmas 18 and 19 assume that the Gaussians have the same covariance, which clearly does not suit us (even in reduced space, where the covariances are closer to spherical). We address this by noting that the original proofs rely on assigning labels according to minimum Euclidean distance between x and the μk and assuming the worst case of spherical Gaussians with variance λmax. Instead, we assign labels according to the Mahalanobis distances between y and the projected , which obviates the assumption of equal covariances.
Importantly, we also quantify the influence of d in the bound for m, since Dasgupta’s result makes no assumption of low-rank covariances. If d > 4/(ε2) log(T2)/2δ, then we can use for discrimination between different Gaussians the same measurements that we would need anyway, in order to obtain a d-dimensional projection of x in the correct hyperplane. The converse is also true when d < 4/(ε2) log(T2)/2δ.
B. Block-Coherence Result
We now turn to our second result. We begin by writing the following concentration inequality, which plays a major role in many CS results for sub-Gaussian random measurements:
| (23) |
For Gaussian measurements, we have C0(ε) = ε2/4 − ε3/6.
Our second main theorem is a modification of [30, Theorem 3.3], which is itself a modification of [31, Theorem 5.2]:
Theorem 2: Let Ψ ∈ ℝN × (d+1)T be a dictionary with block-coherence μB and subcoherence ν such that . Let Φ ∈ ℝm × N be a measurement matrix whose elements satisfy the concentration inequality (23). Then, for any signal θ that is block 1-sparse in Ψ with block size d + 1, and for given δ ∈ (0, 1) and τ > 0, the linear transformation , U ∈ ℝm × (d+1)T verifies the RIP with constant δ, with probability at least 1 − e−τ, if
| (24) |
We thus have
| (25) |
C. Proof of Theorem 2
The proof is similar to that in [30], which follows the same method as [31] but, due to block-sparsity, has a drastically reduced number of hyperplanes to search over. For sparsity level S, usual CS bounds depend on , which is the number of S-dimensional subspaces in ℝN. For the mixture of rank-d-covariance Gaussians case, however, there exist only T possible combinations of (d + 1)-sized dictionary atom blocks—i.e., the sparsity is highly structured. In addition, we use results from [25] that ensure exact recovery is possible for certain sparsity levels that depend on the block coherence of Ψ.
Following [31], we invoke the Johnson–Lindenstrauss (JL) lemma and the concentration bound (23). First, we consider subdictionary ΨΛ with |Λ| = d + 1. We cover the unit sphere ℝd with a finite set Q of points such that ‖q‖ = 1 for all q ∈ Q and, for all θ : ‖θ‖ = 1, we have minq∈Q ‖θ − q‖ ≤ δ/4. It is known that there exists such a Q with |Q | ≤ (12/δ)d+1. We can apply (23) with ε = (δ/2) to all points ΨΛq and get
| (26) |
with probability at least 1 − 2(12/δ)d+1 e−mC0(δ/2), by the union bound. Defining γ as the smallest number such that ‖UΛθ‖2 ≤ (1 + γ)‖θ‖2 for all θ supported on Λ, it is proved in [31] that i) γ < δ ∈ (0, 1) and ii) (1 − γ)‖θ‖2 ≤ ‖UΛθ‖2 ≤ (1 + γ)‖θ‖2.
We now apply the union bound over all valid subdictionaries—in our case, there are T of them. Thus, the probability that (25) will fail is less than
| (27) |
Now we require that 2Te−mC0(δ/2)+(d+1)log((12)/(δ)) ≤ e−τ and we get the result that if
| (28) |
for given and δ ∈ (0, 1) and τ > 0, then with probability at least 1 − e−τ the composite matrix U has the restricted isometry property with the prescribed δ. The constant C0(δ/2), for a Gaussian measurement matrix, is equal to δ2/16 − δ3/48.
All that is left to do is to relate d with the block-coherence μB and subcoherence ν, in order to guarantee stable and efficient recovery in the same sense as [32]. We take a different route than [33], who reaches the result d ≤ 1/16μ using standard coherence. Instead, we can get much less strict limitations on d by using the result in [28] for block-coherence, where we need the condition .With L = 1, it is straightforward to obtain
| (29) |
(which for ν = 0 reduces to ). This completes the proof.
We point out that condition (29) can be checked for the MFA model learned through the procedure described in Section II, by explicitly computing the quantities ν and μB. Moreover, it is also possible to incorporate (29) directly into the learning process, without necessitating a posteriori verification. For instance, one might employ a rejection sampling step in tandem with the Gibbs sampler to preclude any solutions that would violate (29).
D. Discussion on the Significance of the Bounds
Theorems 1 and 2 allow us to establish expectations regarding the comparative performance of manifold-based CS, and more generally of MFA-based CS, versus traditional sparsity-based CS. For comparison, and assuming sparsity level S (i.e., ‖θ‖0 ≤ S), the current best sparsity-based CS bounds on the number of measurements are O(S log(N/S)) for the case of a subgaussian measurement matrix Φ, and O(S log4(N/S) when Φ is drawn from an orthobasis ensemble, assuming maximum incoherence with the sparsity dictionary [34].
In contrast, our separability and block-sparsity bounds are max(d, O(log T2)) and O(log(2T)) + O(d + 1) respectively, and they have no dependence on the ambient dimension N, while the dependence on d (which play a role analogous to the sparsity S) remains no worse than linear. While the dependence on log N is, in both our results, essentially replaced by a dependence on log T, this works to our considerable advantage because we expect T ≪ N by many orders of magnitude. This is consistent with the experimental results we show in the following section, where our reconstruction quality is comparable to that of the best sparsity-based CS algorithms, but using a much smaller fraction of the measurements.
Furthermore, our bounds relate to recent results in [4] for manifold CS, where the sample complexity bound is also linear in d and logarithmic in the manifold volume and condition number. If one notes the fact that these parameters tend to be larger for more geometrically complex manifolds and that, likewise, such manifolds typically require more Gaussians in our MFA model, i.e., a larger value for the parameter T, we can see that T therefore plays an interesting and perhaps unsuspected geometric role.
V. Experimental Results With Real Data
A. Digit Data
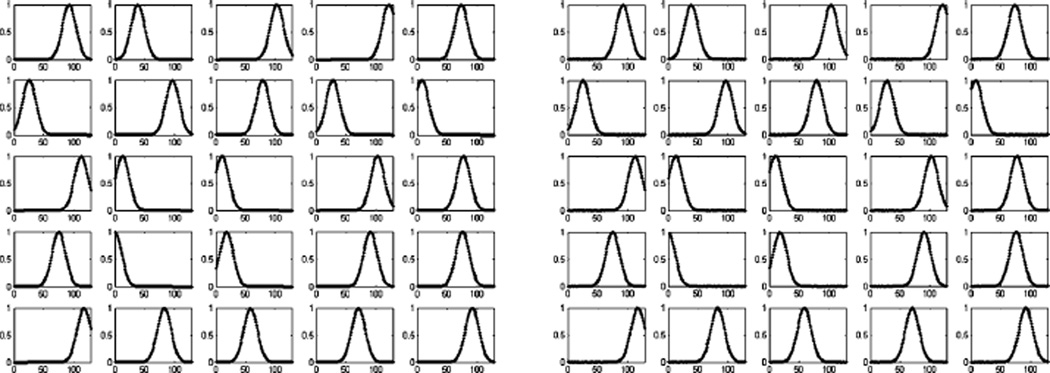
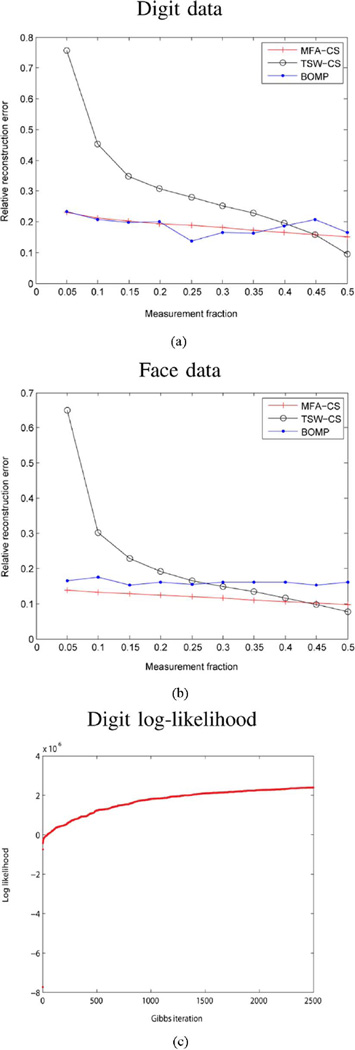
The MNIST digit dataset is commonly used for manifold learning. In this experiment, the training set contains 10 000 images (1000 images for each digit “0” through “9”, each image of size 28 × 28). We train the nonparametric MFA model without using the digit labels. The training results are depicted in Fig. 8. The model uses 25 clusters [Fig. 8(a)] and for each cluster the subspace dimension is around 10 [Fig. 8(b)–(c)]. We then use 100 testing images for compressive sensing and reconstruction. The performance is shown in Fig. 9(a) and examples of the reconstruction are given in Fig. 10. With very few measurements we can obtain a reasonable reconstruction. In these and all subsequent examples the components of Φ are drawn i.i.d. from 𝒩(0, 1). As seen in Fig. 9(a), our method slightly outperforms the BOMP algorithm [25] which, like ours, exploits block-sparsity structure but [25] is not probabilistic. We have chosen the block-sparsity level of BOMP (the number of active blocks) to be the same as that inferred by our method (this is a difficult parameter to set in the absence of our MFA model). Also, since BOMP does not include a procedure for learning the dictionary in the first place, we supply it with our learned MFA factor loadings and means, concatenated as explained in Section IV, which constitute the dictionary.
Fig. 8.
Training results for the MNIST digit data. (a) The top figure is the cluster occupation probability for each sample, and the bottom figure is the probability of using the clusters. (b) Center for each cluster (χt). (c) Factor usage of each cluster (Δt diag(zt)).
Fig. 9.
(a) CS reconstruction error for the MNIST digit data. The vertical axis denotes relative error as a function of the number of measurements, where the latter is in fraction of the size of the original image. The MFA-CS results correspond to the proposed method, BOMP to Block-OMP [25] and TSW-CS corresponds to the tree-structured wavelet-based CS inversion developed in [35], using a Haar wavelet. (b) Similar plot for the face data. (c) MNIST log-probability plot as a function of the number of Gibbs iterations (for learning the underlying MFA, with this done “offline” with the training data, prior to CS analysis); the corresponding plot for the face data looks very similar, and we omit it for brevity.
Fig. 10.
CS reconstruction results for the MNIST digit data. The left-most column shows the original image (different examples for each row), and the subsequent columns show CS reconstruction as a function of number of measurements, quantified in terms of percent relative to the size of the original image. In each column, the left figure corresponds to the CS inversion method in [35] using a Haar wavelet, and the right subfigure corresponds to the proposed method. The second through eleventh columns correspond to 5% to 50% measurements, in increments of 5%.
In Figs. 9(a) and 10, CS reconstruction results are also made with the tree-based wavelet CS inversion algorithm developed in [35]. While the method in [35] does not explicitly exploit the properties of the manifold, it does exploit structure in the wavelet coefficients of typical images and has demonstrated state-of-the-art performance relative to almost all other existing CS algorithms (see [35] for details on those comparisons). The comparative results in Fig. 9(a) and 10 show the benefits of leveraging the manifold information, particularly for a small number of measurements, as predicted by the theory in Section IV. As a trade-off, the wavelet-based method outperforms both our algorithm and BOMP for the high-measurement regime. This is due to approximation error, since the wavelet basis is a complete basis for the observation space, while our learned dictionary is under-complete and much smaller. For many CS applications, we are most interested in the low-measurement-number regime.
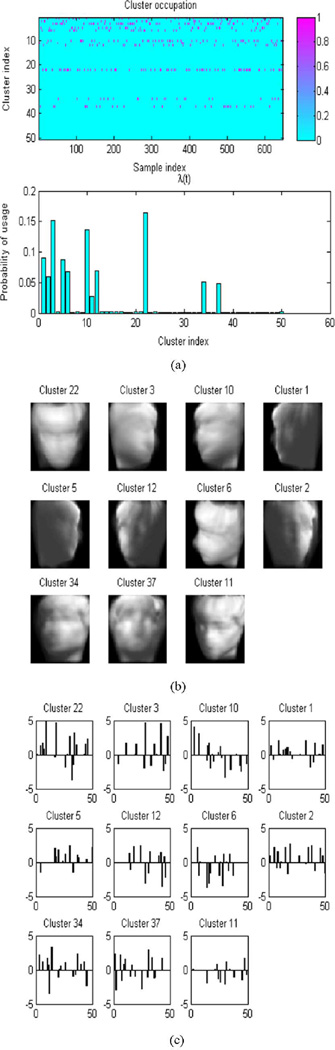
B. Face Data
As our last example, we show results on the face dataset used in the Isomap paper [5]. The faces are from the same subject but with different pose and illumination. We use 648 training images, each with dimension 64 × 64. The training results are depicted in Fig. 11. The model uses 11 clusters [Fig. 11(a)] and for each cluster the subspace dimension is around 13 [Fig. 11(a)–(b)]. We then use 20 (distinct) testing images for compressive sensing and reconstruction. The performance is shown in Fig. 9(b) and examples of the reconstruction are given in Fig. 12. Note again the excellent relative performance, particularly for a small number of measurements, as compared to the state-of-the-art CS algorithm developed in [35] (that exploits wavelet structure, but not manifold information) and to the BOMP algorithm [25], which is not probabilistic. In all examples, note that the wavelet-based approach in [35] eventually provides slightly better performance as the fractional number of measurements becomes large. The MFA-based CS approach is exploiting more prior knowledge than the wavelet-based approach, and therefore it is most advantageous for a relatively small number of CS measurements. The additional prior assumptions about the support of the signals of interest, associated with using the MFA, may introduce small errors with a large number of CS measurements (due to errors in the MFA model). The wavelet-based approach, which makes fewer assumptions about underlying model structure, yields slightly better results in this large-sample regime, but much worse results based on a relatively small number of CS measurements.
Fig. 11.
Training result for the face data. (a) The top figure is the cluster occupation probability for each sample, and the bottom figure is the probability of using the clusters. (b) Cluster centers (χt). (c) Factor usage (Δt diag(zt)) of each cluster.
Fig. 12.
CS reconstruction results for the face data. The left-most column shows the original image (different examples for each row), and the subsequent columns show CS reconstruction as a function of number of measurements, quantified in terms of percent relative to the size of the original image. In each column, the left figure corresponds to the CS inversion method in [35] using a Haar wavelet, and the right subfigure corresponds to the proposed method. The second through eleventh columns correspond to 5% to 50% measurements, in increments of 5%.
VI. Conclusion
In this paper, we have proposed a nonparametric Bayesian framework to learn MFA models for manifolds, with the advantage of inferring the number of clusters and the subspace dimensions simultaneously from the data. Furthermore, we have shown how this nonparametric MFA can be used to construct dictionaries for compressive sensing of signals from the learned manifold, taking advantage of the block-sparsity of such signals. The CS reconstruction estimate can be efficiently obtained in closed form, using Bayes’ rule.
In addition, we have derived theoretical bounds on the number of necessary random measurements in terms of easily computable quantities. It should be noted that Theorem 2 applies to a broader class of signals then the MFA considered here. Any signal living on a union of hyperplanes can be reconstructed with the same guarantees. In fact, the bound is likely loose, since we do not use the fact that the first nonzero coordinate of θ, corresponding to the mean in subdictionary block Ψ[t], is always ‖μt‖. We conjecture that the dependence on d + 1 can be reduced to d. Thus, Theorem 2 is actually true for a superset of the union of hyperplanes considered. Nevertheless, care should be exercised in verifying low dictionary block-coherence and subcoherence, so that the required sparsity level is high enough to be useful.
Interestingly, if we interpret the mixture as a manifold model, the covariance rank d is the intrinsic dimension, which plays the same role in the bounds as the sparsity S, as noted by [3] and [4]. In the same vein, the number of T components is related to the manifold condition number introduced by Nyogi et al. [36] and used by Wakin and Baraniuk—it models the complexity and curvature of the manifold. The present work thus complements prior results on CS for manifolds by introducing a general-purpose reconstruction algorithm, filling an existing gap.
Biographies

Minhua Chen was born in Wuhan, China, on October 25, 1982. He received the B.S. and M.S. degree in electrical engineering at Tsinghua University, Beijing, China, in 2004 and 2006 respectively. He received the Ph.D. degree at Duke University, Durham, NC, in 2009.
His research interest is in machine learning and signal processing.

Jorge Silva received the E.E., M.Sc., and Ph.D. degrees in electrical and computer engineering from the Instituto Superior Técnico (IST), Lisbon, Portugal, in 1993, 1999, and 2007, respectively.
He was a Researcher at the Instituto de Engenharia de Sistemase Computadores (INESC) from 1993 to 1996, under a PRAXIS XXI award and at the Instituto de Sistemas e Robótica (ISR), Lisbon, Portugal, from 2003 to 2007. He was a Teaching Assistant and later Adjunct Professor at the Instituto Superior de Engenharia de Lisboa (ISEL) from 1996 to 2007. In the same period, he did consulting and research and development work for major Portuguese utility and transportation companies. He is currently a Research Scientist at Duke University, Durham, NC, where he is working on statistical models for very high-dimensional data. His research interests include manifold learning, kernel methods, nonlinear prediction, and filtering and computer vision.

John Paisley received the B.S., M.S., and Ph.D. degrees in electrical and computer engineering from Duke University, Durham, NC, in 2004, 2007, and 2010, respectively.
He is currently a Postdoctoral Research Assistant in the Computer Science Department at Princeton University, Princeton, NJ. His research interests include Bayesian models and machine learning.

Chunping Wang received the B.S. and M.S. degrees in electrical engineering from Tsinghua University, Beijing, China, in 2001 and 2003, respectively.
She is currently a graduate student in the Department of Electrical and Computer Engineering at Duke University, Durham, NC, focusing on machine learning and data mining. Her research interests include learning with incomplete data, multitask learning, and collaborative filtering.

David Dunson is a Professor of Statistical Science at Duke University, Durham, NC. His research focuses on the development and application of novel Bayesian statistical methods motivated by high-dimensional and complex data sets. A particular emphasis is on nonparametric Bayesian methods that avoid assumptions, such as normality and linearity, and on latent factor models that allow dimensionality reduction in massive-dimensional settings. Recent projects have developed sparse latent factor models that scale to massive dimensions and improve performance in predicting disease and other phenotypes based on high-dimensional and longitudinal biomarkers. Related methods can be used for combining high-dimensional data from different sources and for massive-dimensional variable selection.
Dr. Dunson is a Fellow of the American Statistical Association and of the Institute of Mathematical Statistics. He received the 2007 Mortimer Spiegelman Award for the top public health statistician, the 2010 Myrto Lefkopoulou Distinguished Lectureship at Harvard University, and the 2010 COPSS Presidents’ Award for the top statistician under 41.
Lawrence Carin (SM’96–F’01) was born in Washington DC on March 25, 1963. He received the B.S., M.S., and Ph.D. degrees in electrical engineering at the University of Maryland, College Park, in 1985, 1986, and 1989, respectively.
In 1989, he joined the Electrical Engineering Department at Polytechnic University, Brooklyn, NY, as an Assistant Professor, and became an Associate Professor there in 1994. In September 1995, he joined the Electrical Engineering Department at Duke University, Durham, NC, where he is currently the William H. Younger Professor of Engineering. He is a co-founder of Signal Innovations Group, Inc. (SIG), a small business, where he serves as the Director of Technology. His current research interests include signal processing, sensing, and machine learning. He has published over 200 peer-reviewed papers.
Dr. Carin is a member of the Tau Beta Pi and Eta Kappa Nu honor societies.
Appendix
As illustrated in Section II, the model likelihood can be expressed as
where Ãtk denotes the kth column of matrix Ãt and Δtk denotes the kth diagonal element in the diagonal matrix Δt. The Gibbs sampling inference algorithm can be derived as follows.
- Sample the cluster index ti from
with Σt defined in Section II-B and . After normalization across t, p(t(i) = t | –) becomes a Multinomial distribution. Due to the low-dimensional subspace structure of Σt, the computation of 𝒩(xi; μt, Σt) can be made more efficient by using the Matrix inversion lemma. Sample the DP concentration parameter η from .
Sample the DP stick weight υt from p(υt | –) Beta(υt; 1 + ∑i:t(i)=t 1, η + ∑i:t(i)>t 1) (t = 1,2,…,T − 1) and υT = 1.
- Sample the factor score ŵi from p(ŵi | t(i), –) = 𝒩( ŵi; ξt(i), Λt(i)) with
- . Thus, we can sample zt and Δt from
where Sample the Bernoulli parameter πt from p(πtk | –) = Beta(πtk; a/K + ztk, b(K − 1)/K + 1 − ztk).
Sample the precision parameter τtk from if ztk =1; otherwise set τtk = 1. The algorithm also works if τtk is always fixed to 1 without update.
- Sample the mean vector for each cluster μt from p(μt | –) = 𝒩(μt; ζt, Γt) where
- Sample the factor loading matrix Ãt from
where Ãjt denotes the jth row of matrix and Ãt and - Sample the precision of the additive noise αt from p(αt | –) = Gamma(αt; gt, ht) with
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Minhua Chen, Electrical and Computer Engineering Department, Duke University, Durham, NC 27708-0291 USA.
Jorge Silva, Electrical and Computer Engineering Department, Duke University, Durham, NC 27708-0291 USA.
John Paisley, Electrical and Computer Engineering Department, Duke University, Durham, NC 27708-0291 USA.
Chunping Wang, Electrical and Computer Engineering Department, Duke University, Durham, NC 27708-0291 USA.
David Dunson, Statistics Department, Duke University, Durham, NC 27708-0291 USA.
Lawrence Carin, Email: lcarin@ee.duke.edu, Electrical and Computer Engineering Department, Duke University, Durham, NC 27708-0291 USA.
References
- 1.Candès E, Wakin M. An introduction to compressive sampling. IEEE Signal Process. Mag. 2008;vol. 25(no. 2):21–30. [Google Scholar]
- 2.Baraniuk R, Cevher V, Duarte M, Hegde C. Model-based compressive sensing. 2008 Preprint. [Google Scholar]
- 3.Baraniuk R, Wakin M. Random projections of smooth manifolds. Found. Comput. Math. 2009;vol. 9(no. 1):51–77. [Google Scholar]
- 4.Wakin M. Manifold-based signal recovery and parameter estimation from compressive measurements. 2008 Preprint. [Google Scholar]
- 5.Tenenbaum J, Silva V, Langford J. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;vol. 290(no. 5500):2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- 6.Tipping M, Bishop C. Mixtures of probabilistic principal component analyzers. Neural Comput. 1999;vol. 11(no. 2):443–482. doi: 10.1162/089976699300016728. [DOI] [PubMed] [Google Scholar]
- 7.Ghahramani Z, Hinton G. The em algorithm for mixtures of factor analyzers. Toronto, ON, Canada: Univ. of Toronto; 1997. Tech. Rep. CRG-TR-96-1. [Google Scholar]
- 8.Sethuraman J. A constructive definition of the Dirichlet prior. Statistica Sinica. 2001;vol. 2:639–650. [Google Scholar]
- 9.Paisley J, Carin L. Nonparametric factor analysis with beta process priors. Proc. ACM 26th Annu. Int. Conf. Machine Learning; New York, NY. 2009. [Google Scholar]
- 10.Brand M. Charting a manifold. Adv. Neural Inf. Process. Syst. 2003:985–992. [Google Scholar]
- 11.Ghahramani Z, Beal M. Variational inference for Bayesian mixtures of factor analysers. Adv. Neural Inf. Process.Systems. 2000;vol. 12:449–455. [Google Scholar]
- 12.Utsugi A, Kumagai T. Bayesian analysis of mixtures of factor analyzers. Neural Comput. 2001;vol. 13(no. 5):993–1002. doi: 10.1162/08997660151134299. [DOI] [PubMed] [Google Scholar]
- 13.Hinton G, Dayan P, Revow M. Modeling the manifolds of images of handwritten digits. IEEE Trans. Neural Netw. 1997;vol. 8(no. 1):65–74. doi: 10.1109/72.554192. [DOI] [PubMed] [Google Scholar]
- 14.Kannan A, Jojic N, Frey B. Fast transformation-invariant component analysis. Int. J. Comput. Vis. 2008;vol. 77(no. 1):87–101. [Google Scholar]
- 15.Verbeek J. Learning nonlinear image manifolds by global alignment of locally linear models. IEEE Trans. Pattern Anal. Mach. Intell. 2006;vol. 28(no. 8):1236–1250. doi: 10.1109/TPAMI.2006.166. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson TS. A Bayesian analysis of some nonparametric problems. Ann. Stat. 1973;vol. 1(no. 2):209–230. [Google Scholar]
- 17.Thibaux R, Jordan M. Hierarchical beta processes and the Indian buffet process; Proc. Int. Conf. Artificial Intelligence Statistics; 2007. [Google Scholar]
- 18.Antoniak C. Mixtures of Dirichlet processes with applications to Bayesian nonparametric problems. Ann. Stat. 1974:1152–1174. [Google Scholar]
- 19.Dasgupta S. Ph.D. dissertation. Berekely: Univ. of California: 2000. Learning probability distributions. [Google Scholar]
- 20.Ishwaran H, James LF. Gibbs sampling methods for stick-breaking priors. J. Amer. Stat. Assoc. 2001;vol. 96:161–173. [Google Scholar]
- 21.Tipping M. Sparse Bayesian learning and the relevance vector machine. J. Mach. Learn. Res. 2001;vol. 1:211–244. [Google Scholar]
- 22.Jain S, Neal R. A split-merge Markov chain Monte Carlo procedure for the Dirichlet process mixture model. J. Comput. Graphic. Stat. 2000 [Google Scholar]
- 23.Papaspiliopoulos O, Roberts G. Retrospective Markov chain Monte Carlo methods for Dirichlet process hierarchical models. Biometrika. 2008;vol. 95(no. 1):169–186. [Google Scholar]
- 24.Blei D, Jordan M. Variational inference for Dirichlet process mixtures. Bayesian Anal. 2006;vol. 1(no. 1):121–144. [Google Scholar]
- 25.Eldar Y, Kuppinger P, Bölcskei H. Compressed sensing of block-sparse signals: Uncertainty relations and efficient recovery. 2009 Arxiv preprint arXiv:0906.3173. [Google Scholar]
- 26.Jasra A, Holmes C, Stephens D. Markov chain Monte Carlo methods and the label switching problem in Bayesian mixture modeling. Stat. Sci. 2005;vol. 20:50–67. [Google Scholar]
- 27.Stephens M. Dealing with label switching in mixture models. J. Roy. Stat. Soc. Series B. 2000;vol. 62:795–809. [Google Scholar]
- 28.Eldar Y, Bolcskei H. Block-sparsity: Coherence and efficient recovery. 2008 Arxiv preprint arXiv:0812.0329. [Google Scholar]
- 29.Eldar Y, Mishali M. Robust recovery of signals from a union of subspaces. 2008 preprint. [Google Scholar]
- 30.Blumensath T, Davies M. Sampling theorems for signals from the union of linear subspaces. IEEE Trans. Inf. Theory. 2009;vol. 55(no. 4):1872–1882. [Google Scholar]
- 31.Baraniuk R, Davenport M, DeVore M, Wakin R. A simple proof of the restricted isometry property for random matrices. Construct. Approxim. 2008;vol. 28(no. 3):253–263. [Google Scholar]
- 32.Tropp J. Greed is good: Algorithmic results for sparse approximation. IEEE Trans. Inf. Theory. 2004;vol. 50(no. 10):2231–2242. [Google Scholar]
- 33.Rauhut H, Schnass K, Vandergheynst P. Compressed sensing and redundant dictionaries. IEEE Trans. Inf. Theory. 2008;vol. 54(no. 5):2210–2219. [Google Scholar]
- 34.Rudelson M, Vershynin R. On sparse reconstruction from Fourier and Gaussian measurements. Commun. Pure Appl. Math. 2008;vol. 61(no. 8) [Google Scholar]
- 35.He L, Carin L. Exploiting structure in wavelet-based Bayesian compressive sensing. IEEE Trans. Signal Process. 2009 Sep;vol. 57(no. 9):3488–3497. [Google Scholar]
- 36.Niyogi P, Smale S, Weinberger S. Finding the homology of submanifolds with high confidence from random samples. Discrete Comput. Geomet. 2008;vol. 39(no. 1):419–441. [Google Scholar]