Abstract
Keratoconus is a thinning corneal dystrophy that begins in the early teenage years and ultimately requires cornea transplantation to restore vision. Here we conducted a highly sensitive mass spectrometric analysis of the epithelium and the stroma from keratoconus and normal donor corneas. We identified a total of 932 and 1,157 proteins in the consolidated data of the epithelium and stroma, respectively. Technical replicates showed strong correlations (≥ 0.88) in levels of all common proteins, indicating very low technical variations in the data. Analysis of the most increased (≥ 1.5 fold) and decreased (≤ 0.8 fold) proteins in the keratoconus corneal epithelial protein extracts identified proteins related to dermal diseases, inflammation, epithelial stratification and mesenchymal changes. Increased proteins included keratins 6A, 16 and vimentin, while the iron transporter lactotransferrin was decreased. The keratoconus stromal proteome suggest endoplasmic reticular stress, oxidative stress and widespread decreases in many extracellular matrix proteoglycan core proteins, lumican and keratocan, collagen types I, III, V and XII. Marked increase in apoptosis and endocytosis-related proteins suggest degenerative changes in keratocytes, the resident cells of the stroma. This is the most comprehensive proteome analysis of the cornea that highlights similarities of keratoconus with other neurodegenerative diseases.
1. Introduction
Keratoconus (KC) is a corneal dystrophy that affects 1/1300 individuals worldwide. The weakening cornea undergoes progressive ectatic thinning and causes severe astigmatism, scarring, and ultimately loss of vision in one out of five patients who require cornea transplantation to restore vision [1]. Certain severe types of rare syndromic keratoconus are associated with other phenotypes such as skeletal defects, sensorineural deafness, Ehlers-Danlos syndrome and Leber Congenital Amaurosis [2]. Isolated keratoconus involving the cornea primarily is often manifested in the early teenage years and progresses over a decade or two. Clinical investigative methods include slit lamp biomicroscopy, confocal imaging and video keratography to determine corneal topography, clarity and cellular properties [3-5]. Vision can be improved by contact lenses or INTACs, which are plastic polymer corneal inserts to reshape the cornea, while riboflavin and UVA cross linking may be used to strengthen the stromal collagen extracellular matrix.
Keratoconus is a complex and heterogeneous disease with the likely involvement of multiple genes and environmental factors. In vivo confocal images show abnormal features in every layer of the cornea; cellular hyper reflectivity, abnormal epithelial and stromal keratocyte shapes. By histology the corneal epithelium shows regional thickening and disruptions in the underlying basement membrane and thinning of the stroma. Analyses of the tear fluid showed abnormal levels of TH1, TH2 and TH17 cytokines, suggesting certain immune dysregulation in this disease as well [6, 7]. Transmission electron microscopy and X-ray diffraction studies of the cornea elucidated abnormal distribution and orientation of collagen lamellae [8, 9]. Earlier studies of selected proteins indicated increases in epithelial proteins such as desmoglein, EMP3 and S100A2 [10] and reduced basement membrane collagen type XII [11]. Biochemical approaches identified heterogeneity in glycosaminoglycan content, stromal proteoglycans decorin, biglycan and the keratan sulfate proteoglycans [12, 13] and decreased hydroxyproline content indicative of reduced collagen [14]. With technical advances in proteomic approaches, a few studies have begun to investigate global proteomic changes in the epithelium and the stroma of keratoconus corneas. These include studies of the corneal epithelium and the stroma by 2D gel electrophoresis [15, 16] and shotgun proteomics of untagged samples [17]. In the current study we used a robust and more sensitive iTRAQ labeling and Nano-Electrospray Ionization Liquid Chromatography Tandem Mass Spectrometry approach to investigate the epithelial and stromal proteome from normal donor and keratoconus corneas. To the best of our knowledge the current study has established the most extensive proteome coverage of the corneal epithelium and the stroma. Relative quantification, based on differential isobaric labels of normal and keratoconus corneal extracts, has identified more than 1.5 fold changes in 277 and 459 proteins in the epithelium and stroma, respectively.
2. Materials and Methods
2.1. Materials
Isobaric peptide labeling (4-plex iTRAQ multiplex kit) was obtained from AB Sciex (Framingham, MA ,USA). Sequencing grade modified porcine trypsin was from Promega. KH2PO4, acetonitrile, KCl, formic acid, guanidine-HCl, sodium acetate, Tris-HCl, trifluoroacetic acid (TFA) and NaCl were from Sigma.
2.2. Human corneas
All KC corneas were obtained from patients seen at the Cornea Service of the Wilmer Eye Institute of Johns Hopkins University according to an approved IRB protocol entitled Genotypic and Phenotypic Assessment of Keratoconus (NA_00006544). Donor cornea tissues used as controls were obtained from autopsy cases without any history of corneal disease, collected by Tissue Banks International (Baltimore, MD). Clinical diagnosis of KC was performed by trained corneal specialists based on corneal topography, along with the presence of the clinical signs. All corneas were procured within 24 hour after surgery or death.
2.3. Protein preparation
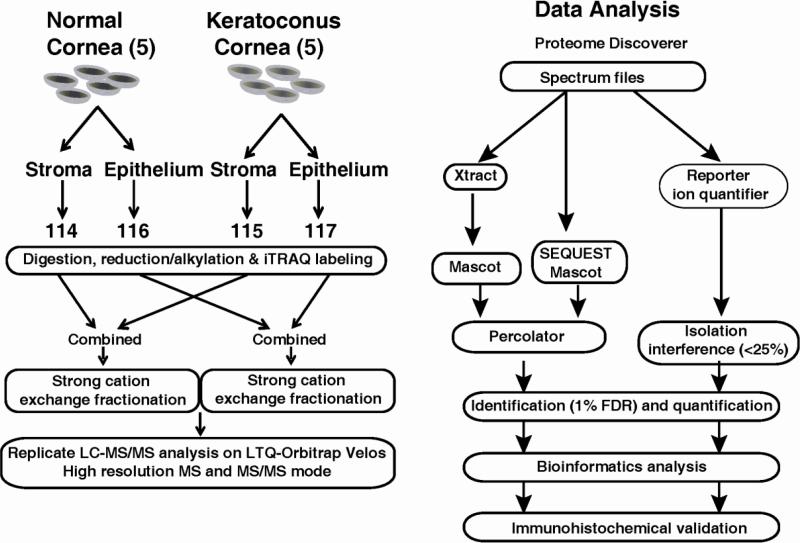
Five corneas from patients with KC and normal autopsy were used for MS analysis. Fig. 1 shows the sequence of events, from cornea acquisition, extraction, iTRAQ labeling of peptides, liquid chromatography and mass spectrometry to data analysis. The epithelium and stroma were separated from each cornea for protein extraction. The corneal samples were frozen in liquid nitrogen, and homogenized by 2.0 ml Biomasher (USA Scientific). After adding 150μL of 0.5% SDS, samples were centrifuged at 15,000g for 30s, sonicated for 1m and then centrifuged at 15,000g for 10m. The supernatant was kept and the protein concentration was measured by Bradford assay kit (Bio-Rad). The proteins from each group were normalized by the total amount, which was also confirmed by SDS-PAGE.
Fig. 1.
Outline of sample preparation and iTRAQ labeling for LC-MS/MS and Data Analysis.
2.4. iTRAQ labeling
Trypsin digestion derived peptides from the epithelium and stroma of pooled keratoconus and normal cornea samples were labeled using 4-plex iTRAQ reagents (AB Sciex) as described earlier [18] (Fig. 1). Briefly, 70μg proteins of each group were reduced using 2μL TCEP at 60°C for 30m; and treated with 1μL of cysteine blocking agent at room temperature for 15 minutes. The samples were then treated with sequencing grade modified porcine trypsin (Promega) (20: 1 protein ratio) overnight at 37°C. The resulting peptides, in a volume of 35 μl each, from normal stroma, KC stroma, normal epithelium and KC epithelium were labeled with 114, 115, 116, and 117 iTRAQ reagents (in 70 μl absolute ethanol), respectively. The labeled peptides were dried and reconstituted in solvent A (5 mM potassium phosphate buffer, pH2.85, in 25% acetonitrile) strong cation exchange (SCX) buffer, combined and fractionated on a Polysulfoethyl A SCX column (PolyLC, USA), using an Agilent 1100 HPLC system. The peptides were eluted using the following buffer gradient: Solvent A and Solvent B (5mM KH2PO4, 30% acetonitrile, 350mM KCl, pH 2.7) linear gradient of 100% in 50 minutes. Each fraction was desalted by C18 stage-tip (3M emphore , C18 Octadecyl extraction disks) and then eluted with 0.1% TFA in 60% acetonitrile. Dry extracted peptides were re-suspended in 7 μL 0.1% formic acid for LC-MS/MS analysis.
2.5. Nanoflow electrospray ionization tandem mass spectrometry analysis
Tandem mass spectrometry analysis of the iTRAQ labeled peptides were carried out on the LTQ-Orbitrap Velos (Thermo Scientific, Waltham, MA) attached to Eksigent (Eksigent, Dublin, CA ) nano-flow 2D liquid chromatography system and an Agilent 1200 microwell plate auto sampler. Peptides were enriched on a 2 cm trap column (YMC gel ODS-A S-10μm), fractionated on Magic C18 AQ, 5μm, 100Å (Michrom Bioresources, Freemont, CA), 75μm × 15 cm column and electrosprayed through a 15 μm emitter (PF3360-75-15-N-5, New Objective). Reversed-phase solvent gradient consisted of solvent A (0.1% formic acid) with increasing levels of solvent B (0.1% formic acid, 90% acetonitrile) over a period of 70 minutes. LTQ orbitrap Velos was set at 2.0 kV spray voltage, full MS survey scan range of 350-1800 m/z, data dependent HCD MS/MS analysis of top 10 precursors with minimum signal of 2000, isolation width of 1.2, 30s dynamic exclusion limit and normalized collision energy of 40. Precursor and the fragment ions were analyzed at 30,000 and 15,000 resolutions, respectively. Two LC-MS/MS technical replicates were carried out on the epithelial and the stromal extracts two analyze correlation between expression levels of proteins with confident peptide ratios (<25% co-isolation impurities).
2.6. MS data analysis
Peptide sequences were identified from isotopically resolved masses in MS and MS/MS spectra extracted with and without deconvolution using Thermo Scientific Xtract software. The data was analyzed using Proteome Discoverer 1.3 (Thermo Scientific) software configured with Mascot and Sequest search nodes. Data was searched against Refseq version 46, human entries with oxidation on methionine and proline, iTRAQ4plex on tyrosine, deamidation on residues N and Q as different variable modifications and iTRAQ4plex on N-terminus and lysine residue, methylthio on cysteine residue as different fixed modifications. Mass tolerances on precursor and fragment masses were set to 15 ppm and 0.03 Da, respectively. Percolator node was used for peptide validation and 1% false discovery rate cutoff was used to filter the data. The peptide and protein data were consolidated using proteome discoverer grouping option and by selecting top one ranked and high confidence peptides. Protein ratios were normalized on protein median values and peptides with >25% isolation interference was excluded from the protein quantification to avoid potential interference of reporter ions from contaminant peaks. The data was further analyzed using the Ingenuity Pathway Analysis (IPA) software to get insights into disease and biological processes. Proteins altered by ≥ 1.5 or ≤ 0.8 in the keratoconus sample compared to the pooled control were used for IPA. Proteins reduced by ≤ 0.8 were converted to negative fold change before analysis by IPA. This software uses a cumulative knowledge base to identify diseases, molecular and cellular pathways, physiological systems and canonical pathways significantly associated with the experimental dataset using right-tailed Fisher Exact Test. A p value ≤ 0.05 (www.ingenuity.com) was considered significant.
3. Results
3.1. Sample description
Patient and normal donor corneas were comparable in ethnicity, with the mean age of the patient pool being younger than the donor pool (Table 1). Two patients had atopy, and three were contact lens wearers, and duration of disease ranged from 7-32 years.
Table. 1.
Keratoconus (KC) and Control (N) Samples
| Sample | Group | Eye | Age | Gender | Ethnicity | Diagnosis (years) | Contact lens | Atopy |
|---|---|---|---|---|---|---|---|---|
| 160 | KC | OS | 39 | M | African American | 10 | No | Yes |
| 166 | KC | OD | 34 | F | Caucasian | 8 | No | No |
| 167 | KC | OD | 25 | M | Hispanic | 7 | Yes | No |
| 239 | KC | OD | 39 | M | African American | 22 | Yes | Yes |
| 241 | KC | OD | 47 | M | Caucasian | 32 | Yes | No |
| 66260 | N | 45 | F | Caucasian | n/a | n/a | n/a | |
| A54397 | N | 64 | F | African American | n/a | n/a | n/a | |
| 66261 | N | 70 | F | Caucasian | n/a | n/a | n/a | |
| 66482 | N | 75 | F | Caucasian | n/a | n/a | n/a | |
| 66470 | N | 77 | M | Caucasian | n/a | n/a | n/a |
OD: oculus dexter; OS: oculus sinister
3.2. Total proteins detected in the corneal epithelium and stroma
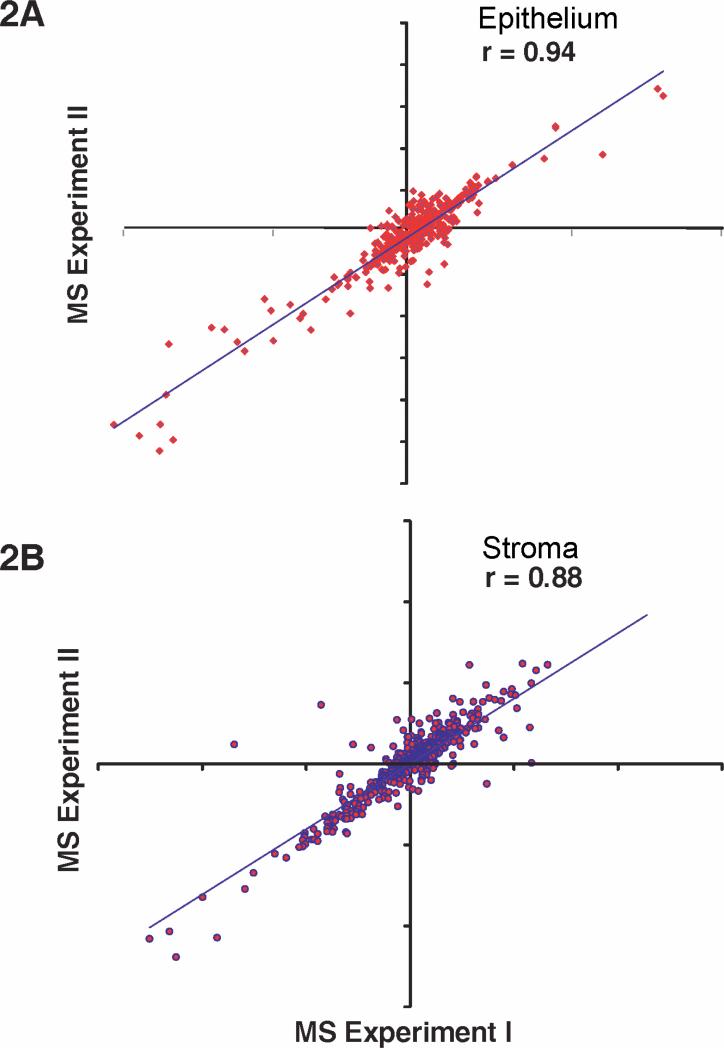
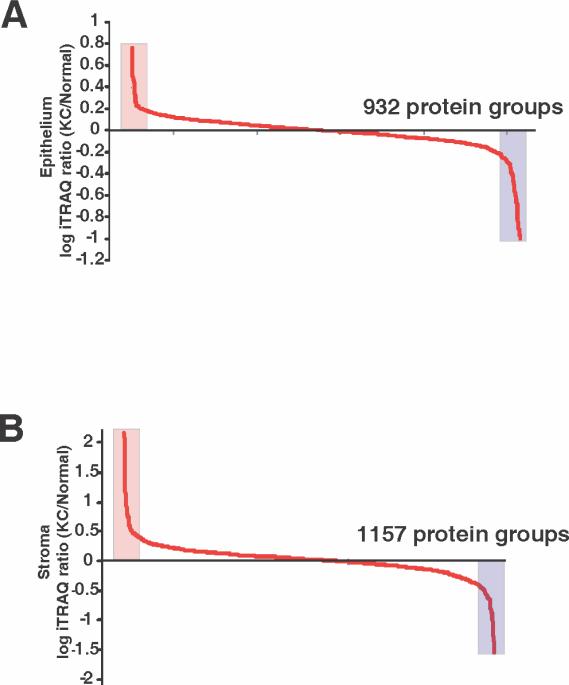
We conducted a quantitative mass spectrometry of the epithelial and the stromal extracts in duplicate. The raw data and Magellan storage files for the data described in this manuscript may be downloaded from ProteomeCommons.org, Tranche, https://proteomecommons.org/tranche/, using the following hash codes: jXHvMx/BkHaM8VvQxVEcBSsRYrcyjy/oKPt7Xhvwq8JUmIRrTL6OVm3TRTQei/doWkxNcyImCRFlmk0VtDP2XcSfNcoAAAAAAAAFwg== The technical replicates showed strong concordance, r = 0.94 and r = 0.88 for the epithelium and stroma, between levels of all common proteins with robust peptide ratios (<25% co-isolation impurities), indicating very low technical variations in the data (Fig. 2). A total of 932 and 1,157 proteins were detected in the consolidated data of the epithelium and stroma, respectively (Fig. 3).
Fig. 2.
Correlation between technical replicates. The expression levels of proteins (KC/Normal) in epithelium and the stroma were compared by linear regression from two technical replicates. All proteins with confident peptide ratios (with <25% co-isolation impurities) in common between replicate experiments were compared. There were 480 and 560 proteins for the epithelium and the stroma, respectively, comprising 14 LC-MS/MS analyses.
Fig. 3.
Overall proteins detected in the epithelium (A) and the stroma (B). A log ratio of fold increase and decrease shows 932 and 1,157 significant proteins in the epithelium and the stroma, respectively.
We selected cut-off values of ≤ 0.8 fold and ≥ 1.5 fold for further study of the differential KC proteome. Although these cut-off values are somewhat low, we selected these for the following reasons. Proteins with previously identified associations with keratoconus, such as A2M (alpha-2 macroglobulin) was decreased by 0.74 fold, while SOD2 (superoxide dismutase 2) was increased by 1.53 fold, indicating that many meaningful changes would reside within this range. Secondly, many extracellular matrix (ECM) proteins are long- lived in the cornea. Therefore, their turnover is expected to be very low and the differential proteome in keratoconus to show small changes. For example, ECM proteins, podocan (0.8 fold), opticin (0.73 fold), collagen type VIII (0.8 fold), were only slightly reduced in the KC cornea. However, even small changes in ECM proteins can have significant biological consequences on corneal thickness, biomechanical strength and architectural differences in the ECM that we did not want to miss by setting high cut off values.
In the epithelial pool 58 proteins were increased ≥ 1.5 fold and 219 were decreased ≤ 0.8 fold in the KC (277 in all) compared to the control pool. In the stromal pool 139 were increased by ≥ 1.5 fold and another 279 decreased by ≤ 0.8 fold compared to the control pool. Overall there were no major differences in the distribution of different categories of proteins in the epithelial versus the stromal extracts (Supplementary Fig. 1). The exception was the extracellular category - the stroma had a larger percent of proteins that belonged to this functional group. This was expected, because compared to the epithelium, the stroma has a larger proportion of connective tissue proteins than the epithelium.
3.3. Differential epithelial proteome in KC
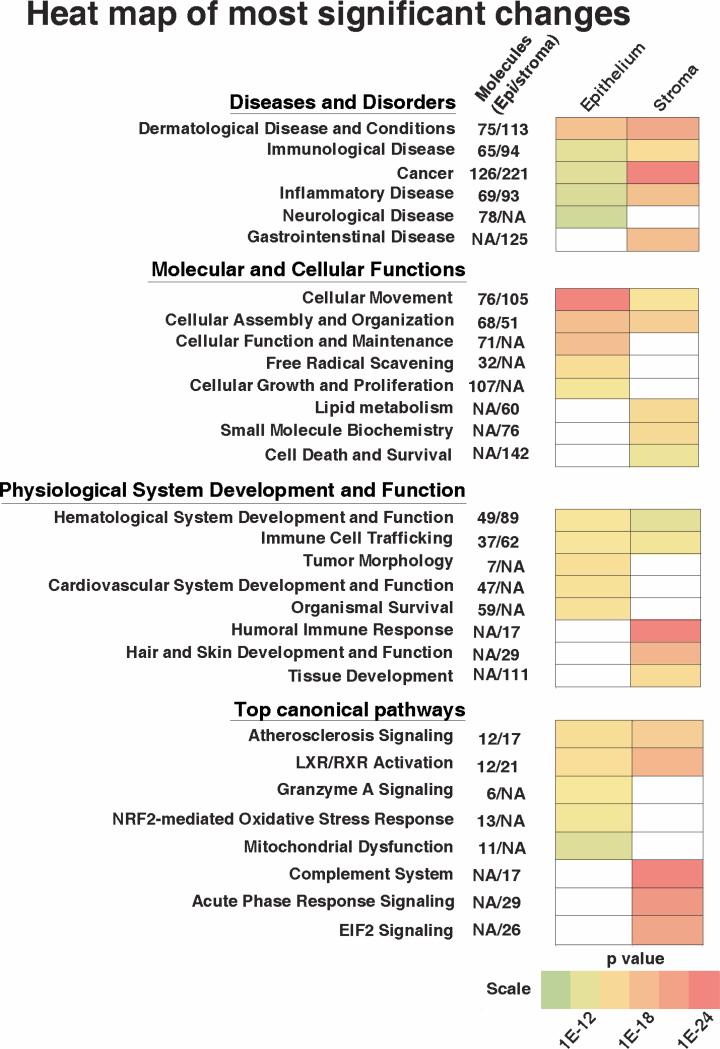
The differential protein profiles were further analyzed using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems) to identify proteins that have known associations with diseases, molecular and cellular functions, physiological systems development and function and canonical pathways. Based on the distribution of proteins that relate to these categories, the IPA software ranks networks and categories, with the smallest p-values as most significant to KC, with the least likelihood of being found together by mere chance alone. Analysis of 277 most changed proteins in the KC epithelial pool identified several categories as most significant that are presented as a heat map of p values (Fig. 4). In the disease category, proteins associated with dermatological diseases and conditions were most significantly changed in the KC epithelial proteome (Supplementary Table 1). In the molecular and cellular functions category, the most significant groups were 1) cellular movement, 2) cellular assembly and organization, 3) cellular function and maintenance, 4) free-radical scavenging and 5) cellular growth and proliferation. In the physiological system development and functional category no one group out of six stood out more than the other. The top 5 canonical pathways associated with the KC epithelium were, 1) atherosclerosis signaling, 2) LXR/RXR activation, 3) granzyme A signaling, 4) NRF2-mediated oxidative stress and 5) mitochondrial dysfunction. The stroma showed marked differences in its association with these categories compared to the epithelium, and these are discussed later.
Fig. 4.
Heat map of most significant changes in the KC epithelium and the stroma. The IPA software classified the protein changes into four categories. Within each category certain groups were significantly changed in the KC epithelium or the stroma as indicated by the colors while no color designates lack of correlation with that tissue.
The dermatological diseases category underscores epithelial dysfunction and contained proteins related to dermatitis, atopy, psoriasis and epithelial ulcerations, such as increased pro-inflammatory proteins S100A8, S100A9, LGALS1, LGALS3, LGALS7 and decreased integrins (ITGA6 and ITGB4). In addition, type I cytokeratin KRT16, and type II cytokeratin KRT6A were increased by 5 fold in KC. These are usually co-expressed in many types of stratified epithelia, and their increase in the KC epithelium is concordant with the epithelial thickening reported by others. Epithelial dysfunction is further supported by decreased basement membrane proteins. The top 20 proteins with the largest changes in KC epithelium are shown in Table 2. The increases include KRT6A, KRT16, HBB, VIM, CALM3 and CRIP1. A two-fold increase in vimentin, a class III intermediate filament protein, usually expressed in mesenchymal cells, suggests epithelial to mesenchymal transformation as an underlying phenomenon in keratoconus. Decreases in the KC epithelium were largely related to epithelial anchoring integrity such as basement membrane and sub-epithelial collagen types VII and XII (COL7A1, COL12A1), all three chains of collagen type VI (COL6A1, COL6A2 and COL6A3) and iron transporter LTF.
Table 2.
Top 10 increased and decreased proteins in KC epithelium
| Gene Symbol | Protein | KC/N |
|---|---|---|
| KRT6A | Keratin 6A | 5.9 |
| KRT16 | Keratin 16 | 5.8 |
| HBB | Hemoglobin, beta | 3.2 |
| CRIP1 | Cysteine-rich protein 1 (intestinal) | 3.1 |
| APOBEC | Apolipoprotein B polypeptide-like | 3.0 |
| PLLP | Plasmolipin | 2.8 |
| S100A8 | S100 calcium binding protein A8 | 2.5 |
| VIM | Vimentin | 2.3 |
| CALML3 | Calmodulin-like 3 | 2.1 |
| RAB25 | RAB25, member RAS oncogene family | 1.8 |
| COL6A3 | Collagen, type VI, alpha 3 | −9.9 |
| COL1A1 | Collagen, type I, alpha 1 | −9.5 |
| LTF | Lactotransferrin | −9.1 |
| COL1A2 | Collagen, type I, alpha 2 | −9.0 |
| ARPP21 | cAMP-regulated phosphoprotein, 21 kD | −8.1 |
| COL7A1 | Collagen, type VII, alpha 1 | −7.9 |
| COL6A2 | Collagen, type VI, alpha 2 | −7.0 |
| COL6A1 | Collagen, type VI, alpha 1 | −6.6 |
| COL12A1 | Collagen, type XII, alpha 1 | −5.4 |
| HSPG2 | Heparan sulfate proteoglycan 2 | −4.7 |
3.4. The stromal proteome
Analysis of 411 proteins present at the level of ≥ 1.5 fold increases and ≤ 0.8 fold decreases in the keratoconus stromal pool identified cancer related proteins as most significant under Diseases and disorders category. These proteins in general relate to cell proliferation and cell death. In the Molecular and cellular functions category, 5 groups were identified as more or less equally significant to the KC stroma: cellular movement, cellular assembly and organization, lipid metabolism, small molecule biochemistry, and cell death and survival. The top canonical pathways that were most significant to the KC stroma were, complement system, acute phase response, EIF2 signaling/protein synthesis, LXR/RXR and atherosclerosis related signaling (Fig. 4). Interestingly, association of the complement pathway, acute phase response and lipid metabolism were specific to the stroma and not the epithelium in KC.
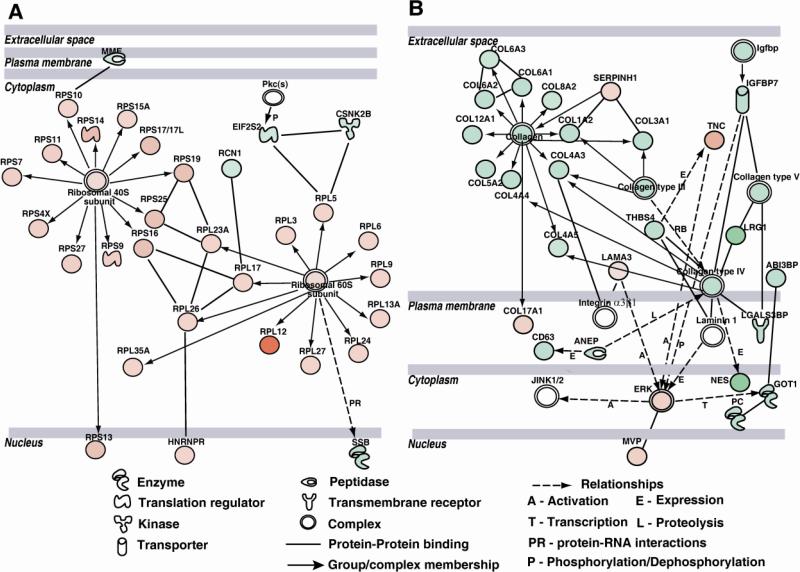
The differential stromal proteome could be linked to several networks, but the most striking were networks 1 and 3 in their stark contrasts (Fig.5). The first network contains many endoplasmic reticulum (ER)- and translation-function related proteins that were increased (Fig. 5A); whereas the second network contains ECM proteins that were all decreased (Fig. 5B). The elongation initiation factor subunit (EIF2S2), MME (matrix metalloendopeptidase), Ca-binding RCN1, and CSNK2B that localize to the ER and Golgi to regulate translation were decreased in KC, suggesting a block in translation initiation. On the other hand, several 40S and 60S ribosomal proteins were all increased indicating accumulations of ribosomal proteins and ER stress (Fig. 5A). The ECM network is comprised of several fibrillar collagen types I, III and V, major constituents of the corneal stroma, that were all decreased in KC. All three chains of collagen type VI were decreased in the KC stromal pool; collagen type VI is widely present in many types of connective tissues including the pericellular matrix, its mutations have been associated with Bethlem myopathies, and its reduction in KC may contribute to keratocyte (stromal cell) dysfunction. Collagen type XII is a fibril associated collagen that is normally present in the anterior corneal stroma and known to interact with stromal proteoglycans, fibrillar collagens and cell surface integrins, and thus implicated in epithelial anchoring to the underlying stroma. Earlier immunofluorescence staining of keratoconus corneas reported a decrease in collagen XII in the sub-epithelial stroma. Its reduction confirmed by our proteomic study underscores its potential role in the weakening of the keratoconus cornea.
Fig. 5.
Protein networks in the stroma. A: Translation-related and ribosomal proteins network. This network shows decreased elongation initiation factor subunit and increased accumulation of ribosomal proteins suggesting potential endoplasmic reticular stress in KC. B: ECM proteins network. Decrease in multiple ECM collagens in the KC stroma that suggest perturbation of the interstitial ECM and the basement membrane.
The top ten increased and decreased proteins in the KC stroma are shown in Table 3. AP2B1and AP1B1 levels were highly increased in KC; these function in clathrin-mediated endocytosis, protein sorting and trafficking. IGLL1/5 is a member of the immunoglobulin superfamily and it was significantly elevated in the KC stroma. CCAR1, a perinuclear phosphoprotein regulates E3 ubiquitin mediated protein degradation and cellular apoptosis. Its marked elevation in the KC stroma also speaks of abnormal protein synthesis/degradation, cellular stress and apoptosis [19]. CRYBB2, CRYBB1 and CRYG5 are major crystallins of the lens that regulate lens transparency; however, we detected these in the cornea and they were markedly reduced in the KC stroma. NES (Nestin), known to localize with intermediate filaments and to be expressed in stem/progenitor cells of various types, was also reduced in KC may be indicative of progenitor cell deficiency.
Table 3.
Top 10 increased and decreased proteins in KC Stroma
| Gene Symbol | Protein | KC/N |
|---|---|---|
| AP2B1 | Adaptor-related Protein complex 2, beta 1 subunit | 50.9 |
| IGLL1/IGLL5 | Immunoglobulin Lambda-ike polypeptide 1 | 40.4 |
| CCAR1 | Cell division cycle and apoptosis regulator 1 | 8.7 |
| RPL12 | Ribosomal protein L12 | 8.5 |
| HAAO | 3-hydroxyanthranilate 3,4-dioxygenase | 7.8 |
| AP1B1 | Adaptor-related protein complex 1, beta 1 subunit | 7.3 |
| HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H1 (H) | 6.5 |
| SERPINB9 | Serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 5.4 |
| TACSTD2 | Tumor-associated calcium signal transducer 2 | 5.1 |
| MFAP4 | Microfibrillar-associated protein 4 | 4.1 |
| CRYBB2 | Crystallin, beta B2 | −18.2 |
| CA1 | Carbonic anhydrase I | −14.6 |
| LCN1 | Lipocalin 1 | −13.6 |
| CRP | C-reactive protein, pentraxin-related | −10.1 |
| CRYGS | Crystallin, gamma S | −8.4 |
| UQCRC2 | Ubiquinol-cytochrome c reductase core protein II | −7.1 |
| HBB | Hemoglobin, beta | −6.3 |
| CRYBB1 | Crystallin, beta B1 | −5.3 |
| NES | Nestin | −5.2 |
| PRR4 | Proline rich 4 (lacrimal) | −5.1 |
Other remarkable decreases in the KC stroma that were not among the top ten, but are major structural components of the corneal stroma are proteoglycans, lumican , decorin, biglycan and keratocan that associate with fibrillar collagens to make up the biomechanically strong, refractive and transparent ECM of the cornea.
3.5. Changes in hydroxylated proteins
We also analyzed the presence of hydroxylated peptides in the normal control and keratoconus corneal proteome (Supplementary Table 2). A majority of the collagens showed reduced hydroxyproline containing peptides in KC. For example, for Collagen type V, 45 peptides were detected, and most were decreased in KC. In the stroma, we detected multiple hydroxyproline peptides in collagen types I, III, VI, XII and XVIII. In all cases the ratio of hydroxyproline containing peptides in KC compared to control stroma was reduced. Proline and lysine residues are hydroxylated in collagens in the rough endoplasmic reticulum, mediated enzymatically by prolyl/lysyl hydroxylases and additional chaperone proteins. Hyroxylation occurs co-translationally mostly, before trimerization of collagens. Hydroxyprolines are important for inter-chain H-bonding and stabilization of the triple helix, while hydroxylysines are further glycosylated for appropriate hydration and collagen organization [20]. Over and under hydroxylation of collagens can occur due to defects in the enzymes, chaperones or other functional stress of the ER. Several osteogenesis imperfect types are associated with changes in hydroxylation of collagens and associated connective tissue abnormalities [21]. In the KC proteome we found that there was a global decrease in hydroxylated peptides which is consistent with the total proteomic data that shows decreases in many collagens. Altered hydroxylation in KC could relate to the general oxidative stress and ER dysfunction reported for KC and supported by our global proteomic data. Earlier studies have suggested reduced collagen cross-linking and reduced hydroxyproline content [22] without definitive quantification as we have presented in this study. Thus, localized under hydroxylation of lysines and prolines in the cornea could contribute to collagen structural anomalies and biomechanical weaknesses as seen in keratoconus, further relating this condition to other connective tissue disorders.
3.6. Predicted upstream regulators of the KC proteome
Based on proteins changed in the KC epithelium and the stroma, the IPA software predicted a set of upstream regulators. One caveat to keep in mind before considering these upstream regulators is that, unlike gene expression data which can be directly correlated to upstream microRNA, transcription factor or growth factor regulators, the protein changes could be the outcome of regulated expression or degradation of the proteins. In any case, the analysis identified several growth factors, kinase and transcription factors as potential regulators of the epithelium and stroma (Table 4). A majority of regulatory factors were predicted to be down regulated in the KC epithelium. The differential proteomes of the epithelium and the stroma predicted down regulation of TGFβ1 in both corneal layers of KC patients, concordant with this, the cytoplasmic signal inhibitor SMAD7 was predicted to be activated. Our earlier immunohistochemistry had shown a slightly increased staining of TGFβ2 and pSMAD2 in the KC epithelium [23].
Table 4.
Predicted upstream regulators in KC epithelium and stroma.
| Epithelium | Stroma | ||||
|---|---|---|---|---|---|
| Regulator | Activation Z-Score | P - value | Regulator | Activation Z-Score | P - value |
| SMAD7 | 2.020 | 4.09E-06 | MYCN | 4.061 | 2.30E-18 |
| NFE2L2 | −4.233 | 1.82E-09 | SMAD7 | 2.555 | 1.38E-04 |
| TP53 | −3.119 | 2.90E-12 | SPDEF | 2.121 | 6.25E-05 |
| VEGFA | −2.961 | 8.12E-04 | WISP2 | 2.000 | 5.67E-03 |
| NFkB | −2.782 | 1.91E-02 | FGF2 | −3.531 | 2.56E-05 |
| FGF2 | −2.511 | 3.53E-04 | CEBPB | −3.364 | 1.67E-06 |
| XBP1 | −2.470 | 5.03E-05 | HNF1A | −3.201 | 1.15E-09 |
| INSR | −2.429 | 6.01E-04 | CHUK | −3.116 | 7.26E-03 |
| MAPK14 | −2.279 | 1.09E-03 | IKBKB | −2.717 | 4.68E-03 |
| EPAS1 | −2.194 | 1.63E-02 | MTPN | −2.630 | 1.53E-03 |
| LEP | −2.129 | 1.67E-03 | P38 MAPK | −2.520 | 5.89E-03 |
| SP1 | −2.093 | 1.84E-04 | TGFβ1 | −2.314 | 9.72E-17 |
| TGFβ1 | −2.086 | 2.83E-12 | CREBBP | −2.158 | 4.09E-02 |
| PPARGC1A | −2.021 | 9.99E-04 | TGFβ2 | −2.138 | 7.38E-03 |
Based on target proteins changed in the KC stroma, four and ten regulators were predicted as activated and repressed in KC, respectively. A widespread decrease in several collagens and associated proteoglycans (BGN, COL1A1, COL1A2, COL3A1, COL6A1, COL6A3, DCN, LUM and TGF BI) predicted activation of SMAD7, SPDEF, and the estrogen receptor group as upstream regulators. Activation of MYCN indicative of growth dysregulation was suggested by decreases in PTK2, IGFBP7, TIMP2, COL1A1, COL5A2, B2M and increases in multiple ribosomal proteins. Inhibition of the CEBPB transcription factor which regulates transcription of acute phase response and inflammatory and anti-apoptotic genes, was suggested by decreased levels of SERPINA1, ORM1, MMP3, LBP, CD14, FTL, FBLN1, CRP and HP and increased CTSC.
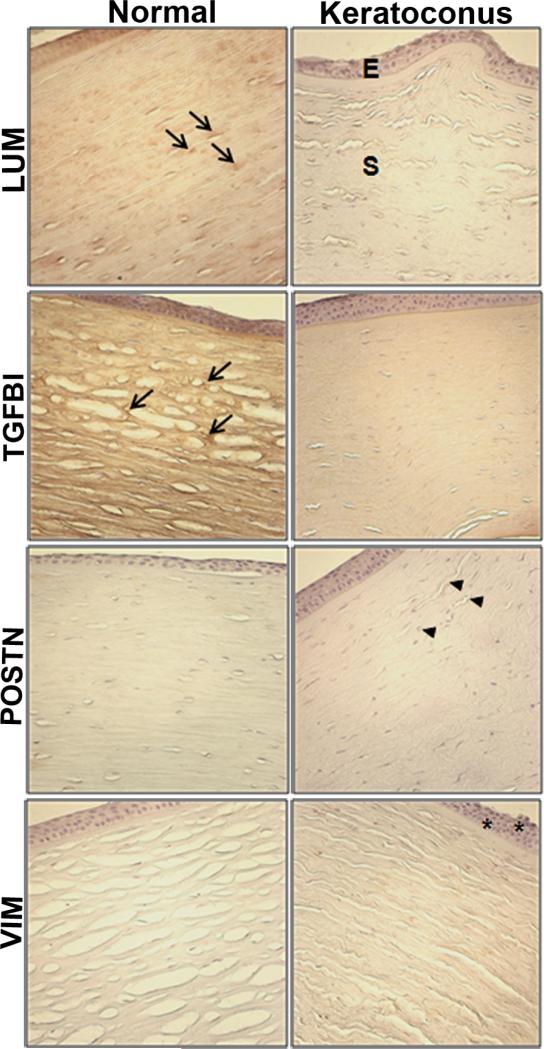
3.7. Immunohistology of selected proteins
We performed immunohistochemistry of paraffin embedded corneal sections to validate selected protein changes in KC (Fig. 6). Concordant with the proteomics data (0.6 KC/N fold change for LUM and 0.4 KC/N for TGFBI), immunostaining for LUM, and TGFBI (TGF β Inducible protein or keratoepithelin) showed decreased staining of the KC stroma. POSTN (periostin) showed increased staining of the KC stroma compared to the control in accordance with its 2.5 fold increase in the KC stroma. The MS data had shown an increase in VIM (vimentin) in the KC epithelial proteome with no change in the stroma. This is reflected in the IHC data as well: stronger staining of the KC epithelium with no change in staining of the stroma compared to control corneas.
Fig. 6.
Immunohistochemistry (IHC) of selected proteins in normal donor and KC corneal sections. Paraffin-embedded corneas were sectioned and immunostained for the proteins indicated. E: epithelium, S: stroma. Arrows show increased staining for lumican(LUM) and TGFBI (TGFβ inducible protein) in the stroma of control donor corneas. Periostin (POSTN) IHC was increased in the stroma of KC corneas (arrowheads). Vimentin (VIM) IHC showed a slight increase in the KC epithelium (asterisks) without any changes in the stroma as seen in the proteome.
4. Discussions and conclusions
Using a highly sensitive isobaric peptide labeling approach on proteins extracted from normal donor and keratoconus corneas we identified a total of 932 and 1,157 proteins in the corneal epithelium and the stroma, respectively. By far this is the largest set of proteins detected, and quantified in the human cornea. A previous study used 2-D gel electrophoresis on the keratoconus epithelium and detected 200-500 spots, and 19 differentially expressed proteins [16]. Another recent study used label-free shot gun proteomic approach to identify 104 epithelial and 44 stromal proteins [17]. Among proteins detected in our system are those known to be present in the cornea, such as Collagen types I, III, V, VI, XII, proteoglycans lumican, keratocan, decorin and biglycan in the stroma. Known epithelial proteins detected in our study include cytokeratin 3, 12, Mucin 16, thioredoxin, transketolase, gelsolin and glutathione S-transferase. A few of the proteins reported as altered in KC by earlier studies were also similarly changed in our study. For example, Collagen XII reported as decreased by earlier immunohistochemistry [11] was found to be decreased in the KC stromal proteome in our study. Transketolase and TGFBI (keratoepithelin) were reported as decreased in the KC in the earlier proteome study [17]; we also found decreased TKT and TGFBI in our keratoconus proteome.
It has been hypothesized that mitochondrial oxidative stress, increased production of reactive oxygen and nitrogen species, increased proteolytic activities and ECM degradation are major underpinnings in keratoconus pathogenesis [14, 24, 25] [26] [27]. Our proteome analysis of the KC epithelium identified atherosclerosis signaling, granzyme signaling, NRF2 mediated oxidative stress and mitochondrial dysfunction as altered canonical pathways in keratoconus. In addition, the most elevated proteins in the KC epithelium included cytoskeletal proteins, encoded by KRT6A, KRT16, VIM and S100A8 that have been linked to dermatological diseases such as psoriasis, inflammation and epithelial-mesenchymal transition. The KC stromal proteome included a broad decrease in many structural collagens and proteoglycans, a minor increase in proteases, altered apoptosis related proteins and complement components that suggest abnormal lipid metabolism, complement functions and cell death as major keratoconic processes.
Taken together, this in depth proteome analysis of the KC epithelium and the stroma speak of a degenerative process in keratoconus with abnormal mitochondrial functions, lipid metabolism, changes in innate immune functions, and increased cell death. These are events echoed in degenerative diseases such as age related macular degeneration [28] and Parkinson's disease [29]. Finally, altered proteins identified in the KC epithelium and the stroma will help to narrow down candidate genes and potential upstream regulators for isolated keratoconus types.
Supplementary Material
Significance.
This study provides, to our knowledge, the most comprehensive proteomic analysis of the vision threatening disease keratoconus, which affects a significant portion of the US and global populations. Using iTRAQ and LC/MS/MS, we have identified significant changes in the human corneal epithelium and stromal proteome that correlate to in vivo clinical findings. The protein changes identified will lead to molecular insights into disease pathogenesis and provide candidate genes for genetic studies of keratoconus.
Highlights.
The largest set (1591 proteins) of corneal proteins identified by LC-MS/MS.
Dermatological disease related proteins were most changed in the KC epithelium.
Apoptosis-related proteins were elevated in the KC stroma.
Several fibrillar collagens and proteoglycans were decreased in the KC stroma.
Our results suggest novel KC candidate genes for further genetic testing.
Acknowledgements
We thank Xiaojun Feng, Ph.D. for her assistance with immunohistochemistry. We thank Robert Cole, Ph.D (Director, Mass Spectrometry and Proteomics Facility of the Johns Hopkins School of Medicine) for technical assistance with mass spectrometry. This study was funded in part by EY11654 (SC) and the Openshaw Keratoconus Research Funds from the Wilmer Eye Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Romero-Jimenez M, Santodomingo-Rubido J, Wolffsohn JS. Keratoconus: a review. Cont Lens Anterior Eye. 2010;33:157–66. doi: 10.1016/j.clae.2010.04.006. quiz 205. [DOI] [PubMed] [Google Scholar]
- 2.Ihalainen A. Clinical and epidemiological features of keratoconus genetic and external factors in the pathogenesis of the disease. Acta Ophthalmol Suppl. 1986;178:1–64. [PubMed] [Google Scholar]
- 3.Hollingsworth JG, Efron N, Tullo AB. In vivo corneal confocal microscopy in keratoconus. Ophthalmic Physiol Opt. 2005;25:254–60. doi: 10.1111/j.1475-1313.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 4.Efron N, Hollingsworth JG. New perspectives on keratoconus as revealed by corneal confocal microscopy. Clin Exp Optom. 2008;91:34–55. doi: 10.1111/j.1444-0938.2007.00195.x. [DOI] [PubMed] [Google Scholar]
- 5.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 6.Jun AS, Cope L, Speck C, Feng X, Lee S, Meng H, et al. Subnormal cytokine profile in the tear fluid of keratoconus patients. PLoS One. 2011;6:e16437. doi: 10.1371/journal.pone.0016437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lema I, Duran JA. Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology. 2005;112:654–9. doi: 10.1016/j.ophtha.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 8.Akhtar S, Bron AJ, Salvi SM, Hawksworth NR, Tuft SJ, Meek KM. Ultrastructural analysis of collagen fibrils and proteoglycans in keratoconus. Acta Ophthalmol. 2008;86:764–72. doi: 10.1111/j.1755-3768.2007.01142.x. [DOI] [PubMed] [Google Scholar]
- 9.Meek KM, Tuft SJ, Huang Y, Gill PS, Hayes S, Newton RH, et al. Changes in collagen orientation and distribution in keratoconus corneas. Invest Ophthalmol Vis Sci. 2005;46:1948–56. doi: 10.1167/iovs.04-1253. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen K, Heegaard S, Vorum H, Birkenkamp-Demtroder K, Ehlers N, Orntoft TF. Altered expression of CLC, DSG3, EMP3, S100A2, and SLPI in corneal epithelium from keratoconus patients. Cornea. 2005;24:661–8. doi: 10.1097/01.ico.0000153556.59407.69. [DOI] [PubMed] [Google Scholar]
- 11.Cheng EL, Maruyama I, SundarRaj N, Sugar J, Feder RS, Yue BY. Expression of type XII collagen and hemidesmosome-associated proteins in keratoconus corneas. Curr Eye Res. 2001;22:333–40. doi: 10.1076/ceyr.22.5.333.5491. [DOI] [PubMed] [Google Scholar]
- 12.Funderburgh JL, Funderburgh ML, Rodrigues MM, Krachmer JH, Conrad GW. Altered antigenicity of keratan sulfate proteoglycan in selected corneal diseases. Invest Ophthalmol Vis Sci. 1990;31:419–28. [PubMed] [Google Scholar]
- 13.Funderburgh JL, Hevelone ND, Roth MR, Funderburgh ML, Rodrigues MR, Nirankari VS, et al. Decorin and biglycan of normal and pathologic human corneas. Invest Ophthalmol Vis Sci. 1998;39:1957–64. [PubMed] [Google Scholar]
- 14.Critchfield JW, Calandra AJ, Nesburn AB, Kenney MC. Keratoconus: I. Biochemical studies. Exp Eye Res. 1988;46:953–63. doi: 10.1016/s0014-4835(88)80047-2. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen K, Vorum H, Fagerholm P, Birkenkamp-Demtroder K, Honore B, Ehlers N, et al. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp Eye Res. 2006;82:201–9. doi: 10.1016/j.exer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava OP, Chandrasekaran D, Pfister RR. Molecular changes in selected epithelial proteins in human keratoconus corneas compared to normal corneas. Mol Vis. 2006;12:1615–25. [PubMed] [Google Scholar]
- 17.Joseph R, Srivastava OP, Pfister RR. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res. 2011;92:282–98. doi: 10.1016/j.exer.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Shao H, Chaerkady R, Chen S, Pinto SM, Sharma R, Delanghe B, et al. Proteome profiling of wild type and lumican-deficient mouse corneas. J Proteomics. 2011 doi: 10.1016/j.jprot.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puliyappadamba VT, Wu W, Bevis D, Zhang L, Polin L, Kilkuskie R, et al. Antagonists of anaphase-promoting complex (APC)-2-cell cycle and apoptosis regulatory protein (CARP)-1 interaction are novel regulators of cell growth and apoptosis. J Biol Chem. 2011;286:38000–17. doi: 10.1074/jbc.M111.222398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birk DE, Bruckner P. Collagen suprastructures. Top Curr Chem. 2005;247:185–205. [Google Scholar]
- 21.Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Bron AJ. Keratoconus. Cornea. 1988;7:163–9. [PubMed] [Google Scholar]
- 23.Engler C, Chakravarti S, Doyle J, Eberhart CG, Meng H, Stark WJ, et al. Transforming growth factor-beta signaling pathway activation in Keratoconus. Am J Ophthalmol. 2011;151:752–9. e2. doi: 10.1016/j.ajo.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown D, Chwa M, Escobar M, Kenney MC. Characterization of the major matrix degrading metalloproteinase of human corneal stroma. Evidence for an enzyme/inhibitor complex. Exp Eye Res. 1991;52:5–16. doi: 10.1016/0014-4835(91)90123-v. [DOI] [PubMed] [Google Scholar]
- 25.Kenney MC, Brown DJ. The cascade hypothesis of keratoconus. Cont Lens Anterior Eye. 2003;26:139–46. doi: 10.1016/S1367-0484(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 26.Fini ME, Yue BY, Sugar J. Collagenolytic/gelatinolytic metalloproteinases in normal and keratoconus corneas. Curr Eye Res. 1992;11:849–62. doi: 10.3109/02713689209033483. [DOI] [PubMed] [Google Scholar]
- 27.Maguen E, Rabinowitz YS, Regev L, Saghizadeh M, Sasaki T, Ljubimov AV. Alterations of extracellular matrix components and proteinases in human corneal buttons with INTACS for post-laser in situ keratomileusis keratectasia and keratoconus. Cornea. 2008;27:565–73. doi: 10.1097/ICO.0b013e318165b1cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012;75:26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abou-Sleiman PM, Muqit MM, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat Rev Neurosci. 2006;7:207–19. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.