Abstract
Background:
MicroRNAs are noncoding regulatory RNAs strongly implicated in carcinogenesis, cell survival, and chemosensitivity. Here, microRNAs associated with chemoresistance in ovarian carcinoma, the most lethal of gynaecological malignancies, were identified and their functional effects in chemoresistant ovarian cancer cells were assessed.
Methods:
MicroRNA expression in paclitaxel (PTX)-resistant SKpac sublines was compared with that of the PTX-sensitive, parental SKOV3 ovarian cancer cell line using microarray and qRT–PCR. The function of differentially expressed microRNAs in chemoresistant ovarian cancer was further evaluated by apoptosis, cell proliferation, and migration assays.
Results:
Upregulation of miR-106a and downregulation of miR-591 were associated with PTX resistance in ovarian cancer cells and human tumour samples. Transfection with anti-miR-106a or pre-miR-591 resensitized PTX-resistant SKpac cells to PTX by enhancing apoptosis (23 and 42% increase), and inhibited their cell migration (43 and 56% decrease) and proliferation (64 and 65% decrease). Furthermore, ZEB1 was identified as a novel target gene of miR-591, and BCL10 and caspase-7 were target genes of miR-106a, as identified by immunoblotting and luciferase assay.
Conclusion:
MiR-106a and miR-591 have important roles in conferring PTX resistance to ovarian cancer cells. Modulation of these microRNAs resensitizes PTX-resistant cancer cells by targeting BCL10, caspase-7, and ZEB1.
Keywords: ovarian cancer, paclitaxel resistance, microRNAs, miR-106a, miR-591
Ovarian cancer is the leading cause of gynaecological cancer-related deaths. More than 70% of patients with ovarian cancer are diagnosed at an advanced stage of disease, when the median overall survival (OS) is <45 months (Winter et al, 2007). Cytoreductive surgery followed by adjuvant therapy with platinum- and taxane-based chemotherapy is the current standard treatment for advanced ovarian cancer (Cannistra, 2004). Despite a multimodality treatment strategy, the overall cure rate is only 30%, as most patients with advanced disease ultimately face recurrence. The main cause of recurrence is the development of resistance to first-line chemotherapeutic agents. Therefore, it is important to identify markers that predict a patient's responsiveness to chemotherapy, which may allow for the development of targeted therapies for overcoming chemoresistance.
At the molecular level, a number of genes and pathways involving drug transport, drug metabolism, and apoptosis have been identified that may have a role in the responsiveness to cancer chemotherapeutics. However, epigenetic, genetic, and postgenetic modifications regulate gene expression. MicroRNAs (miRNAs), noncoding regulatory RNAs of 21–25 nucleotides (Ambros, 2004), are critical regulators of post-transcriptional gene expression.
The miRNAs are transcribed as long RNA precursors (primary miRNAs), which are processed to yield mature miRNAs ∼25 nucleotides in length by Drosha-Pasha/DGCR8 and Dicer. Mature miRNAs are incorporated into the RNA-induced silencing complex and then target the 3′ untranslated region (3′-UTR) of a specific mRNA by base pairing, leading to translational repression or mRNA degradation (Bartel, 2004). MicroRNAs regulate the expression of up to 70% of human genes, implying a potential role for miRNAs in the regulation of nearly every genetic pathway (Lewis et al, 2005; Esquela-Kerscher and Slack, 2006). Therefore, understanding the regulatory role of miRNAs may lead to a better understanding of the molecular events involved in diverse biological processes (Hwang and Mendel, 2007), and could lead to the development of drug targets for therapeutic intervention.
Recent evidence has indicated that altered miRNA levels are related to the response to chemotherapeutic agents as well as oncogenesis. Previous studies have revealed that miR-21 and miR-16 could have a role in the chemotherapeutic response of cholangiocarcinoma and gastric carcinoma cells, respectively (Meng et al, 2006; Xia et al, 2008). With respect to ovarian cancers, a few studies have reported several miRNAs associated with drug resistance in ovarian cancer cells or tissues; however, most of these studies assessed only the expression profiles of miRNAs without any functional validation (Nam et al, 2008; Sorrentino et al, 2008; Eitan et al, 2009). In this study, the purpose of our investigation was to find miRNAs related with paclitaxel (PTX) resistance in ovarian cancer using a high-throughput miRNA microarray and quantitative real-time reverse transcriptase PCR (qRT–PCR), and found that differential expression of miRNAs-106a and -591 in PTX-resistant ovarian cancer sublines (SKpac cells) compared with their parental cell line (SKOV3). Furthermore, we demonstrated that the regulation of miR-106a and miR-591 in ovarian cancer cells affects sensitivity to PTX, cancer cell migration and proliferation, and that ZEB1, BCL10, and caspase-7 are direct target genes of miR-106a and miR-591.
Materials and methods
Cell lines and tissue samples
The human ovarian carcinoma cell line SKOV3 was obtained from American Type Culture Collection (Manassas, VA, USA). Six different PTX-resistant sublines (SKpac-8, -10, -11, -12, -16, and -17) were generated from the parent cell line (SKOV3) by continuous exposure of a stepwise, escalating concentration of PTX over a period of 12 months. SKpac cells were 365.5-fold more resistant to PTX (IC50=7.8 μℳ) than the SKOV3 cell line (IC50=22 nℳ). All cell lines were maintained in McCoy's 5A medium (Gibco/Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum, 100 IU ml−1 penicillin, and 50 μg ml−1 streptomycin in a humidified atmosphere containing 5% CO2 at 37 °C.
Fresh tissue samples from 39 ovarian serous tumours, including six serous benign tumours and 33 malignant serous carcinomas, were obtained at the time of surgery from patients who had undergone oophorectomies for ovarian epithelial tumours. Samples were immediately frozen in liquid nitrogen and stored at −80 °C. The patient's samples were divided into two groups, chemosensitive and chemoresistant, according to the responsiveness to the first-line chemotherapy based on the NCCN guidelines. Informed consent was obtained from each patient prior to surgery and this study was approved by the Ethical Committee of Bundang CHA Medical Center.
MicroRNA extraction and miRNA microarray
For the selection of the candidate miRNAs, the miRNA expression profiles for four PTX-resistant SKpac cell lines (SKpac-8, -10, -11, and -12) and the parent ovarian cancer cell line (SKOV3) were evaluated and compared using a bead-based miRNA microarray (Human-miRNA-V1 Bead Chips; Illumina, San Diego, CA, USA) containing 739 human precursor and mature miRNA oligonucleotide probes. MicroRNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions.
Microarray analysis was conducted following the manufacturer's recommendations. After hybridisation, the arrays were imaged at a resolution of 0.8 μm using a BeadArray Reader. Raw data were extracted using software provided by the manufacturer (Illumina BeadStudio v3.1.3, Gene Expression Module v3.3.8). The selected gene signal values were log-transformed and normalised via the quintile method. Comparative gene expression between SKpac cells and SKOV3 cells was expressed as the fold change. All data analyses and visualisations of differentially expressed genes were conducted using ArrayAssist (Stratagene, La Jolla, CA, USA).
Quantitative real-time PCR with serous carcinoma cell lines and tissue samples
For the validation of miRNA microarray findings, qRT–PCR was conducted on SKOV3 cells, SKpac cells (SKpac-8, -10, -11, -12, -16, and -17), and 39 ovarian tumour samples, including 6 serous benign tumours and 33 malignant serous carcinomas. Total RNA was extracted from fresh tissues and cell lines using TRIzol reagent (Invitrogen). The stem-loop qRT–PCR for mature miRNAs was conducted using a Bio-Rad (Redmond, WA, USA) CFX96 Real-Time PCR Detection System. All PCR reactions were run in triplicate and gene expression relative to RNU48 was calculated using the comparative threshold (Ct) method (2−ΔΔCt).
Transfection of anti-miR inhibitors and pre-miR precursors
Chemically modified RNA-based anti-miR miRNA inhibitors (miR-106a and -96) and pre-miR miRNA precursors (miR-512, -591, -200c, and -203) were purchased from Ambion (Applied Biosystems, Foster City, CA, USA). To transfect cells, 60 pmol of miRNA precursor or 100 pmol of miRNA inhibitor was diluted in 250 μl of serum-free McCoy's 5A media with 5 μl of lipofectamine 2000 (Invitrogen). We used mirVana miRNA inhibitor (for anti-miR transfection) or mirVana miRNA mimic (for pre-miR transfection) (Ambion) for the negative control. To validate the efficiency of transfection, miRNA expression was examined by qRT–PCR.
TUNEL assay
Cells (2 × 107) were fixed with 75% ethanol for 2 h at −20 °C. They were incubated in 0.1% Triton X-100 and 0.1% sodium citrate for 2 min on ice. Apoptotic cells were analysed using the In Situ Cell Death Detection kit (Roche, Mannheim, Germany) and detected by fluorescence-activated cell sorting (FACS) using a flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Colony-forming assay
Cells were seeded at 1 × 105 cells per well in six-well plates. The next day, cells were transfected with miRNA inhibitors or precursors and incubated for 48 h. Transfected cells were then replated at 300 cells per well in a gelatin-coated six-well culture dish. After 14 days, colonies were fixed with 4% paraformaldehyde for 10 min and then visualised using hematoxylin and counted. Groups of >50 cells were scored as colonies.
Cell migration assay
Cell migration was evaluated using the Oris Cell Migration Assay kit (Platypus Technologies, Madison, WI, USA). Cells were plated (2.5 × 105 cells per well) in six-well plates. Twenty-four hours later, cells were transfected with miRNA inhibitors or precursors and incubated for an additional 48 h. Transfected cells were then replated at 2.5 × 104 cells per well in a collagen-coated migration well. The next day, the stoppers were removed to create a detection zone. After 20 h, cells were visualised using hematoxylin, and were counted under a microscope.
Immunoblotting
Cells were lysed in RIPA buffer (Biotech, Seoul, Korea) and immunoblotting was performed. Primary antibodies were incubated overnight at 4 °C as follows: β-actin, 1 : 10 000 (Cell Signaling Technologies, Danvers, MA, USA); ARID4B, 1 : 100 (Abnova, Taipei, Taiwan); RB1, 1 : 500 (Abnova); HIPK3, 1 : 500 (Abcam, Cambridge, UK); ZEB1, 1 : 500 (Abcam); MYLIP, 1 : 1000 (Abgent, San Diego, CA, USA); p21, 1 : 1000 (Epitomics, Burlingame, CA, USA); Caspase-7, 1 : 1000 (Abnova), and BCL10,1 : 1000 (Epitomics).
qRT–PCR array for apoptosis gene
An apoptosis qRT–PCR array (Real-Time Primers, Elkins Park, PA, USA) was used to analyse the expression of apoptosis-related genes. For first-strand cDNA synthesis, 1 μg of total RNA was reverse-transcribed in a final reaction mix of 20 μl using the iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer's instructions. The qRT–PCR was performed on a CFX96 (Bio-Rad) using SYBR Green PCR Master Mix (Bio-Rad) and universal cycling conditions (10 min at 95 °C, 15 s at 95 °C, and 1 min 60 °C for 49 cycles). The fold change in gene expression for all the genes was calculated using the Ct method (2−ΔΔCt).
Luciferase assay
The luciferase assay was performed to assess whether miR-591 could directly repress ZEB1, and whether miR-106a directly represses caspase-7 or BCL10 in ovarian cancer cells. The 3′-UTRs of ZEB1, caspase-7, and BCL10 were amplified by PCR from genomic DNA of SKpac cells. The putative binding sites for miR-591 within the 3′-UTR of ZEB1, and for miR-106a within 3′-UTRs of caspase-7 or BCL10 were identified using the Targetscan algorithm (targetscan.org). The wild-type or mutated binding sites were cloned separately into the Nhel and Xhol sites of the pGL3-control vector (Ambion). The pGL3-control (100 ng) and pRL-TK plasmids (5 ng; for normalisation) were transfected into SKpac cells seeded in 24-well plates (3 × 104 cells per well). Synthetic pre-miR-591 or pre-miR-106a (Ambion) of 20–60 nℳ was added to the above reactions. Luciferase activity was measured after 48 h on an Infinite 200pro series luminometer (Tecan Group, Zurich, Switzerland) using the Dual-Luciferase reporter assay system (Promega, Mannheim, Germany) according to the manufacturer's instructions. All experiments were performed in triplicate and normalised to Renilla luciferase activity.
Statistical analysis
The significant differences between groups were determined using the Student's t-test. Statistical analysis to determine the significant differences in the expression of each miRNA between groups of chemosensitive and chemoresistant ovarian carcinomas was conducted using the non-parametric Kruskal–Wallis test. The survival curves of ovarian cancer patients were estimated via the Kaplan–Meier method, and the resulting curves were compared using the log-rank test. P-values of <0.05 were considered statistically significant. Statistical analysis was performed using the SAS statistics software package (SAS Enterprise Guide 4.1).
Results
MiRNA expression patterns by microarray in PTX-resistant ovarian cancer cells
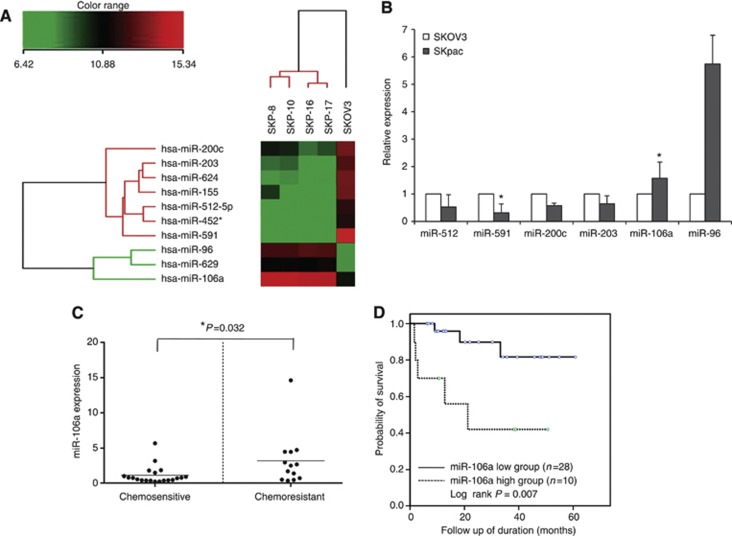
MiRNA expression in four PTX-resistant cell lines (SKpac-8, -10, -11, and -12) was compared with that of the parent SKOV3 ovarian cancer cell line. The expression profiles were determined using a bead-based miRNA microarray containing 739 human precursor and mature miRNA oligonucleotide probes. Ten miRNAs (three upregulated and seven downregulated) were determined to be deregulated by >10-fold in PTX-resistant SKpac cells as compared with the parent, PTX-sensitive SKOV3 cells (Table 1), and a tree was generated via hierarchical clustering analysis (Figure 1A). Of these 10 miRNAs, 6 miRNAs were statistically different: miR-96 and miR-106a were significantly upregulated, and miR-591, miR-512-5p, miR-203, and miR-200c were significantly downregulated (P<0.05). The microarray data were prepared according to minimum information about a microarray experiment (MIAME) recommendations and deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The GEO accession number for the platform is GSE43598.
Table 1. Significantly altered miRNAs by microRNA microarray in PTX-resistant SKpac cells compared with parent SKOV3 cells.
| miRNA | Fold | s.d. | P-value | Chromosome | Putative target genes |
|---|---|---|---|---|---|
| miR-96 |
20.69 |
0.25 |
0.00 |
7q32.2 |
CCNG1, RAB35, FOXO1, CASP2, SOX5, ABCD1, PPP1R9B, ATG7, RASA1, CCND2 |
| miR-106a |
16.65 |
0.54 |
0.02 |
Xq26.2 |
RB1, VEGFA, HIPK3, CDKN1A/p21, ARID4B, Mylip, BCL10, CASP7 |
| miR-629 |
10.6 |
1.25 |
>0.05 |
15q23 |
TP53INP2, PERP, CFLAR, RASSF8, TNFRSF10D, PTPRB, CARD8, RAB12, ZEB1, AKTIP |
| miR-591 |
−327 |
0.25 |
0.00 |
7q21.3 |
ZEB1, MOXD1, CXorf41, RANBP9, PGGT1B, SPARC |
| miR-624 |
−41.22 |
6.07 |
>0.05 |
14q12 |
PPP6R3, CASP10, RRAS2, IL20, RAB22A, SMAD2, PTPN22, JAG2, CADM2, KLF4, APAF1 |
| miR-512-5p |
−37.22 |
0.67 |
0.00 |
19q13.42 |
BCL2L2, TERT, CTNNB1, CDK6, TAB3, CEND1, CDK3, NOTCH3, CCNL2 |
| miR-155 |
−40.76 |
21.31 |
>0.05 |
21q21.3 |
MYB, FOS, MAPK14, FGF7, WEE1, RAPH1, RAB3B, TP53INP1, KLF9, CARD11, RHEB |
| miR-203 |
−25.13 |
1.51 |
0.05 |
14q32.33 |
RASAL2, MAP3K13, NFYA, JMY, TCF4, SRC. ZEB1, SMAD9, BCL7A, BCL11B, FOXK1 |
| miR-452 |
−16.97 |
2.97 |
>0.05 |
Xq28 |
BAG4, ITGA9, RAB3B, CDK6, TBK1, RHOU, NOTCH2NL, TNF, IL3, TNFAIP8, CASD1, SMAD4 |
| miR-200c | −12.8 | 0.69 | 0.02 | 12p13.31 | ZEB1, ZEB2, LRP1B, WIPF1, PTPRZ1, ELAVL2, DUSP1, USP25 |
Figure 1.
(A) Hierarchical clustering of miRNA expression profiles by miRNA microarray. Unsupervised hierarchical clustering analysis of miRNAs that exhibited a statistically significant (P<0.05) increase or decrease in PTX-resistant SKpac cells compared with PTX-sensitive SKOV3 cells. The level of miRNA expression is color-coded. (B) Validation by qRT–PCR of candidate miRNAs that showed significantly different expression by miRNA microarray (see A). The bar graph shows the expression of each microRNA in SKpac cells (SKpac-8, -10, -11, -12, -16, and -17) relative to the average expression level in SKOV3 cells (*P<0.05). (C) Expression of miR-106a by qRT–PCR in chemosensitive and chemoresistant ovarian cancer tissues. Relative expression levels are shown normalised to benign tumours. The inner bar in each graph represents the mean value. The expression level in the chemoresistant ovarian cancers was significantly higher (3.2-fold than that in benign tissue) than in the chemosensitive cancers (1.12-fold than that in benign tissue; P=0.032). (D) Kaplan–Meier OS curve. The higher expression group of miR-106a (more than two-fold than that of benign tumour) demonstrated a significantly worse OS (P=0.007) in patients with ovarian serous carcinomas.
Real-time RT–PCR validation of the six significantly different miRNAs in chemoresistant ovarian cancer cells and tissues and their clinical implication
Microarray data were first validated by qRT–PCR in ovarian cancer cells and tissues. Comparing the expression levels of the six differentially expressed miRNAs stated above by qRT–PCR confirmed that miR-106a (1.6-fold) and miR-96 (5.7-fold) were upregulated in SKpac cells compared with that of SKOV3 cells, whereas miR-591 (0.32-fold), miR-512 (0.53-fold), miR-203 (0.64-fold), and miR-200c (0.58-fold) were downregulated (Figure 1B).
To further investigate whether the above-identified miRNAs are associated with chemoresistant ovarian cancers ex vivo, qRT–PCR was performed on 6 benign serous tumours and 33 serous carcinomas (of which 20 were chemosensitive and 13 were chemoresistant). The miRNA expression in both chemoresistant and chemosensitive carcinomas was compared with that of benign tumours. Of the six miRNAs examined, miR-106a was clearly increased in 69% (9 out of 13 cases) of chemoresistant carcinomas (mean value: 3.2-fold), whereas it was increased only in 25% (5 out of 20 cases) of chemosensitive carcinomas (mean value: 1.12-fold) (P=0.032; Figure 1C). Expression of the remaining miRNAs did not significantly differ between these two groups (Supplementary Figure S1).
We also performed the correlation of patient's survival and miRNAs expression to evaluate their clinical impact. The patients were divided into two groups, the higher-expressing group and the lower-expressing group, by the cutoff value of two-fold expression relative to that in the benign serous tumours. Follow-up was available for all 33 patients with serous carcinomas and the mean follow-up of the study population was 29 months (range: 2–80 months). Eight patients (24%) died of disease during the follow-up period. A statistically significant difference in OS was noted (Figure 1D) between the higher-expressing and lower-expressing groups of miR-106a (OS: 50% vs 87%, P=0.007).
Manipulation of miR-106a and miR-591 increased PTX-induced apoptosis in SKpac cells
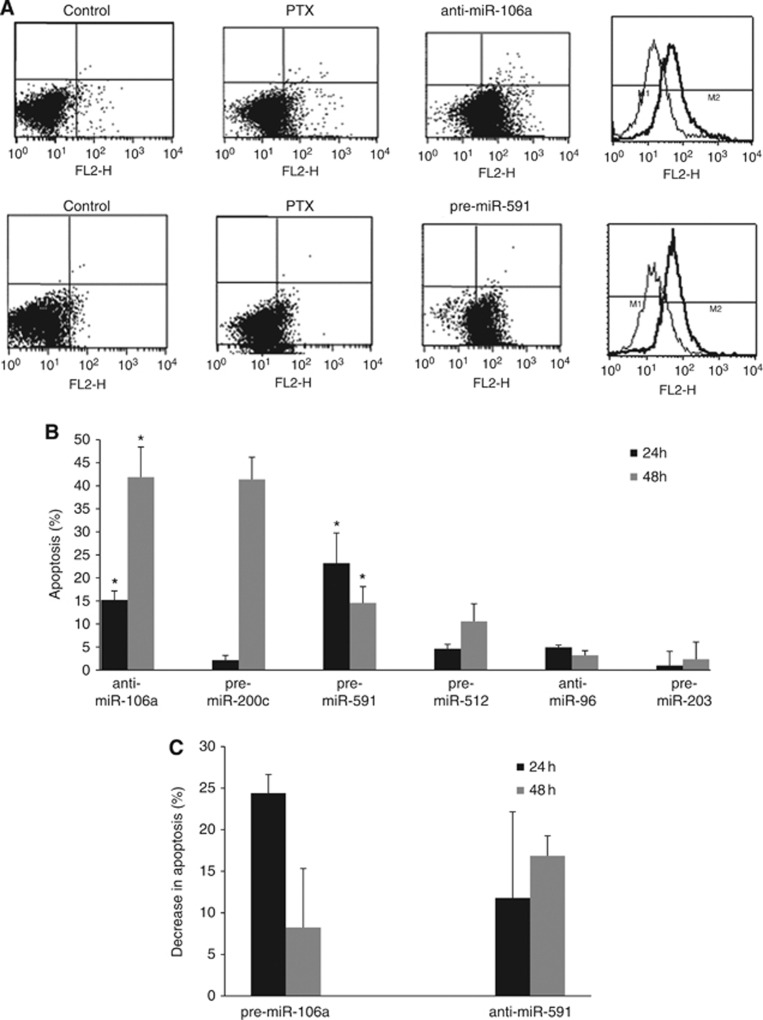
To assess whether miRNA modulation would affect the chemosensitivity of PTX-resistant SKpac cells, PTX-induced apoptosis was examined by TUNEL assay. SKpac cells (SKpac-10, -16 and -17) were transfected with precursors or inhibitors of the six significantly deregulated miRNAs and treated with 80 nℳ PTX. Apoptosis was evaluated by flow cytometry and compared with that of PTX-treated control miRNA-transfected cells. More than 90% of the endogenous miRNA expression was downregulated by the inhibitor, and a >20 000-fold increase in miRNA expression was induced by the precursors (Supplementary Figure S2). PTX-induced apoptosis increased by 15% and 23% at 24 h, and 42% and 15% at 48 h after transfection with anti-miR-106a and pre-miR-591, respectively (P<0.05; Student's t-test), compared with that of control miRNA-transfected SKpac cells (Figure 2A and B). No significant differences were observed in response to transfection with the other miRNA precursors (miR-512 and miR-203) or inhibitor (miR-96), except with pre-miR-200c at 48 h. To confirm whether miR-106a and miR-591 have a direct function in the development of PTX resistance, a gain-of-function approach was used in PTX-sensitive parental SKOV3 cells, which express relatively low levels of miR-106a and high level of miR-591. A TUNEL assay revealed that SKOV3 cells transfected with pre-miR-106a and anti-miR-591 prior to PTX treatment exhibited a marked decrease in apoptosis (8–25%) compared with PTX-treated, control miRNA-transfected cells (Figure 2C).
Figure 2.
TUNEL assay in SKpac cells after transfection of anti-miR-106a or pre-miR-591. (A) Representative graphs of TUNEL assay. Transfection anti-miR-106a or pre-miR-591 markedly increases apoptosis of PTX-resistant SKpac cells following 80 nℳ PTX treatment. (B) The graph represents the mean increase in apoptosis after transfection of each miRNA following PTX treatment compared with PTX-treated, control miRNA-transfected cells (*P<0.05). (C) The graph represents the mean decrease in PTX (20 nℳ)-induced apoptosis in chemosensitive parental SKOV3 cells after transfection with pre-miR-106a, or anti-miR-591. The graph represents the mean±standard error of triplicate experiments.
Alteration of apoptosis-related gene expression by miR-106a and miR-591
To determine which genes or pathways are involved in the regulation of apoptosis by these miRNAs, a qRT–PCR array was performed before and after transfection of anti-miR-106a and pre-miR-591 in SKpac cells (SKpac-10, -16 and -17). Of 84 apoptosis-related genes, 14 pro-apoptotic genes were significantly increased after transfection of anti-miR-106a or pre-miR-591 (Table 2), including members of the TNF ligand and receptor families, the caspase family, DNA damage-associated genes, and BCL10. All of these genes were downregulated in the PTX-resistant SKpac cells compared with the chemosensitive SKOV3 cells, and they were statistically significant except FADD and TNFSF9. The dominant pro-apoptotic genes were TNFRSF10A for anti-miR-106a (30.68-fold), and caspase-8 for pre-miR-591 (9.15-fold). Anti-apoptotic genes were not significantly altered by manipulation of these miRNAs.
Table 2. Apoptosis-related genes significantly altered (>1.5-fold) by modulation of miR-106a, and miR-591.
| |
|
Fold change |
||
|---|---|---|---|---|
| Gene symbol | Gene | Anti-miR106aa (P-value) | Pre-miR-591a (P-value) | SKpac/SKOV3b (P-value) |
|
Groups 1–3: TNF ligand, TNFR, and TRAF families | ||||
| FADD | Fas(TNFRSF6)-associated via death domain | 3.82 (0.047) | — | −1.24 (0.903) |
| LTBR | Lymphotoxin beta receptor (TNFR superfamily member 3) | — | 4.12 (0.045) | −14.21 (0.030) |
| TANK | TRAF family member-associated NFKB activator | 2.16 (0.010) | — | −6 (0.005) |
| TNFRSF10A | Tumour necrosis factor receptor superfamily, member 10a | 30.68 (0.029) | 2.50 (0.048) | −33.42 (0.042) |
| TNFRSF10B | Tumour necrosis factor receptor superfamily, member 10b | 2.46 (0.021) | — | −3.93 (0.031) |
| TNFSF9 | Tumour necrosis factor(ligand) superfamily, member 9 | 2.66 (0.017) | — | −2.14 (0.149) |
| TNFSF13 |
Tumour necrosis factor(ligand) superfamily, member 13 |
1.73 (0.010) |
— |
−15.14 (0.002) |
|
Group 4: Bcl-2 family | ||||
| BCL10 |
B-cell CLL/lymphoma 10 |
5.32 (0.014) |
2.22 (0.000) |
−4.28 (0.050) |
|
Group 5: Caspase and CARD families | ||||
| CASP3 | Caspase-3, apoptosis-related cysteine protease | — | 3.70 (0.027) | −6.56 (0.000) |
| CASP6 | Caspase-6, apoptosis-related cysteine protease | 1.96 (0.026) | 2.24 (0.023) | −2.75 (0.048) |
| CASP7 | Caspase-7, apoptosis-related cysteine protease | 2.73 (0.034) | — | −1.91 (0.021) |
| CASP8 |
Caspase-8, apoptosis-related cysteine protease |
— |
9.15 (0.005) |
−5.80 (0.041) |
|
Group 6: DNA damage | ||||
| TP53 | Tumour protein p53 | 2.94 (0.000) | — | −3.07 (0.003) |
| TP73L | Tumour protein p73-like | 1.57 (0.021) | — | −9.44 (0.016) |
Abbreviation: TRAF=TNF receptor-associated factor.
Mean fold change compared with the mean level of SKpac cells before transfection of each miRNA.
Expression fold of SKpac cells in comparison to SKOV3 cells.
Manipulation of miR-106a and miR-591 impaired cell migration and proliferation in PTX-resistant SKpac cells
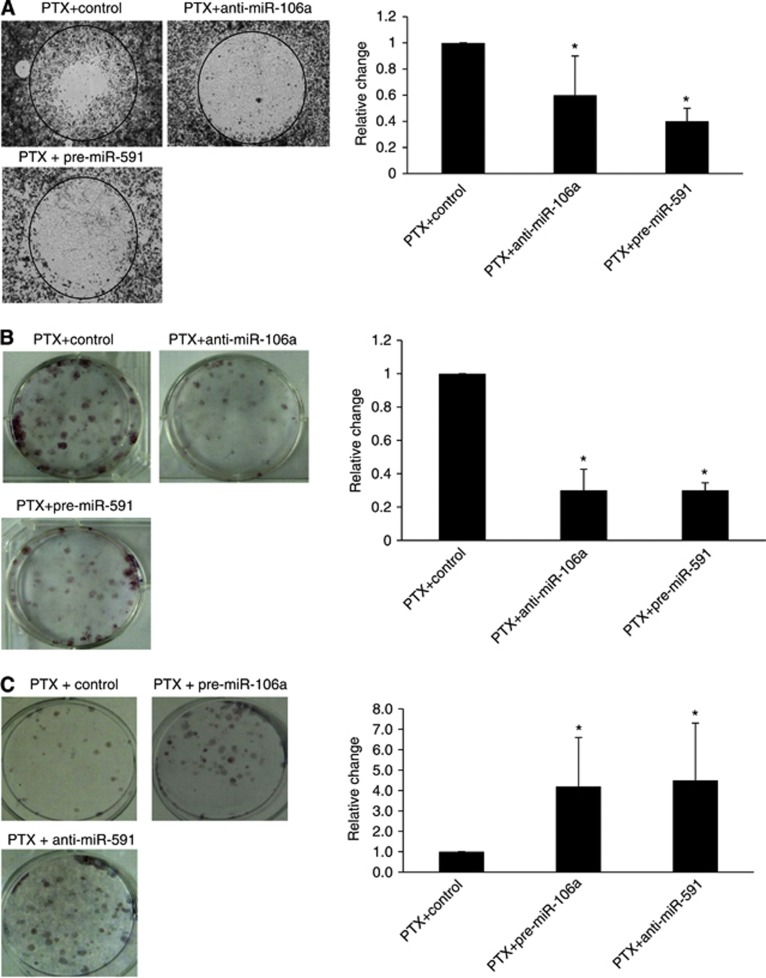
To determine if modulation of the candidate miRNAs renders SKpac cells less migratory, an Oris Cell Migration Assay was performed on SKpac cells (SKpac-10,16, 17). The number of migrating cells decreased by 43% and 56% in response to PTX treatment in cells transfected with anti-miR-106a and pre-miR-591, respectively, compared with PTX-treated, control miRNA-transfected cells (Figure 3A).
Figure 3.
Cell migration and colony-forming assay. (A) Cell migration across collagen-coated wells was assessed using the Oris Cell Migration Assay kit. Migration of PTX-treated (80 nℳ) SKpac cells transfected with pre-miR-591 or anti-miR-106a was markedly decreased compared with that of contol mRNA-transfected cells. The graph represents the mean±standard error of triplicate experiments (*P<0.05). (B) The mean number of colonies formed by SKpac cells transfected with anti-miR-106a or pre-miR-591 decreased compared with PTX-treated, control miRNA-transfected cells. The graph represents the mean±standard error of triplicate experiments (*P<0.05). (C) Colony formation by PTX-treated SKOV3 cells transfected with pre-miR-106a or anti-miR-591 was markedly increased (4–4.5-fold) compared with PTX-treated, control miRNA-transfected cells (*P<0.05).
To determine if these miRNAs were involved in regulating the inhibition of cell growth by PTX, a colony-forming assay was performed. Transfection with anti-miR-106a and pre-miR-591 led to a 64% and 65% decrease, respectively, in the number of colonies formed by SKpac cells at 48 h, compared with PTX-treated, control miRNA-transfected cells (Figure 3B, P<0.05, t-test).
The function of these miRNAs was confirmed using a gain-of-function approach with PTX-sensitive SKOV3 cells. The number of colonies formed by PTX-treated SKOV3 cells transfected with pre-miR-106a or anti-miR-591 was increased by 4–4.5-fold compared with PTX-treated, control miRNA-transfected cells (Figure 3C).
Identification of potential targets for miR-106a and miR-591 in ovarian cancer cells
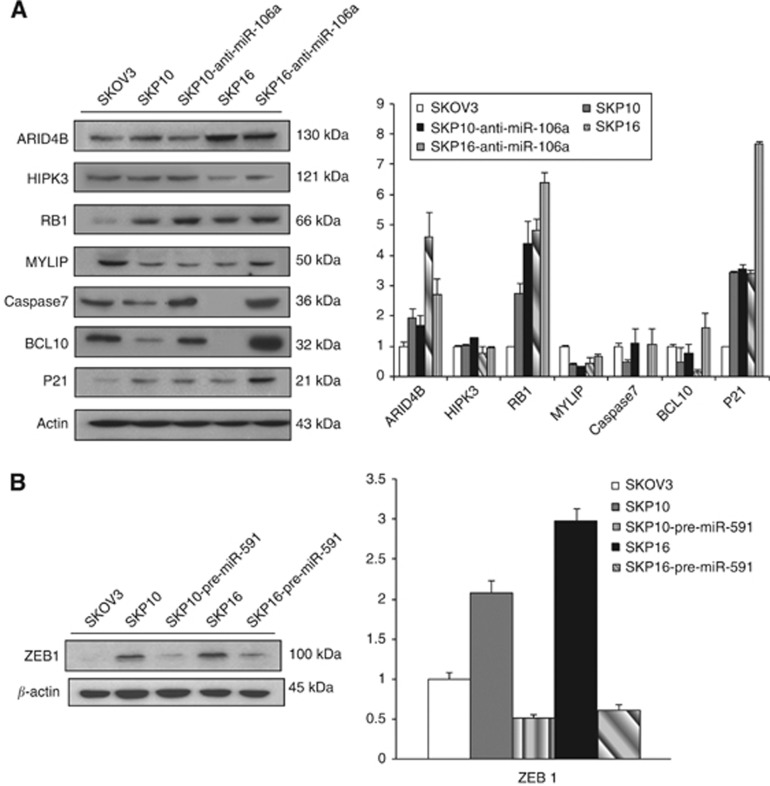
The miRNAs can regulate a large number of target genes, and several databases are available for the prediction of the targets of selected miRNAs. Predicted target genes (Landais et al, 2007; Ivanovska et al, 2008) were identified (Table 1) via Target Scan (http://www.targetscan.org) and PicTar-Vert (http://pictar.mdc-berlin.de). To assess whether these miRNAs directly regulate the putative target genes, the genes related with oncogenesis were selected, and protein expression levels were examined by immunoblotting. The protein levels of RB1, p21, caspase-7 and BCL10 showed a 1.5–2-fold increase after treatment with anti-miR-106a, whereas HIPK3 and MYLIP were not significantly altered (Figure 4A). After treatment with pre-miR-591, ZEB1 level decreased to 0.2-fold, which was 3-fold high in SKpac cells compared with parent SKOV3 cell (Figure 4B).
Figure 4.
Immunoblotting for proteins of putative target genes of miR-106a and miR-591. (A,B) Protein expression of cancer-related, putative target genes of miR-106a (A) and miR-591 (B) was shown by immunoblotting in SKOV3 cells, chemoresistant SKpac cells (SKpac-10 and -16), and SKpac cells transfected with anti-miR-106a or pre-miR-591. The graph represents the relative fold of each protein compared to the level of parental SKOV3 cells. Protein bands were quantitated by densitometric analysis and normalised to β-actin levels. Representative experiment repeated twice with similar results. The protein levels of RB1, p21, BCL10, and Caspase-7 increased two-fold after treatment with anti-miR-106a. ZEB1 expression decreased to 0.2-fold after transfection of pre-miR-591.
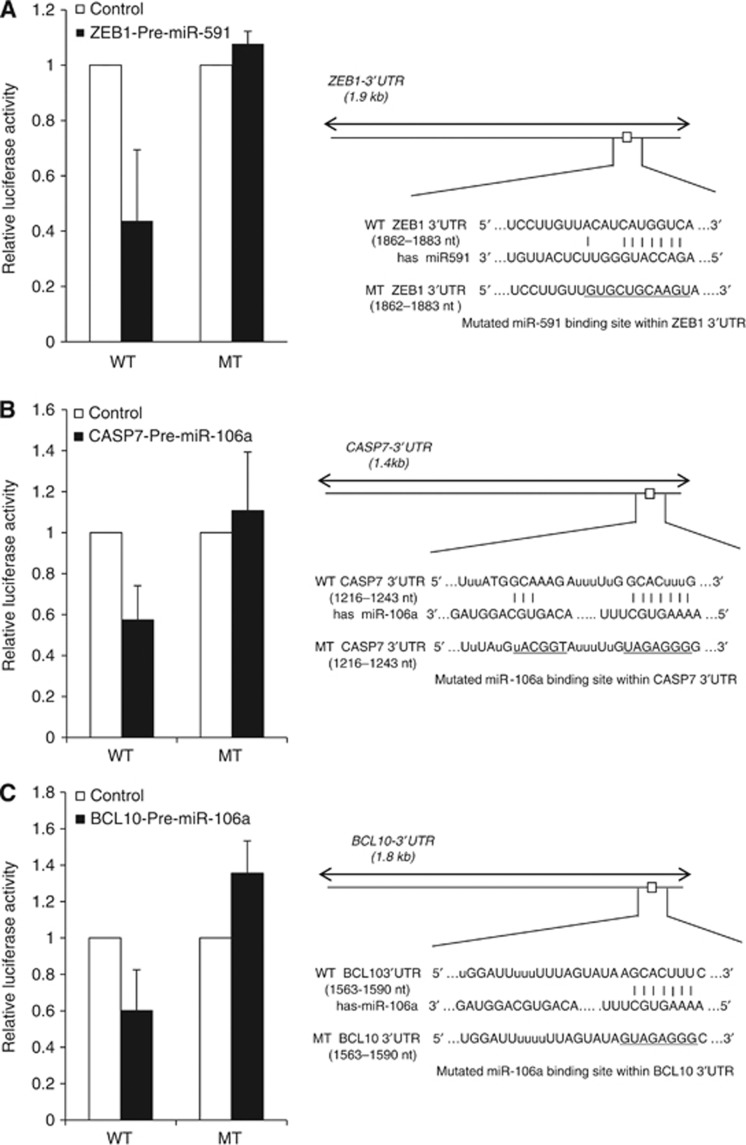
We performed luciferase assay to determine whether miR-591 could directly repress ZEB1, and whether miR-106a directly represses caspase-7 or BCL10 in ovarian cancer cells. The luciferase activity of the reporter construct containing the wild-type ZEB1 3′-UTR was repressed by pre-miR-591 (50%), whereas this miRNA had no effect on the luciferase activity of reporter constructs containing mutant ZEB1 3′-UTR (Figure 5A). The luciferase activity of the reporter construct containing the wild-type caspase-7 or BCL10 3′-UTR was repressed by pre-miR-106a (42% and 40%), whereas this miRNA had no effect with mutant caspase-7 or BCL10 3′-UTR (Figure 5B and C). These results implicate that ZEB1 is a direct target gene of miR-591, and caspase-7 and BCL10 are direct target genes of miR-106a.
Figure 5.
Luciferase assays for putative target genes of miR-106a and miR-591. The wild-type or mutated binding sites were cloned separately into the pGL3-control vector. The pGL3-control (100 ng) and pRL-TK plasmids (5 ng) were transfected, and synthetic pre-miR-591 or pre-miR-106a was added into SKpac cells. All experiments were performed in triplicate and normalised to Renilla luciferase activity. (A) The luciferase activity of reporter construct containing the wild-type ZEB1 3′-UTR was repressed by pre-miR-591 (50%), whereas this miRNA had no effect on the luciferase activity of reporter constructs containing mutant ZEB1 3′-UTR. (B) The luciferase activity of reporter construct containing the wild-type caspase-7 3′-UTR was repressed by pre-miR-106a (42%), whereas this miRNA had no effect on the luciferase activity with mutant caspase7 3′-UTR. (C) The luciferase activity of reporter construct containing the wild-type BCL10 3′-UTR was repressed by pre-miR-106a (40%), whereas this miRNA had no effect on the luciferase activity with mutant BCL10 3′-UTR. MT, mutant type; WT, wild type.
Discussion
Chemoresistance is one of the most important obstacles to the successful treatment of ovarian cancer (Stordal et al, 2007). However, many evidences indicate that the mechanisms responsible for chemoresistance are likely to be multifaceted and extremely intricate (Stordal et al, 2007; Tumbarello et al, 2012).
MicroRNAs are a recently discovered class of small noncoding RNAs encoded by the genomes of a wide range of multicellular organisms. Accumulating evidence suggests that miRNAs can act as regulators of chemosensitivity (Meng et al, 2006; Si et al, 2007; Blower et al, 2008) in addition to regulating oncogenes or tumour suppressors (Esquela-Kerscher and Slack, 2006; Hwang and Mendell, 2007). With respect to ovarian cancer, the upregulation of miR-214 was reported to promote the survival of ovarian cancer cells and induce resistance to cisplatin (Yang et al, 2008). A recent study using high-throughput analysis of the miRNA profile in PTX-resistant cells (Sorrentino et al, 2008) demonstrated an association between some miRNAs and resistance to specific compounds for chemotherapy. Another study using a miRNA microarray with ovarian cancer tissue identified seven miRNAs differentially expressed in platinum-resistant ovarian tumours (Eitan et al, 2009). However, these previous studies only compared the expression profiles of drug-resistant and drug-sensitive cells or tissues, without functional validation documenting the cellular consequences induced by targeted inactivation or re-expression of candidate miRNAs.
In the present study, we screened and selected putative miRNAs affecting PTX resistance in ovarian cancer using a miRNA microarray and qRT–PCR, and found that the upregulation of miR-106a and downregulation of miR-591 were associated with PTX resistance. We confirmed that the modulation of these miRNAs in PTX-resistant ovarian cancer cells resensitized the cells to PTX, through analyses of apoptosis, cell migration, and colony formation. This was supported by a gain-of-function approach showing that the experimental upregulation of miR-106a or downregulation of miR-591 in chemosensitive parental SKOV3 cells led to decrease sensitivity to PTX.
The oncogenic potential of miR-106a has been reported in colon, pancreas, and prostate carcinomas in a large-scale miRNA microarray analysis (Volinia et al, 2006). However, the oncogenic function of miR-106a in ovarian cancer has not yet been reported, and its downregulation has been previously connected to platinum resistance (Sorrentino et al, 2008; Boren et al, 2009). In the present study, however, we observed for the first time that chemoresistance and poor survival of the patient were significantly associated with upregulation of miR-106a, and that experimental downregulation of miR-106a increased the apoptotic, anti-proliferative, and anti-migratory effects of PTX on PTX-resistant ovarian cancer cells.
A role for miR-591 in oncogenesis had not been described until recently, when it was reported that miR-591 suppresses the growth of orthotopic neuroblastoma xenografts (Shohet et al, 2011). The present study also supports that miR-591 is a tumour suppressor, by showing that the restoration of miR-591 increased apoptosis and impaired cancer cell proliferation and migration. In addition, we found that ZEB 1 is a direct target gene of miR-591, by immunoblotting and luciferase assay. The ZEB factors are transcriptional repressors that induce the epithelial–mesenchymal transition (Nakahata et al, 2010; Ohashi et al, 2010; Takeyama et al, 2010) by suppressing the expression of many epithelial genes, including E-cadherin (Vandewalle et al, 2009). The acquisition of mesenchymal phenotype is critically associated with chemoresistance (Adam et al, 2009; Ahmed et al, 2010; Marchini et al, 2012), as well as tumour cell motility and invasion. Taken together, the restoration of miR-591 reverses the sensitization of drug-resistant ovarian cancers to PTX, and reduces cancer cell migration and proliferation, probably by the repression of its target genes, ZEB1. This study is the first to report that miR-591 is associated with PTX resistance, and that ZEB1 is a direct target of miR-591.
As apoptotic cell death is an important factor determining the sensitivity of cancer cells to chemotherapeutic drugs, we also examined which genes or pathways were involved in the regulation of apoptosis by these miRNAs. The TNF ligand/receptor and caspase families were significantly increased by modulation of these miRNAs, with the highest upregulation of TNFRSF10A (miR-106a) and caspase-8 (miR-591). Interestingly, most of these genes were significantly downregulated in PTX-resistant SKpac cells, which showed high miR-106a and low miR-591 expression compared with the chemosensitive SKOV3 cells, suggesting that these genes might be regulated by these miRNAs directly, or indirectly. Among these genes, BCL10 and caspase-7 were direct target genes of miR-106a according to the Target Scan (http://www.targetscan.org), which was confirmed by immunoblotting and luciferase assay. The other genes were likely to be indirectly regulated by these miRNAs. This result is in accordance with the previous study demonstrating that protein expressions of BCL-2 and BCL10 were significantly reduced in PTX-resistant ovarian cancer cells induced by long-term erythropoietin treatment (Solar et al, 2008), and that inactivation of BCL10 was not related with gene mutations in ovarian cancer (Bertoni et al, 1999). Taken together, it is postulated that BCL10 is regulated by miR-106a in ovarian cancer cells, and the chemoresistance associated with miR-106a may be induced by direct regulation of BCL10 and/or caspase-7.
Overall, we found that miR-106a and miR-591 have a critical role in the development of PTX resistance in ovarian cancer cells, and that the regulation of these miRNAs can resensitize PTX-resistant cancer cells through the increase of many pro-apoptotic genes of TNF ligand/receptor and caspase families, and by directly targeting ZEB1, BCL10, and caspase-7. Our results suggest that the modulation of these miRNAs may be a promising therapeutic strategy for overcoming PTX resistance in ovarian cancer.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2009-0070433 and NRF-2012-R1A1B3004095).
Author Contributions
Jin Heong Huh and Tae Heon Kim: searched the target scan for microRNAs, and data analysis; Kwangil Kim: microarray analysis; Ji-Ae Song: western blotting; Yoon Jung Jung: luciferase assay; Joo Yeon Jeong: apoptosis TUNEL assay; Mi Jung Lee: PCR array; Yoo Kyong Kim: real-time RT–PCR; Dong Heon Lee: data analysis; Hee Jung An: study design and writing.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, Bar-Eli M, Dinney C. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15 (16:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Abubaker K, Findlay J, Quinn M. Epithelial mesenchymal transition and cancer stem cell-like phenotypes facilitate chemoresistance in recurrent ovarian cancer. Current Cancer Drug Targets. 2010;10 (3:268–278. doi: 10.2174/156800910791190175. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431 (7006:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116 (2:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bertoni F, Luminari S, Intini D, Carobbio S, Codegoni AM, Spataro V, Neri A. Analysis of BCL-10 gene mutations in ovarian cancer cell lines. Ann Oncol. 1999;10 (10:1259. doi: 10.1023/a:1008309631480. [DOI] [PubMed] [Google Scholar]
- Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, Weinstein JN, Sadee W. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008;7 (1:1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- Boren T, Xiong Y, Hakam A, Wenham R, Apte S, Chan G, Kamath SG, Chen DT, Dressman H, Lancaster JM. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol Oncol. 2009;113 (2:249–255. doi: 10.1016/j.ygyno.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351 (24:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- Eitan R, Kushnir M, Lithwick-Yanai G, David MB, Hoshen M, Glezerman M, Hod M, Sabah G, Rosenwald S, Levavi H. Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecol Oncol. 2009;114 (2:253–259. doi: 10.1016/j.ygyno.2009.04.024. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6 (4:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96 (Suppl:R40–R44. [PubMed] [Google Scholar]
- Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, Linsley PS, Cleary MA. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell Biol. 2008;28 (7:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67 (12:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120 (1:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Marchini S, Fruscio R, Clivio L, Beltrame L, Porcu L, Nerini IF, Cavalieri D, Chiorino G, Cattoretti G, Mangioni C, Milani R, Torri V, Romualdi C, Zambelli A, Romano M, Signorelli M, Giandomenico SD, D'Incalci M. Resistance to platinum-based chemotherapy is associated with epithelial to mesenchymal transition in epithelial ovarian cancer. Eur J Cancer. 2012;49 (2:520–530. doi: 10.1016/j.ejca.2012.06.026. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130 (7:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- Nakahata S, Yamazaki S, Nakauchi H, Morishita K. Downregulation of ZEB1 and overexpression of Smad7 contribute to resistance to TGF-beta1-mediated growth suppression in adult T-cell leukemia/lymphoma. Oncogene. 2010;29 (29:4157–4169. doi: 10.1038/onc.2010.172. [DOI] [PubMed] [Google Scholar]
- Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14 (9:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB, Kalabis J, Vega ME, Kalman RA, Nakagawa M, Klein-Szanto AJ, Herlyn M, Diehl JA, Rustgi AK, Nakagawa H. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70 (10:4174–4184. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohet JM, Ghosh R, Coarfa C, Ludwig A, Benham AL, Chen Z, Patterson DM, Barbieri E, Mestdagh P, Sikorski DN, Milosavljevic A, Kim ES, Gunaratne PH. A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res. 2011;71 (11:3841–3851. doi: 10.1158/0008-5472.CAN-10-4391. [DOI] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26 (19:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Solar P, Feldman L, Jeong JY, Busingye JR, Sytkowski AJ. Erythropoietin treatment of human ovarian cancer cells results in enhanced signaling and a paclitaxel-resistant phenotype. Int J Cancer. 2008;122 (2:281–288. doi: 10.1002/ijc.23071. [DOI] [PubMed] [Google Scholar]
- Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111 (3:478–486. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Stordal B, Pavlakis N, Davey R. A systematic review of platinum and taxane resistance from bench to clinic: an inverse relationship. Cancer Treatment Rev. 2007;33 (8:688–703. doi: 10.1016/j.ctrv.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Takeyama Y, Sato M, Horio M, Hase T, Yoshida K, Yokoyama T, Nakashima H, Hashimoto N, Sekido Y, Gazdar AF, Minna JD, Kondo M, Hasegawa Y. Knockdown of ZEB1, a master epithelial-to-mesenchymal transition (EMT) gene, suppresses anchorage-independent cell growth of lung cancer cells. Cancer Lett. 2010;296 (2:216–224. doi: 10.1016/j.canlet.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello DA, Temple J, Brenton JD. ss3 integrin modulates transforming growth factor beta induced (TGFBI) function and paclitaxel response in ovarian cancer cells. Mol Cancer. 2012;11:36. doi: 10.1186/1476-4598-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66 (5:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103 (7:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter WE, 3rd, Maxwell GL, Tian C, Carlson JW, Ozols RF, Rose PG, Markman M, Armstrong DK, Muggia F, McGuire WP. Prognostic factors for stage III epithelial ovarian cancer: a Gynecologic Oncology Group Study. J Clin Oncol. 2007;25 (24:3621–3627. doi: 10.1200/JCO.2006.10.2517. [DOI] [PubMed] [Google Scholar]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123 (2:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68 (2:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.