Abstract
Background:
Oesophageal adenocarcinomas often show resistances to chemotherapy (CTX), therefore, it would be of high interest to better understand the mechanisms of resistance. We examined the expression of heat-shock proteins (HSPs) and glucose-regulated proteins (GRPs) in pretherapeutic biopsies of oesophageal adenocarcinomas to assess their potential role in CTX response.
Methods:
Ninety biopsies of locally advanced adenocarcinomas before platin/5-fluorouracil (FU)-based CTX were investigated by reverse phase protein arrays (RPPAs), immunohistochemistry (IHC) and quantitative RT–PCR.
Results:
CTX response strongly correlated with survival (P=0.001). Two groups of tumours with specific protein expression patterns were identified by RPPA: Group A was characterised by low expression of HSP90, HSP27 and p-HSP27(Ser15, Ser78, Ser82) and high expression of GRP78, GRP94, HSP70 and HSP60; Group B exhibited the inverse pattern. Tumours of Group A were more likely to respond to CTX, resulting in histopathological tumour regression (P=0.041) and post-therapeutic down-categorisation from cT3 to ypT0–T2 (P=0.040). High HSP60 protein (IHC) and mRNA expression were also associated with tumour down-categorisation (P=0.016 and P=0.004).
Conclusion:
Our findings may enhance the understanding of CTX response mechanisms, might be helpful to predict CTX response and might have translational relevance as they highlight the role of potentially targetable cellular stress proteins in the context of CTX response.
Keywords: oesophageal adenocarcinoma, chemotherapy response, heat-shock proteins, glucose-regulated proteins, reverse phase protein array (RPPA)
Adenocarcinoma of the oesophagus is a very aggressive tumour with increasing incidence, particularly in countries of the Western world. For locally advanced tumours, neoadjuvant chemo(radio)therapy is now frequently utilised as an integral part of a multimodal treatment regimen, and this treatment provides a survival benefit when compared with surgery alone (Lordick et al, 2004a; Sjoquist et al, 2011; van Hagen et al, 2012). However, approximately half of the patients show no obvious response to chemotherapy (CTX). Therefore, the pretherapeutic identification of non-responding patients would be valuable and help patients avoid inefficient therapy, toxic side effects and costs. For oesophageal adenocarcinoma, several studies have concentrated on the analysis of CTX-related markers (Fareed et al, 2009). Despite increasing knowledge regarding the molecular background of this cancer entity, no valid biomarkers have been identified to predict prognosis or CTX response in clinical routine.
Previously, we demonstrated that the regulation and expression of several molecular chaperones, such as heat-shock proteins (HSPs) and glucose-regulated proteins (GRPs), may have an important impact on the biology of this tumour with respect to prognosis (Langer et al, 2008a; Slotta-Huspenina et al, 2012a) and response to CTX (Langer et al, 2008b). In this study, we further elucidated the role of these regulatory proteins in the context of CTX response.
We performed a tissue-based, comprehensive expression analysis of the HSPs HSP60, HSP70, HSP90 and HSP27, including the phosphorylated forms p-HSP27(Ser15), p-HSP27(Ser78) and p-HSP27(Ser82), and the GRPs GRP94 and GRP78 in a well-characterised collection of pretherapeutic biopsies of patients with oesophageal adenocarcinoma. These patients were treated with platin/5-FU neoadjuvant CTX before surgery (Lordick et al, 2004a; Bader et al, 2008). We analysed the expression of these molecules using reverse phase protein array (RPPA) technology, which allows quantitative analysis of protein expression from formalin-fixed, paraffin-embedded tissue and, in addition, by immunohistochemistry (IHC) and quantitative real-time PCR (qRT–PCR). The results of the expression studies were then correlated with tumour response to neoadjuvant CTX treatment.
Materials and methods
Ethics Statement
All patients provided informed written consent, and the study was approved by the Ethics Committee of the Technische Universität München, Munich, Germany (No. 2056/08).
Patient characteristics and tissue specimens
Formalin-fixed and paraffin-embedded tumour samples from 90 patients with locally advanced oesophageal adenocarcinomas (cT3-T4) were investigated. These patients were treated between 1995 and 2009 in the Department of Surgery at Klinikum Rechts der Isar der Technischen Universität München, Germany. The median age of the patients was 62 (range 33–82) years. Eight female (8.9%) and 82 male patients (91.1%) were included in the study. Histopathological examination of the diagnostic biopsies revealed adenocarcinoma with a tumour differentiation grade two (G2, moderately differentiated) in 40 cases (46%) and three (G3, poorly differentiated) in 47 cases (54%). For three cases, no valid tumour differentiation grade could be assigned due to unrepresentative material.
Preoperative CTX
Patients were treated with a cisplatin/oxaliplatin- and 5-FU-based CTX (PLF/OLF scheme) without additional radiation or postoperative CTX according to previously published protocols, including the MUNICON trial (Lordick et al, 2004a, 2007). Thirty patients with an age of ⩽60 years and good health status were also given paclitaxel (T-PLF/OLF; Lordick et al, 2007; Bader et al, 2008). Briefly, a total of 50 mg m−2 cisplatin was scheduled to be given as a 1-h intravenous (i.v.) infusion on days 2, 16 and 30. For patients with a glomerular filtration rate of <60 ml kg−1 min−1, oxaliplatin (85 mg m−2 over 2 h) replaced cisplatin. Moreover, 500 mg m−2 leucovorin was applied i.v. over 2 h on days 2, 9, 16, 23, 30 and 37, followed by a 2000 mg m−2 i.v. infusion of 5-FU over 24 h. Paclitaxel consisted of 80 mg m−2, given as a 3-h i.v. infusion on days 1, 15 and 29. Seventy-one patients received at least one cycle (61 patients two cycles, 10 patients one cycle). Following the schedule of the MUNICON trial, 19 patients had early therapy abort after 2 weeks due to metabolic non-response.
Surgery
A right abdominothoracic approach using an intrathoracic anastomosis and two-field lymphadenectomy (Ivor Lewis procedure) or a transhiatal approach with cervical anastomosis was performed. Surgery was conducted 2–3 weeks after CTX completion.
Histopathological evaluation of tumour regression
The histopathological response following neoadjuvant CTX was investigated, and the procedure was conducted as previously described with a highly standardised work up of the corresponding resection specimens (Becker et al, 2003; Langer et al, 2009). The tumour regression grading (TRG=tumour regression grade) was based on an estimation of the percentage of vital tumour tissue in relation to the macroscopically determined tumour bed, which was completely worked up histologically. The following system of tumour regression was used: grade 1, complete (0% residual tumour); grade 2, subtotal and partial tumour regression (1–50% residual tumour per tumour bed); and grade 3, minimal or no tumour regression (>50% residual tumour per tumour bed). The slides were reviewed separately by the two pathologists (KB and RL). In case of disagreement, both pathologists reviewed the specimen under a double-headed microscope and reached a consensus diagnosis. According to this grading system, 7 patients (7.8%) had complete tumour regression (TRG 1), and 30 patients (33.3%) had subtotal or partial tumour regression (TRG 2). Fifty-three patients (58.9%) demonstrated no response to neoadjuvant CTX (TRG 3), among them 18 patients with metabolic non-response and early therapy abort. Postoperative tumours were classified as ypT0 in 7 cases (7.8%), ypT1 in 12 cases (13%) and ypT2 in 19 cases (21.1%). Therefore, a tumour down-categorisation from cT3 to ypT0–T2 was observed in 38 cases (42%). Postoperative histopathological findings (TRG, UICC classification and tumour differentiation) are presented in Table 1. Regarding subgroups of patients according to the CTX scheme applied (PLF/OLF vs T-PLF/OLF), there was no difference between the groups in terms of response rate (37% vs 41%) and down-categorisation (40% vs 41%).
Table 1. Postoperative histopathology findings following neoadjuvant chemotherapy.
| Factor | Number of patients | % |
|---|---|---|
| |
90 |
100 |
|
Tumour regression grade (TRG) | ||
| TRG 1 | 7 | 7.8 |
| TRG 2 | 30 | 33.3 |
| TRG 3 |
53 |
58.9 |
|
UICC ypT category | ||
| ypT0 | 7 | 7.8 |
| ypT1 | 12 | 13.3 |
| ypT2 | 19 | 21.1 |
| ypT3 |
52 |
57.8 |
|
Lymph node metastases | ||
| Absent | 31 | 34.4 |
| Present |
59 |
65.6 |
|
Distant Metastases | ||
| Absent | 75 | 83.3 |
| Present |
15 |
16.7 |
|
Tumour grading | ||
| G2 | 40 | 44.4 |
| G3 |
47 |
52.2 |
|
Resection status | ||
| R0 | 74 | 82.2 |
| R1 | 16 | 17.8 |
Abbreviation: uicc=Union for International Cancer Control.
Follow-up
Overall survival (OS) was calculated from the day of surgery to death. One patient died within 1 month after surgery and was excluded from survival analysis. Three patients were lost to follow-up. Forty-three of the 86 remaining patients (50%) died with a median OS of 33 months (95% CI 23–42). The medium follow-up time for surviving patients was 40.2 months (range 5.7–134.6).
Protein extraction and antibodies
Protein extractions were performed as previously described (Wolff et al, 2011b). Briefly, tumour tissue from three 10-μm sections was deparaffinised (addition of xylene twice for 10 min) and rehydrated (using 100%, 90% and 70% ethanol for 5 min each). The tumour tissue was microdissected to obtain a percentage of tumour tissue of at least 80% haemorrhagic and necrotic areas were excluded. Approximately 0.5 cm2 of tissue from three 10-μm-thick sections was processed in 100 μl of extraction buffer (EXB Plus, Qiagen GmbH, Hilden, Germany). Protein concentrations were determined using the Bradford protein assay according to the manufacturer's instructions (Bio-Rad, Hercules, CA, USA). Lysates were probed for β-actin by western blot to verify protein extraction success and the suitability of the material for RPPA analysis. Before performing RPPA for HSP/GRP expression analysis, all antibodies were validated by western blot using protein extracts of formalin-fixed paraffin-embedded tissues. A detailed list of the antibodies used is provided in Table 2.
Table 2. Antibodies used for immunohistochemistry (IHC) and western blot (WB)/RPPA analysis.
| Protein | Antibody | Distributor | IHC Dilution | WB/RPPA |
|---|---|---|---|---|
| HSP27 |
#2402 |
Cell Signaling, Danvers, MA, USA |
1 : 250 |
1 : 1000 |
| Phospho-HSP(Ser15) |
#ab39399 |
Abcam, Cambridge, UK |
1 : 500 |
1 : 1000 |
| Phospho-HSP(Ser78) |
#2405 |
Cell Signaling, Danvers, MA, USA |
ND |
1 : 1000 |
| Phospho-HSP(Ser82) |
#2401 |
Cell Signaling, Danvers, MA, USA |
ND |
1 : 1000 |
| HSP60 |
#ab46798 |
Abcam, Cambridge, UK |
1 : 2000 |
1 : 2000 |
| HSP70 |
#ab17850 |
Abcam, Cambridge, UK |
1 : 1 |
1 : 50 |
| HSP90 |
#ab1429 |
Abcam, Cambridge, UK |
1 : 100 |
1 : 200 |
| GRP78 |
#ab32618 |
Abcam, Cambridge, UK |
1 : 1000 |
1 : 1000 |
| GRP94 | #sc1794 | Santa Cruz Biotechnology Inc., CA, USA | 1 : 5000 | 1 : 500 |
Abbreviations: GRP=glucose-related protein; HSP=heat-shock protein; ND= not done; RPPA=reverse phase protein array.
RPPAs and quantitative protein analysis
RPPAs were generated using the Calligrapher MiniArrayer (Bio-Rad) (Gulmann et al, 2006). Three replicates per lysate and dilution (undiluted, 1 : 2, 1 : 4, 1 : 8, 1 : 16, buffer) were applied to a nitrocellulose-coated glass slide (Grace Bio-Labs, Bend, OR, USA), yielding a total of 18 data points per sample. Peroxidase blocking was performed according to the manufacturer's instructions (Dako, Glostrup, Denmark). Immunodetection was conducted similar to western blot analysis. To estimate the total protein content, arrays were stained in parallel with Sypro Ruby Protein Blot Stain (Molecular Probes, Eugene, OR, USA) according to the manufacturer's instructions. The TIFF images of the antibody-stained slides and Sypro Ruby-stained slides were analysed with MicroVigene 3.5.0.0 software (VigeneTech, Carlisle, MA, USA). The MicroVigene signal-intensity points were calculated by the integral of a logistic four-point fit model that was matched optimally to the 18 data points.
IHC
IHC staining included antigen retrieval, antibody incubation and detection and was performed as described previously (Bauer et al, 2012). A detailed list of the antibodies and dilutions is provided in Table 2. According to the results of a previously published study (Slotta-Huspenina et al, 2012a), we only analysed p-HSP27Ser15 from the group of pHSPs; p-HSP27Ser78 and p-HSP27 Ser82 had been shown to exhibit only very focal immunoreaction, which cannot be considered as representative of whole tumoural expression. We counted at least 500 unequivocally identifiable tumour cells. Expression was assessed based on the intensity of cytoplasmic immunostaining and the percentage of stained tumour cells. The intensity was scored as 0 (negative), 1 (weak staining), 2 (moderate staining) or 3 (strong staining). The percentage of positive tumour cells was scored as 0 (none), 1 (<10%), 2 (10–50%), 3 (51–80%) or 4 (>80%). Multiplication of the scores for intensity and percentage resulted in a semiquantitative immunoreactive score (IRS) ranging from 0 to 12. Two independent observers (JSH and ED) evaluated the tumour staining. Differences were discussed at a double-header microscope to achieve a final consensus.
Quantitative real-time PCR
Microdissection, RNA extraction and cDNA synthesis were performed as described previously with minor modifications (Specht et al, 2001). Following tissue preparation, the microdissected tumour tissue was transferred into a sterile 1.5-ml tube containing RNA lysis buffer. Lysis was conducted at 60 °C for 24 h until the tissue was completely solubilised. RNA was purified by phenol and chloroform extraction, followed by precipitation with an equal volume of isopropanol in the presence of 20 μl of 2 ℳ sodium acetate (pH 4.0) and 2 μl of 10 mg ml−1 glycogen at −20 °C. The RNA pellet was washed once in 70% ethanol, dried and resuspended in 20 μl of RNase-free water. One microgram of RNA was transcribed into cDNA using Superscript II reverse transcriptase (Invitrogen, Darmstadt, Germany) and 250 ng of random hexamers (Roche Diagnostics, Penzberg, Germany) in accordance with the manufacturer's recommendations; the final volume of the reaction was 20 μl. Gene expression was quantified using RealTime ready single assays (Roche Diagnostics) for the target genes GRP94 (ID 100489), HSP27 (ID 100497), HSP70 (ID 110730), GRP78 (ID 110805), HSP90 (ID 138013) and HSP60 (ID 137175) and for the housekeeping genes PPIA (ID 102088), ALAS1 (ID 102108) and ACTB (ID 101125), as described previously (Slotta-Huspenina et al, 2012a). Housekeeping genes were selected in a previous study using the RealTime ready Reference Gene Panel (Roche Diagnostics), which contains 19 different reference genes to facilitate the identification of the most suitable genes from 8 different carcinoma samples. Using GeNorm software (Biogazelle, Zwijnaarde, Belgium), the reference genes PPIA, ALAS1 and ACTB were demonstrated to be stably expressed in all the analysed tissues. qRT–PCR was performed in triplicate with the LightCycler 480 Instrument using LightCycler 480 Probes Master (Roche Diagnostics) and 10 ng of cDNA per well. Thermal cycler conditions included 45 cycles at 95 °C for 10 s, 60 °C for 30 s and 72 °C for 1 s. Relative mRNA expression was calculated by the ΔΔCt method using the LightCycler 480 Software and an efficiency-corrected algorithm with standard curves and triple normalisation to the PPIA, ALAS1 and ACTB reference genes.
Statistical analysis
Unsupervised hierarchical clustering was performed using Cluster and TreeView software. Following log transformation and center to median calculations, average hierarchical clustering was performed using Spearman rank correlation (Eisen et al, 1998). SPSS statistical software (IBM SPSS statistics version 21, Ehningen, Deutschland) was used for additional statistical analyses. Associations between the groups of patients are provided in cross tabs, and differences were determined using the χ2-test. Comparisons between groups were performed using non-parametric tests (Mann–Whitney U test, Kruskal Wallis test and Spearman rank correlation). Survival analyses were performed using Kaplan–Meier estimates and log-rank tests. All tests were two-sided, and the significance level was set to P<0.05.
Results
Tumour regression correlates with patients' survival
According to the CTX response classification described above, TRG was prognostic (P=0.001), and complete responders (TRG 1, N=7) and partial responders (TRG 2, N=30) had improved survival (median not reached in both the groups) compared with non-responders (TRG 3, N=53), who had a median OS of 22 months (95% CI 11–33 months). Therefore, patients with complete and partial tumour regression (TRG 1 and 2) were classified as responders, and patients without tumour regression (TRG 3) were classified as non-responders according to their prognosis. Response was associated with postoperative tumour down-categorisation from cT3 to ypT0–T2 (P<0.001), which was observed in 38 patients (42.2%). Patients with tumour down-categorisation had significantly improved outcome (median not reached) compared with patients with ypT3 tumours (N=52); the median OS of patients with ypT3 tumours was only 25 months (95% CI 17–34 months; P=0.007). A better tumour differentiation grade (G2 vs G3) in pretherapeutic biopsies was significantly associated with tumour down-categorisation (P=0.001). Regarding the subgroups of patients according to the CTX scheme applied (PLF/OLF vs T-PLF/OLF), the association between histopathological response/down-categorisation and OS hold true for the PLF/OLF (P=0.002/0.020) and in trend for the smaller T-PLF/OLF-treated patients (P=0.017/0.20).
Expression analysis of HSPs and GRPs by RPPA in pretherapeutic biopsies identifies two patient groups that differ in response to CTX
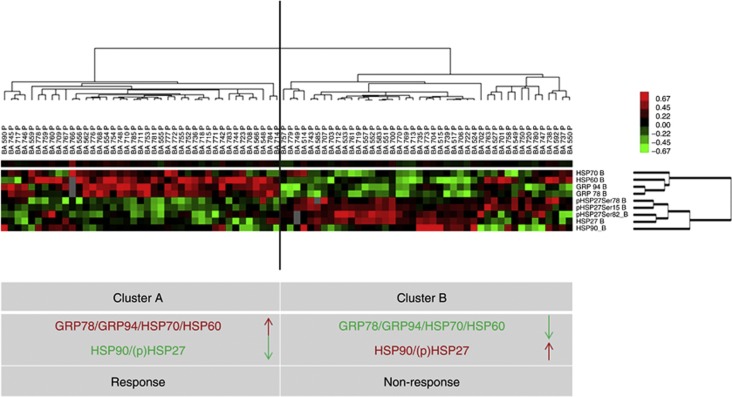
RPPA was performed on 81 pretherapeutic biopsies; in 9 cases, protein lysates could not be further analysed due to limited amounts of extracted proteins. Representative RPPA results for the proteins analysed are provided in Supplementary file S1. All HSP and GRP proteins, including the phosphorylated forms of HSP27, could be detected and were expressed at various levels in tumour biopsies. The median quantitative protein expression (protein/SyproRuby) levels were as follows: HSP27: 164 (range 55–462), p-HSP27(Ser15): 224 (range 52–848), p-HSP27(Ser78): 319 (range 105–1274), p-HSP27(Ser82): 121 (range 32–477), HSP60: 346 (range 87–151), HSP70, 298 (range 110–670), HSP90: 162 (range 75–360), GRP94: 316 (range 94–897), and GRP78: 491 (range 122–1444). In general, there was a significant positive correlation between the protein expression levels of HSP60, HSP70, GRP78 and GRP94 (P<0.001) and between HSP90, HSP27 and p-HSP27(Ser15, Ser78, Ser82) (P<0.001). Unsupervised hierarchical clustering was conducted to generate a tumour-specific protein expression pattern for better understanding of the relationship between the HSP/GRP proteins. This approach is similar to one that was recently published by our group (Slotta-Huspenina et al, 2012a). Two groups of tumours (Group A and Group B) could be clearly distinguished by their specific HSP/GRP protein expression patterns (Figure 1). Group A consisted of 38 tumours characterised by low expression of HSP90, HSP27 and p-HSP27(Ser15, Ser78, Ser82) and high expression of HSP60, HSP70, GRP78 and GRP94. Group B consisted of 43 tumours and displayed an inverse pattern; these cases had low expression levels of HSP60, HSP70, GRP78 and GRP94 and high expression levels of HSP90, HSP27 and p-HSP27(Ser15, Ser78, Ser82). The median expression levels of the single proteins in the two tumour groups are listed in Table 3. Next, we determined whether the identified HSP/GRP protein patterns correlated with CTX responses, particularly with respect to histopathological tumour regression and tumour down-categorisation. Tumours with HSP/GRP-pattern A were more likely to respond to neoadjuvant CTX (Table 4). There was a significant association with better tumour regression (P=0.041) as well as a post-treatment down-categorisation (P=0.040). Moreover, patients with pretherapeutic HSP/GRP-pattern A were more likely to be free of distant metastases (P=0.023). No significant association was observed between the HSP/GRP protein expression patterns and other histopathological characteristics, which included lymph node status (P=0.075) and tumour differentiation (P=0.132). Analysing subgroups of patients according to the CTX regime applied, significant association between HSP/GRP expression pattern and down-categorisation after CTX hold true for the PLF/OLF-treated patients (P=0.035); however, regarding the small subgroup of patients who additionally were treated with Paclitaxel (T-PLF/OLF), there was only a trend similar between GSP/GRP patterns and response/down-categorisation, which did not reach statistical significance (see Supplementary file 2).
Figure 1.
Unsupervised hierarchical cluster analysis of 81 oesophageal adenocarcinomas based on the expression of HSP27, HSP60, HSP70, HSP90, phosphorylated forms of HSP27 (Ser78, Ser82, Ser15), GRP78 and GRP94 measured by RPPAs. This analysis identified two groups of tumours with specific HSP/GRP protein expression patterns (A and B). The hallmark of Group A was high expression levels of HSP60, HSP70, GRP78 and GRP94 and low expression of HSP27, including its phosphorylated form (p-HSP27) and HSP90. Group B showed the inverse of Group A. Cluster colour key: ed – upregulated; green – downregulated; black – unchanged; grey – missing.
Table 3. Median protein expression levels of reverse phase protein arrays (RPPAs) in tumours with HSP/GRP-patterns A and B identified by unsupervised hierarchical cluster analysis.
| Protein | HSP/GRP-pattern A | HSP/GRP-pattern B | P-value | ||
|---|---|---|---|---|---|
|
Median protein expression level (min–max) | |||||
| HSP60 |
560 (188–1508) |
↑ |
290 (87–749) |
↓ |
<0.001 |
| HSP70 |
331 (109–597) |
↑ |
283 (143–671) |
↓ |
0.069 |
| HSP90 |
148 (77–286) |
↓ |
184 (75–361) |
↑ |
0.003 |
| GRP78 |
621 (149–1444) |
↑ |
402 (123–941) |
↓ |
<0.001 |
| GRP94 |
429 (115–899) |
↑ |
253 (95–810) |
↓ |
<0.001 |
| HSP27 |
122 (56–255) |
↓ |
223 (65–463) |
↑ |
<0.001 |
| p-HSP27 (Ser15) |
142 (52–357) |
↓ |
312 (109–766) |
↑ |
<0.001 |
| p-HSP27 (Ser78) |
216 (106–566) |
↓ |
392 (172–1275) |
↑ |
<0.001 |
| p-HSP27 (Ser82) | 82 (33–239) | ↓ | 190 (52–477) | ↑ | <0.001 |
Abbreviations: GRP=glucose-related protein; HSP=heat-shock protein.
Table 4. Association of HSP/GRP patterns A and B (RPPA) with histopathological response (TRG) and ypT category.
| HSP/GRP-pattern A | HSP/GRP-pattern B | Total | P-value | |
|---|---|---|---|---|
| Response | 20 | 13 | 33 | 0.041 |
| Non-response |
18 |
30 |
48 |
|
| Total |
38 |
43 |
81 |
|
| |
|
|
|
|
| Down-categorisation | 21 | 14 | 35 | 0.040 |
| No down-categorisation |
17 |
29 |
46 |
|
| Total | 38 | 43 | 81 |
Abbreviations: GRP=glucose-related protein; HSP=heat-shock protein; RPPA=reverse phase protein array; TRG=tumour regression grade.
Responder: TRG 1/2, Non-responder: TRG 3; Down-categorisation: ypT0–T2, no down-categorisation: ypT3.
Immunoreactivity scores for HSP60 and HSP27 correlate with tumour down-categorisation
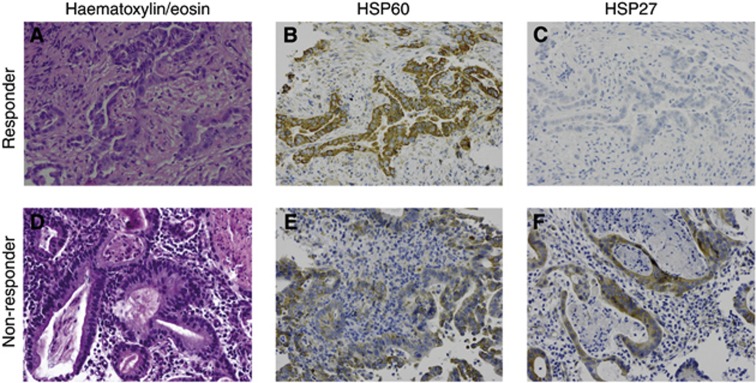
IHC analysis was performed on 82 biopsies according to the criteria given above. For eight cases, tumour cell content was not sufficient for reliable interpretation. Staining intensity scores (IRS) ranged from 0 to 12 for HSP27, HSP60, HSP90, GRP94 and GRP78 and from 0 to 9 for p-HSP27(Ser15) and HSP70. The expression of the single proteins was compared in responders and non-responders. High HSP60 immunoreactivity (cutoff=median=IRS 9) in pretherapeutic biopsies was associated with tumour down-categorisation (P=0.016), and low HSP27 immunoreactivity (cutoff=median=IRS 3) was associated with both down-categorisation (P=0.043) and tumour regression (P=0.012). For the other proteins, no associations were observed between IHC and CTX response. Similar results were obtained when analysing the subgroups of PLF/OLF- and T-PLF/OLF-treated patients (see Supplementary file S2). HSP60 and HSP27 staining are shown for two representative cases (responder and non-responder) in Figure 2.
Figure 2.
IHC staining for HSP60 and HSP27 in pretherapeutic biopsies of two oesophageal adenocarcinoma cases. (A–C) a CTX responder (No.745, TRG 1) and (D–F) a non-responder (No.772, TRG 3), are shown ( × 200): (A) haematoxylin/eosin; (B) very strong HSP60 expression; (C) no HSP27 expression; (D) haematoxylin/eosin; (E) weak HSP60 expression; and (F) strong HSP27 expression.
mRNA levels of HSP60 and HSP70 correlate with tumour down-categorisation
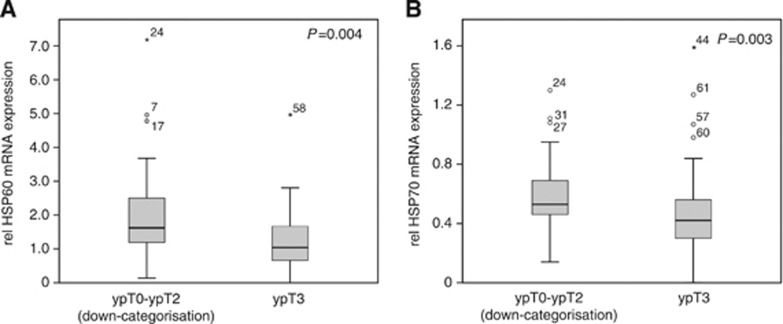
Quantitative real-time PCR analysis was performed on 88 biopsies; in two cases, mRNA extraction was not successful. mRNA from the target genes was detectable at various levels in cancer biopsies from all analysed patients. The relative median mRNA expression levels (ratio of target gene/housekeeping genes) were as follows: HSP27: 0.47 (range 0.1–0.75), HSP60: 1.26 (0.01–7.12), HSP70: 0.48 (0.01–1.59), HSP90: 0.45 (0.01–2.39), GRP78: 0.24 (0.01–1.85), and GRP94: 0.47 (0.01–1.24). The expression level of single HSP/GRP mRNAs was compared between responders and non-responders, but we found no association between any of the mRNAs and histopathological tumour regression. However, higher pretherapeutic HSP60 mRNA levels (P=0.004) and higher HSP70 mRNA levels (P=0.003) were significantly associated with tumour down-categorisation (median HSP60 in yT0-2 tumours:1.62 vs yT3:1.02, median HSP70 in yT0-2 tumours: 0.53 vs yT3: 0.42) (Figure 3). This holds true for the T-PLF/OLF subgroup of patients (P=0.019 and 0.035; see Supplementary file S2).
Figure 3.
Box plot analysis of HSP60 and HSP70 mRNA expression in pretherapeutic biopsies of oesophageal adenocarcinoma. Box plots are illustrating the relative mRNA expression of (A) HSP60 and (B) HSP70 in 88 pretherapeutic biopsies of patients with tumour down-categorisation (ypT0–T2, N=38) and without down-categorisation (ypT3, N=50) after neoadjuvant chemotherapy.
Correlation between RPPA analysis, qRT–PCR and IHC
The two groups identified by RPPA analysis (Groups A and B) exhibited significantly different mRNA expression levels for GRP78 (median relative expression for Group A: 0.30 and for Group B: 0.22, P=0.034) and GRP94 (median relative expression for Group A: 0.56 and Group B: 0.41, P=0.050). HSP60 may also be differentially expressed in these two groups (median relative expression for Group A: 1.45 and for Group B: 1.23, P=0.077). For HSP27, a significant correlation was observed between protein levels (RPPA) and mRNA levels (r=0.335; P=0.002). For HSP70 and HSP90, no correlation was observed between RPPA and qRT–PCR results.
With IHC, tumours with HSP/GRP protein expression pattern A (Group A) had significantly higher IRSs for HSP60 (P=0.010) and a trend for lower p-HSP27 (Ser15) scores (P=0.083). For HSP90, HSP70, GRP94 and GRP78, no correlation was observed between RPPA and IHC.
Between IHC reactive scores and mRNA levels, a significant correlation was detected for HSP90 (P=0.007). For all other analysed proteins, no correlation was observed between IHC staining intensity and mRNA levels.
Discussion
Expression of HSPs and GRPs can be induced in cells following exposure to different insults, allowing cells to survive stress conditions. These proteins act as molecular chaperones either by helping refold damaged proteins or by assisting in their elimination. HSPs and GRPs also have an important role in the regulation of apoptosis (Garrido et al, 2006; Powers et al, 2009). Recently, studies have reported an association of HSPs and GRPs with cancer and demonstrated their important roles in cancer biology. Molecular chaperones have been suggested to influence tumour growth and differentiation and may have a major impact on the clinical outcome of patients (Ciocca and Calderwood, 2005; Calderwood et al, 2006; Fu and Lee, 2006; Lee, 2007; Khalil et al, 2011). Given the important cytoprotective role of HSPs in cancer cell physiology, it is not surprising that HSP expression also has been linked to radio- and CTX resistance (Fu and Lee, 2006; Kuramitsu et al, 2012).
Here, we present a tissue-based study reporting the role of HSPs and GRPs in the CTX response of oesophageal adenocarcinomas. We investigated the expression of the best-known HSPs and GRPs at the protein level by IHC, at mRNA level and by RPPA. We analysed pretherapeutic biopsies of a well-characterised sample collection of oesophageal adenocarcinoma patients. These patients were homogenously treated by neoadjuvant CTX and tumour resection according to the standardised protocols (Lordick et al, 2004a, 2007). Tumour response to neoadujvant treatment was assessed in a highly standardised and elaborated manner by determination of histopathological tumour regression grade (TRG) and tumour stage (Becker et al, 2003; Langer et al, 2009). We identified a specific protein expression pattern by RPPA exhibiting high levels of GRP78, GRP94, HSP70 and HSP60 and low levels of HSP90, HSP27 and p-HSP27(Ser15, Ser78, Ser82); this expression pattern was significantly associated with response to neoadjuvant CTX. In a recently published study on primary resected oesophageal adenocarcinomas, we described a similar HSP/GRP expression pattern (high levels of GRP78, GRP94, HSP60 and a low level of p-HSP27) that was associated with poor prognosis and clinical outcome (Slotta-Huspenina et al, 2012a). This underlines the relevance of the findings of the current study. Both results taken together suggest that the combination of highly expressed GRP78, GRP94, HSP70 and HSP60 and low p-HSP27 expression may, on the one hand, be preferentially found in tumours with an aggressive biological behavior and, on the other hand, in tumours with a higher degree of sensitivity to CTX. However, for the cases analysed in the present study, we were unable to demonstrate that this HSP/GRP expression pattern impacted the prognosis, as the most important and best prognostic factor for OS was TRG, which was histomorphologically assessed in tumour resection specimens after neoadjuvant CTX. Another morphological-based criterium, better tumour differentiation grade using the grading system outlined by the WHO classification of tumours Sobin et al. (2010), highly correlated with post-treatment down-categorisation from cT3 to ypT0–T2 in our cohort. Although there are certain limitations regarding representativity and objectivity in the estimation of histopathological grading (Dikken et al, 2012), currently there are no validated molecular biomarkers in oesophageal adenocarcinomas to predict CTX response. Nevertheless, there is more evidence that the revealed protein pattern is of high relevance in this context. Previously, our group used a proteomic screening approach to explore potential predictive proteins in a small cohort of oesophageal adenocarcinoma patients treated with neoadjuvant CTX (Langer et al, 2008b). In that study, high pretherapeutic expression levels of GRP78, GRP94, HSP60 and HSP27 were associated with response to neoadjuvant CTX. This protein combination coincided with the pattern of the current study (high GRP78, GRP94, HSP70, HSP60 and low HSP90, (p)HSP27) except for one protein – HSP27. The discrepancy of the prediction potential of this marker might be explained by several points: (1) The present study was conducted on a larger sample collection of 90 patients, in contrast to the small sample set of 34 patients included in the former screening study. (2) Different methods for protein and mRNA expression analysis were used in the previous study, and (3) the definition of response was based on the metabolic response assessed by PET – whereas in the current study, it was based on histopathologic tumour regression and down-categorisation (T-stage), which have been shown to be of greater prognostic value in the past few years. However, the divergent findings with respect to HSP27 of the proteomic analysis cannot be fully explained. Moreover, we are aware that both histopathological regression and metabolic response are only surrogate markers associated with improved survival and cannot guarantee recurrence-free long-term survival, because it is supposed to be influenced by multiple clinical, histopathological and molecular factors.
In accordance with our observations in primary resected oesophageal adenocarcinomas, the most striking result from this study was that the specific prognostic protein expression pattern could only be detected by RPPA and subsequent clustering analysis. However, expression levels of single HSP and GRP proteins were not predictive when analysed individually.
RPPA is a new, high-throughput technology and a powerful tool for the molecular characterisation of paucicellular material. RPPA can also be used for the challenging analysis of small numbers of cells of human biopsies (Paweletz et al, 2001; Agari et al, 2012) and samples from formalin-fixed, paraffin-embedded tissue (Wolff et al, 2011a). A benefit of RPPA is the ability to simultaneously quantify different proteins in several biological samples at the same time and under the same experimental conditions, generating patient-specific ‘protein patterns' and signaling networks. Hierarchical cluster analysis of expression data then allows to identify similarities in overall protein expression patterns and to stratify patients based on their molecular level. However, this approach also has some disadvantages: expression of individual proteins becomes less relevant as the clustering process progresses and an incorrect assignment made early in the process cannot be corrected (Tamayo et al, 1999). Using RPPA and cluster analysis, we identified a prognostic HSP/GRP pattern in oesophageal adenocarcinomas in a former study (Slotta-Huspenina et al, 2012a). Now we found a similar HSP/GRP expression pattern that was associated with response to CTX.
Interestingly, high HSP60 and low HSP27 expression assessed by IHC were also significantly associated with CTX response. Thus, HSP60 and HSP27 may serve as surrogate markers for the underlying protein expression profile and may serve as clinically useful predictors for CTX response.
Moreover, as for high HSP60 mRNA expression an association to CTX response could be demonstrated as well, these additional results underline the possible important role of HSP60 in the context of CTX response in oesophageal adenocarcinomas. However, the use of HSP/GRP expression as a molecular predictor for CTX response is not clinically applicable in its current form yet and needs to be validated in independent case collections or in prospective studies.
Although the sample size of the present study is appropriate for the identification of predictive biomarkers and is comparable with other publications in this field, we also face the problem with regard to a homogenous cytotoxic treatment, as others do (Fareed et al, 2009). Most anticancer regimes consist of a combination of various drugs, thus hampering the analysis of substance-specific resistance mechanism in vivo. In this study, we investigated a collective of patients who were all treated with a platin/5-FU-based CTX (PLF/OLF) according to standardised protocols (Lordick et al, 2004b, 2007); additional application of paclitaxel in a subgroup of patients did not show a significant bias with regards to the outcome of the patients, the rate of CTX response and the impact on the molecular findings. We suppose that these findings support the idea of a general cellular sensitivity towards cytotoxic therapy that is unspecific, substance independent and is mainly caused by differential expression profiles of molecular chaperones. The result of our study may not only improve the molecular understanding of CTX resistance but may also impact the development of new molecular targeted substances in the era of personalised treatment. Recently, novel therapeutic agents that inhibit HSPs have been developed and have already emerged as powerful anti-tumoural agents in preclinical settings alone or in combination with other drugs in gastrointestinal and non-gastrointestinal malignancies (Moser et al, 2009; Jego et al, 2010; Dickson et al, 2013).
In summary, using RPPA technology we were able to detect a specific HSP/GRP protein expression pattern in oesophageal adenocarcinomas that was associated with response to neoadjuvant CTX but not with prognosis. Additional results from IHC and qRT–PCR experiments suggest an important role for HSP60 in CTX response. Our findings may be helpful to predict CTX response. Moreover, these findings may facilitate our understanding of CTX response mechanisms and may also serve as a foundation for the development of strategies to overcome CTX resistance.
Acknowledgments
We thank Mrs Melitta Winkler, Ruth Wichnalek and Christina Schott for their excellent technical assistance.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Agari Y, Sakamoto K, Kuramitsu S, Shinkai A. Transcriptional repression mediated by a TetR family protein, PfmR, from Thermus thermophilus HB8. J Bacteriol. 2012;194 (17:4630–4641. doi: 10.1128/JB.00668-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader FG, Lordick F, Fink U, Becker K, Hofler H, Busch R, Siewert JR, Ott K. Paclitaxel in the neoadjuvant treatment for adeno carcinoma of the distal esophagus (AEG I). A comparison of two phase II trials with long-term follow-up. Onkologie. 2008;31 (7:366–372. doi: 10.1159/000135515. [DOI] [PubMed] [Google Scholar]
- Bauer K, Nitsche U, Slotta-Huspenina J, Drecoll E, von Weyhern CH, Rosenberg R, Hofler H, Langer R. High HSP27 and HSP70 expression levels are independent adverse prognostic factors in primary resected colon cancer. Cell Oncol (Dordr) 2012;35 (3:197–205. doi: 10.1007/s13402-012-0079-3. [DOI] [PubMed] [Google Scholar]
- Becker K, Mueller JD, Schulmacher C, Ott K, Fink U, Busch R, Bottcher K, Siewert JR, Hofler H. Histomorphology and grading of regression in gastric carcinoma treated with neoadjuvant chemotherapy. Cancer. 2003;98 (7:1521–1530. doi: 10.1002/cncr.11660. [DOI] [PubMed] [Google Scholar]
- Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31 (3:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10 (2:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MA, Okuno SH, Keohan ML, Maki RG, D'Adamo DR, Akhurst TJ, Antonescu CR, Schwartz GK. Phase II study of the HSP90-inhibitor BIIB021 in gastrointestinal stromal tumors. Ann Oncol. 2013;24 (1:252–257. doi: 10.1093/annonc/mds275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikken JL, Coit DG, Klimstra DS, Rizk NP, van Grieken N, Ilson D, Tang LH. Prospective impact of tumor grade assessment in biopsies on tumor stage and prognostic grouping in gastroesophageal adenocarcinoma: relevance of the seventh edition American Joint Committee on Cancer Staging Manual revision. Cancer. 2012;118 (2:349–357. doi: 10.1002/cncr.26301. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95 (25:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fareed KR, Kaye P, Soomro IN, Ilyas M, Martin S, Parsons SL, Madhusudan S. Biomarkers of response to therapy in oesophago-gastric cancer. Gut. 2009;58 (1:127–143. doi: 10.1136/gut.2008.155861. [DOI] [PubMed] [Google Scholar]
- Fu Y, Lee AS. Glucose regulated proteins in cancer progression, drug resistance and immunotherapy. Cancer Biol Ther. 2006;5 (7:741–744. doi: 10.4161/cbt.5.7.2970. [DOI] [PubMed] [Google Scholar]
- Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5 (22:2592–2601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- Gulmann C, Sheehan KM, Kay EW, Liotta LA, Petricoin EF., 3rd Array-based proteomics: mapping of protein circuitries for diagnostics, prognostics, and therapy guidance in cancer. J Pathol. 2006;208 (5:595–606. doi: 10.1002/path.1958. [DOI] [PubMed] [Google Scholar]
- Jego G, Hazoume A, Seigneuric R, Garrido C. Targeting heat shock proteins in cancer. Cancer Lett. 2010;332 (2:275–285. doi: 10.1016/j.canlet.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Khalil AA, Kabapy NF, Deraz SF, Smith C. Heat shock proteins in oncology: Diagnostic biomarkers or therapeutic targets. Biochim Biophys Acta. 2011;1816 (2:89–104. doi: 10.1016/j.bbcan.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Kuramitsu Y, Wang Y, Taba K, Suenaga S, Ryozawa S, Kaino S, Sakaida I, Nakamura K. Heat-shock protein 27 plays the key role in gemcitabine-resistance of pancreatic cancer cells. Anticancer Res. 2012;32 (6:2295–2299. [PubMed] [Google Scholar]
- Langer R, Feith M, Siewert JR, Wester HJ, Hoefler H. Expression and clinical significance of glucose regulated proteins GRP78 (BiP) and GRP94 (GP96) in human adenocarcinomas of the esophagus. BMC Cancer. 2008a;8:70. doi: 10.1186/1471-2407-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer R, Ott K, Feith M, Lordick F, Siewert JR, Becker K. Prognostic significance of histopathological tumor regression after neoadjuvant chemotherapy in esophageal adenocarcinomas. Mod Pathol. 2009;22 (12:1555–1563. doi: 10.1038/modpathol.2009.123. [DOI] [PubMed] [Google Scholar]
- Langer R, Ott K, Specht K, Becker K, Lordick F, Burian M, Herrmann K, Schrattenholz A, Cahill MA, Schwaiger M, Hofler H, Wester HJ. Protein expression profiling in esophageal adenocarcinoma patients indicates association of heat-shock protein 27 expression and chemotherapy response. Clin Cancer Res. 2008b;14 (24:8279–8287. doi: 10.1158/1078-0432.CCR-08-0679. [DOI] [PubMed] [Google Scholar]
- Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67 (8:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- Lordick F, Ott K, Krause BJ, Weber WA, Becker K, Stein HJ, Lorenzen S, Schuster T, Wieder H, Herrmann K, Bredenkamp R, Hofler H, Fink U, Peschel C, Schwaiger M, Siewert JR. PET to assess early metabolic response and to guide treatment of adenocarcinoma of the oesophagogastric junction: the MUNICON phase II trial. Lancet Oncol. 2007;8 (9:797–805. doi: 10.1016/S1470-2045(07)70244-9. [DOI] [PubMed] [Google Scholar]
- Lordick F, Stein HJ, Peschel C, Siewert JR. Neoadjuvant therapy for oesophagogastric cancer. Br J Surg. 2004a;91 (5:540–551. doi: 10.1002/bjs.4575. [DOI] [PubMed] [Google Scholar]
- Lordick F, Weber WA, Stein HJ, Schuhmacher C, Beer A, Hennig M, Ott K, Peschel C, Schwaiger M, Siewert JR. Individualized neoadjuvant treatment strategy in adenocarcinoma of the esophago-gastric junction (AEG): Interim report on the MUNICON trial. J Clin Oncol (Meeting Abstracts. 2004b;22 (14_suppl:4060. [Google Scholar]
- Moser C, Lang SA, Stoeltzing O. Heat-shock protein 90 (Hsp90) as a molecular target for therapy of gastrointestinal cancer. Anticancer Res. 2009;29 (6:2031–2042. [PubMed] [Google Scholar]
- Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin IE, Liotta LA. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20 (16:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- Powers MV, Clarke PA, Workman P. Death by chaperone: HSP90, HSP70 or both. Cell Cycle. 2009;8 (4:518–526. doi: 10.4161/cc.8.4.7583. [DOI] [PubMed] [Google Scholar]
- Sjoquist KM, Burmeister BH, Smithers BM, Zalcberg JR, Simes RJ, Barbour A, Gebski V. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol. 2011;12 (7:681–692. doi: 10.1016/S1470-2045(11)70142-5. [DOI] [PubMed] [Google Scholar]
- Slotta-Huspenina J, Berg D, Bauer K, Wolff C, Malinowsky K, Bauer L, Drecoll E, Bettstetter M, Feith M, Walch A, Hofler H, Becker KF, Langer R. Evidence of prognostic relevant expression profiles of heat-shock proteins and glucose-regulated proteins in oesophageal adenocarcinomas. PLoS One. 2012a;7 (7:e41420. doi: 10.1371/journal.pone.0041420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobin L, Gospodarowicz ML, Wittekind Ch. TNM Classification of Malignant Tumors. John Wiley & Sons: New York, NY, USA; 2010. [Google Scholar]
- Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158 (2:419–429. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96 (6:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ, Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, Cuesta MA, Blaisse RJ, Busch OR, ten Kate FJ, Creemers GJ, Punt CJ, Plukker JT, Verheul HM, Spillenaar Bilgen EJ, van Dekken H, van der Sangen MJ, Rozema T, Biermann K, Beukema JC, Piet AH, van Rij CM, Reinders JG, Tilanus HW, van der Gaast A. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366 (22:2074–2084. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- Wolff C, Schott C, Malinowsky K, Berg D, Becker KF. Producing reverse phase protein microarrays from formalin-fixed tissues. Methods Mol Biol. 2011a;785:123–140. doi: 10.1007/978-1-61779-286-1_9. [DOI] [PubMed] [Google Scholar]
- Wolff C, Schott C, Porschewski P, Reischauer B, Becker KF. Successful protein extraction from over-fixed and long-term stored formalin-fixed tissues. PLoS One. 2011b;6 (1:e16353. doi: 10.1371/journal.pone.0016353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.