Abstract
Background:
Mutation of the RB1 gene is necessary but not sufficient for the development of retinoblastoma. The nature of events occurring subsequent to RB1 mutation is unclear, as is the retinal cell-of-origin of this tumour.
Methods:
Gene expression profiling of 21 retinoblastomas was carried out to identify genetic events that contribute to tumorigenesis and to obtain information about tumour histogenesis.
Results:
Expression analysis showed a clear separation of retinoblastomas into two groups. Group 1 retinoblastomas express genes associated with a range of different retinal cell types, suggesting derivation from a retinal progenitor cell type. Recurrent chromosomal alterations typical of retinoblastoma, for example, chromosome 1q and 6p gain and 16q loss were also a feature of this group, and clinically they were characterised by an invasive pattern of tumour growth. In contrast, group 2 retinoblastomas were found to retain many characteristics of cone photoreceptor cells and appear to exploit the high metabolic capacity of this cell type in order to promote tumour proliferation.
Conclusion:
Retinoblastoma is a heterogeneous tumour with variable biology and clinical characteristics.
Keywords: retinoblastoma, retina, photoreceptor, chromosome 6p, chromosome 1q, chromosome 16q
Disruption of the RB pathway as a consequence of RB1 gene mutation or mutation of other pathway components (e.g., D-type cyclins, CDK4 or p16INK4A) is common to most if not all human cancers. Although these mutations are assumed to impact primarily on cell cycle regulation, RB1 has many additional roles, including regulation of chromosome stability, senescence and cellular differentiation (Indovina et al, 2013). Regulation of permanent cell cycle withdrawal and terminal differentiation are especially relevant in a developmental context and may be important in the prototype RB1-associated tumour, retinoblastoma. An understanding of the molecular pathogenesis of retinoblastoma may also shed light on additional roles of pRB in other tumour types which also show high frequencies of RB1 gene mutation/deletion (rather than cyclin/CDK/CDKI mutations), for example, osteosarcoma, small-cell lung cancer, bladder cancer and hepatocellular carcinoma.
Knowledge of the cell-of-origin of retinoblastoma is important to understand the role of RB1 mutation in this tumour. The mammalian retina is composed of six neuronal cell types, rod, cone, horizontal, amacrine, bipolar and ganglion cells, and one glial cell type, Müller glia (Figure 1), all of which are derived from a common retinal progenitor cell (RPC) and arise in an evolutionarily conserved birth order during development. Retinal progenitor cells at a specific stage of development show competence to produce post-mitotic precursor cells, with the potential for terminal differentiation into a limited subset of retinal cell types, for example, early progenitor cells give rise to ganglion and cone precusor cells, while late progenitor cells give rise to Müller glial and bipolar precursor cells (Livesey and Cepko, 2001; Dyer and Bremner, 2005).
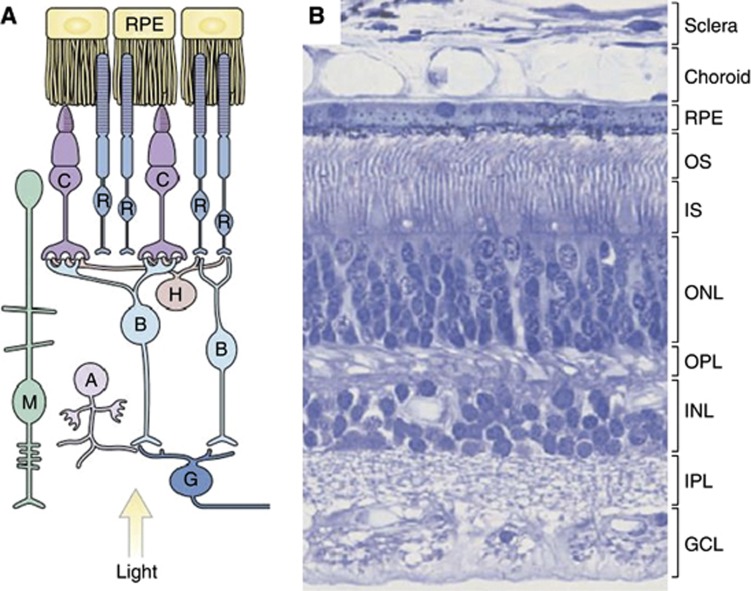
Figure 1.
The human retina. (A) Organisation of the retina. (B) A H&E-stained section of the retina shows the outer and inner segments (OS, IS) of rod and cone photoreceptor cells. Photoreceptor nuclei form the outer nuclear layer (ONL). Nuclei of bipolar, amacrine, horizontal and Müller glial cells form the inner nuclear layer (INL), and the nuclei of ganglion cells form the ganglion cell layer (GCL). The outer plexiform layer (OPL) contains the processes and synaptic terminals of photoreceptors, horizontal and bipolar cells. The inner plexiform layer (IPL) contains the processes and synaptic terminals of bipolar, amacrine and ganglion cells. The processes of Müller glial cells fill all the retinal space not occupied by neurons and blood vessels. (Reproduced from Sung and Chuang (2010)). Abbreviations: A, amacrine cell; B, bipolar cell; C, cone; G, ganglion cell; H, horizontal cell; M, Müller glial cell; R, rod; RPE, retinal pigment epithelium.
Several studies of both human retinoblastoma and mouse retinoblastoma models have sought to define the retinal cell-of-origin of retinoblastoma and to understand why these cells are so susceptible to oncogenic transformation following RB1 mutation. These studies have reached a variety of conclusions, suggesting that retinoblastomas may be heterogeneous in their origin. A detailed immunohistochemical and genetic examination of human retinoblastoma led Xu et al (2009) to suggest, for example, that human retinoblastoma has properties of a cone precursor cell. Investigation of mouse models of retinoblastoma have generally implicated other cell types, however, including horizontal, amacrine and Müller glial precursors (Chen et al, 2004; Ajioka et al, 2007; Pajovic et al, 2011). A further study concluded that multiple cell type-specific developmental programs, including those of amacrine, horizontal, photoreceptor and progenitor cells, are co-expressed in individual human retinoblastoma cells (McEvoy et al, 2011).
A general finding from studies of both human and murine retinoblastoma is that secondary mutations in addition to loss of RB1 are required for tumour development. It has been reported, for example, that loss of RB1 in humans induced non-proliferative retinoma and that increasing genome instability was correlated with progression to retinoblastoma (Dimaras et al, 2008). The observation of recurrent chromosomal alterations including 1q and 6p gain and 16q loss in up to 50% of all retinoblastomas suggests that genes on these chromosomes may contribute to tumour development and progression. A number of genes of interest have been highlighted, for example, MDM4 and KIF14 on 1q, E2F3 on 6p, CDH11 and RBL2/p130 on 16q (Corson and Gallie, 2007). However, the significance of many additional genes that show altered patterns of expression in association with these chromosomal alterations (e.g., CENPF, DEK, KIFC1) remains to be determined (Corson and Gallie, 2007).
In this study, we have used gene expression profiling to gain further insight into the molecular pathways which drive retinoblastoma tumorigenesis and their relationship to retinal developmental processes.
Materials and methods
Patient samples
Frozen tissue from 21 retinoblastomas, enucleated without prior treatment, was used for microarray analyses and qRT-PCR. Tissue was obtained from dissected globes and was divided for RNA and DNA extraction. Histopathology was assessed (blinded to genetic data) on tumour tissue, which was formalin-fixed and paraffin-embedded at the time of diagnosis. Five to 10 tissue sections were assessed according to the guidelines of the International Retinoblastoma Staging Working Group (Sastre et al, 2009). Patients were all treated at the Birmingham's Children Hospital, UK, and consent was obtained for tissue banking for ethically approved research. The study was approved by the local Research Ethics Committee.
Array-CGH
DNA was prepared from retinoblastomas and peripheral blood lymphocytes from 10 normal healthy individuals using phenol/chloroform extraction (normal DNAs were then pooled for further processing). DNA was sonicated (100–1000 bp), purified (Qiagen Ltd, Crawley, UK) and then amplified in a two part procedure using primer A (5′-GTTTCCCAGTCACGGTCNNNNNNNNN-3′) for initial linear amplification, followed by primer B (5′-GTTTCCCAGTCACGGTC-3′) (Wang et al, 2003) for 30 cycles of amplification with the inclusion of aminoallyl-dUTP to faciliate post amplification labelling. Fragmentation and labelling of 7.5 μg of amplified DNA was performed using the GeneChip WT dsDNA Terminal Labeling kit (Affymetrix UK Ltd, High Wycombe, UK). Hybridization to Affymetrix GeneChip Human Promoter 1.0R tiling arrays was carried out according to the standard Affymetrix procedures.
Gene expression arrays
Total RNA was isolated from retinoblastoma samples using Trizol extraction (Life Technologies Ltd, Paisley, UK), and additional purification was carried out using the Qiagen RNeasy system (Qiagen). RNA integrity was assessed using the Agilent Bioanalyzer 2100 (Agilent Technologies UK Ltd, Wokingham, UK). First- and second-strand cDNA synthesis, labelling and hybridisation to Affymetrix Human Gene 1.0ST arrays were performed according to the standard Affymetrix protocol. Affymetrix data were extracted, normalised and summarised using the robust multi-average method implemented in the Affymetrix Expression Console. CEL files from the same Affymetrix array platform (Human Gene 1.0ST) were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) for normal adult retina (GSM607947, GSM607948) and 96-day human fetal retina (GSM460264), and were normalised and summarised in parallel with retinoblastoma samples.
Array data analysis
CEL files from Affymetrix GeneChip Human Promoter 1.0R Tiling Arrays were analysed for copy number alterations using Partek Genomic Suite (PGS). Data were first log2 transformed and quantile normalised (with adjustment for GC content and probe sequence). Using the normal sample as baseline, copy number estimates were then obtained for each of 4.2 million probes on the array and segmentation applied with parameters set at: minimum number of genomic markers: 10, P-value threshold: 0.01 and signal to noise: 1.0.
Principal component and hierarchical clustering (HC) analysis of expression data from Affymetrix Human Gene 1.0ST arrays was carried out using PGS. Default parameters were used for principal component analysis (PCA; dispersion matrix: correlation; eigenvector scaling: normalised) and elipsoids were plotted at 2 standard deviations from the centroid of each group. Hierarchical clustering metrics used were Pearson correlation (distance measurement) and average linkage clustering. For other analyses, summarised Affymetrix data were imported into MultiExperiment Viewer (Saeed et al, 2006). Differentially expressed genes were identified using SAM (Significance Analysis of Microarrays) with a q-value or false discovery cutoff at 5%. Functional annotation of lists of differentially expressed genes was carried out using the Database for Annotation, Visualisation and Integrated Discovery (DAVID) (Huang et al, 2009). Annotation categories searched included GOTERM_BP (Biological Process), GOTERM_MF (Molecular Function) and pathways from the Biocarta, Kegg and Reactome databases. Hierarchical clustering of the reduced data set (retinal-associated genes) was performed using the GenePattern Software Suite (Reich et al, 2006). Geneset enrichment analysis (GSEA) was carried out using canonical pathways included in the Molecular Signatures Database (v3.0) of the GSEA software package (Subramanian et al, 2005). Data for GSEA was preprocessed so that only a single probeset for each gene (that with the highest expression) was included.
Chromosome 1q, 6p and 16q differentially expressed genes (fold change ⩾1.5, q⩽0.05) were defined as potentially cancer related on the basis of encoding proteins of known function and citations in the literature, implicating involvement in cancer or in cellular regulatory processes.
Real-time RT-PCR
One microgram of total RNA was used for cDNA synthesis using the QuantiTect Reverse Transcription kit (Qiagen) according to the manufacturer's protocol. Quantitative RT-PCR was performed in 1 × SYBR Green PCR mixture (Applied Biosystems, Warrington, UK), 1 μl diluted cDNA (1 : 5) and 0.5 μℳ gene-specific primers (Supplementary Table S7) designed using Primer 3 (http://frodo.wi.mit.edu) software. The reactions were performed in triplicate and β-actin was used as endogenous normalisation control.
Results
Retinoblastoma is a heterogeneous tumour
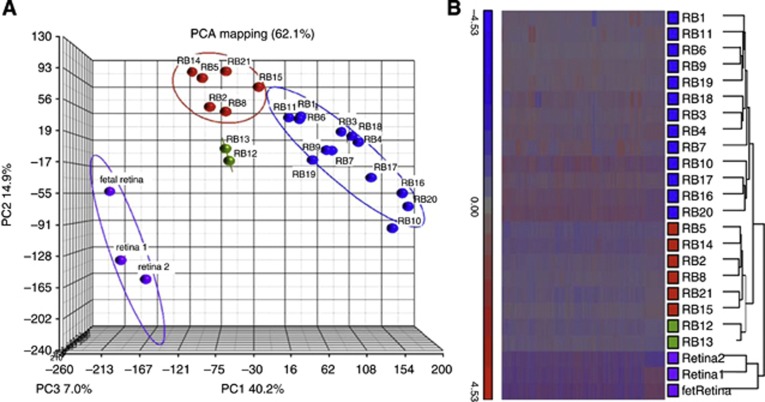
Initial exploratory analysis of the gene expression profiles of 21 retinoblastomas (Table 1) was carried out to assess heterogeneity. Principal component analysis showed a clear separation between normal retina and retinoblastoma, and in addition indicated a further sub-division of retinoblastomas into two main groups with 13 and 6 members, respectively (Figure 2A). A further two samples (group 3) were subsequently shown to be more similar to normal retina (see below). This grouping was also shown in a HC analysis (Figure 2B).
Table 1. Characteristics of retinoblastomas.
| |
|
|
|
|
|
|
|
|
|
Chromosome copy number alterationsf |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RB ID | Histological differentiation | Types of rosettes | Extent of rosettesa | Apoptosisb | Choroid invasionc | Choroid invasion depth | Optic nerve invasiond | Scleral invasion | Genetic groupe | 1q gain | 6p gain | 16q loss |
| RB1 |
Poor |
FW, N |
1 |
2 |
2 |
D |
3 |
0 |
1 |
+ |
− |
+ |
| RB3 |
Poor |
N |
1 |
2 |
0 |
S |
1 |
0 |
1 |
+ |
+ |
− |
| RB4 |
Poor |
N |
2 |
2 |
1 |
D |
3 |
0 |
1 |
− |
+ |
+ |
| RB6 |
Well |
FW |
3 |
2 |
0 |
S |
0 |
0 |
1 |
− |
(+)g |
− |
| RB7 |
Poor |
− |
0 |
2 |
2 |
D |
3 |
0 |
1 |
+ |
− |
+ |
| RB9 |
Poor |
− |
0 |
2 |
2 |
D |
4 |
0 |
1 |
− |
− |
− |
| RB10 |
Well |
FW, N |
4 |
2 |
0 |
S |
0 |
0 |
1 |
− |
+ |
− |
| RB11 |
Poor |
FW, P |
1 |
2 |
0 |
S |
2 |
0 |
1 |
− |
+ |
− |
| RB16 |
Intermed |
FW, N, P |
2 |
2 |
2 |
D |
3 |
0 |
1 |
ND |
ND |
ND |
| RB17 |
Poor |
N |
1 |
2 |
1 |
D |
3 |
0 |
1 |
− |
− |
− |
| RB18 |
Poor |
P |
0 |
2 |
2 |
D |
1 |
0 |
1 |
+ |
+ |
+ |
| RB19 |
Poor |
FW, N, P |
1 |
2 |
2 |
D |
1 |
0 |
1 |
− |
+ |
− |
| RB20 |
Well |
FW |
3 |
2 |
0 |
S |
0 |
0 |
1 |
− |
+ |
− |
| RB2 |
Well |
FW |
4 |
1 |
1 |
D |
3 |
0 |
2 |
− |
− |
− |
| RB5 |
Well |
FW, N |
3 |
1 |
0 |
S |
0 |
0 |
2 |
− |
− |
− |
| RB8 |
Well |
FW |
3 |
1 |
2 |
D |
1 |
1 |
2 |
− |
− |
− |
| RB14 |
Well |
FW, N |
4 |
1 |
0 |
S |
0 |
0 |
2 |
ND |
ND |
ND |
| RB15 |
Intermed |
FW |
2 |
2 |
0 |
S |
0 |
0 |
2 |
− |
− |
− |
| RB21 |
poor |
FW, N, P |
1 |
2 |
0 |
S |
0 |
0 |
2 |
− |
− |
− |
| RB12 |
Well |
FW, N |
3 |
1 |
1 |
D |
2 |
0 |
3 |
− |
− |
− |
| RB13 | Well | FW, N | 4 | 1 | 0 | S | 0 | 0 | 3 | − | − | − |
Abbreviations: FW=Flexner-Wintersteiner (characteristic of retinoblastoma); N=Homer-Wright rosettes (associated with tumours of neural origin); ND=not determined; P=pallisading; S, D=superficial, deep choroidal invasion.
Extent of rosettes: 0, none; 1, <10% 2, 10–25% 3, 25–50% 4, >50%.
Apoptosis: 1, low (patchy distribution); 2, high (observed over whole tumour area).
Choroid invasion: 0, none; 1, localised; 2, extensive.
Optic nerve invasion: 0, none; 1, pre-laminar; 2, intra-laminar; 3, retro-laminar; 4, to resection margin.
Genetic group–see text.
+/−: Presence/absence of chromosome copy number alteration; nd: not done.
Gain restricted to ∼3 Mb within 6p21.3.
Figure 2.
Retina and retinoblastoma gene expression data. (A) Principal component analysis shows separation of retinoblastoma samples into three groups, group 1 (blue symbols), group 2 (red symbols) and group 3 (green symbols). Normal adult retina and fetal retina are from http://www.ncbi.nlm.nih.gov/geo/. (B) Hierarchical clustering (HC) of the same samples. Colour coding shows concordance with PCA.
Functional annotation of differentially expressed genes in retina and retinoblastma (fold change ⩾2.0, q⩽0.05; Supplementary Table S1) indicated that, overall, the most highly enriched gene ontology (GO) categories were those associated with photoreceptor differentiation/maintenance and phototransduction in normal adult retina, and with cell cycle and mitosis in retinoblastoma (Table 2). More detailed examination of the differences between the three groups, using pairwise comparisons (Supplementary Tables S2–S4), identified significant enrichment of GO categories relating to mitosis, spindle function, DNA replication and cytokinesis in groups 1 and 2 relative to group 3 (Table 3). Group 2 was additionally characterised by GO categories relating to photoreceptor development and differentiation, while in both groups 2 and 3 phototransduction was also highlighted (Table 4).
Table 2. Gene ontology of genes differentially expressed (a) in normal adult retina and (b) retinoblastoma.
| Cluster | GO term | Fold enrichment | P-value |
|---|---|---|---|
|
(a) | |||
| Annotation cluster 1 | Enrichment score: 14.46 | ||
| GOTERM_BP_FAT | GO:0007601∼visual perception | 3.32 | 0.0000 |
| GOTERM_BP_FAT | GO:0050953∼sensory perception of light stimulus | 3.32 | 0.0000 |
| GOTERM_BP_FAT |
GO:0007600∼sensory perception |
1.07 |
0.3013 |
| Annotation cluster 2 | Enrichment score: 8.82 | ||
| GOTERM_BP_FAT | GO:0008104∼protein localisation | 1.65 | 0.0000 |
| GOTERM_BP_FAT | GO:0045184∼establishment of protein localisation | 1.65 | 0.0000 |
| GOTERM_BP_FAT |
GO:0015031∼protein transport |
1.64 |
0.0000 |
| Annotation cluster 3 | Enrichment score: 7.20 | ||
| GOTERM_BP_FAT | GO:0007602∼phototransduction | 5.64 | 0.0000 |
| GOTERM_BP_FAT | GO:0009583∼detection of light stimulus | 5.01 | 0.0000 |
| GOTERM_BP_FAT | GO:0009582∼detection of abiotic stimulus | 3.29 | 0.0000 |
| GOTERM_BP_FAT | GO:0009581∼detection of external stimulus | 2.89 | 0.0000 |
| GOTERM_BP_FAT |
GO:0051606∼detection of stimulus |
2.37 |
0.0000 |
| Annotation cluster 4 | Enrichment score: 6.15 | ||
| GOTERM_BP_FAT | GO:0046530∼photoreceptor cell differentiation | 5.58 | 0.0000 |
| GOTERM_BP_FAT | GO:0042461∼photoreceptor cell development | 5.43 | 0.0000 |
| GOTERM_BP_FAT | GO:0001754∼eye photoreceptor cell differentiation | 5.32 | 0.0000 |
| GOTERM_BP_FAT |
GO:0042462∼eye photoreceptor cell development |
5.12 |
0.0000 |
| Annotation cluster 5 | Enrichment score: 5.25 | ||
| GOTERM_MF_FAT | GO:0043167∼ion binding | ||
| GOTERM_MF_FAT | GO:0046872∼metal ion binding | 1.17 | 0.0000 |
| GOTERM_MF_FAT | GO:0043169∼cation binding | 1.16 | 0.0000 |
| GOTERM_MF_FAT | GO:0043169∼cation binding | 1.16 | 0.0000 |
|
(b) | |||
| Annotation cluster 1 | Enrichment score: 185.65 | ||
| GOTERM_MF_FAT | GO:0004984∼olfactory receptor activity | 9.23 | 0.0000 |
| GOTERM_BP_FAT | GO:0007606∼sensory perception of chemical stimulus | 7.95 | 0.0000 |
| GOTERM_BP_FAT | GO:0007608∼sensory perception of smell | 8.35 | 0.0000 |
| KEGG_PATHWAY | hsa04740:Olfactory transduction | 6.36 | 0.0000 |
| GOTERM_BP_FAT | GO:0007600∼sensory perception | 4.96 | 0.0000 |
| GOTERM_BP_FAT | GO:0007186∼G-protein-coupled receptor protein signalling pathway | 4.09 | 0.0000 |
| GOTERM_BP_FAT | GO:0050890∼cognition | 4.46 | 0.0000 |
| GOTERM_BP_FAT |
GO:0050877∼neurological system process |
3.50 |
0.0000 |
| Annotation cluster 2 | Enrichment score: 20.53 | ||
| GOTERM_BP_FAT | GO:0000279∼M phase | 3.44 | 0.0000 |
| GOTERM_BP_FAT | GO:0022403∼cell cycle phase | 2.94 | 0.0000 |
| GOTERM_BP_FAT | GO:0022402∼cell cycle process | 2.30 | 0.0000 |
| GOTERM_BP_FAT |
GO:0007049∼cell cycle |
2.07 |
0.0000 |
| Annotation cluster 3 | Enrichment score: 19.54 | ||
| GOTERM_BP_FAT | GO:0000280∼nuclear division | 3.68 | 0.0000 |
| GOTERM_BP_FAT | GO:0007067∼mitosis | 3.68 | 0.0000 |
| GOTERM_BP_FAT | GO:0000087∼M phase of mitotic cell cycle | 3.62 | 0.0000 |
| GOTERM_BP_FAT | GO:0048285∼organelle fission | 3.54 | 0.0000 |
| GOTERM_BP_FAT |
GO:0000278∼mitotic cell cycle |
2.64 |
0.0000 |
| Annotation cluster 4 | Enrichment score: 7.86 | ||
| GOTERM_BP_FAT | GO:0051321∼meiotic cell cycle | 3.46 | 0.0000 |
| GOTERM_BP_FAT | GO:0051327∼M phase of meiotic cell cycle | 3.41 | 0.0000 |
| GOTERM_BP_FAT |
GO:0007126∼meiosis |
3.41 |
0.0000 |
| Annotation cluster 5 | Enrichment score: 5.79 | ||
| GOTERM_BP_FAT | GO:0065004∼protein-DNA complex assembly | 3.67 | 0.0000 |
| GOTERM_BP_FAT | GO:0031497∼chromatin assembly | 3.56 | 0.0000 |
| GOTERM_BP_FAT | GO:0006334∼nucleosome assembly | 3.41 | 0.0000 |
| GOTERM_BP_FAT | GO:0034728∼nucleosome organisation | 3.20 | 0.0000 |
| GOTERM_BP_FAT | GO:0006333∼chromatin assembly or disassembly | 2.63 | 0.0000 |
| GOTERM_BP_FAT | GO:0006325∼chromatin organisation | 1.07 | 0.4285 |
Abbreviation: GO=gene ontology.
Genes with at least two-fold (q⩽0.05) differential expression in normal adult retina vs retinoblastoma were analysed using DAVID (http://david.abcc.ncifcrf.gov). The top five most highly enriched clusters of GO terms are presented. The group enrichment score used to rank biological significance is the geometric mean (in −log scale) of member's P-values in a corresponding annotation cluster. GOTERM_BP: Biological Process. GOTERM_MF: Molecular function. G0_FAT: includes a subset of the Gene Ontology term set created in order to filter the broadest terms so that they do not overshadow the more specific terms.
Table 3. Gene ontology of genes differentially expressed in (a) group 1 and (b) group 2, both relative to group 3.
| Cluster | GO Term | Fold enrichment | P-value |
|---|---|---|---|
|
(a) | |||
| Annotation cluster 1 | Enrichment score: 34.87 | ||
| GOTERM_BP_FAT | GO:0000279∼M phase | 8.99 | 0.0000 |
| GOTERM_BP_FAT | GO:0022403∼cell cycle phase | 7.49 | 0.0000 |
| GOTERM_BP_FAT | GO:0007067∼mitosis | 10.46 | 0.0000 |
| GOTERM_BP_FAT | GO:0000280∼nuclear division | 10.46 | 0.0000 |
| GOTERM_BP_FAT | GO:0000087∼M phase of mitotic cell cycle | 10.28 | 0.0000 |
| GOTERM_BP_FAT | GO:0048285∼organelle fission | 10.05 | 0.0000 |
| GOTERM_BP_FAT | GO:0022402∼cell cycle process | 5.82 | 0.0000 |
| GOTERM_BP_FAT |
GO:0000278∼mitotic cell cycle |
7.11 |
0.0000 |
| Annotation cluster 2 | Enrichment score: 29.41 | ||
| GOTERM_MF_FAT | GO:0004984∼olfactory receptor activity | 9.12 | 0.0000 |
| GOTERM_BP_FAT | GO:0007608∼sensory perception of smell | 7.74 | 0.0000 |
| GOTERM_BP_FAT | GO:0007606∼sensory perception of chemical stimulus | 7.17 | 0.0000 |
| KEGG_PATHWAY | hsa04740:Olfactory transduction | 5.66 | 0.0000 |
| GOTERM_BP_FAT | GO:0007600∼sensory perception | 4.29 | 0.0000 |
| GOTERM_BP_FAT | GO:0050890∼cognition | 3.82 | 0.0000 |
| GOTERM_BP_FAT | GO:0007186∼G-protein-coupled receptor protein signalling pathway | 3.39 | 0.0000 |
| GOTERM_BP_FAT | GO:0050877∼neurological system process | 2.99 | 0.0000 |
| GOTERM_BP_FAT |
GO:0007166∼cell surface receptor linked signal transduction |
2.15 |
0.0000 |
| Annotation cluster 3 | Enrichment score: 10.49 | ||
| GOTERM_BP_FAT | GO:0065004∼protein-DNA complex assembly | 11.36 | 0.0000 |
| GOTERM_BP_FAT | GO:0031497∼chromatin assembly | 10.80 | 0.0000 |
| GOTERM_BP_FAT | GO:0006334∼nucleosome assembly | 10.62 | 0.0000 |
| GOTERM_BP_FAT | GO:0034728∼nucleosome organisation | 9.60 | 0.0000 |
| GOTERM_BP_FAT | GO:0006333∼chromatin assembly or disassembly | 7.40 | 0.0000 |
| GOTERM_BP_FAT | GO:0034621∼cellular macromolecular complex subunit organisation | 3.55 | 0.0000 |
| GOTERM_BP_FAT | GO:0034622∼cellular macromolecular complex assembly | 3.69 | 0.0000 |
| GOTERM_BP_FAT |
GO:0006325∼chromatin organisation |
2.73 |
0.0001 |
| Annotation cluster 4 | Enrichment score: 8.65 | ||
| GOTERM_BP_FAT | GO:0051327∼M phase of meiotic cell cycle | 7.67 | 0.0000 |
| GOTERM_BP_FAT | GO:0007126∼meiosis | 7.67 | 0.0000 |
| GOTERM_BP_FAT |
GO:0051321∼meiotic cell cycle |
7.52 |
0.0000 |
| Annotation CLUSTER 5 | Enrichment score: 8.27 | ||
| GOTERM_BP_FAT | GO:0006974∼response to DNA damage stimulus | 3.90 | 0.0000 |
| GOTERM_BP_FAT | GO:0006281∼DNA repair | 4.47 | 0.0000 |
| GOTERM_BP_FAT | GO:0033554∼cellular response to stress | 2.66 | 0.0000 |
|
(b) | |||
| Annotation cluster 1 | Enrichment score: 66.78 | ||
| GOTERM_BP_FAT | GO:0022403∼cell cycle phase | 10.48 | 0.0000 |
| GOTERM_BP_FAT | GO:0000279∼M phase | 11.91 | 0.0000 |
| GOTERM_BP_FAT |
GO:0022402∼cell cycle process |
8.34 |
0.0000 |
| Annotation cluster 2 | Enrichment score: 53.36 | ||
| GOTERM_BP_FAT | GO:0000278∼mitotic cell cycle | 9.83 | 0.0000 |
| GOTERM_BP_FAT | GO:0007067∼mitosis | 13.57 | 0.0000 |
| GOTERM_BP_FAT | GO:0000280∼nuclear division | 13.57 | 0.0000 |
| GOTERM_BP_FAT | GO:0000087∼M phase of mitotic cell cycle | 13.33 | 0.0000 |
| GOTERM_BP_FAT |
GO:0048285∼organelle fission |
13.04 |
0.0000 |
| Annotation cluster 3 | Enrichment score: 21.83 | ||
| GOTERM_BP_FAT | GO:0006281∼DNA repair | 7.23 | 0.0000 |
| GOTERM_BP_FAT | GO:0006974∼response to DNA damage stimulus | 6.13 | 0.0000 |
| GOTERM_BP_FAT |
GO:0033554∼cellular response to stress |
4.29 |
0.0000 |
| Annotation cluster 4 | Enrichment score: 14.86 | ||
| GOTERM_BP_FAT | GO:0007126∼meiosis | 10.47 | 0.0000 |
| GOTERM_BP_FAT | GO:0051327∼M phase of meiotic cell cycle | 10.47 | 0.0000 |
| GOTERM_BP_FAT |
GO:0051321∼meiotic cell cycle |
10.26 |
0.0000 |
| Annotation cluster 5 | Enrichment score: 13.93 | ||
| GOTERM_BP_FAT | GO:0065004∼protein-DNA complex assembly | 12.82 | 0.0000 |
| GOTERM_BP_FAT | GO:0031497∼chromatin assembly | 12.33 | 0.0000 |
| GOTERM_BP_FAT | GO:0034728∼nucleosome organisation | 11.04 | 0.0000 |
| GOTERM_BP_FAT | GO:0006334∼nucleosome assembly | 11.66 | 0.0000 |
| GOTERM_BP_FAT | GO:0006333∼chromatin assembly or disassembly | 8.45 | 0.0000 |
| GOTERM_BP_FAT | GO:0034622∼cellular macromolecular complex assembly | 4.40 | 0.0000 |
| GOTERM_BP_FAT | GO:0006325∼chromatin organisation | 3.21 | 0.0000 |
Abbreviation: GO=gene ontology.
Genes with at least two-fold (q⩽0.05) differential expression in group 1 or group 2 vs group 3 were analysed using DAVID (http://david.abcc.ncifcrf.gov). The top five most highly enriched clusters of GO terms are presented.
Table 4. Gene ontology of genes differentially expressed in (a) group 2 and (b) group 3 relative to group 1.
| Cluster | GO term | Fold enrichment | P-value |
|---|---|---|---|
|
(a) | |||
| Annotation cluster 1 | Enrichment score: 9.60 | ||
| GOTERM_BP_FAT | GO:0050953∼sensory perception of light stimulus | 8.45 | 0.0000 |
| GOTERM_BP_FAT | GO:0007601∼visual perception | 8.45 | 0.0000 |
| GOTERM_BP_FAT | GO:0050890∼cognition | 2.28 | 0.0000 |
| GOTERM_BP_FAT | GO:0007600∼sensory perception | 2.33 | 0.0000 |
| GOTERM_BP_FAT |
GO:0050877∼neurological system process |
1.98 |
0.0001 |
| Annotation cluster 2 | Enrichment score: 5.52 | ||
| GOTERM_BP_FAT | GO:0007602∼phototransduction | 19.07 | 0.0000 |
| GOTERM_BP_FAT | GO:0009583∼detection of light stimulus | 16.13 | 0.0000 |
| GOTERM_BP_FAT | GO:0009582∼detection of abiotic stimulus | 9.68 | 0.0000 |
| GOTERM_BP_FAT | GO:0009581∼detection of external stimulus | 8.50 | 0.0000 |
| GOTERM_BP_FAT | GO:0009416∼response to light stimulus | 5.47 | 0.0000 |
| GOTERM_BP_FAT | GO:0051606∼detection of stimulus | 5.33 | 0.0001 |
| GOTERM_BP_FAT | GO:0009314∼response to radiation | 3.78 | 0.0003 |
| GOTERM_BP_FAT |
GO:0009628∼response to abiotic stimulus |
2.74 |
0.0008 |
| Annotation cluster 3 | Enrichment score: 4.19 | ||
| GOTERM_BP_FAT | GO:0042462∼eye photoreceptor cell development | 18.88 | 0.0000 |
| GOTERM_BP_FAT | GO:0001754∼eye photoreceptor cell differentiation | 17.98 | 0.0000 |
| GOTERM_BP_FAT | GO:0042461∼photoreceptor cell development | 15.73 | 0.0000 |
| GOTERM_BP_FAT | GO:0046530∼photoreceptor cell differentiation | 15.10 | 0.0000 |
| GOTERM_BP_FAT |
GO:0048592∼eye morphogenesis |
5.47 |
0.0047 |
| Annotation cluster 4 | Enrichment score: 3.06 | ||
| GOTERM_BP_FAT | GO:0048858∼cell projection morphogenesis | 3.60 | 0.0001 |
| GOTERM_BP_FAT | GO:0032990∼cell part morphogenesis | 3.44 | 0.0002 |
| GOTERM_BP_FAT | GO:0032989∼cellular component morphogenesis | 2.38 | 0.0043 |
| GOTERM_BP_FAT |
GO:0000902∼cell morphogenesis |
2.47 |
0.0044 |
| Annotation cluster 5 | Enrichment score: 1.96 | ||
| GOTERM_BP_FAT | GO:0010324∼membrane invagination | 2.86 | 0.0084 |
| GOTERM_BP_FAT | GO:0006897∼endocytosis | 2.86 | 0.0084 |
| GOTERM_BP_FAT | GO:0016044∼membrane organisation | 2.15 | 0.0183 |
|
(b) | |||
| Annotation cluster 1 | Enrichment score: 20.08 | ||
| GOTERM_BP_FAT | GO:0050953∼sensory perception of light stimulus | 5.17 | 0.0000 |
| GOTERM_BP_FAT | GO:0007601∼visual perception | 5.17 | 0.0000 |
| GOTERM_BP_FAT |
GO:0007600∼sensory perception |
1.59 |
0.0001 |
| Annotation cluster 2 | Enrichment score: 10.44 | ||
| GOTERM_BP_FAT | GO:0007602∼phototransduction | 10.40 | 0.0000 |
| GOTERM_BP_FAT | GO:0009583∼detection of light stimulus | 8.80 | 0.0000 |
| GOTERM_BP_FAT | GO:0009582∼detection of abiotic stimulus | 5.81 | 0.0000 |
| GOTERM_BP_FAT | GO:0009581∼detection of external stimulus | 5.10 | 0.0000 |
| GOTERM_BP_FAT | GO:0051606∼detection of stimulus | 3.78 | 0.0000 |
| GOTERM_BP_FAT |
GO:0009416∼response to light stimulus |
3.36 |
0.0000 |
| Annotation cluster 3 | Enrichment score: 6.40 | ||
| GOTERM_BP_FAT | GO:0031644∼regulation of neurological system process | 3.14 | 0.0000 |
| GOTERM_BP_FAT | GO:0051969∼regulation of transmission of nerve impulse | 3.15 | 0.0000 |
| GOTERM_BP_FAT |
GO:0050804∼regulation of synaptic transmission |
3.16 |
0.0000 |
| Annotation cluster 4 | Enrichment score: 6.03 | ||
| GOTERM_BP_FAT | GO:0048858∼cell projection morphogenesis | 2.73 | 0.0000 |
| GOTERM_BP_FAT | GO:0032990∼cell part morphogenesis | 2.62 | 0.0000 |
| GOTERM_BP_FAT | GO:0030030∼cell projection organisation | 2.24 | 0.0000 |
| GOTERM_BP_FAT | GO:0048667∼cell morphogenesis involved in neuron differentiation | 2.63 | 0.0000 |
| GOTERM_BP_FAT | GO:0031175∼neuron projection development | 2.41 | 0.0000 |
| GOTERM_BP_FAT | GO:0048812∼neuron projection morphogenesis | 2.50 | 0.0000 |
| GOTERM_BP_FAT |
GO:0007409∼axonogenesis |
2.40 |
0.0001 |
| Annotation cluster 5 | Enrichment score: 4.88 | ||
| GOTERM_BP_FAT | GO:0008104∼protein localisation | 1.67 | 0.0000 |
| GOTERM_BP_FAT | GO:0045184∼establishment of protein localisation | 1.65 | 0.0000 |
| GOTERM_BP_FAT | GO:0015031∼protein transport | 1.62 | 0.0000 |
Abbreviation: GO=gene ontology.
Genes with at least two-fold (q⩽0.05) differential expression in group 2 or group 3 vs group 1 were analysed using DAVID (http://david.abcc.ncifcrf.gov). The top five most highly enriched clusters of GO terms are presented.
The conclusion drawn from these results is that groups 1 and 2 appear to have the greatest proliferative potential, but these groups differ in the extent of photoreceptor differentiation. The expression profile of group 2 retinoblastomas appears to be most consistent with derivation from a retinal cone photoreceptor lineage. Group 1 retinoblastomas may derive either from a different retinal lineage or from an early uncommitted cell type, for example, a RPC.
Retinoblastomas show different patterns of retinal gene expression
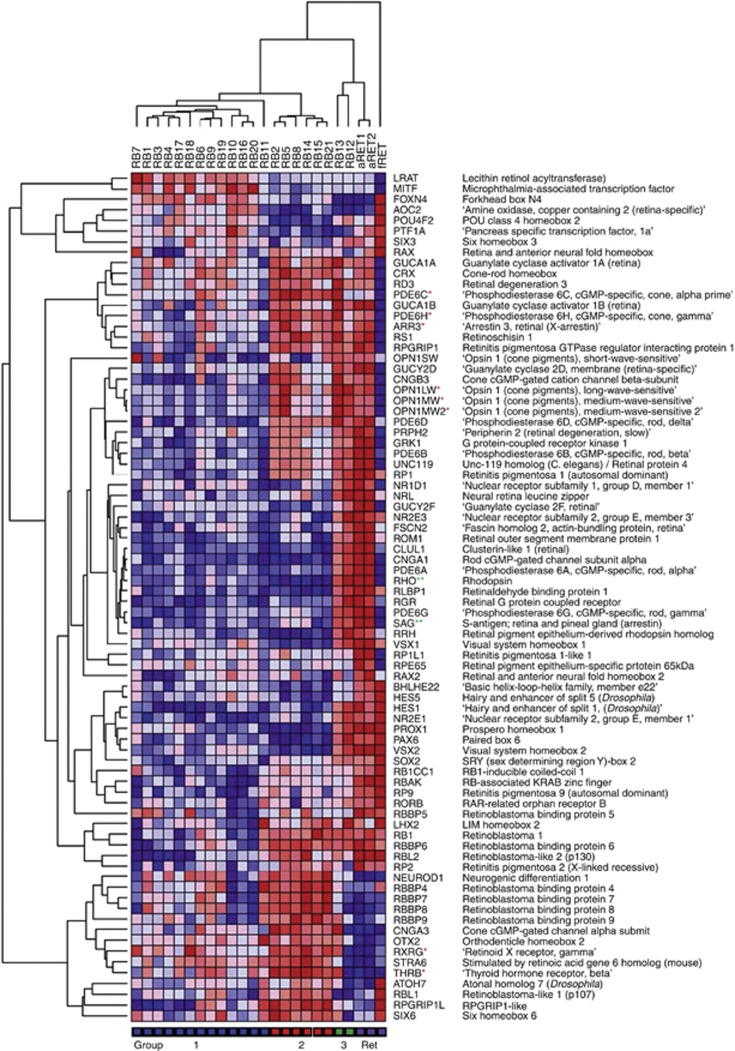
To investigate the histogenesis of retinoblastoma groups in more detail, data were extracted for all genes on the array associated with retinal development and function. Genes were identified using the Affymetrix gene description; also included were genes listed by Hennig et al (2008), which function in the regulation of photoreceptor gene expression and genes listed by Byerly and Blackshaw (2009) with roles in vertebrate retinal development. Unsupervised HC of tumours, based on 80 retina-associated genes, produced an identical grouping to that obtained with PCA and with HC of the complete data set (Figure 3), and provided information about the expression of genes associated with different retinal cell types in each of the tumour groups.
Figure 3.
Hierarchical clustering of retina and retinoblastoma based on 80 genes associated with retinal development and function. Abbreviations: aRET, adult retina; fRET, fetal retina. *Cone-enriched genes; **Rod-enriched genes.
The expression of cone photoreceptor-enriched transcription factors, RXRG and THRB, was of particular interest in view of the reported cone precursor origin of retinoblastoma (Xu et al, 2009). The expression of RXRG and THRB was increased 1.4- and 1.5-fold, respectively, in group 2 retinoblastomas relative to group 1, and was three- to five-fold increased relative to normal adult retina. Furthermore, the high expression in group 2 of downstream genes encoding cone opsins (OPN1MW, OPN1LW), cone arrestin (ARR3) and cone phosphodiesterases (PDE6C, PDE6H) is consistent with the development of group 2 retinoblastomas from a cone lineage cell. The fact that there was little or no expression of either rod rhodopsin (RHO) or rod arrestin (SAG) in this group confirms that the observed gene expression profile is not a consequence of the inclusion of normal retinal tissue. Expression of RHO and SAG were 170- and 50-fold higher, respectively, in normal retina, where rods outnumber cones by ∼20 : 1. In contrast, the two group 3 samples (RB12 and RB13) showed high-level expression of both cone and rod genes (e.g., OPN1MW/LW, RHO, NR2E3), as well as several genes characteristic of other retinal neuronal and glial cell types (HES1, PAX6, PROX1, VSX2, RLBP1/CRALBP) (Livesey and Cepko, 2001; Cid et al, 2010; Joly et al, 2011). Their similarity to the normal retinal samples (Figure 3) is suggestive of inclusion of some normal retinal tissue, and these two samples were not analysed further.
Although the expression of RXRG and THRB was moderately reduced rather than absent in group 1, a much greater decrease (six- to eight-fold) in the expression of OPN1MW/LW genes relative to group 2 suggests that group 1 retinoblastomas do not complete the cone differentiation programme and may be arrested at an earlier stage of development. It was noted that some group 1 retinoblastomas also showed variable upregulation (⩽1.5-fold) of a number of genes encoding transcription factors implicated more generally in eye and retinal development, particularly in the development of retinal horizontal and amacrine cells, for example, FOXN4, PTFIA and MITF (Fujitani et al, 2006). These results suggest that group 1 retinoblastomas have the potential for at least the early stages of differentiation of more than one retinal cell type.
To further validate these results, quantitative RT-PCR was carried out for selected genes characteristic of each of the three groups. In the case of all genes tested (FOXN4, ARR3, RXRG, RHO, NR2E3 and PAX6), expression patterns were in agreement with the array results (Supplementary Figure S1).
Histopathology and cytogenetics of retinoblastoma sub-types
We next examined whether the genetic classification of retinoblastomas into different groups, with differing levels of cone photoreceptor differentiation, was related to histopathological or cytogenetic characteristics. A detailed histopathological review of all retinoblastomas (carried out by a reviewer without knowledge of the genetic data) showed relatively good agreement between genetic and histological definitions of photoreceptor differentiation (Table 1). Four out of six (66.7%) group 2 retinoblastomas were classified as well differentiated on the basis of histological features (the extent of photoreceptor-like Flexner-Wintersteiner rosettes), compared with only 3 out of 13 (23.1%) group 1 retinoblastomas. However, these figures may under-estimate the extent of correlation between genetic and histopathological data, as tissue used for genetic analysis was representative of only a small part of the tumour. It is also of interest that group 1 retinoblastomas showed an increased frequency of both post-laminar optic nerve invasion (PLONI, 6 out of 13, 46.1%) and deep choroid invasion (DCI, 8 out of 13, 61.5%) compared with group 2 (1 out of 6, 16.7% and 2 out of 8, 25.0% for PLONI and DCI respectively), although these differences did not reach statistical significance (Table 1). Patients with these features are considered to be at higher risk of extra-ocular tumour spread (Shields et al, 2006). Surprisingly, the frequency of apoptotic cells was also significantly higher in group 1 retinoblastomas (P=0.004).
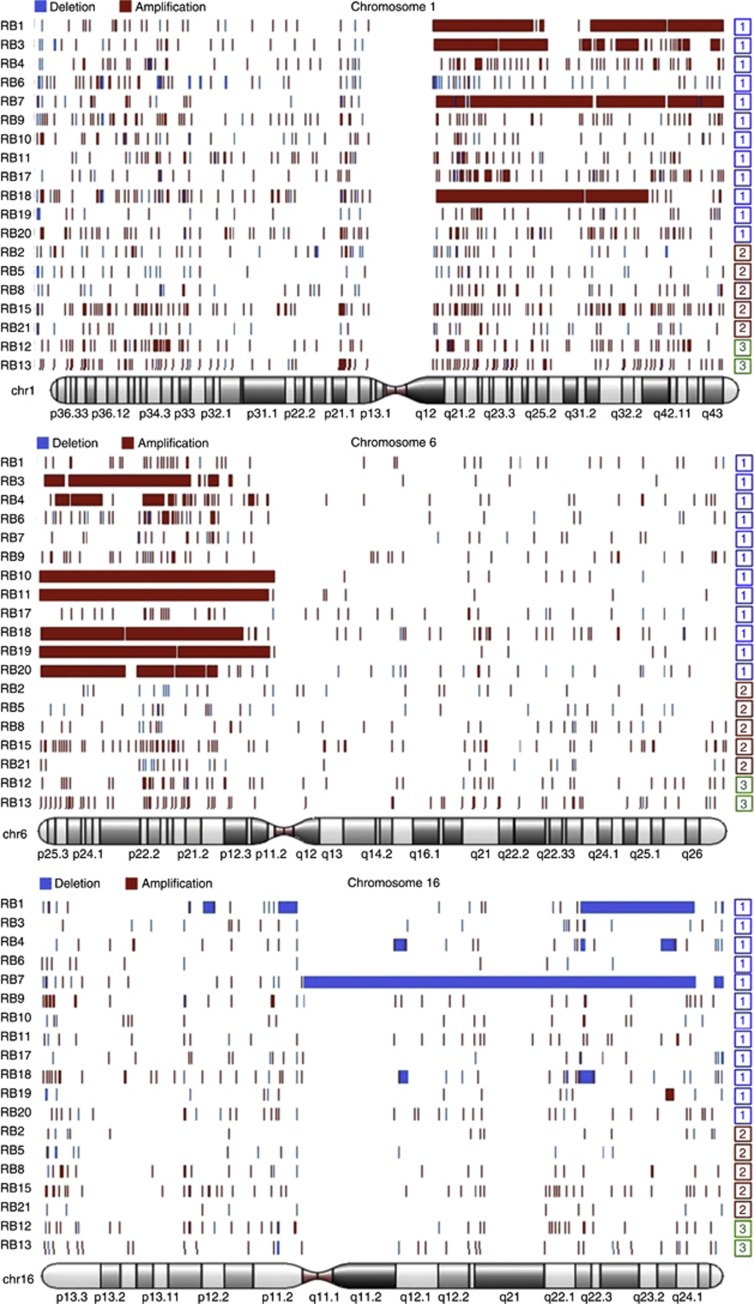
The occurrence of cytogenetic alterations most frequently reported to be associated with adverse clinical features was also investigated, that is, gain of chromosomes 1q and 6p and loss of chromosome 16q. Array-CGH of 19 samples for which material was available showed that these alterations tended to occur together and were significantly more frequent in group 1 retinoblastomas (P=0.003 for any alteration; P=0.029 for 6p gain) (Table 1, Figure 4). Overall, these results lend further weight to the classification of retinoblastoma into two distinct groups with differing genetic and clinical characteristics.
Figure 4.
Array-CGH of retinoblastomas. The chromosomes with the most frequent copy number alterations were chromosomes 1, 6 and 16. These alterations were restricted to group 1 retinoblastomas.
Gene expression changes associated with recurrent chromosome alterations
We next asked whether the altered pattern of differentiation in group 1 retinoblastomas might be related to the expression of genes on chromosomes 1q, 6p and 16q. SAM analyses identified a total of 555 genes mapping to these chromosomal regions, which showed significantly altered expression (fold change ⩾1.5; FDR q⩽0.05) in retinoblastomas with the relevant chromosomal alteration (Supplementary Table S5). A subset of 56 protein-coding genes had functions which potentially could contribute to retinoblastoma tumorigenesis (based on involvement in other tumour types or in cell regulatory processes) or which are relevant to retinal/photoreceptor development and function (Table 5).
Table 5. Differentially expressed genes on chromosomes 1q, 6p and 16q.
| Gene symbol | Gene name | Cyto band | Fold change | q (%) |
|---|---|---|---|---|
|
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
1q21.2 |
2.1 |
1.3 |
|
HDGF |
Hepatoma-derived growth factor |
1q23.1 |
1.7 |
0.0 |
|
VANGL2 |
Vang-like 2 (van gogh, Drosophila) |
1q23.2 |
1.5 |
1.3 |
|
NUF2 |
NUF2, NDC80 kinetochore complex component |
1q23.3 |
2.7 |
0.8 |
|
CENPL |
Centromere protein L |
1q25.1 |
1.6 |
0.9 |
|
CACYBP |
Calcyclin-binding protein |
1q25.1 |
1.6 |
0.7 |
|
ASPM |
Asp (abnormal spindle) homolog, |
1q31 |
2.6 |
2.0 |
|
KIF14 |
Kinesin family member 14 |
1q32.1 |
2.3 |
1.0 |
|
NR5A2 |
Nuclear receptor subfamily 5, group A, member 2 |
1q32.1 |
1.8 |
0.5 |
|
YOD1 |
YOD1 OTU deubiquinating enzyme 1 |
1q32.2 |
1.5 |
0.0 |
|
NEK2 |
NIMA (never in mitosis gene a)-related kinase 2 |
1q32.3 |
2.0 |
4.1 |
|
CENPF |
Centromere protein F (mitosin) |
1q41 |
2.4 |
2.7 |
|
PRIM2 |
Primase, DNA, polypeptide 2 |
6p11.2 |
1.7 |
0.4 |
|
MCM3 |
Minichromosome maintenance complex component 3 |
6p12 |
1.7 |
0.3 |
|
BMP5 |
Bone morphogenetic protein 5 |
6p12.1 |
1.7 |
0.0 |
|
TFAP2D |
Transcription factor AP-2 delta |
6p12.1 |
1.6 |
0.2 |
|
OPN5 |
Opsin 5 |
6p12.3 |
2.0 |
0.0 |
|
RUNX2 |
Runt-related transcription factor 2 |
6p21 |
1.7 |
0.0 |
|
UHRF1BP1 |
UHRF1-binding protein 1 |
6p21 |
1.5 |
0.0 |
|
CDC5L |
CDC5 cell division cycle 5-like |
6p21 |
1.5 |
0.8 |
|
STK38 |
Serine/threonine kinase 38 |
6p21 |
1.9 |
0.1 |
|
USP49 |
Ubiquitin-specific peptidase 49 |
6p21 |
1.8 |
0.1 |
|
POLH |
Polymerase (DNA directed), eta |
6p21.1 |
1.7 |
0.1 |
|
FOXP4 |
Forkhead box P4 |
6p21.1 |
1.5 |
0.1 |
|
PIM1 |
Pim-1 oncogene |
6p21.2 |
1.7 |
0.0 |
|
TEAD3 |
TEA domain family member 3 |
6p21.2 |
1.6 |
0.0 |
|
DAXX |
Death-domain associated protein |
6p21.3 |
1.6 |
0.0 |
|
KIFC1 |
Kinesin family member C1 |
6p21.3 |
2.2 |
0.0 |
|
EHMT2 |
euchromatic histone-lysine N-methyltransferase 2 |
6p21.31 |
1.5 |
0.1 |
|
FANCE |
Fanconi anaemia, complementation group E |
6p21.31 |
1.8 |
0.0 |
|
E2F3 |
E2F transcription factor 3 |
6p22 |
1.5 |
0.2 |
|
JARID2 |
Jumonji, AT rich interactive domain 2 |
6p22.3 |
1.5 |
0.2 |
|
DEK |
DEK oncogene |
6p22.3 |
1.5 |
0.4 |
|
CDKAL1 |
CDK5 regulatory subunit associated protein 1-like 1 |
6p22.3 |
1.7 |
0.1 |
|
NUP153 |
Nucleoporin 153 kDa |
6p22.3 |
1.6 |
0.1 |
|
SOX4 |
SRY-box 4 |
6p22.3 |
1.6 |
0.1 |
|
RNF182 |
Ring finger protein 182 |
6p23 |
1.5 |
0.2 |
|
PRPF4B |
PRP4 pre-mRNA processing factor 4 homolog B |
6p25.2 |
1.5 |
0.3 |
|
RNF8 |
RING finger protein 8 |
6p31.3 |
1.6 |
0.0 |
|
N4BP1 |
NEDD4-binding protein 1 |
16q12.1 |
2.0 |
1.2 |
|
CHD9 |
Chromodomain helicase DNA-binding protein 9 |
16q12.2 |
3.1 |
0.0 |
|
RBL2 |
retinoblastoma-like 2 (p130) |
16q12.2 |
2.4 |
1.2 |
|
RPGRIP1L |
RPGRIP1-like |
16q12.2 |
2.3 |
2.4 |
|
RSPRY1 |
Ring finger and SPRY domain containing 1 |
16q13 |
2.2 |
0.8 |
|
BBS2 |
Bardet-Biedl syndrome 2 |
16q21 |
3.0 |
0.0 |
|
CNOT1 |
CCR4-NOT transcription complex, subunit 1 |
16q21 |
2.1 |
0.0 |
|
NAE1 |
NEDD8 activating enzyme E1 |
16q22 |
2.0 |
4.2 |
|
PRMT7 |
Protein arginine methyltransferase 7 |
16q22.1 |
1.6 |
1.2 |
|
SF3B3 |
Splicing factor 3b, subunit 3, |
16q22.1 |
1.9 |
1.2 |
|
TERF2 |
Telomeric repeat-binding factor 2 |
16q22.1 |
1.9 |
4.1 |
|
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
16q22.1 |
1.7 |
2.4 |
|
TXNL4B |
Thioredoxin-like 4B |
16q22.2 |
1.9 |
4.1 |
|
CDK10 |
Cyclin-dependent kinase 10 |
16q24 |
1.5 |
0.8 |
|
USP10 |
Ubiquitin-specific peptidase 10 |
16q24.1 |
2.5 |
4.1 |
|
FBXO31 |
F-box protein 31 |
16q24.2 |
1.6 |
1.0 |
| TCF25 | Transcription factor 25 (basic helix-loop-helix)/NULP1 | 16q24.3 | 2.5 | 1.1 |
Chromosome 1q genes identified in retinoblastomas with 1q gain included several with functions in mitosis, particularly centrosome duplication, chromosome segregation and the mitotic spindle checkpoint (ASPM, CENPF, KIF14, NUF2, NEK2). Additional chromosome 1q genes identified have functions in promoting cell cycle progression (CKS1B) and transcriptional regulation during development (NR5A2).
Chromosome 6p genes that showed increased expression in association with 6p gain again emphasised the same functional categories highlighted by 1q genes, for example, mitosis (KIFC1, NUP153), cell cycle progression (E2F3, UHRF1BP1) and developmental transcriptional regulation (FOXP4, SOX4, TEAD3). TEAD3 is a downstream target of the Hippo pathway, and it is of interest that combined inactivation of RB and Hippo pathway kinases has been shown to result in dedifferentiation in the Drosophila retina (Nicolay et al, 2010). Chromosome 6p genes also included several with roles in the epigenetic regulation of gene expression, particularly through modification of chromatin (DAXX, DEK, EHMT2, JARID2, RUNX2, PRPF4B).
Genes which were downregulated in association with 16q loss included RBL2 (p130), which has an essential role in maintaining permanent cell cycle arrest and senescence following events initiated by RB1 (Helmbold et al, 2011). It is of interest that RBL2 expression was reduced not only in retinoblastomas with 16q loss (N=4), but also showed a 2.2-fold decrease overall in group 1 retinoblastomas relative to group 2, which may reflect epigenetic regulation (De Falco and Giordano, 2006). Other 16q genes identified included USP10, which is upregulated in response to DNA damage and which stabilises p53, also FBXO31 and NAE1, which function in ubiquitin pathways to regulate cell cycle progression, cell growth and survival, and CHD9 and PRMT7, which have roles in chromatin remodelling, with the latter also implicated in the DNA damage response (Karkhanis et al, 2012). A further consequence of chromosome 16q loss was the decreased expression of BBS2 and RPGRIP1L, which are required for retinal photoreceptor function.
Geneset enrichment analysis analysis of regulatory pathways in group 1 and group 2 retinoblastomas
Geneset enrichment analysis was used to obtain further information about the regulatory pathways responsible for the differing characteristics of group 1 and 2 retinoblastomas. Genesets enriched in group 1 retinoblastomas included several related to G-protein-coupled receptor-mediated signalling (Supplementary Table S6). There was a strong emphasis on neurotransmitter signalling, particularly involving class A1 (RHO-like) GPCRs, which include the amine ligand-binding receptors (e.g., serotonin, adrenergic and cholinergic receptors) and peptide ligand-binding receptors (hypocretin/orexin, NPY, melanocortin and tachykinin receptors) (Supplementary Figure S2). These results are of interest as although glutamine is the major neurotransmitter within the retina, particularly for photoreceptor, bipolar and ganglion cells, additional neurotransmitters/receptors are a feature of other cell types. Serotonin and NPY, for example, are associated with subsets of amacrine cells (Bagnoli et al, 2003; Ghai et al, 2009), tachykinins with amacrine and ganglion cells, and orexins with horizontal, bipolar, amacrine and ganglion cells (Kolb et al, 1995; Savaskan et al, 2004). This genome-wide analysis of gene expression patterns is consistent with the idea that group 1 retinoblastomas show characteristics of multiple retinal cell types.
In the case of group 2 retinoblastomas, many significantly enriched genesets were relevant to photoreceptor function, particularly mitochondrial function/energy metabolism as well as transcription and RNA splicing (Supplementary Table S6, Supplementary Figure S3). Photoreceptors, especially cones, are among the most metabolically active cells in the body (Eckmiller 2004; Reidel et al, 2011), and normal retinal/photoreceptor functions maintained in group 2 retinoblastomas may promote tumour growth.
Discussion
The aetiology of retinoblastoma and its relationship to retinal histogenesis have been intensively studied. However, as yet there is still relatively little consensus on the cell-of-origin or the molecular mechanisms involved. Our results suggest that retinoblastomas, like most other tumour types, are not a homogeneous group but may be classified into at least two different sub-types based on molecular and histopathological characteristics. Furthermore we suggest that these sub-types may have differing aetiology and origin.
Classification of retinoblastomas
Our results, based on gene expression profiling, are consistent with the existence of two sub-types of retinoblastoma, group 1 retinoblastomas that show features of multiple retinal cell types, and group 2 retinoblastomas that show a distinctive cone photoreceptor expression profile. This sub-division is further supported by the finding that recurrent chromosomal alterations typical of retinoblastoma (1q loss, 6p gain and 16q loss) were restricted to group 1 tumours, and in addition suggest that the genetic mechanisms driving tumorigenesis in each group may also be distinct. It was also noted that adverse histopathological features (PLONI and deep choroid invasion) were more frequent in group 1 retinoblastomas.
Our classification of retinoblastoma is in contrast to two previous reports, which emphasised the overall similarity between retinoblastomas, but nevertheless also provides support for both of these studies (Xu et al, 2009; McEvoy et al, 2011). Xu et al (2009) proposed a cone precursor origin for retinoblastoma (similar to our group 2 tumours), following immunohistochemical detection of cone-specific markers RXRG, THRB and cone arrestin in all 40 retinoblastomas examined. Consistent with this finding, both our group 1 and 2 retinoblastomas expressed genes encoding these proteins, albeit at higher levels in group 2. Similarly, the failure to detect several developmental markers of other retinal cell types (PROX1, CHX10/VSX2, PAX6) in retinoblastoma cells in the study of Xu et al (2009) is also consistent with our observation that these genes were significantly downregulated in both group 1 and 2 retinoblastomas relative to normal adult and normal fetal retina (Figure 3). We conclude, therefore, that the results of Xu et al (2009) do not exclude the possibility that some retinoblastomas also have characteristics of non-cone retinal cell types.
McEvoy et al (2011) concluded from gene expression analyses that retinoblastoma tumour cells express genes associated with multiple retinal cell types. This is similar to our findings for group 1 retinoblastomas. However, further examination of the data of McEvoy et al (2009; GEO:GSE29683) on 52 human retinoblastomas suggests that there is a significant range of expression values for individual retinal cell type-specific genes in this large complex data set (Supplementary Figure S4). These results are not inconsistent with the idea that some retinoblastomas have a predominantly cone-like expression profile.
The retinoblastoma cell-of-origin
The identification of different sub-types of retinoblastoma raises the question as to whether these represent divergent evolution from a common tumour progenitor cell or alternatively if there is a different cell-of-origin in each case. It is important to consider this question in the context of normal retinal development. Retinal progenitor cells progress through a series of competence states during development, giving rise to retinal cell type-specific precursors in a temporally restricted manner, a process which may be regulated at least in part by the sequential expression of retinal transcription factors (Ohsawa and Kageyama, 2008; Agathocleous and Harris, 2009). However, there is considerable overlap in the generation of different cell types and individual progenitor cells from the same developmental time point not only show extensive heterogeneity of gene expression but may also express genes encoding multiple retinal transcription factors within a single cell (Trimarchi et al, 2008). Consistent with these observations, recent evidence suggests that RPCs show a surprising level of stochasticity in cell fate choice, and the observed variability in the number and type of cells resulting from clonal expansion of individual RPCs (a RPC may divide to produce two daughter RPCs (P/P), a RPC and a differentiating cell (P/D) or two differentiating cells (not necessarily identical) (D/D)) fits well with a model in which cells divide or differentiate with fixed probabilities related to the ratios of different cell types in the normal retina (Gomes et al, 2011; He et al, 2012).
Taking this information into consideration, we propose a model for group 1 retinoblastomas in which these tumours arise from a RPC in which the balance between P/P, P/D and D/D divisions is disrupted to favour continued proliferation, but with individual tumour cells showing stochastic regulation of lineage progression and variable patterns of gene expression. The higher level of apoptosis observed in group 1 retinoblastomas, compared with group 2, may reflect a proportion of tumour cells that undergo terminal D/D divisions. This model is consistent with published expression data on single retinoblastoma cells (McEvoy et al, 2011). Examination of this data indicates that single cells obtained following repeated ( × 3) passage of a human retinoblastoma in mice do not show selection for a single retinal lineage but instead display a high level of inter-cell variability in the pattern of transcription factor expression (Supplementary Figure S5). These results support the idea that stochastic regulation of cell fate is an inherent characteristic of these retinoblastomas, as well as of normal retina.
Group 2 retinoblastomas in contrast are characterised by a stable pattern of cone photoreceptor gene expression, including not only genes encoding cone-enriched transcription factors but also multiple genes required for photoreceptor structure and function (Supplementary Table S2). These retinoblastomas might arise in a RPC in which the stochastic regulation of cell fate is strongly biased towards a cone cell lineage. Alternatively, they may originate in an RXRG-expressing precursor cell, primed for cone differentiation, which has failed to undergo cell cycle exit. Continued proliferation may be a consequence of the combined effects of pRB loss and also upregulation of genes responsible for cell cycle progression, for example, cyclin E2 and the mitotic cyclin-dependent kinase, CDK1, which showed 1.9- to 2.1-fold increased expression in group 2 relative to group 1 retinoblastoma and even larger increases relative to fetal and adult retina (results not shown).
The molecular aetiology of group 1 retinoblastomas
The restricted occurrence of chromosome 1q and 6p gain and 16q loss in group 1 retinoblastomas suggests that genes on these chromosomes are relevant to the aetiology of this tumour group. In agreement with previous reports, this study identified several functionally related genes with roles in spindle microtubule organisation and kinetochore/centrosome-mediated mitotic functions, for example, KIF14, CENPF, ASPM, NEK2 and NUF2 on chromosome 1 and KIFC1 on chromosome 6p (Grassmann et al, 2005; Gratias et al, 2005; Orlic et al, 2006; Bowles et al, 2007; Ganguly and Shields, 2010). These genes have particular relevance for group 1 retinoblastomas, as orientation of the mitotic spindle has been shown to influence the distribution of cell-fate determinants to one or both daughter cells, thereby producing symmetric (P/P, D/D) or asymmetric cell divisions (P/D or D/D with different cell types). In the retina, RPCs with horizontally oriented spindles (relative to the retinal pigment epithelium) generate identical daughter cells, while vertically oriented spindles tend to generate two different daughters, and it has been suggested that even slight changes in spindle orientation may influence the outcome of cell division (Cayouette et al, 2006). Proteins such as ASPM have been shown to be critical for the maintenance of symmetric divisions in the developing CNS (Nigg and Stearns, 2011), and it is tempting to speculate that increased expression of ASPM might have a similar function in retinoblastoma, resulting in increased production of RPCs.
Genes on chromosomes 1q, 6p and 16q may also contribute to tumorigenesis through more direct effects on cell cycle regulation. An important consequence of loss of pRB function is aberrant regulation of the G1–S transition and these genes may further potentiate this effect through over-riding downstream cell cycle checkpoints. Of particular interest is the upregulation of the chromosome 1q gene CKS1B, which has an essential role in the SKP2-mediated ubiquitination of a range of cell cycle inhibitory proteins including p27/CDKN1B and also p130/RBL2 (Tedesco et al, 2002; Krishnan et al, 2010; Liberal et al, 2011). CKS1B maps to 1q21, a common region of gain in cancer, and its amplification/overexpression has been reported in many tumours (Wang et al, 2009; Chen et al, 2010; Krishnan et al, 2010). Also of interest is the downregulation of the 16q gene USP10, which de-ubiquinates p53 and is an essential regulator of its stability (Yuan et al, 2010). It has been suggested that loss of USP10 provides an alternative mechanism to antagonise p53 function in tumours such as renal cell carcinoma, which (like retinoblastoma) do not have a high frequency of p53 mutations (Yuan et al, 2010). USP10 was also identified by Gratias et al (2007) in a search for genes mapping to 16q24, which were differentially expressed in retinoblastomas with 16q LOH.
Although investigation of the molecular consequences of chromosome alterations in group 1 retinoblastomas provides clues about pathways which are aberrantly regulated in tumorigenesis, it is also important to identify the primary mitogenic signal(s). Potential candidates include growth factors, neurotrophins and neurotransmitters, all of which have been implicated in the regulation of cell proliferation in the developing retina (Martins and Pearson, 2008). Our GSEA analysis points to a role for neurotransmitter signalling, especially through amine and peptide ligand-binding receptors. This is a significant result as it provides further support for the novel finding of McEvoy et al (2011) that among paediatric solid tumour cell lines, the retinoblastoma cell lines Y79 and WERI1 are uniquely sensitive to broad-acting monoamine receptor inhibitors, and that blockade specifically of amine receptors (but not other major neurotransmitter signalling pathways) reduced retinoblastoma growth and survival. Inhibition of neurotransmitter signalling may therefore have important therapeutic potential in retinoblastoma.
The molecular aetiology of group 2 retinoblastomas
Group 2 retinoblastomas were observed to have a gene expression signature strongly indicative of differentiation along a cone photoreceptor lineage. This is consistent with the observation that human differentiated cone photoreceptors (but not early cone precursors) stain strongly for pRB, suggesting that pRB does not mediate the initial proliferative arrest in these cells (Lee et al, 2006), but may be important for maintaining cell cycle exit and differentiation. A consequence of loss of RB function may be to permit cell cycle re-entry of cells, which otherwise are competent to form mature photoreceptors and have all of the components required for the visual cycle.
Our results suggest that there are two main features of cone photoreceptors which have relevance for tumorigenesis in group 2 retinoblastomas. The first is the extremely high metabolism of this cell type – photoreceptors are among the most metabolically active cells in the body (Reidel et al, 2011). Of particular note in group 2 retinoblastomas was the upregulation of multiple components of the TCA and oxidative phosphorylation (OXPHOS) pathways. Although tumour cells are frequently reported to use aerobic glycolysis in preference to OXPHOS (Warburg effect), there is increasing evidence that OXPHOS and glycolysis cooperate to sustain energy needs in tumorigenesis, with dynamic regulation reflecting the tumour microenvironment (Jose et al, 2011). Our results, and those of Chen et al (2007), suggest that in environments such as the retina and the brain where energy demand is high, tumour cells use enhanced mitochondrial respiratory pathways for energy production. The very high metabolic activity of retinal photoreceptor cells is also paralleled by elevated transcriptional and translational activity, and it is of interest that mutations of several pre-mRNA processing genes (e.g., PRPF3, PRPF31, PRPC8) are associated with retinal disease phenotypes (Cao et al, 2011; Tanackovic et al, 2011). Elevated mRNA processing, observed in group 2 retinoblastomas, may impact on tumorigenesis both through an overall promotion of cell growth and division, but potentially also through a generalised loss of splicing fidelity which is a hallmark of cancer (Ghigna et al, 2008). Overall these results suggest that cellular and metabolic adaptations for efficient photoreceptor function in the normal retina may also contribute to a growth advantage for tumour cells in this microenvironment, and that this may account for the particular sensitivity of cone photoreceptor cells to oncogenic transformation.
Conclusions
In summary, the results of this study suggest that the molecular aetiology of retinoblastoma reflects aberrant regulation of developmental processes required for the highly ordered production of the different retinal cell types from a single progenitor cell. Although we cannot exclude the possibility that retinoblastomas represent a broad spectrum of developmental phenotypes, nevertheless our results suggest that most tumours arise from either a RPC or a committed cone precursor. Our findings are important for the development of novel targeted therapeutics for retinoblastoma and, in addition, strongly suggest that the clinical significance of retinoblastoma molecular heterogeneity merits further investgation.
Acknowledgments
We thank Carol Hitchcott and Susan Cavanagh for co-ordinating sample collection. We also thank families of retinoblastoma patients for consenting to tumour banking. This work was supported by the UK Children's Eye Cancer Trust (CHECT), the Birmingham Children's Hospital Research Foundation and the Birmingham Children's Hospital Department of Oncology.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Ajioka I, Martins RA, Bayazitov IT, Donovan S, Johnson DA, Frase S, Cicero SA, Boyd K, Zakharenko SS, Dyer MA. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell. 2007;131:378–390. doi: 10.1016/j.cell.2007.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnoli P, Dal Monte M, Casini G. Expression of neuropeptides and their receptors in the developing retina of mammals. Histol Histopathol. 2003;18:1219–1242. doi: 10.14670/HH-18.1219. [DOI] [PubMed] [Google Scholar]
- Bowles E, Corson TW, Bayani J, Squire JA, Wong N, Lai PB, Gallie BL. Profiling genomic copy number changes in retinoblastoma beyond loss of RB1. Genes Chromosomes Cancer. 2007;46:118–129. doi: 10.1002/gcc.20383. [DOI] [PubMed] [Google Scholar]
- Byerly M, Blackshaw S. Vertebrate retina and hypothalamus development. Wiley Interdiscip Rev Syst Biol Med. 2009;1:380–389. doi: 10.1002/wsbm.22. [DOI] [PubMed] [Google Scholar]
- Cao H, Wu J, Lam S, Duan R, Newnham C, Molday RS, Graziotto JJ, Pierce EA, Hu J. Temporal and tissue specific regulation of RP-associated splicing factor genes PRPF3, PRPF31 and PRPC8--implications in the pathogenesis of RP. PLoS One. 2011;6:e15860. doi: 10.1371/journal.pone.0015860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M, Poggi L, Harris WA. Lineage in the vertebrate retina. Trends Neurosci. 2006;29:563–570. doi: 10.1016/j.tins.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5:539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, 3rd, Felding-Habermann B. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- Chen L, Chan TH, Guan XY. Chromosome 1q21 amplification and oncogenes in hepatocellular carcinoma. Acta Pharmacol Sin. 2010;31:1165–1171. doi: 10.1038/aps.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid E, Santos-Ledo A, Parrilla-Monge M, Lillo C, Arevalo R, Lara JM, Aijon J, Velasco A. Prox1 expression in rod precursors and Muller cells. Exp Eye Res. 2010;90:267–276. doi: 10.1016/j.exer.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Corson T, Gallie B. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosomes Cancer. 2007;46:617–634. doi: 10.1002/gcc.20457. [DOI] [PubMed] [Google Scholar]
- De Falco G, Giordano A. pRb2/p130: a new candidate for retinoblastoma tumor formation. Oncogene. 2006;25:5333–5340. doi: 10.1038/sj.onc.1209614. [DOI] [PubMed] [Google Scholar]
- Dimaras H, Khetan V, Halliday W, Orlic M, Prigoda N, Piovesan B, Marrano P, Corson T, Eagle R, Squire J, Gallie B. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet. 2008;17:1363–1372. doi: 10.1093/hmg/ddn024. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Bremner R. The search for the retinoblastoma cell of origin. Nat Rev Cancer. 2005;5:91–101. doi: 10.1038/nrc1545. [DOI] [PubMed] [Google Scholar]
- Eckmiller MS. Defective cone photoreceptor cytoskeleton, alignment, feedback, and energetics can lead to energy depletion in macular degeneration. Prog Retin Eye Res. 2004;23:495–522. doi: 10.1016/j.preteyeres.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CV. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Shields CL. Differential gene expression profile of retinoblastoma compared to normal retina. Mol Vis. 2010;16:1292–1303. [PMC free article] [PubMed] [Google Scholar]
- Ghai K, Zelinka C, Fischer AJ. Serotonin released from amacrine neurons is scavenged and degraded in bipolar neurons in the retina. J Neurochem. 2009;111:1–14. doi: 10.1111/j.1471-4159.2009.06270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics. 2008;9:556–570. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes FL, Zhang G, Carbonell F, Correa JA, Harris WA, Simons BD, Cayouette M. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development. 2011;138:227–235. doi: 10.1242/dev.059683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann C, Gratias S, Stephan H, Schuler A, Schramm A, Klein-Hitpass L, Rieder H, Schneider S, Kappes F, Eggert A, Lohmann D. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene. 2005;24:6441–6449. doi: 10.1038/sj.onc.1208792. [DOI] [PubMed] [Google Scholar]
- Gratias S, Rieder H, Ullmann R, Klein-Hitpass L, Schneider S, Bononi R, Kappler M, Lohmann D. Allelic loss in a minimal region on chromosome 16q24 is associated with vitreous seeding of retinoblastoma. Cancer Res. 2007;67:408–416. doi: 10.1158/0008-5472.CAN-06-1317. [DOI] [PubMed] [Google Scholar]
- Gratias S, Schuler A, Hitpass L, Stephan H, Rieder H, Schneider S, Horsthemke B, Lohmann D. Genomic gains on chromosome 1q in retinoblastoma: consequences on gene expression and association with clinical manifestation. Int J Cancer. 2005;116:555–563. doi: 10.1002/ijc.21051. [DOI] [PubMed] [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris WA. How variable clones build an invariant retina. Neuron. 2012;75:786–798. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmbold H, Galderisi U, Bohn W. The switch from Rb1/p105 to Rb2/p130 in DNA damage and cellular senescence. J Cell Physiol. 2011;4:22786. doi: 10.1002/jcp.22786. [DOI] [PubMed] [Google Scholar]
- Hennig A, Peng G-H, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133. doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Indovina P, Marcelli E, Casini N, Rizzo V, Giordano A. Emerging roles of RB family: new defense mechanisms against tumor progression. J Cell Physiol. 2013;228:525–535. doi: 10.1002/jcp.24170. [DOI] [PubMed] [Google Scholar]
- Joly S, Pernet V, Samardzija M, Grimm C. Pax6-positive Muller glia cells express cell cycle markers but do not proliferate after photoreceptor injury in the mouse retina. Glia. 2011;59:1033–1046. doi: 10.1002/glia.21174. [DOI] [PubMed] [Google Scholar]
- Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor's dilemma. Biochim Biophys Acta. 2011;1807:552–561. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Karkhanis V, Wang L, Tae S, Hu YJ, Imbalzano AN, Sif S. Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase delta catalytic subunit gene, POLD1. J Biol Chem. 2012;287:29801–29814. doi: 10.1074/jbc.M112.378281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H, Fernandez E, Ammermuller J, Cuenca N. Substance P: a neurotransmitter of amacrine and ganglion cells in the vertebrate retina. Histol Histopathol. 1995;10:947–968. [PubMed] [Google Scholar]
- Krishnan A, Nair SA, Pillai MR. Loss of cks1 homeostasis deregulates cell division cycle. J Cell Mol Med. 2010;14:154–164. doi: 10.1111/j.1582-4934.2009.00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TC, Almeida D, Claros N, Abramson DH, Cobrinik D. Cell cycle-specific and cell type-specific expression of Rb in the developing human retina. Invest Ophthalmol Vis Sci. 2006;47:5590–5598. doi: 10.1167/iovs.06-0063. [DOI] [PubMed] [Google Scholar]
- Liberal V, Martinsson-Ahlzen HS, Liberal J, Spruck CH, Widschwendter M, McGowan CH, Reed SI. Cyclin-dependent kinase subunit (Cks) 1 or Cks2 overexpression overrides the DNA damage response barrier triggered by activated oncoproteins. Proc Natl Acad Sci USA. 2011;23:23. doi: 10.1073/pnas.1102434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Martins RA, Pearson RA. Control of cell proliferation by neurotransmitters in the developing vertebrate retina. Brain Res. 2008;1192:37–60. doi: 10.1016/j.brainres.2007.04.076. [DOI] [PubMed] [Google Scholar]
- McEvoy J, Flores-Otero J, Zhang J, Nemeth K, Brennan R, Bradley C, Krafcik F, Rodriguez-Galindo C, Wilson M, Xiong S, Lozano G, Sage J, Fu L, Louhibi L, Trimarchi J, Pani A, Smeyne R, Johnson D, Dyer MA. Coexpression of normally incompatible developmental pathways in retinoblastoma genesis. Cancer Cell. 2011;20:260–275. doi: 10.1016/j.ccr.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay B, Bayarmagnai B, Moon N, Benevolenskaya E, Frolov M. Combined inactivation of pRB and hippo pathways induces dedifferentiation in the Drosophila retina. PLoS Genet. 2010;6:e1000918. doi: 10.1371/journal.pgen.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45:72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- Pajovic S, Corson TW, Spencer C, Dimaras H, Orlic-Milacic M, Marchong MN, To KH, Theriault B, Auspitz M, Gallie BL. The TAg-RB murine retinoblastoma cell of origin has immunohistochemical features of differentiated Muller glia with progenitor properties. Invest Ophthalmol Vis Sci. 2011;52:7618–7624. doi: 10.1167/iovs.11-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Leifeld T, Gould J, Lerner J, Tamayo P, Mesirov J. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Reidel B, Thompson JW, Farsiu S, Moseley MA, Skiba NP, Arshavsky VY. Proteomic profiling of a layered tissue reveals unique glycolytic specializations of photoreceptor cells. Mol Cell Proteomics. 2011;10:M110.002469. doi: 10.1074/mcp.M110.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A, Bhagabati N, Braisted J, Liang W, Sharov V, Howe E, Li J, Thiagarajan M, White J, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Sastre X, Chantada GL, Doz F, Wilson MW, de Davila MT, Rodriguez-Galindo C, Chintagumpala M, Chevez-Barrios P. Proceedings of the consensus meetings from the International Retinoblastoma Staging Working Group on the pathology guidelines for the examination of enucleated eyes and evaluation of prognostic risk factors in retinoblastoma. Arch Pathol Lab Med. 2009;133:1199–1202. doi: 10.5858/133.8.1199. [DOI] [PubMed] [Google Scholar]
- Savaskan E, Muller-Spahn F, Meier F, Wirz-Justice A, Meyer P. Orexins and their receptors in the human retina. Pathobiology. 2004;71:211–216. doi: 10.1159/000078675. [DOI] [PubMed] [Google Scholar]
- Shields C, Mashayekhi A, Au A, Czyz C, Leahy A, Meadows A, Shields J. The international classification of retinoblastoma predicts chemoreduction success. J Ophthamol. 2006;113:2276–2280. doi: 10.1016/j.ophtha.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Chuang JZ. The cell biology of vision. Cell Biol. 2010;190:953–963. doi: 10.1083/jcb.201006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G, Ransijn A, Thibault P, Abou Elela S, Klinck R, Berson EL, Chabot B, Rivolta C. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum Mol Genet. 2011;20:2116–2130. doi: 10.1093/hmg/ddr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco D, Lukas J, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2) Genes Dev. 2002;16:2946–2957. doi: 10.1101/gad.1011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS One. 2008;3:e1588. doi: 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, Latreille JP, Wilson RK, Ganem D, DeRisi JL. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XC, Tian J, Tian LL, Wu HL, Meng AM, Ma TH, Xiao J, Xiao XL, Li CH. Role of Cks1 amplification and overexpression in breast cancer. Biochem Biophys Res Commun. 2009;379:1107–1113. doi: 10.1016/j.bbrc.2009.01.028. [DOI] [PubMed] [Google Scholar]
- Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, Liu A, Jhanwar SC, Abramson DH, Cobrinik D. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009;137:1018–1031. doi: 10.1016/j.cell.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.